Abstract
This review is provided in recognition of the extensive contributions of Dr. Robert J. Lefkowitz to the G protein-coupled receptor (GPCR) field and to celebrate his 75th birthday. Since one of the authors trained with Bob in the 80s, we provide a history of work done in the Lefkowitz lab during the 80s that focused on dissecting the mechanisms that regulate GPCR signaling, with a particular emphasis on the GPCR kinases (GRKs). In addition, we highlight structure/function characteristics of GRK interaction with GPCRs as well as a review of two recent reports that provide a molecular model for GRK-GPCR interaction. Finally, we offer our perspective on some future studies that we believe will drive this field.
Keywords: arrestins, GPCR, GRK, phosphorylation, signaling, X-ray crystallography
1. A brief history of GRKs
The history of G protein-coupled receptor kinases (GRKs) really began with the identification of an enzymatic activity in rod membranes that could phosphorylate rhodopsin in a light-dependent manner [1]. This enzyme was called rhodopsin kinase (now GRK1) and it was subsequently purified and found to specifically phosphorylate light-activated rhodopsin [2]. Similar studies in the Lefkowitz lab during the late 70s and early 80s were focused on understanding the mechanisms involved in the loss of responsiveness of β-adrenergic receptor (βAR) signaling following prolonged stimulation with agonist (a process called desensitization). These studies revealed that the βAR underwent a mobility shift on SDS PAGE following agonist treatment [3]. This mobility shift was subsequently shown to be due to phosphorylation of the receptor [4], and additional studies established that at least some of this phosphorylation was due to the cAMP dependent protein kinase (PKA) [5]. In vitro studies demonstrated that PKA could directly phosphorylate the β2AR to a stoichiometry of 2 mol phosphate/mol receptor and that this phosphorylation attenuated receptor coupling to the heterotrimeric G protein Gs [6]. Thus, these early studies identified a mechanism of feedback regulation that involved phosphorylation of the β2AR by PKA, the protein kinase activated by the βAR signaling pathway. This feedback regulation of the β2AR by PKA was termed heterologous desensitization.
While a role for PKA phosphorylation of the β2AR was evident from these early studies, additional studies in the Lefkowitz lab revealed that the β2AR could also be phosphorylated in an agonist-dependent manner in S49 lymphoma cell lines that lacked the ability to activate PKA [7]. This observation led to a search for the enzyme that phosphorylated the β2AR in an agonist-dependent manner and ultimately resulted in the identification of the β-adrenergic receptor kinase or βARK (now called GRK2) [8]. βARK was analogous to rhodopsin kinase, given that both enzymes phosphorylated the active conformation of the receptor, and raised interesting questions about the similarities between phototransduction through rhodopsin and hormonal signaling through the β2AR [9]. Indeed, subsequent studies revealed that βARK could also phosphorylate light activated rhodopsin while rhodopsin kinase could phosphorylate the agonist-occupied β2AR [10]. Additional studies suggested that βARK had broad specificity since activation of multiple receptors promoted its translocation from the cytosol to the plasma membrane [11,12]. Moreover, βARK was also able to directly phosphorylate the α2-adrenergic receptor in vitro [13]. βARK was eventually purified [14] and cloned [15] revealing that it is a 689 amino acid serine/threonine protein kinase that specifically phosphorylates the agonist-occupied form of GPCRs such as the β2AR. Moreover, the cloning studies suggested that βARK is likely a member of a larger family of G protein-coupled receptor kinases [15].
During the course of these studies, another protein that contributes to receptor desensitization was identified. This protein was initially identified in the visual system and was termed S-antigen or 48 kDa protein and later named arrestin by Herman Kühn [16,17]. Arrestin had the interesting property of binding to light activated rhodopsin that had been phosphorylated by rhodopsin kinase and was found to quench phototransduction [16]. Studies in the Lefkowitz lab identified a similar role for an arrestin in desensitizing β2AR signaling in a βARK-dependent manner [18]. These efforts ultimately led to the identification of a non-visual arrestin termed β-arrestin that specifically binds to βARK-phosphorylated β2AR to inhibit receptor interaction with Gs [19]. Thus, these early studies revealed that GRKs play a central role in promoting arrestin binding to agonist-activated GPCRs to turn off receptor activation of heterotrimeric G proteins, a process termed homologous desensitization.
Once βARK was cloned, additional efforts led to the cloning of βARK2 (now called GRK3) [20], rhodopsin kinase [21], IT11 (now called GRK4) [22], GRK5 [23], GRK6 [24] and GRK7 [25,26]. The seven mammalian GRKs contribute to the phosphorylation and regulation of hundreds of G protein-coupled receptors (GPCRs). While GRKs have been extensively reviewed [27–33], here we focus on our current understanding of how GRKs interact with activated GPCRs.
2. Structure/function analysis of GRK-GPCR interaction
2.1. GRK structure
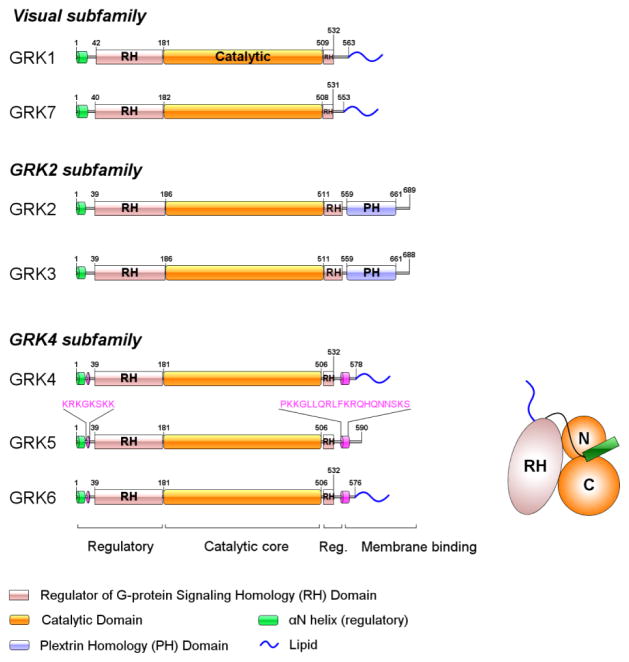
GRKs are serine/threonine protein kinases most related to the AGC kinase subfamily. GRKs have a modular structure with a central catalytic domain that sits within a regulator of G protein signaling homology (RH) domain [34,35] that is bracketed by a short N-terminal α-helical domain (αN-helix) and a variable C-terminal lipid-binding region [36] (Figs. 1 and 2). This basic structure is conserved in all GRKs going back to unicellular eukaryotes and non-metazoan opisthokonts [37]. The C-terminal region mediates membrane localization via prenylation (in GRK1 and 7), palmitoylation (in GRK4 and 6), or direct lipid binding either via a pleckstrin homology (PH) domain (in GRK2 and 3) or a polybasic/hydrophobic domain (in GRK5).
Figure 1. General architecture of GRKs.
GRKs are divided into 3 subfamilies based on sequence homology and are composed of two main domains, Regulator of G protein signaling Homology (RH) and catalytic domains. αN-helix comprising the first ~20 residues plays a regulatory role by bridging the N- and C-lobes of catalytic domain. The C-terminal fragment mediates membrane localization of GRKs. GRK4 subfamily includes two polybasic regions at N- and C-termini, and GRK5 relies on these regions to interact with negatively-charged phospholipids. GRK2 and GRK3 have a PH domain that interacts with acidic phospholipids and Gβγ subunits.
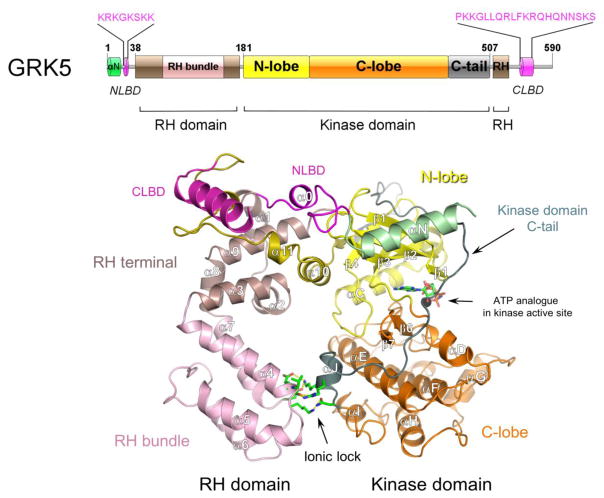
Figure 2. Structure of GRK5.
Crystal structure of GRK5 bound to AMP-PNP (PDB ID 4TND). The RH bundle and terminal subdomains, catalytic C-lobe and N-lobe subdomains, N-terminal lipid binding domain (NLBD) and an ionic lock between the RH and kinase domains are highlighted. Disordered αN-helix (green) and C-terminal lipid binding domain (CLBD) (magenta) were computationally modeled.
To date, all mammalian GRKs except for GRK3 and GRK7 have been crystallized. The first published structure was of GRK2 in complex with Gβγ from the Tesmer laboratory [38]. This structure provided important insight including the observation that the kinase domain is inserted into the RH domain and that contacts between the RH and kinase domains help to maintain the kinase in an inactive, open conformation. The X-ray crystal structure of GRK6 revealed a similar architecture with the RH domain making extensive contacts with the kinase domain, which remains in an open conformation even with a bound ATP analog [39]. The RH domain also forms an extensive dimer interface in GRK6 and while it is unclear whether this has a physiological role, there is evidence that a similar interface in GRK5 plays a role in membrane localization [40]. Interestingly, GRK6 was also crystallized in a more active conformation with a partially closed kinase domain and an extended αN-helix that bridged the kinase domain [41]. The authors proposed that this structure provides potential insight into a conformation similar to GRK bound to a receptor. GRK1 was crystallized next and found to homodimerize using a conserved interface within the RH domain [42]. GRK1 also crystallized in several conformations including some that revealed the C-terminal extension of the kinase domain and one where the αN-helix was observed. Based on the position of the αN-helix close to the hinge and active site tether (AST) of the kinase domain, the authors proposed a conceptual model for GRK1 docking to activated rhodopsin. GRK5 was crystallized in the presence of sangivamycin or AppNHp [43] as well as in complex with a high affinity inhibitor [44]. These studies also revealed that the RH domain helps to maintain the kinase domain in an inactive conformation [43] while inhibitor binding helps to close the kinase domain and partially relieve the structural constraint from the RH domain [44]. Most recently, a GRK4 polymorphism (A486V) that has been implicated in mediating hypertension was also crystallized [45]. In addition to these structures, GRK2 alone [46] as well as GRK2 in complex with Gβγ and Gαq have also been crystallized [47]. Overall, these structures reveal that the RH and kinase domains are in extensive contact with each other and appear to maintain the kinase in an inactive, open conformation. Moreover, these studies and others suggest that the αN-helix appears to stabilize kinase domain closure via a process that may be regulated by GPCR binding [41,42,48].
2.2. Using peptides as substrates to understand GRK function
While there is no clear consensus sequence that has been identified for GRK-mediated phosphorylation, the use of synthetic peptides has provided a number of interesting insights. GRK1 and GRK2 have a preference for phosphorylating peptides containing a serine/threonine with either carboxyl- or amino-terminally localized acidic residues, respectively [49], while GRK5 [50] and GRK6 [51] appear to prefer peptides with a serine/threonine preceded by basic residues. While these studies provide potential insight on the sequence specificity of GRKs, they need to be considered from the perspective that peptides are very poor GRK substrates. For example, the best peptide substrates for GRK2 typically have a Km that is 103–104 fold higher than for a receptor (e.g., 0.2–3 mM for peptides vs. ~0.2 μM for the β2AR) [49,52,53]. Similarly, the receptor is also a much better substrate with a Vmax ~103-fold higher than a peptide [53]. Thus, a GPCR is an ~106 fold better GRK substrate (Vmax/Km) compared to a peptide. Interestingly, recent studies identified a peptide from β-tubulin (DEMEFTEAESNMN) that had a Km of 34 μM as a GRK2 substrate, although it had a Vmax some 104-fold lower than found in other peptide studies raising concerns about the conditions that were used [54,55]. Another interesting twist is that peptides were also used to show that GRK binding to a receptor effectively activates the kinase. This was initially shown for rhodopsin activation of GRK1 where the ability to phosphorylate a peptide was dramatically enhanced by light-activated C-terminally truncated rhodopsin [56]. Similar studies with GRK2 revealed an ~200-fold increase in peptide phosphorylation by either light-activated rhodopsin or agonist-occupied β2AR [57]. One conclusion from these studies is that GRK interaction with a receptor surface enhances the affinity and ultimately the rate of phosphorylation.
2.3. GPCR regions involved in GRK binding
To better understand the process of GRK-mediated phosphorylation of GPCRs, a number of studies have focused on trying to identify the GPCR and GRK regions involved in interaction. Early studies found that peptides synthesized from the intracellular domains of the β2AR could either serve as substrates for GRK2 or, in some cases, inhibitors of GRK2 mediated-phosphorylation [52]. The most potent inhibitor of these peptides was derived from the first intracellular loop (ICL1) of the β2AR and had an IC50 of ~40 μM, although peptides from ICL2 and ICL3 could also inhibit receptor phosphorylation. Interestingly, the Lohse group made a few modifications to the β2AR ICL1 peptide that enhanced inhibition achieving an IC50 of 0.6 μM [58]. This modified peptide was also found to effectively inhibit the ability of GRK3 and GRK5 to phosphorylate the β2AR suggesting that this may be a general region of GRK interaction with the β2AR [58]. An additional strategy to study peptide effects on GPCR-GRK interaction has involved the use of lipidated peptides called pepducins [59]. Interestingly, pepducins from the β2AR ICL1 were able to effectively promote GRK-mediated phosphorylation of the β2AR providing further support for an important role of ICL1 in this process [60]. Taken together, these studies support the notion that multiple regions of the receptor are involved in GRK interaction.
An additional strategy that has been used extensively to try to define the regions most important for GRK interaction is receptor mutagenesis. Some of the early work involved characterization of GRK1 interaction with rhodopsin and revealed that the intracellular loops of the receptor likely serve as an initial platform for GRK docking [61,62]. For example, Shi et al. performed extensive alanine scanning mutagenesis of the three intracellular loops in rhodopsin and evaluated the ability of these mutants to be phosphorylated by GRK1 [61]. These studies found that mutation of residues in ICL1 (Thr62, Val63, and Gln64) resulted in an ~50% increase in phosphorylation while mutations of residues in ICL2 (Arg147, Phe148, Gly149) and ICL3 (Ala233, Ala234, Ala235) led to ~50% and ~80% decreases in phosphorylation, respectively. Thurmond et al. also found a significant role for ICL2 since deletion or replacement of ICL2 resulted in a complete loss of rhodopsin phosphorylation by GRK1 [62]. Deletion or replacement of ICL3 also resulted in a significant decrease in receptor phosphorylation while additional data supported a role for both ICL2 and ICL3 in direct binding of GRK1 [62]. The interaction of the α2A-adrenergic receptor (α2AAR) with GRK2 was also studied extensively since previous studies had demonstrated that GRK2 mediates the phosphorylation of 4 adjacent serines in the α2AAR ICL3 [63]. In these studies, it was shown that glutathione S-transferase (GST) fusion proteins containing either the ICL2 or ICL3 of the α2AAR could directly bind to GRK2 while there was no interaction with ICL1 or C-terminal domain fusion proteins [64]. Truncation mutagenesis identified three discrete regions within the α2AAR ICL3 in GRK2 binding while site-directed mutagenesis supported a role for specific basic residues (Arg225, Lys320, and Lys358) in ICL3 in GRK2-mediated phosphorylation of the α2AAR [64]. Basic residues in ICL2 from the metabotropic glutamate receptor 1 (mGluR1) (Lys691 and Lys692) were also shown to play a role in GRK2 binding [65]. A recent study also implicated a role for ICL3 residues (Leu226 and Val230) in GRK1 mediated phosphorylation of rhodopsin and proposed that GRK1, arrestin and the G protein transducin utilize a similar site of binding on rhodopsin [66]. In addition to these studies, there has been a large amount of work focused on identifying GRK phosphorylation sites on various receptors with the in vivo sites of rhodopsin phosphorylation by GRK1 being first described [67]. Taken together, these studies support a direct role for ICL2 and ICL3 in binding GRKs as well as a role for ICL1 in facilitating binding and/or phosphorylation of the receptor. Moreover, GRK interaction with the receptor intracellular loops likely provides allosteric control of GRK activation through triggering the kinase domain closure required to effectively phosphorylate residues within the GPCR C-terminus and/or ICL3.
2.4. GRK regions involved in GPCR binding and phosphorylation
Numerous studies have also focused on identifying the GRK residues important in mediating the binding and phosphorylation of GPCRs. In addition to the expected critical role of an invariant catalytic lysine in GRK2-mediated receptor phosphorylation and desensitization of the β2AR [68], early studies also implicated a role for the N-terminal region of GRK1 in rhodopsin interaction [69]. These studies demonstrated that an antibody made against a peptide encompassing GRK1 residues 17-34 effectively blocked GRK1-mediated phosphorylation of rhodopsin without affecting phosphorylation of a peptide substrate. Similarly, the Ca2+ binding protein recoverin was found to bind to the N-terminal region of GRK1 and inhibit GRK1-mediated phosphorylation of rhodopsin [70–72]. Additional work on GRK1 and GRK2 used truncation and point mutagenesis to identify an essential role for the N-terminal region in mediating rhodopsin phosphorylation [73]. These studies found that truncation of the N-terminal 15 or 30 amino acids as well as mutation of a conserved glutamic acid (Glu7 in GRK1 and Glu5 in GRK2) effectively inhibited rhodopsin phosphorylation by the GRK without affecting peptide phosphorylation. Importantly, the GRK1 mutants retained their ability to translocate from the cytosol to rod outer segments upon light activation suggesting that they could still bind to rhodopsin. A role for the N-terminal region was also implicated in GRK5 with a highly conserved Leu (Leu3 in GRK5) as well as Thr10 being found important in receptor phosphorylation [74]. Interestingly, these studies also found that an N-terminal peptide from GRK5 (residues 1-14) effectively inhibited GRK5-mediated phosphorylation of rhodopsin while having no effect on the phosphorylation of tubulin by GRK5 or rhodopsin by GRK2. It was proposed that this far N-terminal domain was an amphipathic α-helix that plays an important role in phospholipid binding. Additional studies implicating an important role for this N-terminal α-helix (αN-helix) in receptor phosphorylation was provided by Pao et al. who found that individual mutation of various N-terminal residues in GRK2 (Asp3, Leu4, Leu7/Leu8 and Asp10) effectively disrupted receptor phosphorylation [48]. In addition, a peptide containing the N-terminal 14 residues from GRK2 could form an amphipathic α-helix that selectively inhibited GRK2-mediated phosphorylation of rhodopsin in a non-competitive manner.
Structural studies of GRK1 [42] and GRK6 [41] also provided important insight into the αN-helix. These studies were the first to visualize this region in the crystal structures and, in GRK6, the αN-helix appears to bridge the active site tether of the kinase C-tail and the kinase small lobe and stabilize closure of the kinase domain [41]. A particularly important residue in the αN-helix/kinase interface was Arg190 which makes direct contact with residues in the αN-helix and kinase C-tail. Indeed, the equivalent residue was shown to be important in GRK1, GRK2 and GRK6 function [75,76]. In addition, mutagenesis of the αN-helix in GRK6 identified a number of residues particularly important in receptor phosphorylation including Ile6, Val7, Asn9, and Leu12 [41]. Similarly, Leu6, Glu7, Val9, Val10, Asn12 and Phe15 were found to be critical in mediating rhodopsin phosphorylation by GRK1 [77]. An additional study revealed a close correlation between functionally important residues within the αN-helix of C. elegans GRK-2 (a mammalian GRK2 ortholog) with its in vivo functional role in chemosensory signaling [78]. While it is clear that the GRK αN-helix plays an essential role in mediating GPCR phosphorylation, it is currently unclear whether this region is directly involved in receptor binding or serves as a switch to facilitate receptor phosphorylation.
Additional GRK regions involved in GPCR interaction and/or phosphorylation have also been identified. For example, a proline rich motif just before the nucleotide gate in GRK2 appears to be involved in GPCR binding [79,80]. Multiple studies have also implicated an important role for the RH domain in GPCR interaction. For example, GRK2 RH domain constructs containing residues 1-190 or 45-185 were able to co-immunoprecipitate with the mGluR1a [81] while Asp527 in the RH domain α11 helix was found to play an essential role in GRK2 binding to the mGluR1a [82]. Evolutionary trace analysis and mutagenesis was also used to show an important role for the α3, α9 and α10 helices of the RH domains in GRK5 and GRK6 mediated phosphorylation of the β2AR [83]. Mutation significantly inhibited GPCR phosphorylation though additional studies demonstrated substantial reduction not only of receptor phosphorylation but also GRK5 autophosphorylation and tubulin phosphorylation by α3, α9 and α10 mutants. This might suggest an allosteric effect of these RH domain mutations on the catalytic properties of the kinase domain. Interestingly, peptides from the α3, α9 and α10 helices of GRK5 were also able to inhibit the phosphorylation of rhodopsin by GRK5 with varied specificity for GRK2, GRK6 and GRK7 [84]. There are also a few noteworthy studies that provided a comprehensive mutagenesis analysis of GRKs [76,85]. For GRK2, studies from the Tesmer and Sterne-Marr labs dissected regions involved in kinase activation and GPCR phosphorylation including the αN-helix and the kinase domain AST. The important role of the αN-helix in mediating GPCR phosphorylation has been noted in a number of studies as described above and, here, the authors proposed that residues Leu4, Val7, Leu8, Val11 and Ser12 directly interact with GPCRs while Asp10, Tyr13, Ala16 and Met17 contribute to closure and activation of the kinase domain [85]. Gly475, Val477 and Ile485 in the AST were also implicated in closure and activation of the kinase domain. Taken together, it is evident that multiple residues within the αN-helix, the RH domain and the kinase domain play an essential role in mediating GPCR binding and/or phosphorylation.
3. A model for GRK interaction with GPCRs
Two recent studies used comprehensive integrated approaches to further define the interaction of GRKs with GPCRs. In one study, the interaction of GRK5 with the β2AR was characterized [86] while the other study focused on GRK1 interaction with rhodopsin [87]. The application of structural approaches such as X-ray crystallography and cryo-electron microscopy (cryo-EM) for characterization of GRK-GPCR interaction is limited by the low affinity, high flexibility and requirement of lipids for stable binding to occur. Thus, to help determine the β2AR-GRK5 binding interface, chemical cross-linking coupled with mass spectrometry was utilized [86]. This is a powerful method for providing low-resolution spatial information for protein complexes that are not stable enough or are too heterogeneous for crystallography [88]. The utility of this approach has been demonstrated in many studies of diverse macromolecular complexes including a cannabinoid receptor subtype 2/Gαi complex [89]. Interaction between GRK5 and the β2AR is highly sensitive to addition of agonist and acidic lipids and, under optimal conditions, both proteins appear to stay in a complex as confirmed by size exclusion chromatography. The complex was further stabilized by cross-linking, and then mass-spectrometry was employed to map positions of cross-linked residues at the β2AR-GRK5 binding interface. Three main clusters of intermolecular cross-links were observed: 1) ICL3 of β 2AR was found proximal to membrane-binding domains of GRK5; 2) ICL2 cross-linked with the RH bundle subdomain; and 3) the β2AR C-terminus bearing the sites of GRK phosphorylation cross-linked mainly with the kinase catalytic cleft [86]. Using unambiguous distance restraints derived from the cross-linking data in combination with recently developed computational methods of structural modeling and refinement, a low-resolution three-dimensional model of the β2AR-GRK5 complex was generated (Fig. 3). This model was validated by hydrogen deuterium exchange mass spectrometry (HDX-MS) analysis and suggests large conformational changes in GRK5 upon binding to the β2AR that result in disruption of a transient electrostatic contact between the RH and catalytic domains (ionic lock, Fig. 2) and closure of the catalytic site [86].
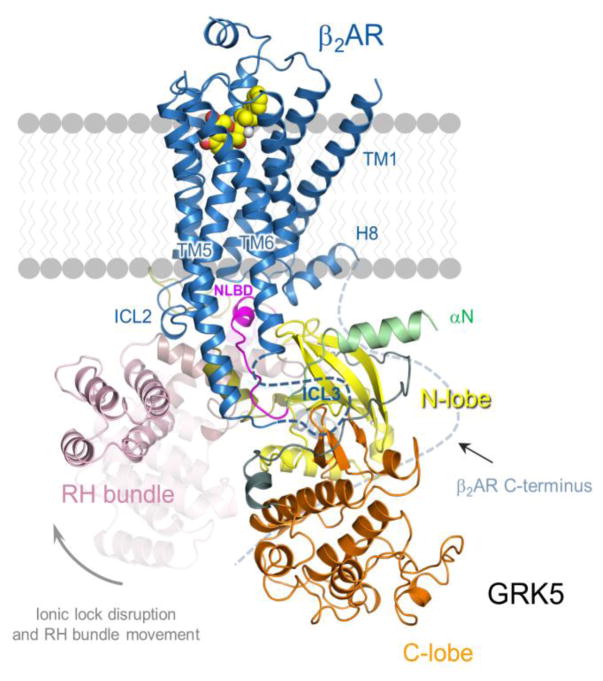
Figure 3. A model for GRK5 interaction with the β2AR.
Structural constraints derived from mass-spectrometry analysis of the cross-linked β2AR-GRK5 complex were applied to crystal structure of the β2AR in complex with Gs (PDB ID 3SN6) and elongated conformation of GRK5 determined by MD simulations of ionic lock disruption in sangivamycin-bound GRK5 (PDB ID 4TNB). The docking model of β2AR-GRK5 complex was refined with full flexibility of protein backbone and residue side chains. This model was further evaluated and validated by HDX-MS studies revealing binding interface based on reduced rate of deuterium uptake of protein regions in the complex as compared to free protein. Accordingly to our model, ICL2 of β2AR is aligned against RH bundle subdomain of GRK5 while ICL1/helix 8 of β2AR is aligned against N-lobe and NLBD of GRK5. The C-terminus of β2AR is shown schematically and is aligned against the kinase catalytic cleft.
A number of important observations became evident from visual inspection of the β2AR-GRK5 model. For example, these results support a role for ICL3 in GRK binding as shown in earlier studies [61,62,64,66]. Interestingly, two lipid-binding domains in GRK5 were found in close proximity to ICL3 by approaching it from internal (N-terminal lipid binding domain, NLBD) and external (C-terminal lipid binding domain, CLBD) sides of TM5 and TM6. This enables the NLBD to occupy the cytoplasmic core of the receptor previously found to accommodate the C-terminal α-helix of Gs and the finger loop of arrestin, important binding determinants of β2AR-Gs [90] and rhodopsin-arrestin complexes [91]. This places the NLBD in the center of the β2AR-GRK5 binding interface. It was also noticed that positioning of GRK5 lipid-binding domains near receptor loops doesn’t preclude their interaction with acidic phospholipids. While the CLBD might be anchored on acidic lipids of the plasma membrane inner leaflet in the vicinity of the GPCR cytoplasmic pocket, binding of the NLBD within predominantly hydrophobic receptor core opens up an interesting interplay between allosteric property of acidic lipids to facilitate receptor activation and GRK5 binding to receptor. In this regard, acidic lipids have recently emerged as direct positive modulators of GPCR activity [92,93]. For instance, phosphatidylglycerol (PG) markedly favored agonist binding and facilitated β2AR activation in high-density lipoparticles [92]. PG can prolong activated state of the receptor presumably via entering the cytosolic transmembrane (TM) pocket of β2AR between TM6 and TM7 [93] thereby preventing TM6 shifting back to its position near TM3 characteristic of the inactive conformation of the receptor. It’s possible that filling the cytoplasmic pocket of the β2AR with an acidic lipid (PG or PIP2) following receptor activation can also facilitate GRK5 binding to the receptor without perturbation of GRK5 basal contacts with phospholipids. Thus, the GRK5 binding pose in the model ensures that GRK5 interactions with the phospholipid bilayer and receptor are not mutually exclusive but rather complementary. Indeed, it’s energetically more favorable to maintain association with phospholipids to enable receptor contact. Cooperation of GRK5 membrane and receptor binding for the formation of the stable complex with β2AR is in agreement with the high rate of β2AR phosphorylation in the presence of both acidic lipids and agonist, whereas agonist alone isn’t capable of effectively driving complex formation.
Another aspect of the β2AR-GRK5 docking model is the orientation of the kinase domain with respect to the β2AR. It occupies a space between the β2AR ICL3 and helix 8/C-terminus to enable the kinase catalytic cleft to be in close proximity to phosphorylation sites on the receptor C-terminus. Moreover, the structural proximity of ICL3 to the GRK5 active site likely helps to account for ICL3 phosphorylation by GRKs observed for some GPCRs [63]. The relative position of the GRK5 N-lobe in the vicinity of helix 8 of the β2AR appears to assist in proper orientation of the GRK5 catalytic domain with respect to the C-terminus of β2AR. This is in agreement with HDX-MS data showing that the complex formation is followed by reduced deuterium uptake of GRK5 N-lobe regions that appear to contact helix 8 in the model [86]. Helix 8 also formed intermolecular contacts with the arrestin finger loop in the crystal structure of rhodopsin and arrestin [91]. Moreover, cryo-EM structure of the calcitonin receptor in complex with Gs demonstrated a contribution of helix 8 to Gs coupling [94]. GRK5 might employ similar sites to interact with the β2AR.
Among the regions involved in β2AR-GRK5 interaction, the surprising role of the RH bundle subdomain of GRK5 was further elucidated. The RH bundle subdomain appears to translocate to ICL2 of the β2AR following large conformational changes in GRK5 triggered by receptor binding. This involves disruption of a network of interactions that establish electrostatic contact between hydrophilic residues of the RH bundle subdomain and kinase domain C-lobe (the ionic lock), followed by RH/kinase domain dissociation. Repositioning of the RH bundle near ICL2 and phospholipid surface requires high plasticity of the enzyme and facilitates complex assembly. High plasticity of GRK5 was initially observed in molecular dynamics (MD) simulations of the ionic lock disruption in GRK5 and confirmed by EM imaging of GRK5 [86]. Transition of GRK5 from a compact “crystallographic” form into an elongated conformation in response to receptor binding might contribute to the increase of GRK5 affinity for β2AR via an induced-fit mechanism where enzyme elongation can help to involve distant regions of GRK5 for β2AR binding thereby increasing the contact surface area between the two proteins. Blocking these conformational changes in GRK5 by placing a disulfide bond between the two domains abolished the ability of GRK5 to phosphorylate GPCR emphasizing the essential role of GRK5 conformational dynamics for GPCR phosphorylation [86].
The high interdomain flexibility of GRK5 observed in MD simulations and EM is partly reminiscent of the domain dynamics in other modular kinases like Src and Abl, which adopt an elongated conformation upon activation [95,96]. While interdomain dynamics in Src are regulated by the phosphorylation state of a specific Tyr residue at the C-terminus of Src, the ionic lock might play a similar role in GRKs, although this seems to be a more transient interaction than that found in Src and Abl. Destabilization of the ionic lock by mutagenesis only moderately enhances kinase activity with an ~60% increase in catalytic efficiency for ATP suggesting that the ionic lock inhibits the basal activity of GRK5 only to some extent. Conversely, stabilization of the ionic lock in GRK5 by an engineered disulfide bond abrogates kinase activation although it can be restored by disruption of the disulfide bond [86]. This resembles the activation mechanism of Src when phosphorylation/dephosphorylation of a specific C-terminal Tyr can switch between inactive and active kinase. In contrast, activation of wild-type GRK5 requires less energy for disruption of relatively weak and transient contact and therefore, wild-type GRK5 is more prone to activation in its basal state. This might be important for GRK function in cells since GRKs also contribute to phosphorylation of non-GPCR substrates in the cytoplasm and nucleus [30,31].
A recent study by He et al. used a genetic approach combined with biochemical assays to identify some key determinants of GRK1 assembly with rhodopsin [87]. To monitor rhodopsin-GRK1 interaction, a proximity-based Tango assay was used. This assay was based on cleavage of a transcriptional activator fused to the C-terminus of rhodopsin by a TEV protease fused to the N-terminus of GRK1. When the two proteins interact, the TEV protease cleaves off the transcriptional activator which enters the nucleus and activates reporter gene transcription. This study revealed that the RH domain of GRK1 is the primary binding site for rhodopsin with critically important residues being mapped on the α3, α9, α10 and the αN-α1 loop of the RH domain. These data were validated using in vitro biochemical assays and partly supported by HDX-MS studies. Interestingly, the receptor interface mapped by the Tango assay correlates well with the functionally important sites identified previously using evolutionary trace analysis [83]. However, this interface is partially capped by packing of the αN-α1 loop and C-terminus on α9 and α3, and therefore, these regions must be displaced to accommodate GPCRs. He et al. also mapped two regions on GRK1 with reduced deuterium uptake in complex with rhodopsin by HDX-MS analysis [87]. In line with the Tango assay data, α9-helix showed decreased HDX while protection of α6-helix of RH bundle subdomain didn’t correlate with results of the Tango assay. Nevertheless, protection of α6-helix is in agreement with RH bundle domain translocation, which plays a role in β2AR-GRK5 interaction [86]. While HDX-MS analysis of β2AR-GRK5 [86] revealed a broader spectrum of protected regions, the rate of HDX decrease captured in β2AR-GRK5 and rhodopsin-GRK1 studies was still markedly lower than observed for β2AR-Gs interaction [97], perhaps reflecting the lower affinity of GPCR-GRK binding as compared to GPCR-Gs coupling.
As previously discussed, the GRK αN-helix was predicted to stabilize the active state of GRKs and to selectively recognize the activated state of a GPCR [41,77]. The αN-helix is accommodated between the N-lobe and RH terminal subdomain in a GRK1/ATP crystal structure [77] whereas it bridges the N- and C-lobes of the kinase domain in a GRK6/sangivamycin structure and contributes to GRK6 activation [41]. This suggests two potential scenarios of GRK docking on a GPCR, if the αN-helix serves as a central element of GPCR-GRK architecture. While we don’t have reliable data regarding the engagement of the GRK5 αN-helix in receptor binding, deuterium uptake of peptide 4–10 derived from the αN-helix of GRK5 was variable preventing unambiguous HDX-MS analysis of its structural dynamics [86]. In addition, truncation of the N-terminal 30 residues didn’t alter the ability of GRK1 to interact with rhodopsin in a Tango assay monitoring direct protein-protein binding [87]. Consistent with this result, an N-terminal 1–30 peptide from GRK1 didn’t interact with receptor in the Tango assay. Since effective GRK phosphorylation of GPCRs depends on the ability of GRKs to interact with phospholipids, dock on receptors and undergo activation, perhaps the αN-helix contributes to all of these processes.
In summary, recent studies of the binding interface and dynamics of GPCR-GRK interaction provide new insight into the role of the RH domain. The RH domain is a distinct element of GRKs that distinguishes them from other protein kinase families. The RH domain can serve as a docking site for GPCRs and helps to control kinase activation via transient contact of the RH bundle and kinase C-lobe subdomains (the ionic lock). The mechanism of ionic lock disruption is dynamically regulated by GPCR binding which provides a molecular basis for GPCR-stimulated activation of GRKs. Together, these studies provide a mechanistic link between GPCR interaction and GRK activation and highlight an important role of the RH domain in this process.
4. Future directions
We have gained significant insight on the structure of GRKs, structure/function analysis of GRK interaction with GPCRs as well as initial insight on how GRK binding to a GPCR results in kinase activation and GPCR phosphorylation. We also know that GRKs are good targets in disease with studies from the Tesmer and Koch groups being particularly insightful on strategies to inhibit GRK2 in cardiovascular disease [31,44,98]. So where do we go from here? Since it is evident that GRKs play a central role in mediating the switch from GPCR interaction with G proteins to GPCR interaction with arrestins, it is critical that we better understand how GRKs target GPCRs and determine the specificity of GRK-GPCR interaction and how interaction mediates the phosphorylation of specific residues on the receptor. Such studies require further investigation of the structures of GPCR-GRK complexes using approaches such as X-ray crystallography, cryo-EM and nuclear magnetic resonance. It will also be important to further understand the conformational dynamics of these interactions using techniques such as double electron-electron resonance spectroscopy, fluorescence resonance energy transfer analysis or similar approaches along with computational tools (MD simulations). These lines of investigation should ultimately enable a better understanding of GPCR regulation by GRKs.
Highlights.
GRKs mediate activation-dependent phosphorylation of GPCRs
GRKs play a central role in switching GPCR signaling from G proteins to arrestins
GRKs bind to multiple intracellular regions of the GPCR
GRK binding promotes significant conformational changes that activate GRK activity
Acknowledgments
JLB would like to thank Bob Lefkowitz for providing a superb training environment and for his support over the years that fostered my scientific development and helped to propel my career as an independent investigator. JLB would also like to thank the many past and present pre-doctoral and postdoctoral fellows who have trained in my lab for their many contributions in dissecting the mechanisms involved in GPCR signaling. This work was supported in part by National Institutes of Health grants R01 GM044944, R01 GM047417, R01 HL136219 and P01 HL114471.
Abbreviations
- αN-helix
N-terminal α-helical domain
- α2AAR
α2A-adrenergic receptor
- AST
active site tether
- βAR
β-adrenergic receptor
- β2AR
β2-adrenergic receptor
- βARK
β-adrenergic receptor kinase
- CLBD
C-terminal lipid binding domain
- EM
electron microscopy
- GPCR
G protein-coupled receptor
- GRK
G protein-coupled receptor kinase
- GST
glutathione-S-transferase
- HDX-MS
hydrogen deuterium exchange mass spectrometry
- ICL
intracellular loop
- MD
molecular dynamics
- mGluR1
metabotropic glutamate receptor 1
- NLBD
N-terminal lipid binding domain
- PG
phosphatidylglycerol
- PH
pleckstrin homology
- PKA
cAMP dependent protein kinase
- RH
Regulator of G Protein Signaling Homology
- TM
transmembrane
Footnotes
Conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kühn H, Dreyer WJ. Light dependent phosphorylation of rhodopsin by ATP. FEBS Lett. 1972;20:1–6. doi: 10.1016/0014-5793(72)80002-4. [DOI] [PubMed] [Google Scholar]
- 2.Shichi H, Somers RL. Light-dependent phosphorylation of rhodopsin. Purification and properties of rhodopsin kinase. J Biol Chem. 1978;253:7040–7046. [PubMed] [Google Scholar]
- 3.Stadel JM, Nambi P, Lavin TN, Heald SL, Caron MG, Lefkowitz RJ. Catecholamine-induced desensitization of turkey erythrocyte adenylate cyclase. Structural alterations in the beta-adrenergic receptor revealed by photoaffinity labeling. J Biol Chem. 1982;257:9242–9245. [PubMed] [Google Scholar]
- 4.Stadel JM, Nambi P, Shorr RG, Sawyer DF, Caron MG, Lefkowitz RJ. Catecholamine-induced desensitization of turkey erythrocyte adenylate cyclase is associated with phosphorylation of the beta-adrenergic receptor. Proc Natl Acad Sci U S A. 1983;80:3173–3177. doi: 10.1073/pnas.80.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sibley DR, Peters JR, Nambi P, Caron MG, Lefkowitz RJ. Desensitization of turkey erythrocyte adenylate cyclase. Beta-adrenergic receptor phosphorylation is correlated with attenuation of adenylate cyclase activity. J Biol Chem. 1984;259:9742–9749. [PubMed] [Google Scholar]
- 6.Benovic JL, Pike LJ, Cerione RA, Staniszewski C, Yoshimasa T, Codina J, Birnbaumer L, Caron MG, Lefkowitz RJ. Phosphorylation of the mammalian beta-adrenergic receptor by cyclic AMP-dependent protein kinase: Regulation of the rate of receptor phosphorylation and dephosphorylation by agonist occupancy and effects on coupling of the receptor to the stimulatory guanine nucleotide regulatory protein. J Biol Chem. 1985;260:7094–7101. [PubMed] [Google Scholar]
- 7.Strasser RH, Sibley DR, Lefkowitz RJ. A novel catecholamine-activated cyclic 3′,5′-phosphate independent pathway for beta-adrenergic receptor phosphorylation in wild-type and mutant S49 lymphoma cells: mechanism of homologous desensitization of adenylate cyclase. Biochemistry. 1986;25:1371–1377. doi: 10.1021/bi00354a027. [DOI] [PubMed] [Google Scholar]
- 8.Benovic JL, Strasser RH, Caron MG, Lefkowitz RJ. Beta-adrenergic receptor kinase: Identification of a novel protein kinase which phosphorylates the agonist-occupied form of the receptor. Proc Natl Acad Sci USA. 1986;83:2797–2801. doi: 10.1073/pnas.83.9.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefkowitz RJ, Benovic JL, Kobilka BK, Caron MG. Receptors and rhodopsin: Shedding new light on an old subject. Trends Pharmacol Sci. 1986;7:444–448. [Google Scholar]
- 10.Benovic JL, Mayor F, Jr, Somers RL, Caron MG, Lefkowitz RJ. Light- dependent phosphorylation of rhodopsin by beta-adrenergic receptor kinase. Nature. 1986;322:867–872. doi: 10.1038/321869a0. [DOI] [PubMed] [Google Scholar]
- 11.Strasser RH, Benovic JL, Caron MG, Lefkowitz RJ. Beta-agonist- and prostaglandin E1-induced translocation of the beta-adrenergic receptor kinase: Evidence that the kinase may act on multiple adenylate cyclase-coupled receptors. Proc Natl Acad Sci USA. 1986;83:6362–6366. doi: 10.1073/pnas.83.17.6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayor F, Jr, Benovic JL, Caron MG, Lefkowitz RJ. Somatostatin induces translocation of the beta-adrenergic receptor kinase and desensitizes somatostatin receptors in S49 lymphoma cells. J Biol Chem. 1987;262:6468–6471. [PubMed] [Google Scholar]
- 13.Benovic JL, Regan JR, Matsui H, Mayor F, Jr, Cotecchia S, Leeb-Lundberg LMF, Caron MG, Lefkowitz RJ. Agonist-dependent phosphorylation of the alpha-2-adrenergic receptor by the beta-adrenergic receptor kinase. J Biol Chem. 1987;262:17251–17253. [PubMed] [Google Scholar]
- 14.Benovic JL, Mayor F, Jr, Staniszewski C, Lefkowitz RJ, Caron MG. Purification and characterization of the beta-adrenergic receptor kinase. J Biol Chem. 1987;262:9026–9032. [PubMed] [Google Scholar]
- 15.Benovic JL, DeBlasi A, Stone WC, Caron MG, Lefkowitz RJ. Primary structure of the beta-adrenergic receptor kinase delineates a potential multigene family of receptor specific kinases. Science. 1989;246:235–240. doi: 10.1126/science.2552582. [DOI] [PubMed] [Google Scholar]
- 16.Pfister C, Chabre M, Plouet J, Tuyen VV, De Kozak Y, Faure JP, Kühn H. Retinal S antigen identified as the 48K protein regulating light-dependent phosphodiesterase in rods. Science. 1985;228:891–893. doi: 10.1126/science.2988124. [DOI] [PubMed] [Google Scholar]
- 17.Wilden U, Wüst E, Weyand I, Kühn H. Rapid affinity purification of retinal arrestin (48 kDa protein) via its light-dependent binding to phosphorylated rhodopsin. FEBS Lett. 1986;207:292–295. doi: 10.1016/0014-5793(86)81507-1. [DOI] [PubMed] [Google Scholar]
- 18.Benovic JL, Kuhn H, Weyand I, Codina J, Caron MG, Lefkowitz RJ. Functional desensitization of the isolated beta-adrenergic receptor by the β-adrenergic receptor kinase: Potential role of an analog of the retinal protein arrestin (48 kDa protein) Proc Natl Acad Sci USA. 1987;84:8879–8882. doi: 10.1073/pnas.84.24.8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. Beta-arrestin: A protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 20.Benovic JL, Onorato JJ, Arriza JL, Stone WC, Lohse M, Jenkins N, Gilbert DJ, Copeland NG, Caron MG, Lefkowitz RJ. Cloning, expression and chromosomal localization of beta-adrenergic receptor kinase 2: A new member of the receptor kinase family. J Biol Chem. 1991;266:14939–14946. [PubMed] [Google Scholar]
- 21.Lorenz W, Inglese J, Palczewski K, Onorato JJ, Caron MG, Lefkowitz RJ. The receptor kinase family: primary structure of rhodopsin kinase reveals similarities to the beta-adrenergic receptor kinase. Proc Natl Acad Sci U S A. 1991;88:8715–8719. doi: 10.1073/pnas.88.19.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambrose C, James M, Barnes G, Lin C, Bates G, Altherr M, Duyao M, Groot N, Church D, Wasmuth JJ, Lehrach H, Housman D. A novel G protein-coupled receptor kinase gene cloned from 4p16.3. Hum Mol Genet. 1992;1:697–703. doi: 10.1093/hmg/1.9.697. [DOI] [PubMed] [Google Scholar]
- 23.Kunapuli P, Benovic JL. Cloning and expression of GRK5: A member of the G protein-coupled receptor kinase family. Proc Natl Acad Sci USA. 1993;90:5588–5592. doi: 10.1073/pnas.90.12.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benovic JL, Gomez J. Molecular cloning and expression of GRK6: A new member of the G protein-coupled receptor kinase family. J Biol Chem. 1993;268:19521–19527. [PubMed] [Google Scholar]
- 25.Hisatomi O, Matsuda S, Satoh T, Kotaka S, Imanishi Y, Tokunaga F. A novel subtype of G-protein-coupled receptor kinase, GRK7, in teleost cone photoreceptors. FEBS Lett. 1998;424:159–164. doi: 10.1016/s0014-5793(98)00162-8. [DOI] [PubMed] [Google Scholar]
- 26.Weiss ER, Raman D, Shirakawa S, Ducceschi MH, Bertram PT, Wong F, Kraft TW, Osawa S. The cloning of GRK7, a candidate cone opsin kinase, from cone- and rod-dominant mammalian retinas. Mol Vis. 1998;4:27. [PubMed] [Google Scholar]
- 27.Pitcher JA, Freedman NJ, Lefkowitz RJ. G protein-coupled receptor kinases. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 28.Krupnick JG, Benovic JL. The role of receptor kinases and arrestins in G protein- coupled receptor regulation. Annu Rev Pharmacol Toxicol. 1998;38:289–319. doi: 10.1146/annurev.pharmtox.38.1.289. [DOI] [PubMed] [Google Scholar]
- 29.Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinases and arrestins. Annu Rev Physiol. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- 30.Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hullmann J, Traynham CJ, Coleman RC, Koch WJ. The expanding GRK interactome: Implications in cardiovascular disease and potential for therapeutic development. Pharmacol Res. 2016;110:52–64. doi: 10.1016/j.phrs.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black JB, Premont RT, Daaka Y. Feedback regulation of G protein-coupled receptor signaling by GRKs and arrestins. Semin Cell Dev Biol. 2016;50:95–104. doi: 10.1016/j.semcdb.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nogués L, Reglero C, Rivas V, Neves M, Penela P, Mayor F., Jr G-protein- coupled receptor kinase 2 as a potential modulator of the hallmarks of cancer. Mol Pharmacol. 2017;91:220–228. doi: 10.1124/mol.116.107185. [DOI] [PubMed] [Google Scholar]
- 34.Siderovski DP, Hessel A, Chung S, Mak TW, Tyers M. A new family of regulators of G-protein-coupled receptors? Curr Biol. 1996;6:211–212. doi: 10.1016/s0960-9822(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 35.Carman CV, Parent J-L, Day PW, Pronin AN, Sternweis PC, Wedegaertner PB, Gilman AG, Benovic JL, Kozasa T. Selective regulation of Gq/11α by an RGS domain in the G protein-coupled receptor kinase, GRK2. J Biol Chem. 1999;274:34483–34492. doi: 10.1074/jbc.274.48.34483. [DOI] [PubMed] [Google Scholar]
- 36.Homan KT, Tesmer JJ. Structural insights into G protein-coupled receptor kinase function. Curr Opin Cell Biol. 2014;27:25–31. doi: 10.1016/j.ceb.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mushegian A, Gurevich VV, Gurevich EV. The origin and evolution of G protein- coupled receptor kinases. PLoS One. 2012;7:e33806. doi: 10.1371/journal.pone.0033806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJG. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gβγ. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 39.Lodowski DT, Tesmer VM, Benovic JL, Tesmer JJG. Crystal structure of G protein-coupled receptor kinase 6 defines the conserved molecular features of the GRK family. J Biol Chem. 2006;281:16785–16793. doi: 10.1074/jbc.M601327200. [DOI] [PubMed] [Google Scholar]
- 40.Xu H, Jiang X, Shen K, Fischer CC, Wedegaertner PB. The regulator of G protein signaling (RGS) domain of G protein-coupled receptor kinase 5 (GRK5) regulates plasma membrane localization and function. Mol Biol Cell. 2014;25:2105–2115. doi: 10.1091/mbc.E13-09-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boguth CA, Singh P, Huang CC, Tesmer JJ. Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 2010;29:3249–3259. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJ. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J Biol Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komolov KE, Bhardwaj A, Benovic JL. Atomic structure of GRK5 reveals distinct structural features novel for G protein-coupled receptor kinases. J Biol Chem. 2015;290:20629–20647. doi: 10.1074/jbc.M115.647297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Homan KT, Waldschmidt HV, Glukhova A, Cannavo A, Song J, Cheung JY, Koch WJ, Larsen SD, Tesmer JJG. Crystal structure of G protein-coupled receptor kinase 5 in complex with a rationally designed inhibitor. J Biol Chem. 2015;290:20649–20659. doi: 10.1074/jbc.M115.647370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen SJ, Parthasarathy G, Darke PL, Diehl RE, Ford RE, Hall DL, Johnson SA, Reid JC, Rickert KW, Shipman JM, Soisson SM, Zuck P, Munshi SK, Lumb KJ. Structure and function of the hypertension variant A486V of G protein-coupled receptor kinase 4. J Biol Chem. 2015;290:23060–23073. doi: 10.1074/jbc.M115.648907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lodowski DT, Barnhill JF, Pyskadlo RM, Ghirlando R, Sterne-Marr R, Tesmer JJ. The role of Gβγ and domain interfaces in the activation of G protein-coupled receptor kinase 2. Biochemistry. 2005;44:6958–6970. doi: 10.1021/bi050119q. [DOI] [PubMed] [Google Scholar]
- 47.Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Snapshot of activated G proteins at the membrane: the Gαq-GRK2-Gβγ complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- 48.Pao CS, Barker B, Benovic JL. Role of the amino terminus of G protein-coupled receptor kinase 2 in receptor phosphorylation. Biochemistry. 2009;48:7325–7333. doi: 10.1021/bi900408g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onorato J, Palczewski K, Regan JW, Caron MG, Lefkowitz RJ, Benovic JL. Role of acidic amino acids in peptide substrates of the beta-adrenergic receptor kinase and rhodopsin kinase. Biochemistry. 1991;30:5118–5125. doi: 10.1021/bi00235a002. [DOI] [PubMed] [Google Scholar]
- 50.Kunapuli P, Onorato JJ, Hosey MM, Benovic JL. Expression, purification and characterization of the G protein-coupled receptor kinase GRK5. J Biol Chem. 1994;269:1099–1105. [PubMed] [Google Scholar]
- 51.Loudon RP, Benovic JL. Expression, purification, and characterization of the G protein-coupled receptor kinase GRK6. J Biol Chem. 1994;269:22691–22697. [PubMed] [Google Scholar]
- 52.Benovic JL, Onorato J, Lohse MJ, Dohlman HG, Staniszewski C, Caron MG, Lefkowitz RJ. Synthetic peptides of the hamster beta-2-adrenergic receptor as substrates and inhibitors of the beta-adrenergic receptor kinase. Br J Clin Pharmacol. 1990;30:3S–12S. doi: 10.1111/j.1365-2125.1990.tb05462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim CM, Dion SB, Onorato JJ, Benovic JL. Expression and characterization of two beta-adrenergic receptor kinase isoforms using the baculovirus expression system. Receptor. 1993;3:39–55. [PubMed] [Google Scholar]
- 54.Asai D, Toita R, Murata M, Katayama Y, Nakashima H, Kang JH. Peptide substrates for G protein-coupled receptor kinase 2. FEBS Lett. 2014;588:2129–2132. doi: 10.1016/j.febslet.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 55.Asai D, Murata M, Toita R, Kawano T, Nakashima H, Kang JH. Role of amino acid residues surrounding the phosphorylation site in peptide substrates for G protein-coupled receptor kinase 2 (GRK2) Amino Acids. 2016;48:2875–2880. doi: 10.1007/s00726-016-2345-6. [DOI] [PubMed] [Google Scholar]
- 56.Palczewski K, Buczyłko J, Kaplan MW, Polans AS, Crabb JW. Mechanism of rhodopsin kinase activation. J Biol Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 57.Chen CY, Dion SB, Kim CM, Benovic JL. Beta-adrenergic receptor kinase: Agonist-dependent receptor binding promotes kinase activation. J Biol Chem. 1993;268:7825–7831. [PubMed] [Google Scholar]
- 58.Winstel R, Ihlenfeldt HG, Jung G, Krasel C, Lohse MJ. Peptide inhibitors of G protein-coupled receptor kinases. Biochem Pharmacol. 2005;70:1001–1008. doi: 10.1016/j.bcp.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Carr R, III, Benovic JL. From biased signaling to polypharmacology: unlocking unique intracellular signaling using pepducins. Biochem Soc Trans. 2016;44:555–561. doi: 10.1042/BST20150230. [DOI] [PubMed] [Google Scholar]
- 60.Carr R, III, Schilling J, Song J, Carter RL, Du Y, Yoo SM, Traynham CJ, Koch WJ, Cheung JY, Tilley DG, Benovic JL. β-arrestin-biased signaling through the β2-adrenergic receptor promotes cardiomyocyte contraction. Proc Natl Acad Sci USA. 2016;113:E4107–4116. doi: 10.1073/pnas.1606267113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi W, Osawa S, Dickerson CD, Weiss ER. Rhodopsin mutants discriminate sites important for the activation of rhodopsin kinase and Gt. J Biol Chem. 1995;270:2112–2119. doi: 10.1074/jbc.270.5.2112. [DOI] [PubMed] [Google Scholar]
- 62.Thurmond RL, Creuzenet C, Reeves PJ, Khorana HG. Structure and function in rhodopsin: peptide sequences in the cytoplasmic loops of rhodopsin are intimately involved in interaction with rhodopsin kinase. Proc Natl Acad Sci U S A. 1997;94:1715–1720. doi: 10.1073/pnas.94.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eason MG, Moreira SP, Liggett SB. Four consecutive serines in the third intracellular loop are the sites for beta-adrenergic receptor kinase-mediated phosphorylation and desensitization of the alpha 2A-adrenergic receptor. J Biol Chem. 1995;270:4681–4688. doi: 10.1074/jbc.270.9.4681. [DOI] [PubMed] [Google Scholar]
- 64.Pao CS, Benovic JL. Structure/function analysis of α2A-adrenergic receptor interaction with G protein-coupled receptor kinase 2. J Biol Chem. 2005;280:11052–11058. doi: 10.1074/jbc.M412996200. [DOI] [PubMed] [Google Scholar]
- 65.Dhami GK, Babwah AV, Sterne-Marr R, Ferguson SS. Phosphorylation-independent regulation of metabotropic glutamate receptor 1 signaling requires g protein-coupled receptor kinase 2 binding to the second intracellular loop. J Biol Chem. 2005;280:24420–24427. doi: 10.1074/jbc.M501650200. [DOI] [PubMed] [Google Scholar]
- 66.Jones Brunette AM, Sinha A, Farrens DL. Evidence that the rhodopsin kinase (GRK1) N-terminus and the transducin Gα C-terminus interact with the same “hydrophobic patch” on rhodopsin TM5. Biochemistry. 2016;55:3123–3135. doi: 10.1021/acs.biochem.6b00328. [DOI] [PubMed] [Google Scholar]
- 67.Ohguro H, Van Hooser JP, Milam AH, Palczewski K. Rhodopsin phosphorylation and dephosphorylation in vivo. J Biol Chem. 270:14259–14262. doi: 10.1074/jbc.270.24.14259. 270. [DOI] [PubMed] [Google Scholar]
- 68.Kong G, Penn R, Benovic JL. A beta-adrenergic receptor kinase dominant negative mutant slows desensitization of the beta-2-adrenergic receptor. J Biol Chem. 1994;269:13084–13087. [PubMed] [Google Scholar]
- 69.Palczewski K, Buczyłko J, Lebioda L, Crabb JW, Polans AS. Identification of the N-terminal region in rhodopsin kinase involved in its interaction with rhodopsin. J Biol Chem. 1993;268:6004–6013. [PubMed] [Google Scholar]
- 70.Gorodovikova EN, Senin II, Philippov PP. Calcium-sensitive control of rhodopsin phosphorylation in the reconstituted system consisting of photoreceptor membranes, rhodopsin kinase and recoverin. FEBS Lett. 1994;353:171–2. doi: 10.1016/0014-5793(94)01030-7. [DOI] [PubMed] [Google Scholar]
- 71.Chen CK, Inglese J, Lefkowitz RJ, Hurley JB. Ca2+-dependent interaction of recoverin with rhodopsin kinase. J Biol Chem. 1995;270:18060–18066. doi: 10.1074/jbc.270.30.18060. [DOI] [PubMed] [Google Scholar]
- 72.Levay K, Satpaev DK, Pronin AN, Benovic JL, Slepak VZ. Localization of the sites for Ca2+-binding proteins on G protein-coupled receptor kinases. Biochemistry. 1998;37:13650–13659. doi: 10.1021/bi980998z. [DOI] [PubMed] [Google Scholar]
- 73.Yu QM, Cheng ZJ, Gan XQ, Bao GB, Li L, Pei G. The amino terminus with a conserved glutamic acid of G protein-coupled receptor kinases is indispensable for their ability to phosphorylate photoactivated rhodopsin. J Neurochem. 1999;73:1222–1227. doi: 10.1046/j.1471-4159.1999.0731222.x. [DOI] [PubMed] [Google Scholar]
- 74.Noble B, Kallal LA, Pausch MH, Benovic JL. Development of a yeast bioassay to characterize G protein-coupled receptor kinases: Identification of an N-terminal region essential for receptor phosphorylation. J Biol Chem. 2003;278:47466–47476. doi: 10.1074/jbc.M308257200. [DOI] [PubMed] [Google Scholar]
- 75.Huang CC, Yoshino-Koh K, Tesmer JJ. A surface of the kinase domain critical for the allosteric activation of G protein-coupled receptor kinases. J Biol Chem. 2009;284:17206–17215. doi: 10.1074/jbc.M809544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sterne-Marr R, Leahey PA, Bresee JE, Dickson HM, Ho W, Ragusa MJ, Donnelly RM, Amie SM, Krywy JA, Brookins-Danz ED, Orakwue SC, Carr MJ, Yoshino-Koh K, Li Q, Tesmer JJ. GRK2 activation by receptors: role of the kinase large lobe and carboxyl-terminal tail. Biochemistry. 2009;48:4285–4293. doi: 10.1021/bi900151g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang CC, Orban T, Jastrzebska B, Palczewski K, Tesmer JJ. Activation of G protein-coupled receptor kinase 1 involves interactions between its N-terminal region and its kinase domain. Biochemistry. 2011;50:1940–1949. doi: 10.1021/bi101606e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wood JF, Wang J, Benovic JL, Ferkey DM. Structural domains required for C. elegans G protein-coupled receptor kinase 2 (GRK-2) function in vivo. J Biol Chem. 2012;287:12634–12644. doi: 10.1074/jbc.M111.336818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gan XQ, Wang JY, Yang QH, Li Z, Liu F, Pei G, Li L. Interaction between the conserved region in the C-terminal domain of GRK2 and rhodopsin is necessary for GRK2 to catalyze receptor phosphorylation. J Biol Chem. 2000;275:8469–8474. doi: 10.1074/jbc.275.12.8469. [DOI] [PubMed] [Google Scholar]
- 80.Gan X, Ma Z, Deng N, Wang J, Ding J, Li L. Involvement of the C-terminal proline-rich motif of G protein-coupled receptor kinases in recognition of activated rhodopsin. J Biol Chem. 2004;279:49741–49746. doi: 10.1074/jbc.M407570200. [DOI] [PubMed] [Google Scholar]
- 81.Dhami GK, Anborgh PH, Dale LB, Sterne-Marr R, Ferguson SS. Phosphorylation-independent regulation of metabotropic receptor signaling by G protein-coupled receptor kinase 2. J Biol Chem. 2002;277:25266–25272. doi: 10.1074/jbc.M203593200. [DOI] [PubMed] [Google Scholar]
- 82.Dhami GK, Dale LB, Anborgh PH, O’Connor-Halligan KE, Sterne-Marr R, Ferguson SS. G protein coupled receptor kinase 2 regulator of G protein signaling homology domain binds to both metabotropic glutamate receptor 1a and Gαq to attenuate signaling. J Biol Chem. 2004;279:16614–16620. doi: 10.1074/jbc.M314090200. [DOI] [PubMed] [Google Scholar]
- 83.Baameur F, Morgan DH, Yao H, Tran TM, Hammitt RA, Sabui S, McMurray JS, Lichtarge O, Clark RB. Role for the regulator of G protein signaling homology domain of G protein-coupled receptor kinases 5 and 6 in beta 2-adrenergic receptor and rhodopsin phosphorylation. Mol Pharmacol. 2010;77:405–415. doi: 10.1124/mol.109.058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baameur F, Hammitt RA, Friedman J, McMurray JS, Clark RB. Biochemical and cellular specificity of peptide inhibitors of G protein-coupled receptor kinases. Int J Pept Res Ther. 2014;20:1–12. doi: 10.1007/s10989-013-9357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beautrait A, Michalski KR, Lopez TS, Mannix KM, McDonald DJ, Cutter AR, Medina CB, Hebert AM, Francis CJ, Bouvier M, Tesmer JJ, Sterne-Marr R. Mapping the putative G protein-coupled receptor (GPCR) docking site on GPCR kinase 2: insights from intact cell phosphorylation and recruitment assays. J Biol Chem. 2014;289:25262–25275. doi: 10.1074/jbc.M114.593178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Komolov KE, Du Y, Duc NM, Betz RM, Rodrigues JPGLM, Leib RD, Patra D, Skiniotis G, Adams CM, Dror RO, Chung KY, Kobilka BK, Benovic JL. Structural and functional analysis of a β2-adrenergic receptor complex with GRK5. Cell. 2017;169:407–412. doi: 10.1016/j.cell.2017.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.He Y, Gao X, Goswami D, Hou L, Pal K, Yin Y, Zhao G, Ernst OP, Griffin P, Melcher K, Xu HE. Molecular assembly of rhodopsin with G protein-coupled receptor kinases. Cell Res. 2017;27:728–747. doi: 10.1038/cr.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rappsilber J. The beginning of a beautiful friendship: Cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mnpotra JS, Qiao Z, Cai J, Lynch DL, Grossfield A, Leioatts N, Hurst DP, Pitman MC, Song ZH, Reggio PH. Structural basis of G protein-coupled receptor-Gi protein interaction: Formation of the cannabinoid CB2 receptor-Gi protein complex. J Biol Chem. 2014;289:20259–20272. doi: 10.1074/jbc.M113.539916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah ST, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dawaliby R, Trubbia C, Delporte C, Masureel M, Van Antwerpen P, Kobilka BK, Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat Chem Biol. 2015;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neale C, Herce HD, Pomès R, Garcia A. Can specific protein-lipid interactions stabilize an active state of the beta 2 adrenergic receptor? Biophys J. 2015;109:1652–1662. doi: 10.1016/j.bpj.2015.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, Danev R, Baumeister W, Miller LJ, Christopoulos A, Kobilka BK, Wootten D, Skiniotis G, Sexton PM. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 2017;546:118–123. doi: 10.1038/nature22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bernadó P, Pérez Y, Svergun DI, Pons M. Structural characterization of the active and inactive states of Src kinase in solution by small-angle X-ray scattering. J Mol Biol. 2008;376:492–505. doi: 10.1016/j.jmb.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 96.Nagar B, Hantschel O, Seeliger M, Davies JM, Weis WI, Superti-Furga G, Kuriyan J. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 97.Chung KY, Rasmussen SGF, Liu T, Li S, DeVree BT, Chae PS, Calinski D, Kobilka BK, Woods VL, Sunahara RK. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waldschmidt HV, Homan KT, Cato MC, Cruz-Rodríguez O, Cannavo A, Wilson MW, Song J, Cheung JY, Koch WJ, Tesmer JJ, Larsen SD. Structure-based design of highly selective and potent G protein-coupled receptor kinase 2 inhibitors based on paroxitine. J Med Chem. 2017;60:3052–3069. doi: 10.1021/acs.jmedchem.7b00112. [DOI] [PMC free article] [PubMed] [Google Scholar]