Abstract
Pyruvate dehydrogenase complex (PDC) deficiency is a major cause of primary lactic acidemia in children. Prompt and correct diagnosis of PDC deficiency and differentiating between specific vs multiple, or secondary deficiencies has important implications for clinical management and therapeutic interventions. Both genetic and enzymatic testing approaches are being used in the diagnosis of PDC deficiency. However, the diagnostic efficacy of such testing approaches for individuals affected with PDC deficiency has not been systematically investigated in this disorder. We sought to evaluate the diagnostic sensitivity and variability of the various PDC enzyme assays in females and males at the Center for Inherited Disorders of Energy Metabolism (CIDEM).
CIDEM data were filtered by lactic acidosis and functional PDC deficiency in at least one cell/tissue type (blood lymphocytes, cultured fibroblasts or skeletal muscle) identifying 186 subjects (51% male and 49% female), about half were genetically resolved with 78% of those determined to have a pathogenic PDHA1 mutation. Assaying PDC in cultured fibroblasts in cases where the underlying genetic etiology is PDHA1, was highly sensitive irrespective of gender; 97% (95% confidence interval [CI]: 90%–100%) and 91% (95% CI: 82%–100%) in females and males, respectively. In contrast to the fibroblast-based testing, the lymphocyte- and muscle-based testing were not sensitive (36% [95% CI: 11%–61%, p = 0.0003] and 58% [95% CI: 30%–86%, p = 0.014], respectively) for identifying known PDC deficient females with pathogenic PDHA1 mutations. In males with a known PDHA1 mutation, the sensitivity of the various cell/tissue assays (75% lymphocyte, 91% fibroblast and 88% muscle) were not statistically different, and the discordance frequency due to the specific cell/tissue used for assaying PDC was 0.15 ± 0.11. Based on this data, a practical diagnostic algorithm is proposed accounting for current molecular approaches, enzyme testing sensitivity, and variability due to gender, cell/tissue type used, and successive repeat testing.
Keywords: Biochemical testing, sensitivity, variability, diagnostic algorithm, pyruvate dehydrogenase complex deficiency, PDHA1.
1. Introduction
The mitochondrial multi-protein pyruvate dehydrogenase complex (PDC) is the primary gateway between glycolysis and the tricarboxylic acid (TCA) cycle, catalyzing oxidative decarboxylation of pyruvate to acetyl-CoA. More than 500 cases of PDC deficiency have been reported, and many others presumably are not reported or diagnosed, so the actual incidence is unknown. This neurometabolic disorder results in significant morbidity and mortality in children. The clinical presentation of PDC deficiency is variable and ranges from fatal congenital lactic acidosis and congenital brain abnormalities to relatively mild ataxia or neuropathy with normal cognitive function and long survival [1–3]. Among subjects with mutations in known PDC genes, this clinical heterogeneity is not correlated with genotype and occurs even with identical mutations [1–3].
The majority of genetically-resolved subjects with functional PDC deficiency have mutations in the chromosome X-linked PDHA1 gene, and less commonly have mutations of the other 4 PDC-specific genes (PDHB, DLAT, PDHX, and PDP1) [1]. Investigations into the molecular etiology of about a third of genetically- unresolved PDC-deficient subjects have identified categories of several other “PDC-associated” genes (an all-inclusive term that includes the PDC-specific genes as well as other genes noted below) whose products are involved with branched-chain amino acid [BCAA] catabolism or other 2-ketoacid dehydrogenase complexes (DLD), thiamine-pyrophosphate cofactor synthesis (TPK), synthesis of lipoate (LIAS, LIPT1, and LIPT2), and iron-sulfur cluster complexes (BOLA3, NFU1, GLRX5, and IBA57), as well as with secondary PDC deficiencies, including fatty acid β-oxidation (ECHS1 and HIBCH) and tricarboxylic acid cycle (SUCLA2) defects where the mechanism of decreased activity is not yet known.[4–7].
The best predictor of survival and cognitive outcome in those affected with PDC deficiency appears to be the age of onset, with neonatal presentations typically associated with early death, and childhood-onset cases (> 4 years of age) associated with better survival and with normal or mild to severe cognitive disability [1]. Typically, the mean age of diagnosis of PDC deficiency is about 45 months with the median being about 20 months [1,8].
Use of ketogenic diets is currently the main therapeutic intervention in PDC deficiency and appears to have a positive effect in the areas of epilepsy, ataxia, sleep disturbance, speech/language development, social functioning, and frequency of hospitalizations [1,8–10]. The efficacy of this therapeutic intervention appears to depend on the disease phenotype and the attainment and maintenance of ketosis [8]. However, administration of a ketogenic diet may not be completely effective and may be harmful (or even lethal) in cases where PDC deficiency is associated with more general impairment of formation of acetyl-CoA (e.g., fatty acid β-oxidation [FAO] or BCAA metabolism defects) or oxidation (e.g., TCA cycle defects or other mitochondrial dysfunction) of acetyl-CoA [4,5,11]. Therefore, prompt and correct diagnosis of this disorder is critical for the long-term positive outcome of a child affected with a specific PDC deficiency.
Both genetic (molecular) and enzymatic (functional) testing approaches are being used in the diagnosis of PDC deficiency. The diagnostic efficacy of each of the above testing approaches for neonates and children affected with PDC deficiency has not been systematically investigated because of the rarity of this disorder. We sought to evaluate the diagnostic sensitivity and variability (for repeat testing, gender and cell/tissue type) of the various biochemical PDC assays available at our clinical reference laboratory and, based on such data, propose an efficient diagnostic strategy for clinical use for this devastating disorder.
2. Materials and methods
All samples referred to the Center for Inherited Disorders of Energy Metabolism (CIDEM) for clinical PDC enzyme testing were de-identified for this study and most were part of the University Hospitals Cleveland Medical Center (UHCMC) IRB-approved Disorders of Pyruvate Metabolism study and/or the Natural History and Advanced Genetic Study of Pyruvate Dehydrogenase Complex Deficiencies (NIH RDCRN NAMDC funded study).
2.1 Molecular confirmation
Some subjects were first identified to have a PDHA1 variant (usually a variant of unknown clinical significance, VUS) and then referred to CIDEM for confirmation of PDC enzyme deficiency. Subjects who were determined to be PDC enzyme deficient first at CIDEM but without a known genetic etiology were enrolled in the IRB-approved studies for molecular confirmation by either 1) Sanger sequencing for the common PDC genes (DLAT, DLD, PDHA1, PDHB, PDHX, and PDP1 performed at CIDEM on a research or clinical basis at UHCMC Center for Human Genetics Laboratory (CHGL), 2) next-generation sequencing (NGS) of 23 genes associated with pyruvate metabolism (i.e., targeted gene panel; TGP) including BOLA3, DLAT, DLD, LIAS, LIPT1, LIPT2, NFU1, PDHA1, PDHB, PDHX, PDK1, PDK2, PDK3, PDK4, PDP1, PDP2, PC, PCK1, PCK2, SLC19A2, SLC19A3, SLC25A19 and TPK1 (performed at UHCMC CHGL), or 3) whole exome sequencing (WES; performed at the Genomics Core Facility of CWRU) [4,5].
2.2 Biochemical confirmation
1-14C-pyruvate-based assay of PDC, both activated-dephosphorylated and inactivated phosphorylated, in disrupted blood lymphocytes, cultured skin fibroblasts and skeletal muscles were as previously described [12,13]. Subjects were assigned to have “low” PDC activity when the determined activity was less than the 3rd percentile of controls [<0.98 (60% of control mean), <1.26 (52% of control mean) and <1.20 (38% of control mean) nmol/min/mg of protein in blood lymphocytes (mean and RR: 1.63 and 0.98–2.72, n = 596), cultured fibroblasts (mean and RR: 2.42 and 1.26–4.42, n = 329) and skeletal muscle (mean and RR: 3.17 and 1.20–6.52, n = 340), respectively]. The clinically validated samples were assayed with control fibroblasts, and citrate synthase and/or dihydrolipoamide dehydrogenase as internal controls (and for blood lymphocytes, an off-site control shipped together to assess sample integrity), and had similar specific activities for the 1-14C labeled pyruvate and count readouts. Most of the fibroblast or muscle PDC assays with low activity were repeated at a separate time with a subculture of fibroblasts or a different aliquot of muscle (see Section 2.3) and the average activity value was used. We usually limit cell culture passage number to ≤10 for enzymatic assays but sometimes the initial passage number of the cultured fibroblasts referred to the lab is not known or reported. We do not rely on fibroblast-based enzymatic data when cell passage numbers are ≥13. Blood lymphocytes were assayed only once for PDC activity because of limited specimen volumes.
2.3 Data filtering and statistical analyses
CIDEM database was sorted by subject gender and whether subject had a 1) known pathogenic PDHA1 mutation determined retrospectively after initial functional PDC testing or 2) was referred from 2009–2017 for functional PDC testing with an already identified PDHA1 variant, predominantly (26/29) categorized as VUS. Independently, CIDEM database was also filtered based on ICD-9 or ICD-10 codes for lactic acidosis, patients with lactic acidosis and functional deficiency in at least one cell/tissue-type tested, with inclusion of all subjects with known pathogenic PDHA1 mutation. The observed cell/tissue type-specific discordance in PDC activity (i.e., low activity in one cell/tissue but normal activity in another cell/tissue) in subjects (specifically males) with completed PDC assays in more than one cell/tissue type were used to determine overall tissue heterogeneity in the PDC assay.
As part of the laboratory standard operating procedure (SOP), if a fibroblast-based PDC activity is low, repeat testing is performed on a different culture (i.e., with one additional passage number) of the same cells usually 1–2 weeks apart depending on cell growth rate. Skeletal muscle-based PDC assays are usually repeated within 1 week per SOP. The discordance frequency for repeat assays (i.e., number of subjects with at least one repeat assay with ≥cutoff value for “low” PDC activity) was determined (Table 1 for fibroblast PDC assays and Supplementary Fig. 1 for skeletal muscle assays). Variability of repeat PDC assays between groups was determined by measuring the difference in the maximum and minimum %PDC activities relative to control mean for each subject and comparing this between the various groups and genders (Table 2 and Supplementary Fig. 1 for repeated fibroblast and skeletal muscle PDC assays).
Table 1.
Discordance frequency with successive repeat fibroblast PDC testing
| Group | Gender | Total subjects | No. of subjects with Fb assay repeated ≥2x (%) | No. of subjects with discordance in Fb assay upon repeat (%) | p value |
|---|---|---|---|---|---|
| PDHA1-Only | M | 34 | 25 (74) | 1 (4) | 0.275 |
| F | 30 | 28 (93) | 3 (11) | ||
|
|
|||||
| All-CIDEM | M | 70 | 43 (61) | 2 (5) | 0.031 |
| F | 78 | 57 (73) | 9 (16) | ||
M, male; F, female. Discordance implies at least one repeat assay with ≥cutoff value for “low” PDC activity. P-values shown were obtained using t-test analysis; for “All-CIDEM” group, application of z-test resulted in a p-value of 0.039.
Table 2.
Variability of successive repeat fibroblast PDC assays between the PDHA1-Only and All-CIDEM groups
| Group | Gender | Total subjects | Percentage of subjects with Fb assay repeated ≥2x | Difference in maximum and minimum % PDC activities per subject relative to control mean activity | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Mean of difference | STD of difference | Range of difference | p value | ||||
| PDHA1-Only | M | 34 | 74 | 10% | 10% | 0–34% | 0.094 |
| F | 30 | 93 | 14% | 11% | 0–42% | ||
|
| |||||||
| All-CIDEM | M | 70 | 61 | 11% | 9% | 0–34% | 0.052 |
| F | 78 | 73 | 16% | 20% | 0–124% | ||
STD, standard deviation; M, male; and F, female.
Control: mean ± STD, range, n value; 2.42 ± 0.88 nmol/min/mg of protein, 1.26–4.42, 329. Because mean and STD are being compared and not proportions, only t-test analysis was used.
95% Confidence intervals were determined using the normal approximation for binomial proportions. For statistical analysis, either the two-sample t-test with un-pooled variances for sample sizes ≤30 or the two-proportion z-test for samples sizes >30 were used, both at a one-tailed significance level of p = 0.05.
3. Results
3.1 Characteristics of groups
3.1.1 Lactic acidotic and functional deficient (All-CIDEM) group
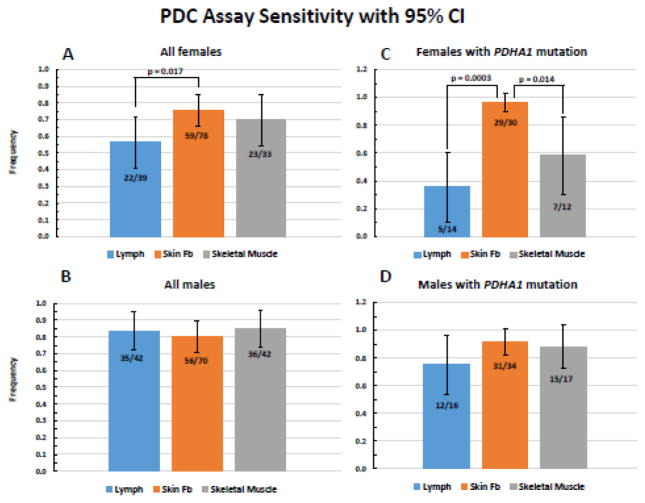
CIDEM data filtered by lactic acidosis and functional PDC deficiency in at least one cell/tissue type identified 186 subjects, about 50% of whom had been genetically tested (either at CIDEM or by the referring physician) and found to have a pathogenic mutation in PDHA1 (38%, 70/186) or a pair of pathogenic mutations in other autosomal PDC-associated genes (11%, 20/186), such as PDHB (6), DLAT (3), DLD (2), PDP1 (1), LIAS (1), LIPT1 (1), ECHS1 (3), SUCLA2 (2), and NFU1 (1) (refer to Table 3 in Bedoyan et al. 2017 for a list of currently known genes associated with functional PDC deficiency [4] and to Table 1 of DeBrosse et al 2012 to determine the frequency of specific PDC gene mutations at that time [1]). The above number of genetically-resolved subjects included the series reported in DeBrosse et al 2012 [1]. The molecular etiologies for the remainder were either unknown or not reported to CIDEM. About 78% (70/90) of the genetically-resolved subjects had a pathogenic mutation in PDHA1, a number comparable to the 74–90% observed by others [1,2,14]. Forty-nine percent (91/186) and 51% (95/186) were female and male, respectively, and 36% (33/91) and 39% (37/95) of females and males, respectively, had an identified pathogenic PDHA1 mutation. Not all subjects were evaluated by enzyme assay in cultured fibroblasts, blood lymphocytes and/or skeletal muscle. The proportion of females and males with low PDC activity in cultured fibroblasts was 0.76 (59/78; 95% CI = 0.66–0.85) and 0.80 (56/70; 95% CI = 0.71–0.89), respectively (Fig. 1A and 1B). This proportion in blood lymphocytes in females and males was 0.56 (22/39; 95% CI = 0.41–0.72) and 0.83 (35/42; 95% CI = 0.72–0.95), respectively (Fig. 1A and 1B). The proportion of females with low PDC activity in lymphocytes (56%, 22/39) was significantly lower compared to that in fibroblasts (76% [59/78], p = 0.017) (Fig. 1A).
Fig. 1.
PDC assay sensitivity for blood lymphocytes, cultured fibroblasts and skeletal muscles. A and B, proportion of subjects with PDC activity deficiency in at least one cell/tissue sample and lactic acidosis (All-CIDEM group). C and D, proportion of subjects with a pathogenic PDHA1 mutation and functional PDC deficiency in at least one cell/tissue type (PDHA1-Only group). 95% Confidence intervals are indicated along with statistical significance (p <0.05) when applicable.
3.1.2 Lactic acidotic, functional deficient with known PDHA1 mutation (PDHA1-Only) group
Among subjects with functional (enzymatic) PDC deficiency (in at least one cell/tissue type) who were retrospectively confirmed to have a pathogenic PDHA1 mutation, and including those referred with a PDHA1 variant and later found to have functional PDC deficiency in at least one cell/tissue type, the proportion of females and males with low PDC activity in cultured fibroblasts was 0.97 (29/30; 95% CI = 0.90–1.03) and 0.91 (31/34; 95% CI = 0.82–1.01), respectively (Fig. 1C and 1D). This proportion in blood lymphocytes for females and males was 0.36 (5/14; 95% CI = 0.11–0.61) and 0.75 (12/16; 95% CI = 0.54–0.96), respectively; and in skeletal muscles was 0.58 (7/12; 95% CI = 0.30–0.86) and 0.88 (15/17; 95% CI = 0.73–1.04), respectively (Fig. 1C and 1D). The proportion of females with pathogenic PDHA1 and low PDC activity in lymphocytes and skeletal muscles were significantly lower than that in fibroblasts (p = 0.0003 and p = 0.014, respectively) (Fig. 1C). Females with a pathogenic PDHA1 mutation and low PDC activity in lymphocytes and skeletal muscle constituted about 1/2 and 2/3 the number found in males for the respective cell/tissue type; 48% (0.36/0.75) for lymphocytes and 66% (0.58/0.88) for skeletal muscles, most likely reflecting random X-inactivation in female tissues. It is unclear why this variability is not also observed in cultured fibroblasts between females and males (0.97 and 0.91, respectively), but we speculate that this may be due to the shared embryonic origin of fibroblasts and nervous system (ectoderm) as compared to muscle and lymphocytes (mesoderm). One way of testing this hypothesis could involve generating induced pluripotent stem cells (IPSC) from patient fibroblasts and then differentiating them to skeletal muscle or nervous system cells for subsequent enzymatic testing of PDC activity. We find robust PDC activity in neuronal progenitor cells (NPC) derived from IPSC originating from fibroblasts allowing for future testing of this hypothesis (data not shown). The frequency of males with a PDHA1 mutation and low PDC activity was not statistically different between lymphocytes, fibroblast and muscle assays (Fig. 1D).
3.1.3 Repeat assay discordance
The low fibroblast PDC activities were comparable in both females and males with a pathogenic PDHA1 mutation; mean ± STD and range in females and males were 0.63 ± 0.25, 0.24–1.10 (n = 29) and 0.57 ± 0.34, 0.00–1.24 (n = 31), respectively (Supplementary Table 1). However, we observed a statistically significant (p = 0.031) increase in frequency of discordance with successive repeat of fibroblast assays in females compared to males in the All-CIDEM but not the PDHA1-Only group (Table 1 and Supplementary Fig. 1A and 1B, and also see Supplementary Fig. 4 in [1]). We observed an almost statistical significance (p = 0.052) for variability in successive repeat PDC assays between gender in the All-CIDEM group with mean, STD and range of difference (between maximum and minimum PDC activities) in repeat culture and assays in females higher compared to males (Table 2).
3.1.4 Cell/tissue type-specific discordance
We assumed that cell/tissue type-specific discordance frequency (i.e., subjects with discordant activities for a sample pair of cells or tissue tested such as lymphocytes vs fibroblasts, lymphocytes vs skeletal muscles, and fibroblasts vs skeletal muscles) could reflect cell/tissue type-specific variability. Because of random X-inactivation in females, we only determined the cell/tissue type-specific discordance frequency in males (37 cases) with a known PDHA1 mutation and this was 0.15 ± 0.11 (Supplementary Fig. 2). The reason for this tissue discordance in males with PDHA1 mutations without evidence of mosaicism remains unexplained [15,16]. The cell/tissue type-specific discordance frequency for All-CIDEM males (95 cases) irrespective of their genetic etiology for PDC deficiency was higher at 0.23 ± 0.08, which may be due to heterogeneity of the underlying genetic etiologies (Supplementary Fig. 2). The tissue discordance frequency for All-CIDEM females (91 cases) was 0.37 ± 0.11 (data not shown), higher than for All-CIDEM males.
3.2 Molecular-first approach
Between 2009 and 2017, 29 subjects (17 male and 12 female) with an identified PDHA1 variant were referred to CIDEM for functional PDC testing (representing a preference for a molecular- first diagnostic approach by the referring physician; with variant classification of 26 VUS, two pathogenic and one probably pathogenic). This was an opportunity for CIDEM to evaluate the pathogenicity of the VUSs. Only about 55% (16/29) showed functional PDC deficiency in at least one cell/tissue type, confirming the pathogenicity of the specific PDHA1 variant (Supplementary Table 2). About 41% (12/29) of subjects had testing completed in more than one cell/tissue type (yellow highlights in Supplementary Table 2). Among the 15 subjects that underwent cultured fibroblast PDC testing, 80% of both females (4/5) and males (8/10) showed low activity. However, among the 20 subjects that underwent blood lymphocyte PDC testing, only 22% (2/9) of females and 45% (5/11) of males showed low activity (Supplementary Table 2).
4. Discussion
Prompt and correct diagnosis of PDC deficiency (and differentiating between specific, generalized, or secondary deficiency) has important implications for clinical management and therapeutic interventions in neonates, infants, young children, and selected adults. Initiation of a therapeutic intervention on the basis of only functional PDC deficiency without follow up genetic resolution of the etiology is not recommended and could be harmful [4,5,11]. Likewise terminating a PDC deficiency work-up following a normal PDC activity assay result in one cell/tissue type without pursuing additional molecular and/or enzymatic testing using another cell/tissue type is ill advised, because of the observed significant cell/tissue type-specific variability in assay outcome irrespective of the underlying genetic etiology of the PDC deficiency (Results 3.1.4).
4.1 Sensitivity and variability
Assaying PDC in cultured fibroblasts in cases where the underlying genetic etiology is PDHA1 (most common), is highly sensitive irrespective of gender; 97% (95% CI: 90%–100%) and 91% (95% CI: 82%–100%) in females and males, respectively. However, we find that about 2/3 of females with a pathogenic PDHA1 mutation will have a normal blood-lymphocyte-based PDC assay. Similarly, the assay using skeletal muscle is not very sensitive either but better than what is observed using lymphocytes. Thus, follow-up genetic and/or enzymatic testing of a normal lymphocyte- or skeletal muscle-based PDC assay result is necessary when suspecting PDC deficiency.
The reason for the observed increase in frequency of discordance with successive repeat fibroblast-based PDC testing in females compared to males in the All-CIDEM but not the PDHA1-Only group remains unexplained (Tables 1 and 2, and Supplementary Fig. 1A and 1B). A similar difference between genders with repeat testing of skeletal muscle (Supplementary Fig. 1C and 1D) cannot be concluded because of the relatively low number of subjects tested. Whether the observed gender-specific variability in repeat testing could be unique to cells of ectodermal origin (e.g., fibroblasts) as compared to those of mesodermal origin (e.g., muscle cells) is also unclear and remains to be determined.
4.2 Diagnostic algorithm
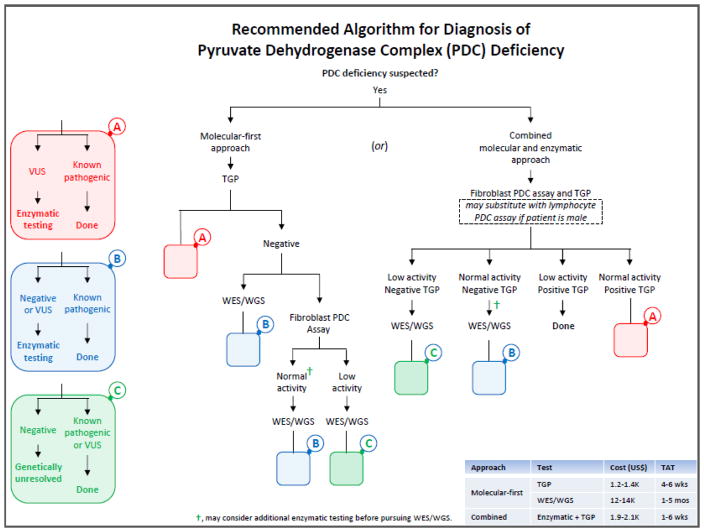
We propose a practical diagnostic approach for when PDC deficiency is suspected for use by pediatricians, neonatologists, neurologists, intensivists, child development specialists, clinical geneticists and biochemical geneticists, who encounter cases with non-specific clinical features such as hypotonia, seizure, developmental delay, ataxia, gait disturbances and/or “cerebral palsy” (Fig. 2). PDC deficiency would be suspected when 1) lactate (L) and pyruvate (P) in plasma and/or CSF are above their respective reference ranges, with normal or slightly elevated L:P ratio (10–25), 2) alanine in plasma and/or CSF is above the reference range, 3) plasma lactate and alanine concentrations are higher in a post-prandial state than a fasting state, 4) an abnormal brain MRI or CT is noted (e.g., ventriculomegaly with microcephaly, corpus callosum abnormality, basal ganglia or midbrain lesions, etc…), and/or 5) an abnormal MR spectrometry (with a significant lactate peak) is noted [17]. Although mutations in PDHA1 constitute the majority of genetically-resolved cases with functional PDC deficiency, mutations in several other genes have been associated with primary (specific and generalized) and secondary PDC deficiencies (see Table 3 in [4]). Therefore, other metabolic screening (or secondary) testing could be implemented early in the diagnostic workup once PDC deficiency is suspected in order to help with differentiating primary from secondary PDC deficiencies and the choice of path in Fig. 2 for diagnostic testing. Other findings suggesting a possible secondary PDC deficiency may include noting elevation(s) in 1) succinyl-, methylmalonyl- and/or propionylcarnitine in plasma and/or urine acylcarnitines, 2) branched-chain amino acids in plasma and/or CSF amino acids, 3) serum methylmalonic acid, and/or 4) methylmalonic acid, propionic acid, branched-chain α-keto or hydroxy acids, 2-methyl-2,3-dihydroxybutyric acid and/or α-ketoglutaric acid in urine organic acids.
Fig. 2.
Recommended algorithm for the diagnosis of PDC deficiency. “Enzymatic testing” is an all-inclusive descriptive term for various enzyme-based testing options noted below (after the initial pursuit of PDC testing in a specific cell/tissue). Enzymatic (functional) testing, when molecular testing is negative or a variant of uncertain significance (VUS) in a specific gene is identified, may include assaying PDC (in a different cell/tissue type than the initial testing), KDC, pyruvate carboxylase (PC), phosphoenolpyruvate carboxykinase (PEPCK), propionyl-CoA carboxylase (PCC), succinyl-CoA ligase (SUCL), branched-chain α-ketoacid dehydrogenase complex (BCKDC), electron complex chain complexes (ETC), and/or oxidative phosphorylation (OxPhos) activities. Please refer to Discussion for when to suspect PDC deficiency and what tests to pursue in a patient with lactic acidosis and hyperalaninemia in order to differentiate between primary and secondary PDC deficiencies. For brevity, left side colored blocks “A” (pink), “B” (blue) and “C” (green) represent steps (algorithmic options) that are repeated (as smaller pink, blue and green blanks) within the algorithm. “Positive TGP” implies identifying a pair of known pathogenic mutations, VUSs or a combination of both in a known autosomal PDC-associated gene; for the X-linked PDHA1 gene, either a single known pathogenic mutation or VUS is expected. Abbreviations used are as follows: TAT, turn-around-time; TGP, targeted gene panel; and WES/WGS, whole exome/genome sequencing.
Targeted gene panel (TGP) testing (i.e., molecular- first approach) may be the first test chosen when suspecting PDC deficiency (whether primary or secondary) (Fig. 2). However, enzymatic (functional) confirmation or testing is ultimately needed if a VUS is identified or if clinical suspicion is high for PDC deficiency (e.g., elevated lactate and pyruvate with normal L/P ratio) despite normal TGP testing or WES/WGS, delaying implementation of specific therapeutic interventions for this complex disorder. Identifying functional PDC deficiency in a patient alone cannot differentiate specific (primary) from secondary PDC deficiencies, which is important to know prior to implementing a therapeutic intervention. An approach using concurrent enzymatic and molecular testing (i.e., combined approach) would be cost effective with shorter turnaround-time (TAT) depending on the cell/tissue type used (e.g., lymphocyte- vs fibroblast PDC assay TATs are ~7 days vs ~4 weeks, respectively). We propose that when suspecting PDC deficiency in a male, a combined blood lymphocyte PDC assay (overall >80% test sensitivity; Fig. 1B) and TGP testing may be the most cost effective and efficient approach with the shortest TAT (Fig. 2). In females, however, the combination of fibroblast-based PDC assay (overall ~76% test sensitivity; Fig. 1A) and TGP testing should be considered instead. As the genetic etiologies of the various PDC deficiencies become known, the number of genes covered on TGPs will likely expand and WES/WGS variant analysis will likely become streamlined, reducing TAT and potentially the cost of molecular testing.
Supplementary Material
Acknowledgments
HKS was in part supported by the Binz Memorial Fund through CWRU School of Medicine. JKB was in part supported by NIH RDCRN 5U54NS078059-05 project NAMDC 7413 grant. We thank Dr. Suzanne D. DeBrosse for critically reading this manuscript. This work was initially inspired by a 13 month-old female patient at our institution who was eventually diagnosed with PDC deficiency.
Abbreviations
- PDC
pyruvate dehydrogenase complex
- CI
confidence interval
- TGP
targeted gene panel
- TAT
turn-around-time
- RR
reference range
- WES
whole exome sequencing
- and WGS
whole genome sequencing
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.DeBrosse SD, Okajima K, Zhang S, Nakouzi G, Schmotzer CL, Lusk-Kopp M, Frohnapfel MB, Grahame G, Kerr DS. Spectrum of neurological and survival outcomes in pyruvate dehydrogenase complex (PDC) deficiency: lack of correlation with genotype. Mol Genet Metab. 2012;107:394–402. doi: 10.1016/j.ymgme.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Patel KP, O’Brien TW, Subramony SH, Shuster J, Stacpoole PW. The spectrum of pyruvate dehydrogenase complex deficiency: clinical, biochemical and genetic features in 371 patients. Mol Genet Metab. 2012;106:385–394. doi: 10.1016/j.ymgme.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quintana E, Gort L, Busquets C, Navarro-Sastre A, Lissens W, Moliner S, Lluch M, Vilaseca MA, De Meirleir L, Ribes A, Briones P, Group PDHW. Mutational study in the PDHA1 gene of 40 patients suspected of pyruvate dehydrogenase complex deficiency. Clin Genet. 2010;77:474–482. doi: 10.1111/j.1399-0004.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- 4.Bedoyan JK, Yang SP, Ferdinandusse S, Jack RM, Miron A, Grahame G, DeBrosse SD, Hoppel CL, Kerr DS, Wanders RJ. Lethal neonatal case and review of primary short-chain enoyl-CoA hydratase (SCEH) deficiency associated with secondary lymphocyte pyruvate dehydrogenase complex (PDC) deficiency. Mol Genet Metab. 2017;120:342–349. doi: 10.1016/j.ymgme.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang X, Bedoyan JK, Demirbas D, Harris DJ, Miron A, Edelheit S, Grahame G, DeBrosse SD, Wong LJ, Hoppel CL, Kerr DS, Anselm I, Berry GT. Succinyl-CoA synthetase (SUCLA2) deficiency in two siblings with impaired activity of other mitochondrial oxidative enzymes in skeletal muscle without mitochondrial DNA depletion. Mol Genet Metab. 2017;120:213–222. doi: 10.1016/j.ymgme.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferdinandusse S, Waterham HR, Heales SJ, Brown GK, Hargreaves IP, Taanman JW, Gunny R, Abulhoul L, Wanders RJ, Clayton PT, Leonard JV, Rahman S. HIBCH mutations can cause Leigh-like disease with combined deficiency of multiple mitochondrial respiratory chain enzymes and pyruvate dehydrogenase. Orphanet J Rare Dis. 2013;8:188. doi: 10.1186/1750-1172-8-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loupatty FJ, Clayton PT, Ruiter JP, Ofman R, Ijlst L, Brown GK, Thorburn DR, Harris RA, Duran M, Desousa C, Krywawych S, Heales SJ, Wanders RJ. Mutations in the gene encoding 3-hydroxyisobutyryl-CoA hydrolase results in progressive infantile neurodegeneration. Am J Hum Genet. 2007;80:195–199. doi: 10.1086/510725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sofou K, Dahlin M, Hallbook T, Lindefeldt M, Viggedal G, Darin N. Ketogenic diet in pyruvate dehydrogenase complex deficiency: short- and long-term outcomes. J Inherit Metab Dis. 2017;40:237–245. doi: 10.1007/s10545-016-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wexler ID, Hemalatha SG, McConnell J, Buist NR, Dahl HH, Berry SA, Cederbaum SD, Patel MS, Kerr DS. Outcome of pyruvate dehydrogenase deficiency treated with ketogenic diets. Studies in patients with identical mutations. Neurology. 1997;49:1655–1661. doi: 10.1212/wnl.49.6.1655. [DOI] [PubMed] [Google Scholar]
- 10.Scholl-Burgi S, Holler A, Pichler K, Michel M, Haberlandt E, Karall D. Ketogenic diets in patients with inherited metabolic disorders. J Inherit Metab Dis. 2015;38:765–773. doi: 10.1007/s10545-015-9872-2. [DOI] [PubMed] [Google Scholar]
- 11.Ferdinandusse S, Friederich MW, Burlina A, Ruiter JP, Coughlin CR, 2nd, Dishop MK, Gallagher RC, Bedoyan JK, Vaz FM, Waterham HR, Gowan K, Chatfield K, Bloom K, Bennett MJ, Elpeleg O, Van Hove JL, Wanders RJ. Clinical and biochemical characterization of four patients with mutations in ECHS1. Orphanet J Rare Dis. 2015;10:79. doi: 10.1186/s13023-015-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerr D, Grahame G, Nakouzi G. Assays of pyruvate dehydrogenase complex and pyruvate carboxylase activity. Methods Mol Biol. 2012;837:93–119. doi: 10.1007/978-1-61779-504-6_7. [DOI] [PubMed] [Google Scholar]
- 13.Chuang DT, Hu CW, Patel MS. Induction of the branched-chain 2-oxo acid dehydrogenase complex in 3T3-L1 adipocytes during differentiation. Biochem J. 1983;214:177–181. doi: 10.1042/bj2140177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperl W, Fleuren L, Freisinger P, Haack TB, Ribes A, Feichtinger RG, Rodenburg RJ, Zimmermann FA, Koch J, Rivera I, Prokisch H, Smeitink JA, Mayr JA. The spectrum of pyruvate oxidation defects in the diagnosis of mitochondrial disorders. J Inherit Metab Dis. 2015;38:391–403. doi: 10.1007/s10545-014-9787-3. [DOI] [PubMed] [Google Scholar]
- 15.Kerr DS, Berry SA, Lusk MM, Ho L, Patel MS. A deficiency of both subunits of pyruvate dehydrogenase which is not expressed in fibroblasts. Pediatr Res. 1988;24:95–100. doi: 10.1203/00006450-198807000-00022. [DOI] [PubMed] [Google Scholar]
- 16.Wexler ID, Hemalatha SG, Liu TC, Berry SA, Kerr DS, Patel MS. A mutation in the E1 alpha subunit of pyruvate dehydrogenase associated with variable expression of pyruvate dehydrogenase complex deficiency. Pediatr Res. 1992;32:169–174. doi: 10.1203/00006450-199208000-00009. [DOI] [PubMed] [Google Scholar]
- 17.Kerr DS, Bedoyan JK. Disorders of Pyruvate Metabolism and the Tricarboxylic Acid Cycle, Chapter 8. In: Sarafoglou K, Hoffmann GF, Roth KS, editors. Pediatric endocrinology and inborn errors of metabolism. 2. New York: McGraw-Hill Education; 2017. pp. 105–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.