Abstract
Rationale
Arrhythmogenic cardiomyopathy (ACM) is caused primarily by mutations in genes encoding desmosome proteins. Ventricular arrhythmias are the cardinal and typically early manifestations, whereas myocardial fibroadiposis is the pathological hallmark. Homozygous DSP (desmoplakin) and JUP (plakoglobin) mutations are responsible for a subset of ACM patients that exhibit cardiac arrhythmias and dysfunction, palmo-planter keratosis, and hair abnormalities (cardiocutaneous syndromes).
Objective
To determine phenotypic consequences of deletion of Dsp in a subset of cells common to the heart and skin.
Methods and Results
Expression of chondroitin sulfate proteoglycan 4 (CSPG4) was detected in epidermal keratinocytes and the cardiac conduction system (CCS). CSPG4pos cells constituted ~ 5.6±3.3% of the non-myocyte cells in the mouse heart. Inducible post-natal deletion of Dsp under the transcriptional control of the Cspg4 locus led to ventricular arrhythmias, atrial fibrillation, atrioventricular conduction defects, and death by 4 months of age. Cardiac arrhythmias occurred early and in the absence of cardiac dysfunction and excess cardiac fibro-adipocytes, as in human ACM. The mice exhibited palmo-plantar keratosis and progressive alopecia, leading to alopecia totalis, associated with accelerated proliferation and impaired terminal differentiation of keratinocytes. The phenotype is similar to human cardiocutaneous syndromes caused by homozygous mutations in DSP.
Conclusions
Deletion of Dsp under the transcriptional regulation of the CSPG4 locus led to lethal cardiac arrhythmias in the absence of cardiac dysfunction or fibroadiposis, palmoplantar keratosis, and alopecia, resembling the human cardiocutaneous syndromes. The findings offer a cellular basis for early cardiac arrhythmias in ACM patients and cardiocutaneous syndromes.
Keywords: Desmoplakin, Neuroglial cells, Arrhythmias, Sudden death, Cardiocutaneous syndrome, desmosome, arrhythmogenic right ventricular cardiomyopathy, desmosome cardiomyopathy
Subject Terms: Animal Models of Human Disease, Mechanisms, Myocardial Biology
INTRODUCTION
Arrhythmogenic cardiomyopathy (ACM) encompasses a group of myocardial diseases, whose cardinal manifestations are ventricular arrhythmias, occurring prior to and independent of systolic dysfunction. 1 Ventricular arrhythmias, manifesting as palpitations, syncope, and sudden cardiac death (SCD), are the most common initial presentation of ACM. 2–4 Atrial arrhythmias are also common and present in up to half of the patients with ACM, typically in those with atrial and right ventricular enlargement. 5, 6 Ventricular arrhythmias occur in the advanced stages of ACM in association with cardiac dysfunction.
The classic form of ACM is arrhythmogenic right ventricular cardiomyopathy (ARVC), which is characterized by progressive fibro-fatty replacement of cardiac myocytes, predominantly but not exclusively in the right ventricle, myocyte atrophy, and apoptosis. 1, 4 Heart failure typically develops in those with long-standing disease, is usually refractory to therapy, and often requires heart transplantation. 1
Mutations in five genes encoding desmosome proteins, namely plakophilin 2 (PKP2), junction protein plakoglobin (JUP), desmoplakin (DSP), desmocollin 2 (DSC2), and desmoglien 2 (DSG2), are the most common known causes of ACM. 7–12 Homozygous mutations in DSP and JUP cause ACM occurring in conjunction with palmo-plantar keratosis, wooly hair and alopecia, which are referred to as cardiocutaneous syndromes. 7, 8,13 Mutations in several others genes, including PLN and LMNA, encoding phospholamban and lamin A/C, respectively, also have been implicated in ACM. 14, 15
Desmosomes are components of the intercalated discs (IDs), which are primarily present in the epithelial cells and involved in cell-to-cell attachment, mechano-transduction, and mechanical integrity of the tissue. 16–19 The mechanisms by which mutations in desmosome proteins lead to ACM and its typical phenotypes of ventricular arrhythmias, excess fibro-adipocytes, or cardiocutaneous syndromes are not fully understood. Deletion of the Dsp gene exclusively in cardiac myocytes in mice leads to cardiac dysfunction, premature death, and mild fibro-adipogenesis. 19 However, cardiac arrhythmias in the myocyte-specific Dsp-deficient mice occur in the context of cardiac dysfunction, but not independent of cardiac dysfunction. 19 The arrhythmic phenotype in the cardiac myocyte-specific deletion of Dsp is in contrast to the human ACM, where cardiac arrhythmias are often the early manifestations, occurring prior to and independent of cardiac dysfunction. 2–4 The dissociation of early cardiac arrhythmias and function might simply reflect potential differential effects of the desmosome protein mutations in stabilizing the ion channel at the IDs and maintaining mechanical integrity of the myocardium. Alternatively, it may indicate possible involvement of non-myocyte cells in the pathogenesis of early cardiac arrhythmias in ACM.
Desmosome proteins are known to be expressed primarily in cardiac myocytes and epithelial cells. They are also expressed, albeit at lower levels in the non-myocyte cardiac cells, such as the mesenchymal cells and fibro-adipocyte progenitors (FAPs). 20, 21 Cells expressing the mutant desmosome proteins, whether myocytes or non-myocyte cells, are expected to be involved in the pathogenesis of ACM. 20, 21
The publicly available RNA sequencing and microarray databases include chondroitin sulfate proteoglycan 4 transcript (Cspg4) among those expressed in the cardiac conduction system (CCS), including the Purkinje fibers and the atrioventricular node (AVN). 22, 23 The CCS, sharing an embryonic origin with cardiac myocytes, also expresses desmosome proteins. 24, 25 CSPG4 is also expressed in the keratinocytes and hair follicle cells, which are known to express DSP 26, 27. Therefore, CSPG4, along with desmosome proteins, might serve as a shared molecular link between early cardiac arrhythmias and the skin phenotype in the cardiocutaneous syndromes. Thus, we determined the phenotypic consequences of deletion of Dsp under the transcriptional regulation of the Cspg4 locus.
METHODS
An expanded Methods section is provided as Online Supplementary Material.
Studies in the animal models conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health and was approved by the Institutional Animal Care and Use Committee. Human tissue use was approved by the Institutional Review Board.
Histology (H&E, Masson trichrome, and Oil Red O staining), immunohistochemistry and immunofluorescence
These techniques were performed as published. 18, 20, 28 Custom-made and commercially available antibodies were used to detect expression of the proteins of interest, including CSPG4 and DSP in isolated cardiac myocytes, non-myocyte cardiac cells, and myocardial sections (Online Table I).
Isolation of human and mouse adult myocytes and non-myocyte cardiac cells
Cardiac myocyte-depleted cell fraction was obtained by collagenase type 2 digestion as published. 20 The tissue samples were digested with collagenase, filtered through a 40 μm cell strainer, and the cells were precipitated by centrifugation. The pelleted cells were re-suspended in a complete medium in the presence of antibiotics, and plated on plates coated with 0.1% gelatin.
Cells expressing CSPG4 were isolated from myocyte-depleted cardiac cells by flow cytometry and Fluorescence-Activated Cell Sorting (FACS), as described, except for using a custom-made rabbit anti CSPG4 antibody. 20
Cspg4-DsRed.T1 reporter mice
Cspg4-DsRed.T1 is a bacterial artificial chromosome (BAC) transgenic reporter mouse line that expresses a red fluorescence protein variant, Ds-Red.T1, under the transcriptional regulation of the Cspg4 locus (Stock No: 008241, Jackson Laboratory). 29
Cspg4-Cre/Esr1*:DspW/F and Cspg4-Cre/Esr1*:DspF/F mice
These mice were generated upon crossing of Dsp floxed mice with the Cspg4-Cre/Esr1* deleter mice. 19, 30 The Dsp gene was deleted in 21-day old mice upon intra-peritoneal injection of tamoxifen.
Quantitative polymerase chain reaction (qPCR)
qPCR was performed as published and the relative normalized values (2^−ΔΔ method, shown as relative to the WT mice) were used to compare the transcript levels. 18, 28
Immunoblotting
Apoptosis
Apoptosis was detected in thin myocardial section by TUNEL assay, as published.19, 31 The number of TUNEL positive cells was quantified in approximately, 20,000 cells per mouse and in 6 mice per group.
Echocardiography
2D, M mode, and Doppler echocardiography was performed 3–6 months old Cspg4-Cre/Esr1*:DspW/F and Cspg4-Cre/Esr1*:DspF/F mice and their corresponding WT controls as published. 18, 20, 28, 32
Electrocardiography and Electrophysiology (EP)
Surface ECG and heart rate were recorded after insertion of 29-gauge needle electrodes subcutaneously into the forelimbs and connecting the leads to an Animal Bio Amp, PowerLab, and the LabChart7 software.
EP studies were performed in isoflurane-anesthetized mice at temperature between 36–37 °C. Intracardiac bipolar atrial and ventricular electrograms were obtained using a 1.1F octapolar catheter (EPR-800; Millar Instruments), as described. 33, 34 EP pacing protocols, including single, double, and burst ventricular pacing protocols were used for ventricular tachycardia (VT) induction at baseline and after injection of isoproterenol (3 mg/kg i.p.). The atrial stimulation protocol to evaluate inducibility of atrial fibrillation was performed with incremental atrial pacing, as described. 34 All pacing protocols were performed a maximum of three times. Non-sustained and sustained VT was defined as reproducible (at least twice) ectopic ventricular rhythms lasting 4 to 9 rapid consecutive ventricular beats and ≥10 beats, respectively. Atrial fibrillation was defined as reproducible rapid and fragmented atrial electrograms with irregular ventricular response for ≥1 second.
Statistical analysis
Data that followed a Gaussian distribution pattern were presented as mean ± SD and were compared between two groups by t test and among multiple groups by ANOVA followed by post-hoc pairwise comparisons. Otherwise, data were presented as the median values and compared by Kruskall-Wallis test, as were the categorical data.
RESULTS
CSPG4 is expressed in CCS and skin keratinocytes
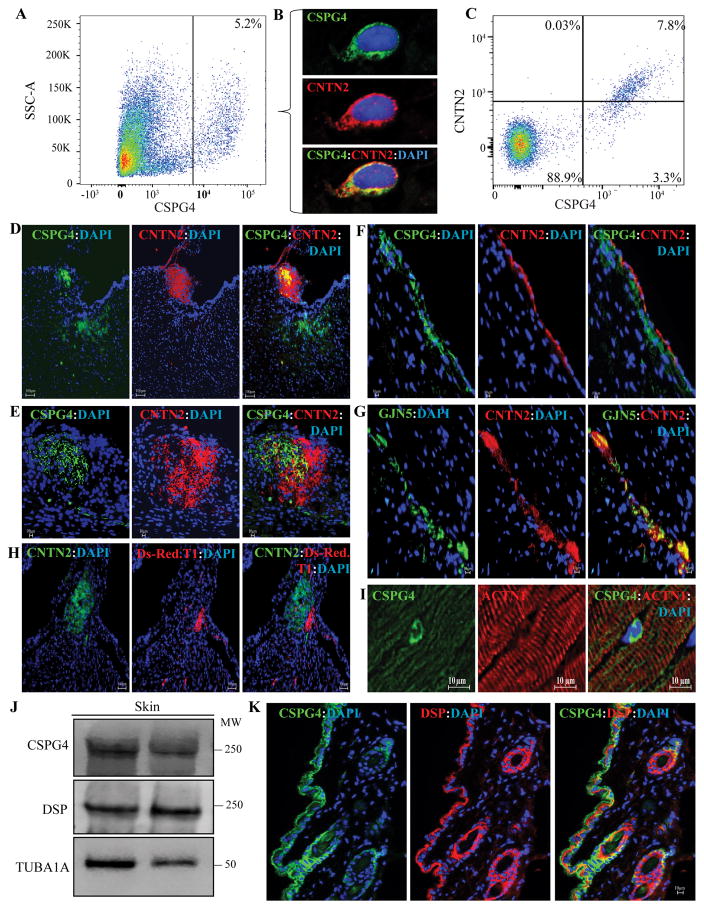
To corroborate the transcriptomic data detecting the Cspg4 transcripts in the heart, non-myocyte fraction of cardiac cells was isolated and sorted against a custom-made anti-CSPG4 antibody. FACS analysis showed that approximately 5.6±3.3% of cardiac non-myocyte cells expressed CSPG4 (Figure 1A, and Online Figure 1). To further corroborate detection of the Cspg4 transcripts in the CCS, co-expression of CSPG4 and contactin 2 (CNTN2), an established CCS marker, in cardiac cells was analyzed by FACS and immunostaining. CSPG4 and CNTN2 were co-expressed in a subset of isolated cardiac non-myocyte cells and about 2/3rd of cells expressing CSPG4 also expressed CNTN2 (Figure 1, panels B and C and Online Figure I).
Figure 1. Detection of expression of CSPG4 in the mouse heart and skin.
A. Flow cytometry plot showing CSPG4pos isolated by FACS from the mouse non-myocyte cardiac cell fraction. CSPG4pos cells represented 5.6±3.3% (N=6) of the non-myocyte cells in the mouse heart. Corresponding controls are shown in Online Figure 1. B. Co-immunostaining of cardiac cells isolated by flow cytometry for the expression of CSPG4 (validation) and cardiac conduction system (CCS) marker contactin 2 (CNTN2). As shown CSPG4 and CNTN2 are co-expressed in the isolated cells. C. Flow cytometry plot showing sorting of cardiac non-myocyte cells against anti CSPG4 and CNTN2 antibodies. As shown, approximately 2/3rd of the cells expressing CSPG4 also expressed CNTN2. D and E. Co-immunofluorescence staining of thin myocardial sections for CSPG4 and CNTN2 showing expression of the CSPG4 protein in in the AV nodal area, identified by the expression of the CCS marker CNTN2. Lower (D) and higher (E) magnifications of the AV nodal area are shown. F and G panels illustrate co-expression of CSPG4 and CNTN2 (Panel F) or GJN5 (connexin 40) and CNTN2 (Panel G) in the CCS, likely representing one of the bundle branches. H. Thin myocardial sections from the Cspg4-DsRedT.1 reporter mouse showing expression of Ds-Red.T.1 protein, a surrogate for CSPG4, in the AV nodal area, which is also identified by the expression of CNTN2. I. Isolated CSPG4pos cells within the myocardium residing between myofibrillar bundles, likely representing a pericytes or a neuroglial type II cells. The panel also shows absence of expression of CSPG4 in cardiac myocytes. J. Immunoblot showing expression of CSPG4 and DSP proteins in the mouse skin tissue along with a corresponding control for the loading condition. K. Thin skin sections showing co-expression and co-localization of CSPG4 and DSP in the epidermis and around hair follicles.
To detect expression and localization of CSPG4 in the heart, thin myocardial sections from adult mice were stained with a custom-made antibody against CSPG4. Expression of CSPG4 was detected in the AV node and the bundle branches, which were identified by the expression of contactin 2 (CNTN2) and/or connexin 40 (GJA5), two well-established CCS markers (Figure 1, panels D–G). 22, 35 To corroborate the immunofluorescence data, thin myocardial sections from the Cspg4-DsRed.T1 reporter mouse hearts were examined under fluorescence microscopy. DsRed.T1 expression, reflecting Cspg4 locus transcriptional activity and hence, an antibody-independent surrogate for CSPG4, was detected in the AV node (Figure 1H). Ds-Red.T.1 expression and localization in the myocardial sections was largely consistent with the detection of CSPG4 expression in the CCS by immunofluorescence staining, albeit less pronounced. Co-localization of CSPG4 and CNTN2 was partial, suggesting expression of CSPG4 in a subset of the CCS cells and cellular heterogeneity of the CCS. In addition to the CCS, CSPG4 was also expressed in a subset of cells within the interstitial tissue in the myocardium, likely representing capillary mural cells or pericytes (Figure 1I). Notably, expression of CSPG4 was not detected in the ventricular myocytes both by immunostaining and by examination of myocardial sections from the Cspg4-DsRed.T1 reporter mice (Figure 1H, and I).
Given the detection of the CSPG4 in capillary mural cells and its known expression in a subset of pericytes 36, isolated cardiac non-myocyte cells were co-stained for CSPG4 and PDGFRB proteins, the latter a marker for pericytes. 37 Expressions of the CSPG4 and PDGFRB proteins were detected in 8.1±10.6% and 2.6±3.3% of the isolated cardiac non-myocyte cells, respectively (Online Figure IIA). Only about 10% of cells expressing PDGFRB also expressed CSPG4. In addition, CSPG4 was not expressed in the endothelial cells, marked by the expression of PECAM1 (Online Figure IIB).
CSPG4 was also strongly expressed in murine back skin, as detected by immunoblotting and immunofluorescence staining (Figure 1J and K). Its expression was predominantly localized to epidermal keratinocytes in the interfollicular epidermis and hair follicles (Figure 1K). As expected, DSP was also expressed throughout the epidermis (Figure 1K). To further corroborate the findings, independent of the antibody performance, expression of CSPG4 was analyzed in the Cspg4-DsRed.T1 mice where DS-RedT.1 reporter protein serves as a surrogate for the expression of CSPG4 protein. DsRed.T1 expression was detected in the skin, predominantly in the interfollicular epidermis and hair follicles (Online Figure III).
Selected ion channel and desmosome proteins are expressed in cardiac CSPG4pos cells
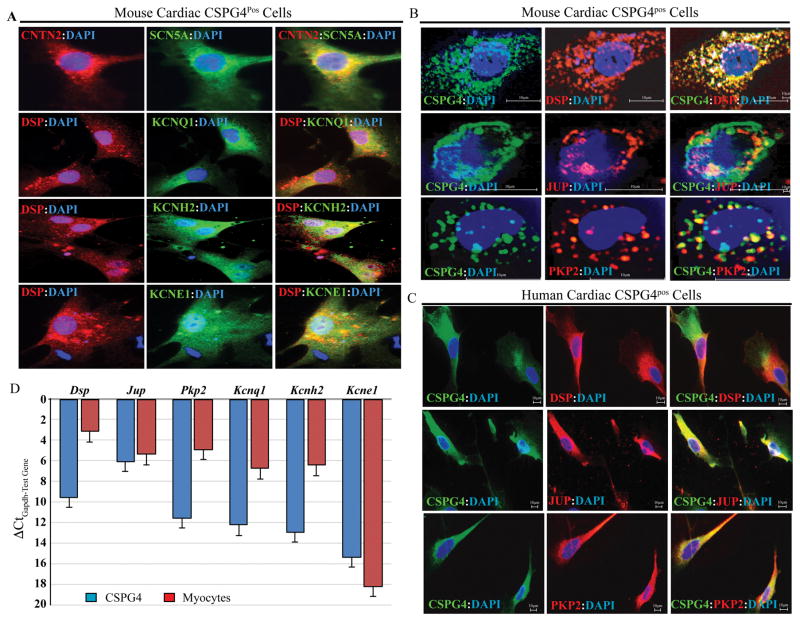
Given that CSPG4 also tagged the CCS, expression of selected ion channel genes was analyzed in the CSPG4pos cells isolated from the heart by FACS. Immunofluorescence staining detected expression of sodium voltage-gated channel α subunit 5 (SCN5A), potassium voltage-gated channel subfamily Q member 1(KCNQ1), potassium voltage-gated channel subfamily H member 2 (KCNH2 or HERG), and potassium voltage-gated channel subfamily E regulatory subunit 1 (KCNE1) in the cardiac CSPG4pos cells (Figure 2A).
Figure 2. Expression of selected ion channel and desmosome proteins in the mouse cardiac CSPG4pos cells.
A. Isolated mouse cardiac CSPG4pos cells were stained for expression of selected ion channels involved in cardiac arrhythmias and either CNTN2 or DSP, based on antibody compatibility. As shown, ion channel proteins SCN5A, KCNQ1, KCNH2, and KCNE1 were expressed in the CSPG4pos cells, along with DSP or CNTN2. B. Immunostaining showing expression of desmosome proteins DSP, JUP and PKP2 in FACS isolated mouse cardiac CSPG4pos cells co-stained with specific antibodies against selected desmosome proteins and CSPG4. C. Cardiac CSPG4pos cells isolated from non-myocyte fraction of cardiac cells were co-stained for the expression of CSPG4 (validation) and desmosome proteins DSP, JUP, and PKP2. As shown, desmospme proteins were expressed in the human cardiac CSPG4pos cells. D. Transcript levels of selected genes encoding desmosome and ion channel proteins in isolated cardiac CSPG4pos cells and cardiac myocytes, as detected by qPCR and depicted as ΔCtGapdh-test gene.
To determine whether cardiac CSPG4pos cells express desmosome proteins, and hence, contribute to the pathogenesis of ACM caused by desmosome protein mutations, mouse cardiac non-myocyte cell fraction was sorted against an anti CSPG4 antibody and the isolated cells were stained for the co-expression of selected desmosome proteins and CSPG4. Figures 2B illustrates expression of selected desmosome proteins, including DSP, JUP, and PKP2 along with the expression of CSPG4 in the FACS-isolated murine cardiac non-myocyte cells.
To extend the findings to the CSPG4pos cells isolated from the human hearts, cardiac non-myocyte cell fraction was isolated from the explanted human hearts not used for cardiac transplantation and co-stained with antibodies against CSPG4 and selected desmosome proteins. As observed in CSPG4pos cells isolated from the mouse heart, desmosome proteins were also co-expressed with CSPG4 in the isolated human cardiac non-myocyte cells (Figure 2C).
To complement the results of immunofluorescence studies, RNA was extracted from isolated cardiac CSPG4pos cells and the transcript levels of selected genes encoding desmosome and ion channel proteins were quantified by qPCR. The results verified expression of Dsp, Jup, Pkp2, Kcnq1, Kcnh2, and Kcne1 in the isolated CSPG4pos cells, albeit the transcripts were less abundant than the corresponding transcripts in the isolated cardiac myocytes (Figure 2D).
Finally, to determine whether cardiac myocytes express CSPG4, isolated cardiac myocytes were co-stained for cardiac actinin α2 (ACTN2) and CSPG4. As shown (Online Figure IVA), CSPG4 was not expressed in cardiac myocytes. Likewise, transcript levels of Myh6 and Actc1 were quantified by qPCR in isolated cardiac CSPG4pos cells. Myh6 and Actc1 transcript levels comprised approximately < 0.01% and 0.04% of those in cardiac myocytes (Online Figure IVB).
Post-natal in vivo deletion of Dsp in the CSPG4pos cells (Cspg4-Cre/Esr1*:DspW/F and Cspg4-Cre/Esr1*:DspF/F mice)
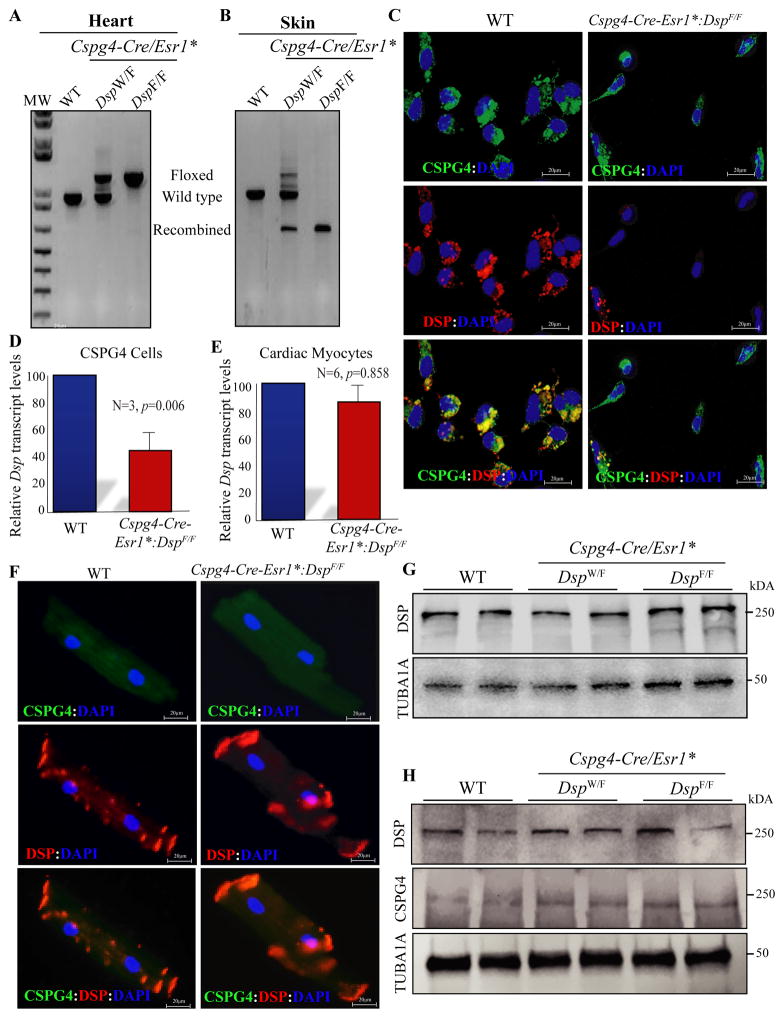
To determine whether deletion of Dsp in CSPG4pos cells induces a cardiac and/or skin phenotype, one or both copies of the Dsp were deleted post-natally (P21) under transcriptional regulation of the Cspg4 locus (Online Figure V). The approach of post-natal deletion reduced potential confounding effects, resulting from the transcriptional activity of the Cspg4 locus in other cell types during cardiac development. Recombination efficiency of the Dsp gene in the heart was low (1.63% in the Cspg4-Cre/Esr1*:DspF/F mice), which is consistent with the small number of cardiac CSPG4pos cells and the absence of transcriptional activity of the Cspg4 locus in other cardiac cells (Figure 3A). Recombination efficiency, however, was almost 100% in the skin in the Cspg4-Cre/Esr1*:DspF/F mice, in accord with the expression of CSPG4 in the keratinocytes and their abundance in the skin (Figure 3B).
Figure 3. Fidelity of the approach to deleting the Dsp gene specifically in the CSPG4pos cells.
A and B. Recombination efficiency in the heart and skin tissues. The floxed, wild type and recombined alleles are identified by size. Recombination efficiency was low in the heart and high in the skin in keeping with the small and high number of cells expressing CSPG4 in the heart and skin, respectively. C. Absence of DSP expression in the CSPG4pos cells isolated from the Cspg4-Cre/Esr1*:DspF/F mouse hearts and the corresponding WT control. The upper immunofluorescence panel shows expression of CSPG4, the middle DSP, and the lower overlay of the two panels in the experimental groups. As shown, DSP was expressed in the CSPG4pos cells isolated from the WT but not from the Cspg4-Cre/Esr1*:DspF/F mouse hearts. D. Quantitative PCR data of the Dsp transcript levels in the CSPG4pos cells isolated from the WT and Cspg4-Cre/Esr1*:DspF/F mouse hearts. Transcript levels of Dsp were reduced by more than 50% in the CSPG4pos cells isolated from Cspg4-Cre/Esr1*:DspF/F mouse hearts, as compared to the WT cells (46.25±10.04%, N=3, p=0.006). E. Quantitative PCR data of the Dsp transcript levels in cardiac myocytes, which were unchanged in cardiac myocytes isolated from Cspg4-Cre/Esr1*:DspF/F mouse hearts as compared to controls (N=6, p=0.858). F. Expression and localization of DSP in cardiac myocytes isolated from the WT and Cspg4-Cre/Esr1*:DspF/F mouse hearts. DSP expression was detected and localized to myocyte ends in both genotypes. The panels also show absence of expression of the CSPG4 protein in cardiac myocytes. G. Expression of DSP protein, detected by immunoblotting, in the cardiac myocytes isolated from the WT, Cspg4-Cre/Esr1*:DspW/F (heterozygous) and Cspg4-Cre/Esr1*:DspF/F (homozygous) mice. As shown, DSP levels were similar among the experimental groups, indicating intactness of DSP in cardiac myocytes. A corresponding blot for tubulin α1 (TUB1A1), as a control for loading conditions, is also shown. H. Expression of DSP, CSPG4, and TUB1A1 in the cardiac protein extracts from two mice per experimental group are shown. DSP protein levels were equal among the groups, in accord with the predominant expression of DSP in cardiac myocytes.
To further confirm deletion of the Dsp gene in the CSPG4pos cells, expression of Dsp transcript and DSP protein were detected in the isolated cardiac CSPG4pos cells. Whereas DSP was expressed in the CSPG4pos cells isolated from the WT mouse hearts, it was not detectable in the CSPG4pos cells isolated from the Cspg4-Cre/Esr1*:DspF/F mouse hearts (Figure 3C). Likewise, Dsp transcript levels were significantly lower in Cspg4-Cre/Esr1*:DspF/F mice when compared to WT (46.3±10.0%, Figure 3D).
To exclude possible fortuitous deletion of Dsp gene in cardiac myocytes, expression and localization of DSP protein were analyzed in the whole heart and isolated cardiac myocytes, respectively. Dsp transcript levels were unchanged in cardiac myocytes isolated from the Cspg4-Cre/Esr1*:DspF/F mouse hearts, as compared to the WT myocytes (Figure 3E). Likewise, DSP localization to the IDs was unaltered in cardiac myocytes isolated from the Cspg4-Cre/Esr1*:DspF/F mouse hearts (Figure 3F). Finally, DSP protein levels in the isolated cardiac myocytes (Figure 3G) as well as in the whole heart (Figure 3H), where myocytes are the predominant cell type expressing DSP, were analyzed by immunoblotting and were not different among the WT, Cspg4-Cre/Esr1*:DspW/F, and Cspg4-Cre/Esr1*ER:DspF/F mice.
Premature death of Cspg4-Cre/Esr1*:DspF/F mice
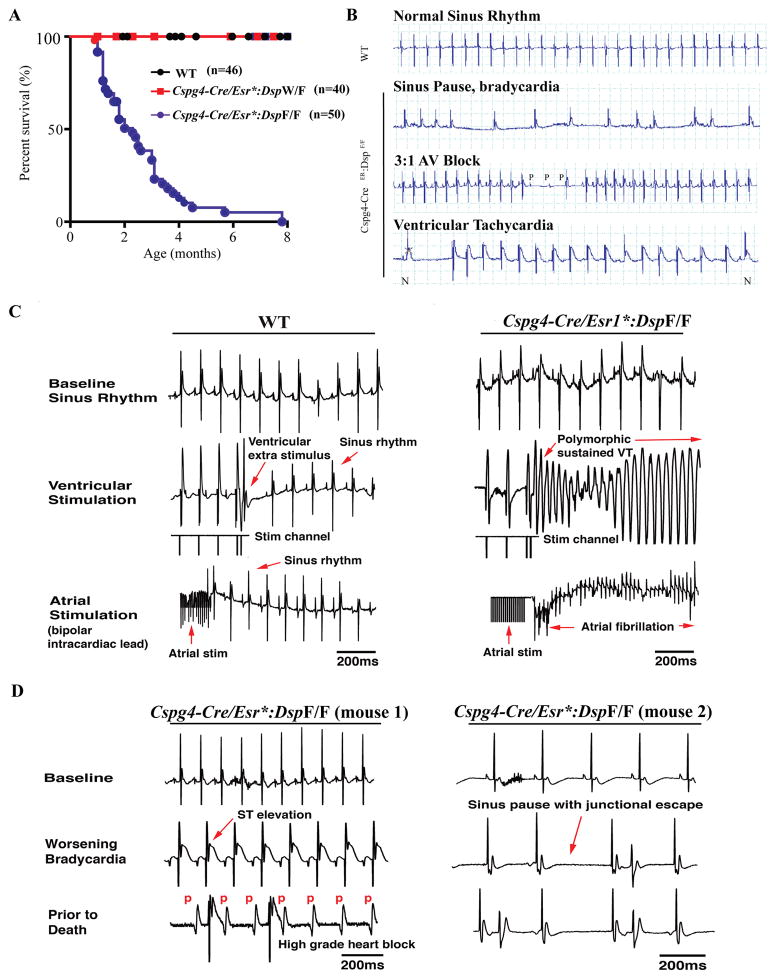
Induced homozygous deletion of Dsp post-natally (day P21) led to a near total mortality within 2–3 months after induction (Figure 4A). The median survival age of the Cspg4-Cre/Esr1*:DspF/F mice was ~30 days post induced deletion of the Dsp gene. Dsp-deficient heterozygous mice had a normal survival up to one year of age (Figure 4A).
Figure 4. Premature death due to cardiac arrhythmias and conduction defects.
A. Kaplan- Meier survival plots showing increased mortality in the Cspg4-Cre/Esr1*:DspF/F mice, with a median survival time of about 30 days post-induced deletion of Dsp and none living past 8 months of age (Chi-squared=152.7, p<0.0001). B. Representative electrocardiographic tracings from WT and Cspg4-Cre/Esr1*:DspF/F mice. Spontaneous episodes of arrhythmias, including ventricular tachycardia, sinus pauses, and advanced AV blocks were detected in the Cspg4-Cre/Esr1*:DspF/F but not the WT mice. Ventricular tachycardia is identified by the change in the QRS morphology, as compared to a normal QRS complex in the same rhythm strip, marked by N, and the ventricular rate. C. Representative tracings from WT and Cspg4-Cre/Esr1*:DspF/F mice showing cardiac rhythm at the baseline and following application of ventricular and atrial electric stimuli. Cardiac rhythm remained sinus after applying ventricular extra stimuli in the WT mice. In contrast, electric stimulation induced sustained polymorphic ventricular tachycardia in the Cspg4-Cre/Esr1*:DspF/F mice. Similarly, atrial stimulation (burst pacing) induced atrial fibrillation in the Cspg4-Cre/Esr1*:DspF/F but not in the WT mice. D. Cardiac rhythm recordings in two Cspg4-Cre/Esr1*:DspF/F mice obtained prior to death. The mice exhibited progressive sinus bradycardia, J point elevation, junctional escape beats, and advanced 3rd degree AV block followed by asystole prior to death.
Cardiac arrhythmias and conduction defects in the Cspg4-Cre/Esr1*:DspF/F mice
To investigate the cause of premature death, cardiac rhythm was monitored for 4 hours per day for 3 consecutive days (a total of 12 h) in each mouse. Implantable telemetric monitoring could not be performed due to a smaller body weight of the Cspg4-Cre/Esr1*:DspF/F mice, which hindered implantation of radio transmitter (Online Figure VI). During cardiac rhythm monitoring the Cspg4-Cre/Esr1*:DspF/F mice exhibited spontaneous high-grade AV block, intermittent atrial fibrillation, and runs of non-sustained VT (Figure 4B). ECG and rhythm phenotypes in the WT and Cspg4-Cre/Esr1*:DspW/F were unremarkable.
To further analyze the propensity of the Dsp-deficient mice to develop cardiac arrhythmias, 1 to 3 months old WT (N=7) and Cspg4-Cre/Esr1*:DspF/F (N=10) mice underwent in vivo EP studies. Cspg4-Cre/Esr1*:DspW/F mice were not studied, as they had normal survival and did not show rhythm abnormalities during cardiac rhythm monitoring. Heart rates at the baseline and conduction intervals were not significantly different between WT and Cspg4-Cre/Esr1*:DspF/F mice, with the exception of the ventricular atrial Wenckebach cycle length at baseline, which was prolonged in the Cspg4-Cre/Esr1*:DspF/F mice (73.3±11.5 msec in the WT and 88.6±13.7 msec in Cspg4-Cre/Esr1*:DspF/F mice, (Online Table II). Intraperitoneal injection of isoproterenol (3 mg/kg) increased the average heart rate equally in the WT and Cspg4-Cre/Esr1*:DspF/F mice.
EP studies were completed in 8 Cspg4-Cre/Esr1*:DspF/F and 7 WT mice. Programmed electrical stimulation evoked ventricular arrhythmias in 2/8 (25%) at the baseline and 5/8 (63%) after isoproterenol injection (3 mg/kg) in the Cspg4-Cre/Esr1*:DspF/F mice (Figure 4C). Likewise, atrial fibrillation was induced in 4/8 (50%) Cspg4-Cre/Esr1*ER:DspF/F mice. Thus, isoproterenol injection induced ventricular arrhythmias and/or atrial fibrillation in 9/10 (90%) of the Cspg4-Cre/Esr1*ER:DspF/F as compared to 1/7 (14%) in the WT mice (p=0.004). Cardiac rhythm was recorded in two Cspg4-Cre/Esr1*:DspF/F mice who died shortly after completion of the pacing protocols. The findings were remarkable for progressive bradycardia with marked ST elevations and progressive heart block preceding death (Figure 4D).
Normal cardiac function in the Cspg4-Cre/Esr1*:DspF/F mice
To determine whether cardiac arrhythmias upon deletion of Dsp in the CSPG4pos cells were associated with cardiac dysfunction or were independent of it, echocardiography was performed in 3-month old Cspg4-Cre/Esr1*:DspF/F mice and age- and sex-matched control WT mice. In addition, echocardiography was also performed in 6 to 10-month old Cspg4-Cre/Esr1*:DspW/F along with matching WT controls. The results, shown in Online Table III and IV, are notable for a normal left ventricular chamber size (when corrected for body weight) and normal cardiac function.
Increased myocardial apoptosis in the Cspg4-Cre/Esr1*:DspF/F mice
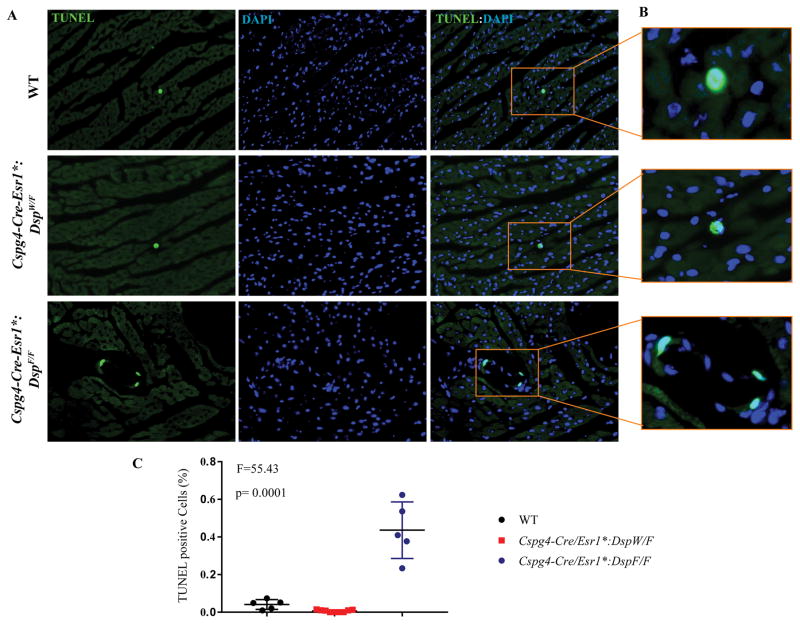
Because apoptosis is a known phenotype of ACM19, approximately 20,000 myocardial cells per mouse and 6 mice per group were examined by TUNEL assay. The number of cells staining positive in the TUNEL assay was increased by approximately 5-fold in the Cspg4-Cre/Esr1*:DspF/F as compared to WT mice (Figure 5).
Figure 5. Increased myocardial apoptosis in the Cspg4-Cre/Esr1*:DspF/F mice, as detected by the TUNEL assay.
A. TUNEL- and the corresponding DAPI-stained thin myocardial sections, showing increased number of TUNEL positive cells in the Cspg4-Cre/Esr1*:DspF/F group. A higher magnification of a TUNEL positive cell in each group and the quantitative data are shown in panels B and C, respectively.
Absence of excess fibro-adipocytes in the heart in the Cspg4-Cre/Esr1*:DspF/F mice
Given that CSPG4 is expressed in a subset of pericytes, because pericytes might be a source of fibro-adipocytes in ACM, and to determine whether cardiac arrhythmias were independent of fibro-adiposis, myocardial histology was examined upon staining of thin myocardial sections with Masson trichrome and Oil Red O. Neither fibrosis, reflected in the collagen volume fraction, nor the number of Oil Red O stained adipocytes was increased in the hearts of Cspg4-Cre/Esr1*:DspW/F and Cspg4-Cre/Esr1*:DspF/F mice as compared to the WT mice (Online Figure VII).
Progressive alopecia and palmo/plantar keratosis in the Cspg4-Cre/Esr1*:DspF/F mice
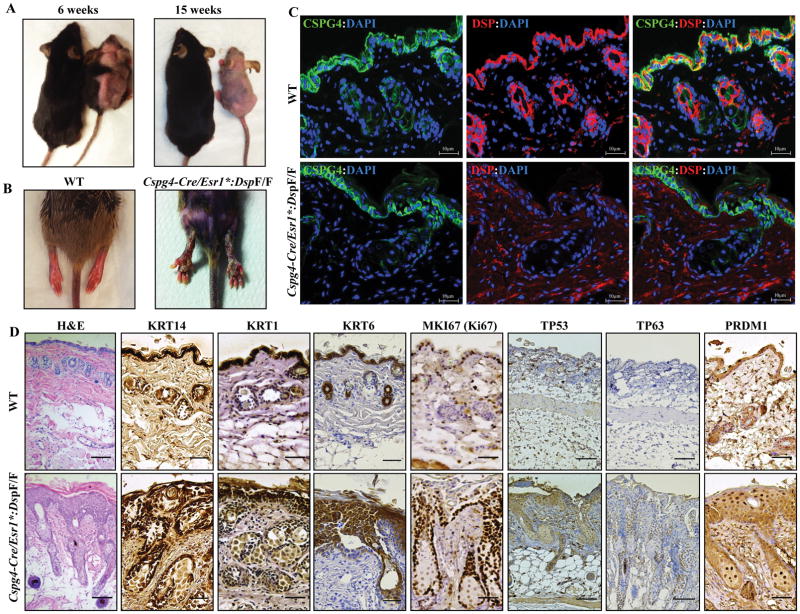
The Cspg4-Cre/Esr1*:DspF/F mice exhibited progressive alopecia starting 2 weeks after induced deletion of the Dsp gene which progressed to alopecia totalis by 4–6 months of age (Figure 6A). Likewise, the mice exhibited severe palmar and plantar keratosis (Figure 6B) resembling the skin phenotypes of human patients with Cavajal and Naxos syndromes, caused by homozygous deletion of DSP and JUP, respectively 7, 8, 38. Immunofluorescence staining of skin section showed effective ablation of DSP in the keratinocytes (Figure 6C), consistent with a very high recombination efficiency (Figure 3B). The Cspg4-Cre/Esr1*:DspW/F mice did not show similar skin phenotypes up to one year of age.
Figure 6. Alopecia totalis, palmoplantar keratosis, increased proliferation and terminal differentiation of epidermal keratinocytes upon deletion of Dsp in the CSPG4pos cells.
A. Representative images of WT and Cspg4-Cre/Esr1*:DspF/F mice illustrating progressive hair loss (alopecia) in the Cspg4-Cre/Esr1*:DspF/F mouse at 6 (left panel) and 15 weeks of age (3 and 12 weeks post tamoxifen-induced deletion of Dsp). B. Representative image showing the presence of severe keratosis on the hind legs of Cspg4-Cre/Esr1*:DspF/F mice. C. Thin skin sections obtained from WT and Cspg4-Cre/Esr1*:DspF/F mice stained for the expression of CSPG4 and DSP proteins. CSPG4 was expressed in the epidermal layer and around hair follicles in both genotypes. Likewise, DSP was expressed and localized to epidermal keratinocytes and hair follicles in the WT but was absent in the Cspg4-Cre/Esr1*:DspF/F mice. D. Immunohistochemical panels of skin tissue from WT and Cspg4-Cre/Esr1*:DspF/F mice showing expression of selected markers for proliferation and differentiation. H&E stained section shows thickened epidermis and perturbed epidermal compaction and sheet formation. KRT14, TP63 and MKI67 (Ki67) stained panels show a hyperproliferative epidermal basal layer, while KRT1, KRT6, TP53 and PRDM1 (BLIMP1) stained panels show impaired terminal differentiation of epidermal keratinocytes.
To delineate the mechanistic basis of alopecia and keratosis and given the role of CSPG4pos cells in renewal of the epidermal keratinocytes 26, 27, 39, skin tissue was stained for markers of cell proliferation, differentiation, inflammation, and apoptosis. Histological analysis of the skin of Cspg4-Cre/Esr1*:DspF/F mice revealed a defect in epidermal sheet formation as well as a hyperplastic interfollicular epidermis (Figure 6D). The Dsp-deficient mice displayed a hyperproliferative and thickened interfollicular epidermal basal layer, as demonstrated by immunohistochemical staining for the proliferation marker Ki67 (MKI67) and basal layer marker keratin 14 (KRT14), tumor protein 53 (TP53), and tumor protein 63 (TP63 or P63) (Figure 6D). To determine whether epidermal differentiation was effected by Dsp-deficiency, skin sections were stained for early differentiation markers KRT1 and KRT6 as well as terminal differentiation marker PRDM1 (PR/SET domain 1). 40 Staining for KRT1, KRT6, and PRDM1 showed strong suprabasal immunoreactivity (Figure 6D). In line with the observed hyperproliferative phenotype, sebaceous glands were enlarged in the skin of Dsp-deficient mice, as assessed by staining for fatty acid synthase (FAS), a sebocyte marker (Online Figure VIII).
Immunohistochemical staining of skin section for the expression of apoptosis markers caspase 3 (CASP3), and BCL2 (BCL2, apoptosis regulator), showed no detectable changes in BCL2 and CASP3 expressions (Online Figure VIII). Similarly, markers of inflammation, including adhesion G protein-coupled receptor F4 (ADGRE4), formerly known as F4/80, a macrophage marker; and tumor necrosis factor alpha (TNFα), a pro-inflammatory cytokine, were unchanged (Online Figure VIII). Likewise, the number of bone marrow-derived hematopoietic cells, identified by the expression of protein tyrosine phosphatase, receptor type C (PTPRC), formerly known as CD45, in the skin were unchanged.
Finally, to determine whether deficiency of DSP in skin keratinocytes impacted localization of JUP, thin skin sections from WT and Cspg4-Cre/Esr1*:DspF/F mice were co-stained for co-expression of KRT14 and JUP. The results showed unaltered localization of JUP in the skin keratinocytes, albeit JUP expression was markedly increased consistent with the increased thickness of epidermis in the Cspg4-Cre/Esr1*:DspF/F mice (Online Figure IX)
DISCUSSION
Cardiac arrhythmias, the cardinal manifestations of ACM, are typically the first manifestations, occurring prior to and in the absence of cardiac dysfunction 1. Cardiac arrhythmias are considered to be the consequence of expression of the mutant or deficiency of the desmosome protein in cardiac myocytes. Experimentally, deletion of genes encoding desmosome proteins in cardiac myocytes in mice leads to cardiac dysfunction but not early cardiac arrhythmias. 19 Cardiac arrhythmias in such models occur concurrently with cardiac dysfunction. 19 The findings of the present study suggest a non-cardiac myocyte origin of the early cardiac arrhythmias in ACM. The findings specifically implicate an origin from cells expressing CSPG4, likely the CCS, which are also known to express desmosome proteins and hence, are expected to be affected in human ACM. 24, 25 Accordingly, deletion of the Dsp gene, specifically in the CSPG4pos cells, which also tags the CCS in the heart, in addition to the capillary mural and neuroglial cells, leads to spontaneous atrial and ventricular arrhythmias and premature mortality. 22, 23 Notably, these arrhythmias occur in the presence of a normal cardiac function and absence of fibro-adiposis. The phenotype simulates the early cardiac arrhythmias in humans with ACM and hence, implicates a distinct cellular basis for early cardiac arrhythmias in ACM.
In accord with the expression of CSPG4 and DSP in skin progenitors and their role in keratinocyte renewal 26, 27, 41, deletion of Dsp specifically in the CSPG4pos cells also led to enhanced proliferation and impaired terminal differentiation of keratinocytes and the clinical phenotype of alopecia totalis and severe keratosis. Strong suprabasal immunoreactivity against KRT1, KRT6 and BLIMP1 suggested initiation but delayed terminal differentiation of keratinocytes in the skin of Dsp-deficient mice. 40 In line with earlier studies, Dsp-deficiency in the epidermal basal layer caused defects in epidermal sheet formation upon mechanical stress. 42 However, no evidence for an inflammatory infiltrate or apoptosis was found. Increased expression of TP53, in the absence of apoptotic epidermal keratinocytes, is in accord with a hyperproliferative epidermal basal layer and impaired (or absent) terminal differentiation in the skin of Cspg4-Cre/Esr1*:DspF/F mice. 43 The observed phenotype, resembling those found in human patients with Carvajal syndrome and Naxos disease 7, 8, 38, defines a cellular basis for the skin phenotype in cardiocutaneous syndromes.
Whereas ventricular and atrial arrhythmias are known phenotypic features of ACM, AV block is not a recognized phenotype. Conduction defect in human ACM typically manifests as an epsilon wave, which also has been reported in PKP2 deficiency. 44 AV block was observed only in the Cspg4-Cre/Esr1*:DspF/F mice but not in heterozygous deletion of DSP, which is the common genotype in human ACM. Similarly, the relevance of progressive bradycardia and AV block preceding death in the DSP-deficient mice to human ACM is unclear. These data along with the previous data point to the role of DSP in regulating cardiac rhythm and conduction. 45 Studies in human patients with homozygous deficiency of DSP would be required to determine whether AV block is also a phenotypic feature of human patients with ACM. Likewise, it is unclear whether atrial fibrillation, a known phenotype of ACM, 5, 6 also occurs prior to and in the absence of cardiac dysfunction in patients with ACM. In addition, the findings in the Cspg4-Cre/Esr1*:DspF/F might be pertinent only to a subset of human patients with the cardiocutaneous syndromes caused by DSP-deficiency but not those caused by missense mutations or those caused by the heterozygous mutations. Finally, phenotypic differences between human ACM and the mouse model are inherent to the model systems, which provide valuable insight into the pathogenesis of the phenotype but typically do not recapitulate the human disease. 46
CSPG4 is also expressed in the mural capillary cells or pericytes and neuroglial cells, which might be a cell source of fibro-adipocytes in ACM. The findings show that deletion of Dsp in the CSPG4pos cells did not result in excess fibro-adipocytes in the heart, hence, excluding these cells, or at least the subset that express CSPG4, as the potential sources of the pathological hallmark of ACM.
The new findings, in conjunction with the previous data, suggest a potential multi-cellular origin of the ACM phenotypes. Accordingly, expression of the mutant proteins, or the deficiency thereof, in cardiac myocyte leads mainly to cardiac dysfunction 19, whereas the effect in cardiac FAPs is excess fibro-adipocytes 20, and that in CSPG4pos cells cardiac arrhythmias. The multi-cellular basis of AC is consistent with the molecular genetics of ACM, as the causal mutations are germ line mutations affecting cell lineages that express the desmosome proteins. Accordingly, each cardiac cell type, expressing the mutant desmosome protein, mainly contributes to a specific phenotype; cardiac myocytes to cardiac dysfunction, CCS to early cardiac arrhythmias, and FAPs to excess fibro-adipocytes.
Several lines of data support fidelity of the Cspg4-Cre mediated deletion of Dsp gene and intactness of expression of DSP in cardiac myocytes. First, by design, Dsp was deleted at post-natal day P21 in order to avoid potential deletion of Dsp in other cell types in which the Cspg4 locus might be transcriptionally active during the development. In support of the experimental approach, the Cspg4 locus, which drives expression of the Cre recombinase, is not transcriptionally active in adult cardiac myocytes, a pre-requisite for the deletion of the Dsp gene. Accordingly, CSPG4 was not expressed in cardiac myocytes and Dsp transcript levels were unchanged in the isolated adult cardiac myocytes. Likewise, DSP protein was expressed and localized to the IDs in the Cspg4-Cre/Esr1*:DspF/F mice. In addition, the DsRed.T1 reporter protein, a surrogate for the transcriptional activity of the Cspg4 locus in the Cspg4:DsRed.T1 reporter mice, was not detected in cardiac myocytes. The DsRed.T1 protein, however, seems to be insoluble and often difficult to detect. Therefore, it might not be the most reliable marker. Therefore, the data were complemented with immunofluorescence staining of isolated cardiac myocytes using custom-made anti CSPG4 antibodies, which did not show expression of CSPG4 in cardiac myocytes. Finally, the Cspg4-Cre/Esr1*:DspF/F mice did not show evidence of cardiac dysfunction, which would be expected upon deletion of Dsp in cardiac myocytes, as reported previously. 19 Consequently, the data indicate that cardiac arrhythmias and conduction defects in the Cspg4-Cre/Esr1*:DspF/F mice are independent of cardiac myocytes and cardiac dysfunction, but rather reflective of deletion of the Dsp gene in non-myocyte cells in the heart, specifically the CCS.
CSPG4 is also expressed in brain tissue and is a marker for neuroglial type II cells. 47 However, neuroglial cells are not known to express desmosome proteins. Likewise, Dsp transcript was not detected by qPCR and its protein by Western Blot in the brain tissue (data not shown). Other epithelial cells, such as the gastrointestinal epithelia cells, as well as vascular pericytes are expected to express CSPG4 and perhaps, even the desmosome proteins. In addition, immunofluorescence staining of non-myocyte cardiac cells showed that heterogeneity of the CSPG4pos cells, a fraction of which also expressed the PDGFRB, a pericyte marker. Deletion of the Dsp gene in such cells and organs might cause concomitant phenotypes, which could confound the findings. However, concomitant phenotypes in organs other than the heart were not discernible at the clinical level, but such organs are not examined in great detail, based on the assumption that unanticipated phenotypes in non-cardiac tissues would not directly relate to cardiac arrhythmias and conduction defects in the Cspg4-Cre/Esr1*:DspF/F mice.
The underpinning mechanism(s) of cardiac arrhythmias in ACM, in general, and early arrhythmias occurring in the presence of a normal left ventricular function, in particular, are not well understood. Desmosomes along with adherens junction are the main constituents of IDs responsible for cell-cell attachment structures and mechanotransduction. In the human heart, they assemble into a single structure, localized to the polar regions of cardiac myocytes, referred to as area composite. 48 However, ID proteins are not exclusive to cardiac myocytes, but are also expressed in the CCS, including His bundles and Purkinje fibers, which is in keeping with shared embryonic origin of CCS and cardiac myocytes. 24 In contrast to cardiac myocytes, however, the ID proteins are not confined to the polar regions, but are rather scattered in cell membrane throughout and involved in lateral cell-cell contacts. 24 In addition to desmosome proteins, the ID protein constituents also include the ion channels, including SCN5A, which is responsible for the Brugada syndrome, long QT3, and familial atrioventricular block. 49–51 Moreover, conductive proteins, such as Connexin 43 (GJA5) and Ankyrin G also localize to the IDs. 52, 53 In accord with these data, CSPG4pos cells express a number of proteins, which are constituents of ion channels in the heart and are involved in cardiac arrhythmias, including SCN5A, KCNQ1, KCNH2, and KCNE1.54 Deficiency of DSP and other desmosome proteins could affect assembly and localization of proteins involved in cardiac conduction affecting the sodium current and conduction velocity, and therefore, predisposing to cardiac conduction defects and arrhythmias. 55, 56 In support of complex interactions between ID protein constituents, mutations in PKP2 have been shown to affect the sodium current and induce a phenotype resembling the Brugada syndrome, likely through interactions with SCN5A. 44 Conversely, mutations in SCN5A have been associated with the ARVC-like phenotype. 57 Moreover, deficiency of desmosome proteins, including DSP, is associated with a reduced expression level of GJA5. 58 Thus, the existing data point to intricate and complex interactions between the conduction system and the desmosome proteins in maintaining normal ion current and conduction in the heart.
In conclusion, the data suggest that deletion of Dsp gene in a subset of cardiac non-myocyte cells and skin keratinocytes, marked by the expression of CSPG4, leads to cardiac arrhythmias, conduction defects, and premature death occurring in the absence of cardiac dysfunction and fibro-adiposis, as in the early stages of human ACM. Likewise, it leads to severe palmoplantar keratosis and alopecia, as observed in the cardiocutaneous syndromes in humans. The data also exclude CSPG4pos cells, which also include a subset of pericytes and neuroglial cells as a cell source of excess fibro-adipocytes in ACM. Collectively, the findings indicate multi-cellular origin of cardiac phenotypes in ACM and provide insights into the pathogenesis of cardiocutaneous syndromes.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Arrhythmogenic cardiomyopathy (ACM) is a primary disease of myocardium caused mostly by mutations in genes encoding desmosome proteins.
Ventricular arrhythmias are the cardinal manifestation of ACM, occurring early and prior to cardiac dysfunction.
Altered functions of ion channels in cardiac myocytes are implicated in the pathogenesis of ventricular arrhythmias in ACM.
A subset of ACM manifests with skin abnormalities occurring in conjunction with cardiomyopathy and is referred to as cardiocutaneous syndromes.
What New Information Does This Article Contribute?
Chondroitin sulfate proteoglycan 4 (CSPG4) is a marker for cardiac conduction system (CCS), tagging a subset of CCS cells.
Cardiac cells expressing CSPG4 also express selected desmosome proteins and ion channels.
Post-natal (P21) inducible conditional deletion of Dsp gene, encoding desmoplakin, under the transcriptional regulation of the Cspg4 locus, i.e., in cells expressing CSPG4, leads to early ventricular and atrial arrhythmias and premature death in the presence of a preserved left ventricular systolic function, as observed in human ACM.
Deletion of Dsp in the CSPG4pos cells also leads to progressive alopecia and severe palmo-plantar keratosis, as observed in cardiocutaneous syndromes in humans.
The findings delineate a cellular basis for early ventricular arrhythmias in ACM, an important cause of sudden cardiac death in the young, particularly in athletes. The present findings along with the previous data point to participation of multiple cell types in the pathogenesis of ACM phenotypes, with CSPG4pos cells contributing to early cardiac arrhythmias, fibro-adipocyte progenitor cells to fibro-adiposis, and cardiac myocytes to cardiac dysfunction and heart failure. Delineation of the molecular and cellular basis of ACM could offer the opportunities to identify new diagnostic tools and novel therapeutic targets in ACM.
Acknowledgments
The authors wish to acknowledge Mr. Alon R. Azares for his technical support with FACS. Dr. Qiong Q Zhou was supported by a scholarship from The Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, China.
SOURCES OF FUNDING
This work was supported in part by grants from NIH, National Heart, Lung and Blood Institute (NHLBI, R01 HL088498 and 1R01HL132401), Leducq Foundation (14 CVD 03), George and Mary Josephine Hamman Foundation, American Heart Association Beginning Grant in Aid (15BGIA25080008 to RL). XHTW was supported by grants from NIH (R01-HL089598, R01-HL091947, R01-HL117641, and R41-HL129570) and American Heart Association (13EIA14560061).
Nonstandard Abbreviations and Acronyms
- ACM
Arrhythmogenic Cardiomyopathy
- ARVC
Arrhythmogenic right ventricular cardiomyopathy
- AVN
Atrioventricular node
- CCS
Cardiac conduction system
- CNTN2
Contactin 2
- CSPG4
Chondroitin sulfate proteoglycan 4
- EPS
Electrophysiological studies
- FACS
Fluorescence-activated cell sorting
- GJA5
Connexin 40
- IDs
Intercalated Disc
- JUP
Junction Plakoglobin
- PDGFRA
Platelet-derived growth factor receptor alpha
- PDGFRB
Platelet-derived growth factor receptor beta
- PKP2
Plakophilin-2
- VT
Ventricular tachycardia
Footnotes
DISCLOSURE
None.
References
- 1.Corrado D, Link MS, Calkins H. Arrhythmogenic right ventricular cardiomyopathy. The New England journal of medicine. 2017;376:61–72. doi: 10.1056/NEJMra1509267. [DOI] [PubMed] [Google Scholar]
- 2.Corrado D, Basso C, Rizzoli G, Schiavon M, Thiene G. Does sports activity enhance the risk of sudden death in adolescents and young adults? Journal of the American College of Cardiology. 2003;42:1959–1963. doi: 10.1016/j.jacc.2003.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Thiene G, Nava A, Corrado D, Rossi L, Pennelli N. Right ventricular cardiomyopathy and sudden death in young people. The New England journal of medicine. 1988;318:129–133. doi: 10.1056/NEJM198801213180301. [DOI] [PubMed] [Google Scholar]
- 4.Corrado D, Basso C, Thiene G, McKenna WJ, Davies MJ, Fontaliran F, Nava A, Silvestri F, Blomstrom-Lundqvist C, Wlodarska EK, Fontaine G, Camerini F. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: A multicenter study. Journal of the American College of Cardiology. 1997;30:1512–1520. doi: 10.1016/s0735-1097(97)00332-x. [DOI] [PubMed] [Google Scholar]
- 5.Chu AF, Zado E, Marchlinski FE. Atrial arrhythmias in patients with arrhythmogenic right ventricular cardiomyopathy/dysplasia and ventricular tachycardia. The American journal of cardiology. 2010;106:720–722. doi: 10.1016/j.amjcard.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, Guo J, Zheng L, Chen G, Ding L, Qiao Y, Sun W, Yao Y, Zhang S. Atrial remodeling and atrial tachyarrhythmias in arrhythmogenic right ventricular cardiomyopathy. The American journal of cardiology. 2016;118:750–753. doi: 10.1016/j.amjcard.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 7.McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman M, Baboonian C, Jeffery S, McKenna WJ. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (naxos disease) Lancet. 2000;355:2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- 8.Alcalai R, Metzger S, Rosenheck S, Meiner V, Chajek-Shaul T. A recessive mutation in desmoplakin causes arrhythmogenic right ventricular dysplasia, skin disorder, and woolly hair. Journal of the American College of Cardiology. 2003;42:319–327. doi: 10.1016/s0735-1097(03)00628-4. [DOI] [PubMed] [Google Scholar]
- 9.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, Towbin JA, Danieli GA. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. American journal of human genetics. 2002;71:1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pilichou K, Nava A, Basso C, Beffagna G, Bauce B, Lorenzon A, Frigo G, Vettori A, Valente M, Towbin J, Thiene G, Danieli GA, Rampazzo A. Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113:1171–1179. doi: 10.1161/CIRCULATIONAHA.105.583674. [DOI] [PubMed] [Google Scholar]
- 11.Awad MM, Dalal D, Cho E, Amat-Alarcon N, James C, Tichnell C, Tucker A, Russell SD, Bluemke DA, Dietz HC, Calkins H, Judge DP. Dsg2 mutations contribute to arrhythmogenic right ventricular dysplasia/cardiomyopathy. American journal of human genetics. 2006;79:136–142. doi: 10.1086/504393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nature genetics. 2004;36:1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 13.Antonov NK, Kingsbery MY, Rohena LO, Lee TM, Christiano A, Garzon MC, Lauren CT. Early-onset heart failure, alopecia, and cutaneous abnormalities associated with a novel compound heterozygous mutation in desmoplakin. Pediatr Dermatol. 2015;32:102–108. doi: 10.1111/pde.12484. [DOI] [PubMed] [Google Scholar]
- 14.van der Zwaag PA, van Rijsingen IA, Asimaki A, Jongbloed JD, van Veldhuisen DJ, Wiesfeld AC, Cox MG, van Lochem LT, de Boer RA, Hofstra RM, Christiaans I, van Spaendonck-Zwarts KY, Lekanne dit Deprez RH, Judge DP, Calkins H, Suurmeijer AJ, Hauer RN, Saffitz JE, Wilde AA, van den Berg MP, van Tintelen JP. Phospholamban r14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: Evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14:1199–1207. doi: 10.1093/eurjhf/hfs119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quarta G, Syrris P, Ashworth M, Jenkins S, Zuborne Alapi K, Morgan J, Muir A, Pantazis A, McKenna WJ, Elliott PM. Mutations in the lamin a/c gene mimic arrhythmogenic right ventricular cardiomyopathy. European heart journal. 2012;33:1128–1136. doi: 10.1093/eurheartj/ehr451. [DOI] [PubMed] [Google Scholar]
- 16.Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: From gene to disease. Circulation research. 2010;107:700–714. doi: 10.1161/CIRCRESAHA.110.223412. [DOI] [PubMed] [Google Scholar]
- 17.Broussard JA, Getsios S, Green KJ. Desmosome regulation and signaling in disease. Cell Tissue Res. 2015;360:501–512. doi: 10.1007/s00441-015-2136-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen SN, Gurha P, Lombardi R, Ruggiero A, Willerson JT, Marian AJ. The hippo pathway is activated and is a causal mechanism for adipogenesis in arrhythmogenic cardiomyopathy. Circulation research. 2014;114:454–468. doi: 10.1161/CIRCRESAHA.114.302810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, Marian AJ. Suppression of canonical wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. The Journal of clinical investigation. 2006;116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombardi R, Chen SN, Ruggiero A, Gurha P, Czernuszewicz GZ, Willerson JT, Marian AJ. Cardiac fibro-adipocyte progenitors express desmosome proteins and preferentially differentiate to adipocytes upon deletion of the desmoplakin gene. Circulation research. 2016;119:41–54. doi: 10.1161/CIRCRESAHA.115.308136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommariva E, Brambilla S, Carbucicchio C, Gambini E, Meraviglia V, Dello Russo A, Farina FM, Casella M, Catto V, Pontone G, Chiesa M, Stadiotti I, Cogliati E, Paolin A, Ouali Alami N, Preziuso C, d’Amati G, Colombo GI, Rossini A, Capogrossi MC, Tondo C, Pompilio G. Cardiac mesenchymal stromal cells are a source of adipocytes in arrhythmogenic cardiomyopathy. European heart journal. 2016;37:1835–1846. doi: 10.1093/eurheartj/ehv579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pallante BA, Giovannone S, Fang-Yu L, Zhang J, Liu N, Kang G, Dun W, Boyden PA, Fishman GI. Contactin-2 expression in the cardiac purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulsmans M, Clauss S, Xiao L, Aguirre AD, King KR, Hanley A, Hucker WJ, Wulfers EM, Seemann G, Courties G, Iwamoto Y, Sun Y, Savol AJ, Sager HB, Lavine KJ, Fishbein GA, Capen DE, Da Silva N, Miquerol L, Wakimoto H, Seidman CE, Seidman JG, Sadreyev RI, Naxerova K, Mitchell RN, Brown D, Libby P, Weissleder R, Swirski FK, Kohl P, Vinegoni C, Milan DJ, Ellinor PT, Nahrendorf M. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169:510–522. e520. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pieperhoff S, Borrmann C, Grund C, Barth M, Rizzo S, Franke WW. The area composita of adhering junctions connecting heart muscle cells of vertebrates. Vii. The different types of lateral junctions between the special cardiomyocytes of the conduction system of ovine and bovine hearts. Eur J Cell Biol. 2010;89:365–378. doi: 10.1016/j.ejcb.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 25.van Weerd JH, Christoffels VM. The formation and function of the cardiac conduction system. Development. 2016;143:197–210. doi: 10.1242/dev.124883. [DOI] [PubMed] [Google Scholar]
- 26.Legg J, Jensen UB, Broad S, Leigh I, Watt FM. Role of melanoma chondroitin sulphate proteoglycan in patterning stem cells in human interfollicular epidermis. Development. 2003;130:6049–6063. doi: 10.1242/dev.00837. [DOI] [PubMed] [Google Scholar]
- 27.Ghali L, Wong ST, Tidman N, Quinn A, Philpott MP, Leigh IM. Epidermal and hair follicle progenitor cells express melanoma-associated chondroitin sulfate proteoglycan core protein. J Invest Dermatol. 2004;122:433–442. doi: 10.1046/j.0022-202X.2004.22207.x. [DOI] [PubMed] [Google Scholar]
- 28.Gurha P, Chen X, Lombardi R, Willerson JT, Marian AJ. Knockdown of plakophilin 2 downregulates mir-184 through cpg hypermethylation and suppression of the e2f1 pathway and leads to enhanced adipogenesis in vitro. Circulation research. 2016;119:731–750. doi: 10.1161/CIRCRESAHA.116.308422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X, Bergles DE, Nishiyama A. Ng2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135:145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- 30.Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, Nishiyama A. Age-dependent fate and lineage restriction of single ng2 cells. Development. 2011;138:745–753. doi: 10.1242/dev.047951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senthil V, Chen SN, Tsybouleva N, Halder T, Nagueh SF, Willerson JT, Roberts R, Marian AJ. Prevention of cardiac hypertrophy by atorvastatin in a transgenic rabbit model of human hypertrophic cardiomyopathy. Circulation research. 2005;97:285–292. doi: 10.1161/01.RES.0000177090.07296.ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruggiero A, Chen SN, Lombardi R, Rodriguez G, Marian AJ. Pathogenesis of hypertrophic cardiomyopathy caused by myozenin 2 mutations is independent of calcineurin activity. Cardiovascular research. 2013;97:44–54. doi: 10.1093/cvr/cvs294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Oort RJ, McCauley MD, Dixit SS, Pereira L, Yang Y, Respress JL, Wang Q, De Almeida AC, Skapura DG, Anderson ME, Bers DM, Wehrens XH. Ryanodine receptor phosphorylation by calcium/calmodulin-dependent protein kinase ii promotes life-threatening ventricular arrhythmias in mice with heart failure. Circulation. 2010;122:2669–2679. doi: 10.1161/CIRCULATIONAHA.110.982298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voigt N, Li N, Wang Q, Wang W, Trafford AW, Abu-Taha I, Sun Q, Wieland T, Ravens U, Nattel S, Wehrens XH, Dobrev D. Enhanced sarcoplasmic reticulum ca2+ leak and increased na+-ca2+ exchanger function underlie delayed afterdepolarizations in patients with chronic atrial fibrillation. Circulation. 2012;125:2059–2070. doi: 10.1161/CIRCULATIONAHA.111.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delorme B, Dahl E, Jarry-Guichard T, Marics I, Briand JP, Willecke K, Gros D, Theveniau-Ruissy M. Developmental regulation of connexin 40 gene expression in mouse heart correlates with the differentiation of the conduction system. Dev Dyn. 1995;204:358–371. doi: 10.1002/aja.1002040403. [DOI] [PubMed] [Google Scholar]
- 36.Ozerdem U, Grako KA, Dahlin-Huppe K, Monosov E, Stallcup WB. Ng2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev Dyn. 2001;222:218–227. doi: 10.1002/dvdy.1200. [DOI] [PubMed] [Google Scholar]
- 37.Cuervo H, Pereira B, Nadeem T, Lin M, Lee F, Kitajewski J, Lin CS. Pdgfrbeta-p2a-creert2 mice: A genetic tool to target pericytes in angiogenesis. Angiogenesis. 2017 doi: 10.1007/s10456-017-9570-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norgett EE, Hatsell SJ, Carvajal-Huerta L, Cabezas JC, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Human molecular genetics. 2000;9:2761–2766. doi: 10.1093/hmg/9.18.2761. [DOI] [PubMed] [Google Scholar]
- 39.Gunnarsson AP, Christensen R, Praetorius J, Jensen UB. Isolating subpopulations of human epidermal basal cells based on polyclonal serum against trypsin-resistant cspg4 epitopes. Experimental cell research. 2017;350:368–379. doi: 10.1016/j.yexcr.2016.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Kretzschmar K, Cottle DL, Donati G, Chiang MF, Quist SR, Gollnick HP, Natsuga K, Lin KI, Watt FM. Blimp1 is required for postnatal epidermal homeostasis but does not define a sebaceous gland progenitor under steady-state conditions. Stem Cell Reports. 2014;3:620–633. doi: 10.1016/j.stemcr.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Q, Zhang H, Liu Y, Adams S, Eilken H, Stehling M, Corada M, Dejana E, Zhou B, Adams RH. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nature communications. 2016;7:12422. doi: 10.1038/ncomms12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasioukhin V, Bowers E, Bauer C, Degenstein L, Fuchs E. Desmoplakin is essential in epidermal sheet formation. Nat Cell Biol. 2001;3:1076–1085. doi: 10.1038/ncb1201-1076. [DOI] [PubMed] [Google Scholar]
- 43.Cottle DL, Kretzschmar K, Schweiger PJ, Quist SR, Gollnick HP, Natsuga K, Aoyagi S, Watt FM. C-myc-induced sebaceous gland differentiation is controlled by an androgen receptor/p53 axis. Cell Rep. 2013;3:427–441. doi: 10.1016/j.celrep.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerrone M, Lin X, Zhang M, Agullo-Pascual E, Pfenniger A, Chkourko Gusky H, Novelli V, Kim C, Tirasawadichai T, Judge DP, Rothenberg E, Chen HS, Napolitano C, Priori SG, Delmar M. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a brugada syndrome phenotype. Circulation. 2014;129:1092–1103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mezzano V, Liang Y, Wright AT, Lyon RC, Pfeiffer E, Song MY, Gu Y, Dalton ND, Scheinman M, Peterson KL, Evans SM, Fowler S, Cerrone M, McCulloch AD, Sheikh F. Desmosomal junctions are necessary for adult sinus node function. Cardiovascular research. 2016;111:274–286. doi: 10.1093/cvr/cvw083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marian AJ. Modeling human disease phenotype in model organisms: “It’s only a model!”. Circulation research. 2011;109:356–359. doi: 10.1161/CIRCRESAHA.111.249409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang W, Zhao N, Bai X, Karram K, Trotter J, Goebbels S, Scheller A, Kirchhoff F. Novel ng2-creert2 knock-in mice demonstrate heterogeneous differentiation potential of ng2 glia during development. Glia. 2014;62:896–913. doi: 10.1002/glia.22648. [DOI] [PubMed] [Google Scholar]
- 48.Franke WW, Borrmann CM, Grund C, Pieperhoff S. The area composita of adhering junctions connecting heart muscle cells of vertebrates. I. Molecular definition in intercalated disks of cardiomyocytes by immunoelectron microscopy of desmosomal proteins. Eur J Cell Biol. 2006;85:69–82. doi: 10.1016/j.ejcb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. Scn5a mutations associated with an inherited cardiac arrhythmia, long qt syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 50.Chen Q, Kirsch GE, Zhang D, Brugada R, Brugada J, Brugada P, Potenza D, Moya A, Borggrefe M, Breithardt G, Ortiz-Lopez R, Wang Z, Antzelevitch C, O’Brien RE, Schulze-Bahr E, Keating MT, Towbin JA, Wang Q. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 51.Schott JJ, Alshinawi C, Kyndt F, Probst V, Hoorntje TM, Hulsbeek M, Wilde AA, Escande D, Mannens MM, Le Marec H. Cardiac conduction defects associate with mutations in scn5a. Nature genetics. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 52.Cohen SA. Immunocytochemical localization of rh1 sodium channel in adult rat heart atria and ventricle. Presence in terminal intercalated disks. Circulation. 1996;94:3083–3086. doi: 10.1161/01.cir.94.12.3083. [DOI] [PubMed] [Google Scholar]
- 53.Sato PY, Coombs W, Lin X, Nekrasova O, Green KJ, Isom LL, Taffet SM, Delmar M. Interactions between ankyrin-g, plakophilin-2, and connexin43 at the cardiac intercalated disc. Circulation research. 2011;109:193–201. doi: 10.1161/CIRCRESAHA.111.247023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cerrone M, Napolitano C, Priori SG. Genetics of ion-channel disorders. Current opinion in cardiology. 2012;27:242–252. doi: 10.1097/HCO.0b013e328352429d. [DOI] [PubMed] [Google Scholar]
- 55.Rizzo S, Lodder EM, Verkerk AO, Wolswinkel R, Beekman L, Pilichou K, Basso C, Remme CA, Thiene G, Bezzina CR. Intercalated disc abnormalities, reduced na(+) current density, and conduction slowing in desmoglein-2 mutant mice prior to cardiomyopathic changes. Cardiovascular research. 2012;95:409–418. doi: 10.1093/cvr/cvs219. [DOI] [PubMed] [Google Scholar]
- 56.Sato PY, Musa H, Coombs W, Guerrero-Serna G, Patino GA, Taffet SM, Isom LL, Delmar M. Loss of plakophilin-2 expression leads to decreased sodium current and slower conduction velocity in cultured cardiac myocytes. Circulation research. 2009;105:523–526. doi: 10.1161/CIRCRESAHA.109.201418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Te Riele AS, Agullo-Pascual E, James CA, Leo-Macias A, Cerrone M, Zhang M, Lin X, Lin B, Sobreira NL, Amat-Alarcon N, Marsman RF, Murray B, Tichnell C, van der Heijden JF, Dooijes D, van Veen TA, Tandri H, Fowler SJ, Hauer RN, Tomaselli G, van den Berg MP, Taylor MR, Brun F, Sinagra G, Wilde AA, Mestroni L, Bezzina CR, Calkins H, Peter van Tintelen J, Bu L, Delmar M, Judge DP. Multilevel analyses of scn5a mutations in arrhythmogenic right ventricular dysplasia/cardiomyopathy suggest non-canonical mechanisms for disease pathogenesis. Cardiovascular research. 2017;113:102–111. doi: 10.1093/cvr/cvw234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lyon RC, Mezzano V, Wright AT, Pfeiffer E, Chuang J, Banares K, Castaneda A, Ouyang K, Cui L, Contu R, Gu Y, Evans SM, Omens JH, Peterson KL, McCulloch AD, Sheikh F. Connexin defects underlie arrhythmogenic right ventricular cardiomyopathy in a novel mouse model. Human molecular genetics. 2014;23:1134–1150. doi: 10.1093/hmg/ddt508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.