Abstract
Rice eating and cooking quality and protein content (PC) are important properties affecting consumers’ preferences, nutrition and health. Linkage QTL mapping and association studies are usually applied to genetically dissect related traits, which could be further facilitated by high density SNP markers and gene annotation based on reference genome to rapid identify candidate genes associated with interested traits. Here, we carried out an association study for apparent amylose content (AC), gel consistency (GC), gelatinization temperature (GT) and PC evaluated in two environments using a diverse panel of 258 accessions from 3 K Rice Genome Project. Wide phenotypic variations were observed in this panel. Genome-wide association study using 22,488 high quality SNPs identified 19 QTL affecting the four traits. Combining gene-based association study and haplotype analyses plus functional annotation allowed us to shortlist nine candidate genes for four important QTL regions affecting AC, GC and GT, including two cloned genes (Wx and ALK), and seven novels. The research suggested that GWAS and gene-based association analysis followed by haplotype analysis is an effective way to detect candidate genes. The identified genes and QTL provided valuable sources for future functional characterization and genetic improvement of rice eating and cooking quality and PC.
Introduction
Rice (Oryza sativa L.) is the staple food for more than half of the world’s population. With the improvement of living standards and increases of diverse demands, rice grain quality has become one of the foremost considerations for rice breeders, producers and consumers. Rice grain quality consists primarily of four components: milling, appearance, eating and cooking, and nutritional qualities. The last two are especially important, as they are related to consumers’ preferences, nutrition and health.
Rice eating and cooking quality (ECQ) is determined mainly by three major physicochemical characteristics, namely, apparent amylose content (AC), gel consistency (GC) and gelatinization temperature (GT). AC is demonstrated to be the most important factor affecting rice ECQ1. AC can be roughly classified into five levels: waxy (1–2%), very low (5–12%), low (12–20%), intermediate (20–25%) and high (>25%)2. Cooked rice kernels with high AC are usually dry, separate, less tender and become hard upon cooling, whereas those with low or intermediate AC are glossy, soft and sticky3. Intermediate AC rice is widely preferred in most rice producing areas of the world since this kind of rice is soft but not too sticky4. Rice cultivars varying in AC could meet the diverse demands for food products and consumers5. GC is a fluid property of rice starch gel, which can be classified into three levels: hard (≤40 mm), medium (41–60 mm) and soft (≥60 mm). Cooked rice with high GC tends to be softer and more elastic. GT is a physical trait responsible for rice cooking time. Usually, GT is estimated as alkali spreading value (ASV) that is assessed by the extent of dispersal of whole milled rice grains in dilute alkali solution (1.7% potassium hydroxide [KOH])6, and could be classified into four groups: high (1–2), high-intermediate (3), intermediate (4–5) and low (6–7)7. Rice kernels with low or intermediate GT need less cooking time which is a desired trait for high quality rice varieties8. As one of the most important parameters of nutritional quality, protein content (PC) is the most abundant constituent in milled rice except starch2. With increasing attention on health, rice with distinct nutritional quality is required to meet the special requirements of consumers.
To facilitate rice high quality breeding, dissecting the genetic basis of rice grain ECQ and PC is useful. The pathway of starch synthesis has been comprehensively studied9. Among these starch synthesis related genes (SSRGs), Wx and ALK are major genes governing AC and GC and GT, respectively1. Other SSRGs, such as AGPlar, BEI, GBSSII, GPT1, ISA2, PUL, SSI, SSIIb, SSIIc, SSIIIa, SSIIIb and SSIVa also have minor effects10,11. However, the pathway of protein synthesis remains unclear to date12. QTL mapping is an effective way to dissect the genetic basis of quantitative traits. Many QTL affecting rice ECQ and PC have been identified through linkage mapping13–16 and association studies17–20, but QTL cloning is still a major challenge to plant geneticists and molecular biologists since the classical strategy using map-based cloning is extremely time-consuming and troublesome21. With the development of technology and reduced costs of genotyping, it’s increasingly easy to obtain genotypic data with millions of SNP markers through genotyping by sequencing (GBS) and high density SNP chips22,23. The high density SNP markers and gene annotation based on high quality reference genomes powerfully facilitate the identification of QTL candidate genes associated with interested traits24. Combining genome-wide association study (GWAS) and gene-based association analysis followed by haplotype analysis is an effective way to identify candidate genes for complex traits including rice grain appearance traits25.
Therefore, the objective of our study is to identify candidate genes affecting rice grain ECQ and PC using GWAS, and gene-based association analysis combining haplotype analysis. A diverse panel consisting of 258 accessions selected from 3 K Rice Genome Project (3 K RGP)26 was evaluated for rice grain ECQ and PC in two environments. GWAS was performed using genome-wide SNPs generated from 3 K RGP by high-throughput sequencing technologies27. Then, for important QTL regions, gene-based association analysis was performed using all available SNP from Rice SNP-Seek Database28. Finally, haplotype analysis was conducted and the phenotype differences among major haplotypes were tested by ANOVA. By this way, numbers of candidate genes governing investigated traits were determined.
Materials and Methods
Plant materials
The materials used in this study comprised 258 rice accessions having similar heading date selected from the 3 K RGP. The detail accession information was described by Wang, et al.25. Roughly, these rice accessions mainly included seven types. Most of them were Xian (indica) (174), followed by Geng (japonica) including temperate Geng (32), tropical Geng (24) and subtropical Geng (14). The remaining were admixture type (7), aus/boro (3) and basmati/sandri (4)25.
Field trials and trait measurements
These accessions were grown in two environments, including Sanya (SY) during Dec 2014 – Apr 2015 and Shenzhen (SZ) during Mar – Jul 2015. In both environments, each accession was planted in a two-row plot with 10 individuals in each row at a spacing of 20 cm × 25 cm with two replicates for each accession. At the maturity (about 35 days after flowering), eight uniform plants in the middle of each plot were bulked harvested and air-dried for three months in the drying houses. Then, around 150 g seeds were dehulled in an electrical dehuller (model JLGJ-45, China) and milled by a desk-top rice miller (JNMJ 6, China). The physicochemical quality traits including AC, GC, GT and PC were analyzed by near infrared spectroscopy (NIRS) using Infratec 1241 Grain Analyzer (FOSS, Denmark) equipped with STM model29,30. About 60 g head milled rice grains of each accession were scanned in duplicate and the average trait value of each accession was used in the following analyses.
Genome-wide marker-trait associations
We carried out GWAS to detect marker-trait associations for all measured traits utilizing 22,488 high quality SNPs and the mean grain quality trait values of the 258 accessions in the two environments. All statistical analyses for GWAS were performed using the SVS software package (SNP & Variation Suite, Version 8.4.0). An EMMAX (Efficient Mixed-Model Association eXpedited)31,32 implementation of the single-locus mixed linear model was applied to the marker dataset. This mixed linear model (MLM) allowed correction for cryptic relatedness and other fixed effects using kinship matrix (K) and population stratification through principle components (Q). The Bonferroni multiple testing correction was applied to identifying significant markers. Significant SNPs affecting the investigated traits were claimed when the test statistics reached P < 1.0 × 10−4 in at least one of the two environments. Our previous study found that the maximum linkage disequilibrium (LD) of the current panel was 0.6225, thus significant SNPs on the same chromosome with LD higher than 0.31 (half of its initial value)25 were delineated into a single QTL.
Gene-based association and haplotype analysis
Gene-based association analysis was carried out to identify candidate genes for important QTL (accounting for over 10% of the trait phenotypic variance). The following four steps were conducted to identify QTL candidate genes. Firstly, we examined all genes located in the 0.31 LD block region of the peak SNP of each important QTL from the Rice Annotation Project Database (RAP-DB). The 0.31 (half of its initial value)25 LD block region means, on average, the LD of markers flanking each peak SNP marker is 0.31. In other words, the average LD of each peak marker with its flanking markers across the population is ≥0.31. Statistically, SNPs beyond the 0.31 LD region centered at the peak SNPs are less likely to contain genes responsible for the detected trait-marker association. Then, all available SNPs located inside of these genes were searched from 32 M SNPs data generated from 3 K RGP in the Rice SNP-Seek Database28. Thirdly, the SNPs with minor allele frequency less than 0.05 and/or missing rate over 20% were removed and the remaining high quality SNPs were used to perform association analyses through MLM using the Q and K applied in GWAS. The threshold was defined as −log10(p) of peak SNP minus 1; Finally, for the genes harboring SNPs with −log10(p) above the threshold, haplotype analysis was carried out, and candidate genes were determined by testing the significant differences among major haplotypes (containing more than 10 samples) through ANOVA. Figure 1 illustrated the technical line of combining GWAS and gene-based association analysis to identify candidate genes.
Figure 1.
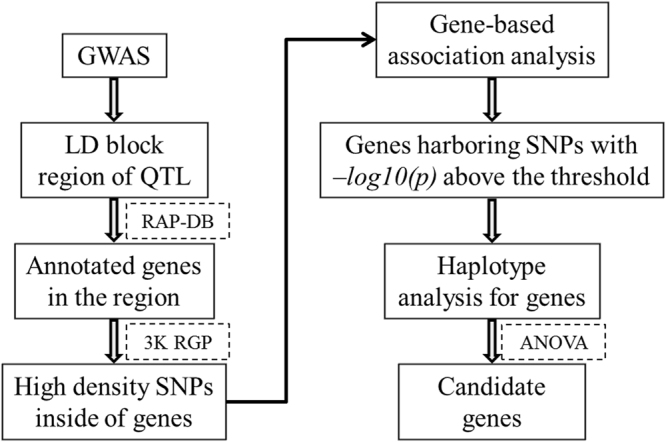
Technical line of combining GWAS and gene-based association analysis to identify candidate genes. RAP-DB: Rice Annotation Project Database. 3 K RGP: 3 K Rice Genome Project.
Results
Trait variations and correlations
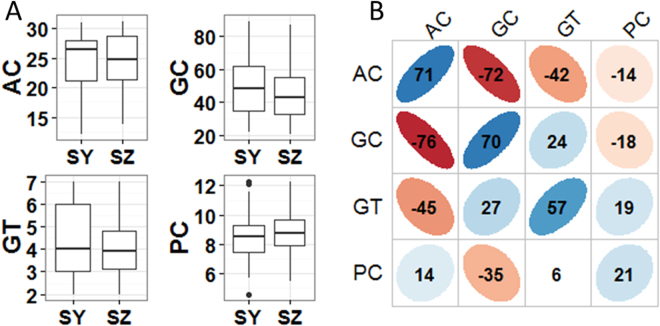
Large variations were observed for all investigated traits in both SY and SZ (Fig. 2A). In SY, AC of the evaluated accessions ranged from 12.1% to 31.1% with a mean of 24.6%. GC varied from 21.9 mm to 89.0 mm with a mean of 50.0 mm. GT was averaged at 4.3 ranging from 2.0 to 7.0. PC ranged from 4.6% to 12.3% with an average of 8.5%. In SZ, AC ranged from 13.7% to 31.3% with a mean of 24.6%. GC varied from 21.0 mm to 87.0 mm with a mean of 45.6 mm. GT was averaged at 4.0 ranging from 2.0 to 7.0. PC ranged from 5.5% to 12.3% with an average of 8.8%. The phenotypic pair-wise correlations were almost similar in both two environments (Fig. 2B). AC was negatively correlated with GC and GT with correlation coefficients (r) of −0.72 (−0.76) and −0.42 (−0.45) in SY (SZ), respectively. Significant positive but weak correlation was observed between GC and GT in SY (r = 0.24) and SZ (r = 0.27). PC was significantly weakly correlated with GC with r being −0.18 and −0.35 in SY and SZ, respectively. The phenotypic correlations between SY and SZ ranged from 0.21 for PC to 0.71 for AC (Fig. 2B).
Figure 2.
(A) Box plots of four investigated traits in two environments. SY: Sanya. SZ: Shenzhen. AC: Apparent Amylose Content. GC: Gel Consistency. GT: Gelatinization Temperature. PC: Protein Content (B) Correlations between four evaluated traits in SY (upper triangular) and SZ (lower triangular). The values were correlation coefficients (r) multiplied by 100. The values on principal diagonal indicated correlations between SY and SZ. The areas and colors of ellipses showed the absolute value of corresponding r. Right and left oblique ellipses indicated positive and negative correlations, respectively. The values without glyphs indicated insignificant at 0.05.
Identification of QTL by GWAS
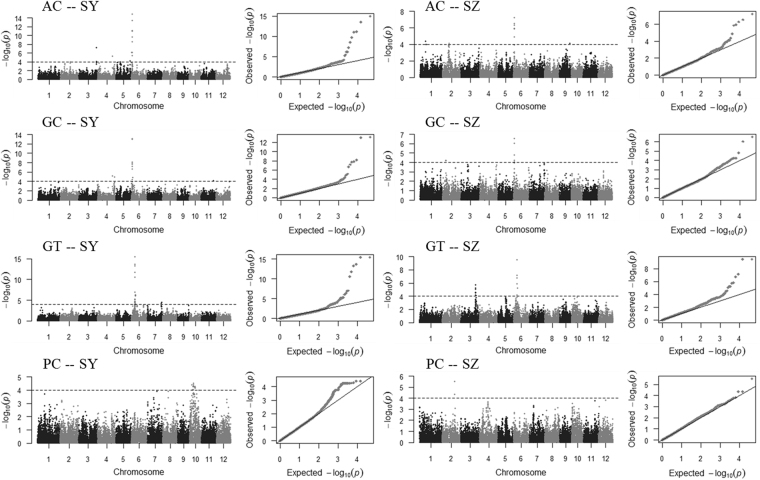
In total, 96 marker-trait associations were detected for the four investigated traits in SY and SZ. For each trait, by delineating significant SNPs on the same chromosome with LD higher than 0.31 into a single QTL, finally 19 QTL were identified (Table 1 and Fig. 3).
Table 1.
QTL identified by GWAS for AC, GC, GT and PC in SY and SZ.
| QTL | Env | Peak | Allelesa | MAFb | p | Effectc | R2 (%)d | Gene/QTL |
|---|---|---|---|---|---|---|---|---|
| qAC1 | SZ | S1_10888508 | C/T | 0.47 | 4.10E-05 | −3.5 | 8.8 | qAC1b |
| qAC2.1 | SZ | S2_12269764 | C/T | 0.24 | 7.50E-05 | −7.1 | 8.5 | |
| qAC2.2 | SY | S2_19252894 | T/G | 0.07 | 9.90E-05 | −3.6 | 6.5 | OsBEIIb |
| qAC3 | SY | S3_33525313 | G/A | 0.08 | 5.90E-08 | −5.6 | 12.9 | qAC3 |
| qAC4 | SY | S4_28862203 | C/T | 0.14 | 5.40E-06 | −5.4 | 8.9 | |
| qAC5 | SY | S5_27617633 | G/T | 0.05 | 6.20E-05 | −4.1 | 7.0 | |
| qAC6 | SY | S6_1746440 | G/A | 0.23 | 1.10E-15 | −8.1 | 31.6 | Wx |
| SZ | S6_1746440 | G/A | 0.24 | 5.90E-08 | −6.9 | 16.3 | ||
| qAC9 | SZ | S9_20798975 | C/T | 0.25 | 7.30E-05 | −6.9 | 8.3 | qAC-9b (9) |
| qGC2 | SZ | S2_6132333 | C/A | 0.05 | 5.70E-05 | 19.3 | 8.2 | qGC-2a |
| qGC4 | SY | S4_28862203 | C/T | 0.14 | 7.50E-06 | 22.4 | 8.7 | |
| qGC6 | SY | S6_1662107 | C/T | 0.23 | 7.00E-14 | 29.4 | 26.2 | Wx |
| SZ | S6_1616444 | G/A | 0.24 | 2.90E-07 | 20.0 | 14.3 | ||
| qGC11 | SY | S11_24266777 | A/T | 0.13 | 7.80E-05 | 16.6 | 6.7 | |
| qGC12 | SY | S12_25629093 | A/G | 0.08 | 8.50E-05 | 15.3 | 6.9 | |
| qGT3 | SZ | S3_29474609 | T/C | 0.49 | 8.70E-05 | −0.8 | 11.9 | |
| qGT6 | SY | S6_6752888 | C/T | 0.31 | 2.56E-16 | 2.8 | 41.0 | ALK |
| SZ | S6_6752888 | C/T | 0.32 | 6.43E-10 | 1.2 | 16.5 | ||
| qGT7 | SY | S7_27788464 | C/G | 0.12 | 3.60E-05 | −1.5 | 7.5 | |
| qPC2 | SZ | S2_24197424 | G/C | 0.07 | 3.00E-06 | 1.6 | 7.6 | |
| qPC10.1 | SY | S10_7659738 | C/T | 0.07 | 2.90E-05 | −2.3 | 8.1 | qPC10 |
| qPC10.2 | SY | S10_17723490 | T/C | 0.29 | 1.00E-04 | −1.4 | 6.7 |
aMajor/Minor allele.
bMAF: Minor allele frequency.
cEffect: Allele effect with respect to the minor allele.
dR2 (%): Phenotypic variance explained.
Figure 3.
Manhattan and Q-Q plots of GWAS for each trait evaluated in SY and SZ. SY: Sanya. SZ: Shenzhen. AC: Apparent Amylose Content. GC: Gel Consistency. GT: Gelatinization Temperature. PC: Protein Content.
For AC, eight QTL were detected in two environments. Four QTL were identified only in SY including qAC2.2, qAC3, qAC4 and qAC5, which accounted for 6.5%, 12.9%, 8.9% and 7.0% of the phenotypic variance, respectively. Three QTL (qAC1, qAC2.1 and qAC9) were detected only in SZ explaining 8.8%, 8.5% and 8.3% of the phenotypic variance, respectively. qAC6 was identified in both SY and SZ, and accounted for 31.6% and 16.3% of the phenotypic variance, respectively (Table 1 and Fig. 3).
Five QTL for GC were identified. These included qGC4, qGC11 and qGC12, detected only in SY which explained 8.7%, 6.7% and 6.9% of the phenotypic variance, respectively, and qGC2 detected only in SZ that accounted for 8.2% of the phenotypic variance. In both SY and SZ, qGC6 was commonly identified and explained 26.2% and 14.3% of the phenotypic variance, respectively (Table 1 and Fig. 3).
For GT, three QTL were detected. qGT7 was identified only in SY and explained 7.5% of the phenotypic variance. qGT3 was detected only in SZ that accounted for 11.9% of the phenotypic variance. qGT6 was identified in both SY and SZ and explained 41.0% and 16.5% of the phenotypic variance, respectively (Table 1 and Fig. 3).
Three QTL affecting PC were detected. These included qPC10.1 and qPC10.2 identified in SY which explained 8.1% and 6.7% of the phenotypic variance, and qPC2 identified in SZ that accounted for 7.6% of the phenotypic variance (Table 1 and Fig. 3).
Candidate genes for important QTL
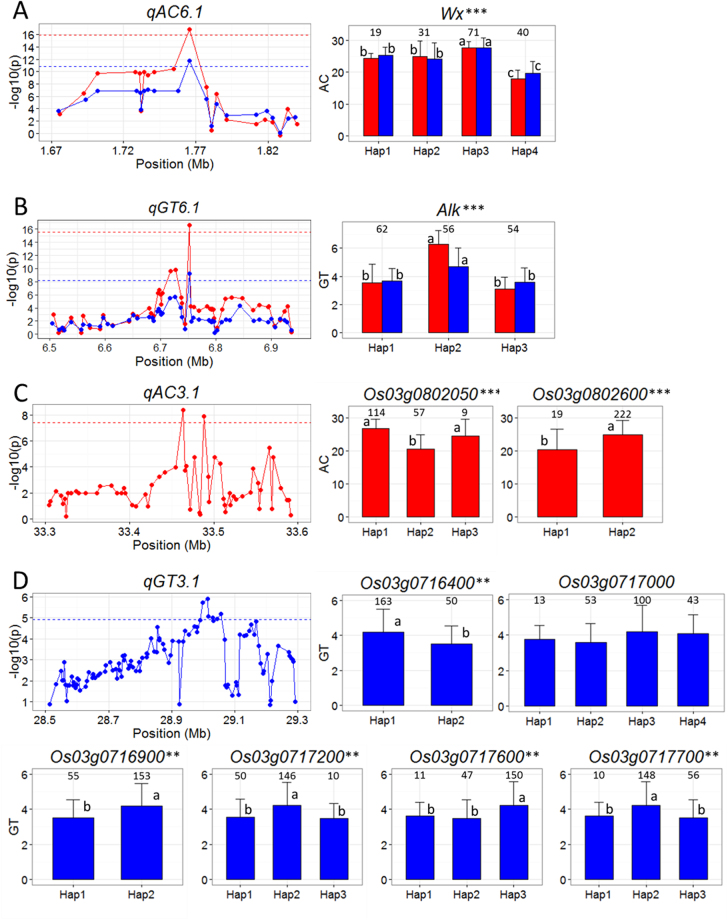
Supplementary Table S1–S4 showed the detailed gene-based association analysis results of the four important QTL regions including qAC6, qGT6, qAC3 and qGT3. The nine candidate genes were shortlisted based on the haplotype analyses (Fig. 4 and Supplementary Table S5).
Figure 4.
(A–D) Gene-based association and haplotypes analysis of targeted genes of related QTL including qAC6 (A), qGT6 (B) qAC3 (C) and qGT3 (D). Each point was a gene indicated by one SNPs having largest –log10(p) value. Dash line showed the threshold to determine significant SNP. The ** and *** suggested significance of ANOVA at p < 0.01 and p < 0.001, respectively. The letter on histogram (a and b) indicated multiple comparisons results at the significant level 0.01. The value on the histogram was the number of individuals of each haplotype. Red and blue color indicated Sanya and Shenzhen environments, respectively.
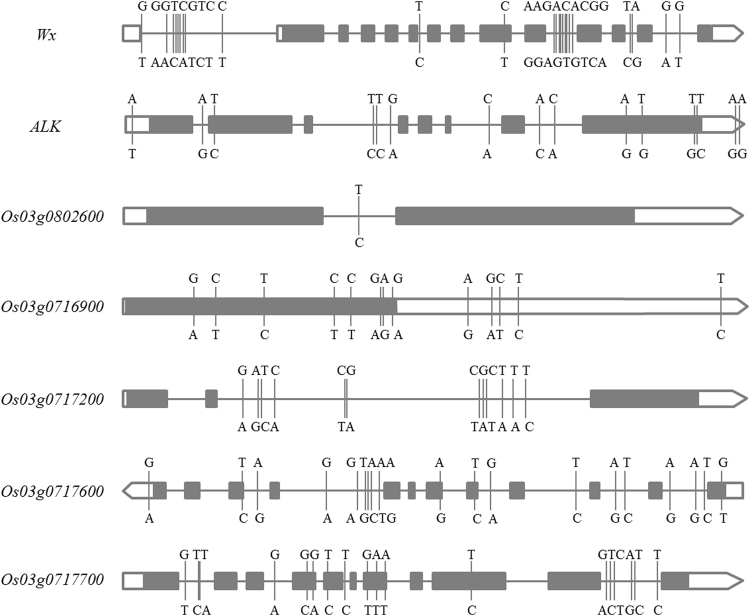
In the region from 1.67 to 1.84 Mb on chromosome 6 harboring qAC6, 143 SNPs of 21 genes were used for association analysis. The peak SNP was S6_1765761 with −log10(p) of 16.9 and 11.8 in SY and SZ, respectively. The well-known gene, Wx (Os06g0133000)33 was found to harbor all SNPs with −log10(p) above the threshold (15.9 and 10.8 in SY and SZ, respectively) (Fig. 4A and Supplementary Table S1). In total, 24 SNPs within the Wx locus were used for haplotype analysis. Of these SNPs, only a synonymous SNP at the 11th position (S6_1768724) of the Wx gene locates in the exon, while all other SNPs locate in the introns (Fig. 5). Four major haplotypes of Wx were detected in the investigated panel. Hap3 had highest AC of 27.6% (27.7%) while Hap4 had lowest AC of 18.0% (19.8%) in SY (SZ) (Table S5).
Figure 5.
Illustration of gene structure and location of SNPs used in haplotype analysis of seven candidate genes. The alleles above (under) the gene structure could increase (decrease) trait values.
In the region from 6.50 to 6.95 Mb on chromosome 6 harboring qGT6, 1,339 SNPs of 53 genes were used for association analyses. The peak SNP was S6_6752888 with −log10(p) of 15.59 and 9.19 in SY and SZ, respectively. Again, all SNPs with −log10(p) above the threshold (14.59 and 8.19 in SY and SZ, respectively) were found to locate in the well-known gene, ALK (Os06g0229800)34,35 (Fig. 4B and Supplementary Table S2), indicating ALK was qGT6. In total, 15 SNPs within the ALK locus were used to analyze haplotypes. These included three SNPs in the 5′UTR and 3′UTR of the gene, two missense mutations in the exons, and the remaining SNPs occurred in the introns (Fig. 5). We noted that the G/A mutation at 10th position of the locus resulted in an amino acid change from Gly to Ser, and the T/C mutation of 13th position changed Phe to Leu. Three major haplotypes of ALK were detected in the evaluated panel with Hap2 having significantly higher GT (6.3 ± 1.0 and 4.7 ± 1.3 in SY and SZ) than Hap1 (3.6 ± 1.3 and 3.7 ± 0.9) and Hap3 (3.1 ± 0.8 and 3.6 ± 1.0) (Table S5).
For qAC3 in the region of 33.3 – 33.6 Mb on chromosome 3, 855 SNPs of 59 genes were used for association analysis. The peak SNP was S3_33463370 (−log10(p) = 8.39). SNPs with −log10(p) above the threshold (7.39) centered around two candidate genes, Os03g0802050 and Os03g0802600 (Fig. 4C and Supplementary Table S3). Os03g0802050 is a gene of unknown function and its gene structure is absent in the RAP-DB. We used 12 SNPs within Os03g0802050 and detected three haplotypes of in the panel. Hap1 and Hap3 had significantly higher AC than Hap2. Os03g0802600 is a putative ATPase with a single SNP (S3_33488536) located in the intron of the gene (Fig. 5) with the Hap2 associated with significantly higher AC.
In the region from 28.50 to 29.30 Mb on chromosome 3 harboring qGT3, 2,351 SNPs of 107 genes were used for association analysis. The peak SNP was S3_29013799 with –log10(p) being 5.92. Six genes harboring SNPs with –log10(p) larger than the threshold (4.92) were identified, including Os03g0716400, Os03g0716900, Os03g0717000, Os03g0717200, Os03g0717600 and Os03g0717700 (Fig. 4D and Supplementary Table S4). Os03g0716400 is a gene of unknown function without functional annotation and gene structure available in RAP-DB. Two major haplotypes consisting of eight SNPs within Os03g0716400 were detected in the rice panel with Hap1 associated with significantly higher GT. Os03g0716900 is a hypothetical gene of unknown function. There were 13 SNPs in this gene, including eight nonsynonymous SNPs in its exons that result in amino acids changes, and five SNPs in 3′UTR of the gene (Fig. 5). We detected two haplotypes at Os03g0716900 in the rice panel with Hap2 having significantly higher GT than Hap1. Os03g0717000 encodes TMK protein precursor. We identified four haplotypes of this gene in the panel but no significant differences for GT were detected among the haplotypes. Os03g0717200 encodes a putative cytochrome b561 family protein. There were 12 SNPs all within the introns of this gene (Fig. 5). We identified three haplotypes of this gene in the panel. Hap2 had significantly higher GT than the other two haplotypes. Os03g0717600 encodes a putative zinc finger (C2H2-type matrin domain containing) protein. There were 19 SNPs within this gene. Three of these SNPs occurred in the exons with a synonymous one plus two nonsynonymous ones causing an amino acid change of from Thr to Ile and another one from Arg into His. The remaining 16 SNPs occurred either in UTR or introns of the gene (Fig. 5). We detected three haplotypes at this gene with Hap3 associated with higher GT. The last candidate of qGT3, Os03g0717700, encodes a putative histidine kinase. We identified three haplotypes consisting of 18 SNPs in this gene in the rice panel. Seven of these SNPs located in the exons with 5 nonsynonymous ones while the remaining 14 SNPs occurred in the introns (Fig. 5). Hap2 had markedly higher GT than Hap1 and Hap3 (Fig. 4D and Table S5).
Discussion
Despite tremendous efforts in the past two decades, resolving individual QTL into their causative genes has been a great challenge in dissecting complex traits at the molecular level. In this respect, QTL mapping using diverse panel populations and GWAS has a much higher resolution than the approach by linkage mapping because mapping populations consisting of random rice accessions used in GWAS have much higher recombination accumulated during their evolutionary history. Indeed, most QTL detected in this study were resolved to relatively few candidate genes, and many of them were adjacent to known SSRGs and/or previously identified QTL for related traits. Four QTL for AC, GC and GT identified in this study were close to the known SSRGs. Of these, qAC6 and qGC6 were the most important ones detected in the same chromosome region in both environments. This QTL region corresponded to the Wx gene (1,765,622 – 1,770,656) encoding GBSSI, a key enzyme for the elongation of amylose chains in amylose biosynthesis well demonstrated previously1,33,36–38. qAC2.2 detected in SY was close to OsBEIIb (19,355,790 – 19,367,127) encoding a kind of starch branching enzyme. One SNP causing a G/C transversion of OsBEIIb was reported to associate with AC39. qGT6, identified in both SY and SZ, was a major QTL affecting GT. It corresponded to ALK encoding SSIIa that mainly affects amylopectin chain-length distribution and alkali disintegration of rice grains34,40. ALK has been demonstrated to be a major gene affecting GT1,38,41,42.
Three other QTL for AC, GC, and PC were adjacent to previously reported QTL for related traits. qAC1 identified in SZ was located in the QTL region flanked by RM572 – RM449 affecting AC reported previously16. qAC3 detected in SY was equivalent to a QTL of the same name flanked by SSR markers RM416 and RM570 reported by Ebadi, et al.43. qAC9 was located in the region of qAC-9b flanked by C609 – C506 detected by Wan, et al.44. qGC2 was mapped very closely to a QTL region flanked by R712-G227 affecting GC reported by Li, et al.45. qPC10.1 detected in SY was in approximate vicinity with qPC10 flanked by RM216 – RM467 detected by Leng, et al.46. These results indicated that the false positives of the QTL identified in this study were very few, if any. The remaining ten QTL identified in this study, including three QTL for AC (qAC2.1, qAC4 and qAC5), three for GC (qGC4, qGC11 and qGC12), two for GT (qGT3 and qGT7) and two (qPC2 and qPC10.2) for PC, were new ones not reported previously. This result would imply that there are many more genes in the rice genome that may have contributed to the tremendous variations for the grain quality traits observed in in the panel.
Our results clearly showed that four large-effect QTL (qAC6, qGT6, qAC3 and qGT3) detected in this study were all previously reported major genes/QTL affecting rice grain quality traits. The candidate gene of qAC6 was Wx 33. It’s reported that Wx a and Wx b had a G/T mutation at 5′ splicing site of intron 1 which caused the inefficiency of GBSS at the posttranscriptional level, while Wx a had higher AC than Wx b 47–49. Here, we found that a SNP at S6_1765761 (the 1st SNP of the haplotype) was the key mutation that resulted in the lowest AC of Hap4 with nucleotide T when compared the other three haplotypes with nucleotide G. Further, we also observed that Hap1 and Hap2 had lower AC than Hap3. Apparently, other mutations in introns of Wx may have also affected its functionality, even though it remains unknown through what unknown mechanism(s) (Fig. 4A and Supplementary Table S5). The candidate gene of qGT6 was ALK 34. Bao, et al.3 reported that the GC/TT polymorphism at 4229/4330 bp of ALK was strongly associated with GT variation. Here, we found that two SNPs at S6_6752887 and S6_6752888 in the exon 8 (the 12th and 13th of the haplotype) were the same GC/TT polymorphism sites which caused higher GT of Hap2. Hap1 and Hap3, which differ at a single SNP at S6_6752357 (the 10th SNP of the haplotype) showed no significant differences for GT. This result suggested that this non-synonymous mutation at S6_6752357 may not necessarily affect the function of ALK (Fig. 4B and Supplementary Table S5).
The above results indicated that gene-based association analysis combining haplotype analysis is an effective way to identify candidate genes for large-effect QTL. We applied this approach to two other large-effect QTL. The first one was qAC3 which explained 12.9% of the phenotypic variance of AC in SY. Ebadi, et al.43 also identified a QTL for AC flanked by RM416 and RM570 in this region. Through gene-based analysis, we were able to shortlist this QTL to two candidate genes (Os03g0802050 and Os03g0802600). In the case of Os03g0802050, the information for functional annotation and gene structure was not available, so that we could not infer the functional variations based on the SNPs locations in the gene. In the case of Os03g0802600 which encodes a putative ATPase, we found a single SNP (S3_33488536) in the intron of the gene was responsible for the phenotypic difference between the two haplotypes (Fig. 5), but the mechanism(s) for why Hap2 (T) had higher AC than Hap1 remains unclear (Fig. 4C).
Another case was qGT3 detected in SZ which accounted for 11.9% of the phenotypic variance. Significant differences in GT were detected between haplotypes of five candidate genes (Os03g0716400, Os03g0716900, Os03g0717200, Os03g0717600 and Os03g0717700), and in all the five cases, significantly increased GT was associated with the major allele(s) (Fig. 4D and Supplementary Table S5), as originally detected in the peak SNP (Table 1). Although, the haplotype differences were insignificant for Os03g0717000 based on ANOVA, the p value was marginal of 0.0517 (Supplementary Table S5), indicating the weak association of this gene with GT when compared with other candidates. For Os03g0716900, Hap2 had significantly higher GT than Hap1, which suggested that the eight missense mutations clustered in the exon affect its function. For Os03g0717600, Hap3 differs from Hap1 and Hap2 at two missense SNPs at S3_29042521 and S3_29042839 (the 10th and 11th SNP of haplotype) in exons 6 and 5. Apparently, these two nonsynonymous SNPs were responsible for the phenotypic differences for GT between the haplotypes. In the case of Os03g0717700, Hap2 had markedly higher GT than Hap1 and Hap3. The five missense mutations at S3_29054380, S3_29054799, S3_29054902, S3_29054985 and S3_29055829 in exons 5, 7 and 9 of this gene were potentially responsible for the QTL (Fig. 4D and Supplementary Table S5). Taken together, additional evidence from gene knockout or gene knockdown by genetic transformation experiments is required to determine which one or ones of these candidate genes are the real causal one for qGT3.
Overall, we identified seven candidates for the two new QTL affecting AC and GT. We realize that the functional inferences of causal genes for the identified QTL based on annotations of gene function and structure may not be sufficient. Currently, further functional validations for these candidates by genetic transformation are in progress to validate the functionalities of the candidate genes on AC and GT. Nevertheless, the current results we presented here provided useful information for genetic validation of the identified QTL candidates and for marker-assisted modification of rice grain quality traits in future breeding.
Conclusion
Considerable genetic variations for four grain quality traits, AC, GC, GT and PC were observed in the current panel. Through GWAS, 19 QTL for the investigated traits were identified. Among them, four QTL were close to SSRGs and five QTL were adjacent to previously identified QTL for related traits. The remaining 10 QTL (three for AC, three for GC, two for GT and two for PC) were novel ones. Nine candidate genes of four important QTL were determined by gene-based association and haplotype analyses, including two known genes (Wx and ALK) and seven novels. These newly identified candidate genes affecting rice grain AC and GT provide valuable information for future functional characterization of these candidates and for MAS-based breeding for improving rice grain ECQ.
Electronic supplementary material
Acknowledgements
This work was funded by the “863” Key Project to JLX (2014AA10A601) from the Chinese Ministry of Science & Technology (http://www.863.gov.cn/); the Shenzhen Peacock Plan (http://www.szsti.gov.cn/) (#: 20130415095710361, Recipient: ZKL); the CAAS Innovative Team Award to JL Xu’s team (http://www.caas.net.cn/), and the Bill & Melinda Gates Foundation (OPP1130530) to ZKL.
Author Contributions
Z.L. and J.X. designed the experiment; X.W., J.Z. and K.C. performed the phenotypic collection; Y.P., X.W., Z.W. and G.Y. performed the analysis and interpretation of the data; X.W., J.X. and J.A. drafted the manuscript; Z.L., J.X. and Y.P. revised the manuscript; all authors approved the final version to be published.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Xiaoqian Wang and Yunlong Pang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17347-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianlong Xu, Email: xujlcaas@126.com.
Zhikang Li, Email: zhkli1953@126.com.
References
- 1.Tian Z, et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA. 2009;106:21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, J. Genes and QTLs for rice grain quality improvement. InTech–Open Science Open Mind, 239–278 (2014).
- 3.Bao J, Shen S, Sun M, Corke H. Analysis of genotypic diversity in the starch physicochemical properties of nonwaxy rice: apparent amylose content, pasting viscosity and gel texture. Starch - Stärke. 2006;58:259–267. doi: 10.1002/star.200500469. [DOI] [Google Scholar]
- 4.Hossaina MS, Singh AK, Fasih-uz-Zaman Cooking and eating characteristics of some newly identified inter sub-specific (indica/japonica) rice hybrids. ScienceAsia. 2009;35:320. doi: 10.2306/scienceasia1513-1874.2009.35.320. [DOI] [Google Scholar]
- 5.Hu P, Zhao H, Duan Z, Linlin Z, Wu D. Starch digestibility and the estimated glycemic score of different types of rice differing in amylose contents. Journal of Cereal Science. 2004;40:231–237. doi: 10.1016/j.jcs.2004.06.001. [DOI] [Google Scholar]
- 6.Little RR, Hilder GB, Dawson EH. Differential effect of dilute alkali on 25 varieties of milled white rice. Cereal Chemistry. 1958;35:111–126. [Google Scholar]
- 7.Juliano, B. In Rice chemistry and technology 443–513 (American Association of Cereal Chemists (AACC) 1985).
- 8.Juliano, B. O. & Villareal, C. Grain quality evaluation of world rices. (Int. Rice Res. Inst., 1993).
- 9.James MG, Denyer K, Myers AM. Starch synthesis in the cereal endosperm. Current opinion in plant biology. 2003;6:215–222. doi: 10.1016/S1369-5266(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 10.Zhou S-R, Yin L-L, Xue H-W. Functional genomics based understanding of rice endosperm development. Current opinion in plant biology. 2013;16:236–246. doi: 10.1016/j.pbi.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Kharabian-Masouleh A, Waters DL, Reinke RF, Ward R, Henry RJ. SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Scientific reports. 2012;2:557. doi: 10.1038/srep00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, et al. Identification of quantitative trait loci responsible for rice grain protein content using chromosome segment substitution lines and fine mapping of qPC-1 in rice (Oryza sativa L.) Molecular Breeding. 2015;35:1–9. doi: 10.1007/s11032-015-0328-z. [DOI] [Google Scholar]
- 13.Tian R, Jiang GH, Shen LH, Wang LQ, He YQ. Mapping quantitative trait loci underlying the cooking and eating quality of rice using a DH population. Molecular Breeding. 2005;15:117–124. doi: 10.1007/s11032-004-3270-z. [DOI] [Google Scholar]
- 14.Wang L, et al. Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theoretical and Applied Genetics. 2007;115:463–476. doi: 10.1007/s00122-007-0580-7. [DOI] [PubMed] [Google Scholar]
- 15.Xu F, et al. QTL mapping for rice grain quality: a strategy to detect more QTLs within sub-populations. Molecular Breeding. 2015;35:1–11. doi: 10.1007/s11032-015-0202-z. [DOI] [Google Scholar]
- 16.Yan B, et al. Analysis of minor quantitative trait loci for eating and cooking quality traits in rice using a recombinant inbred line population derived from two indica cultivars with similar amylose content. Molecular Breeding. 2014;34:2151–2163. doi: 10.1007/s11032-014-0170-8. [DOI] [Google Scholar]
- 17.Huang X, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nature genetics. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature genetics. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 19.Zhao K, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nature communications. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, Zhou L, Ponce K, Ye G. The Usefulness of Known Genes/Qtls for Grain Quality Traits in an Indica Population of Diverse Breeding Lines Tested using Association Analysis. Rice. 2015;8:1–13. doi: 10.1186/s12284-015-0064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, X. et al. New Candidate Genes Affecting Rice Grain Appearance and Milling Quality Detected by Genome-Wide and Gene-Based Association Analyses. Frontiers in plant science7, 10.3389/fpls.2016.01998 (2016). [DOI] [PMC free article] [PubMed]
- 22.He, J. et al. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Frontiers in plant science5, 10.3389/fpls.2014.00484 (2014). [DOI] [PMC free article] [PubMed]
- 23.Chen H, et al. A high-density SNP genotyping array for rice biology and molecular breeding. Molecular plant. 2014;7:541–553. doi: 10.1093/mp/sst135. [DOI] [PubMed] [Google Scholar]
- 24.Yano K, et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nature genetics. 2016;48:927–934. doi: 10.1038/ng.3596. [DOI] [PubMed] [Google Scholar]
- 25.Wang, X. et al. New Candidate Genes Affecting Rice Grain Appearance and Milling Quality Detected by Genome-Wide and Gene-Based Association Analyses. Frontiers in plant science7, 10.3389/fpls.2016.01998 (2017). [DOI] [PMC free article] [PubMed]
- 26.3K RGP. The 3,000 rice genomes project. GigaScience3, 7, 10.1186/2047-217x-3-7 (2014).
- 27.Zheng TQ, et al. Rice functional genomics and breeding database (RFGB): 3 K-rice SNP and InDel sub-database (in Chinese) Chin Sci Bull. 2015;60:367–371. doi: 10.1360/N972014-01231. [DOI] [Google Scholar]
- 28.Alexandrov N, et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic acids research. 2015;43:D1023–1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yun Y-T, et al. QTL Mapping of Grain Quality Traits Using Introgression Lines Carrying Oryza rufipogon Chromosome Segments in Japonica Rice. Rice. 2016;9:62. doi: 10.1186/s12284-016-0135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo Y-J, et al. Effects of allelic variations in starch synthesis-related genes on grain quality traits of Korean nonglutinous rice varieties under different temperature conditions. Breeding Science. 2014;64:164–175. doi: 10.1270/jsbbs.64.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nature genetics. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilhjalmsson BJ, Nordborg M. The nature of confounding in genome-wide association studies. Nature reviews. Genetics. 2013;14:1–2. doi: 10.1038/nrg3382. [DOI] [PubMed] [Google Scholar]
- 33.Wang ZY, et al. The amylose content in rice endosperm is related to the post‐transcriptional regulation of the Waxy gene. The Plant Journal. 1995;7:613–622. doi: 10.1046/j.1365-313X.1995.7040613.x. [DOI] [PubMed] [Google Scholar]
- 34.Gao Z, et al. Map-based cloning of the ALKgene, which controls the gelatinization temperature of rice. Science in China Series C. 2003;46:661. doi: 10.1360/03yc0099. [DOI] [PubMed] [Google Scholar]
- 35.Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y. Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik. 2002;104:1–8. doi: 10.1007/s001220200000. [DOI] [PubMed] [Google Scholar]
- 36.Bao J, et al. QTL mapping for the paste viscosity characteristics in rice (Oryza sativa L.) Theoretical and Applied Genetics. 2000;100:280–284. doi: 10.1007/s001220050037. [DOI] [Google Scholar]
- 37.Fan CC, et al. The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theoretical and Applied Genetics. 2005;110:1445–1452. doi: 10.1007/s00122-005-1975-y. [DOI] [PubMed] [Google Scholar]
- 38.Yang F, et al. Association mapping of starch physicochemical properties with starch synthesis-related gene markers in nonwaxy rice (Oryza sativa L.) Molecular Breeding. 2014;34:1747–1763. doi: 10.1007/s11032-014-0135-y. [DOI] [Google Scholar]
- 39.Lu FH, Park YJ. An SNP downstream of the OsBEIIb gene is significantly associated with amylose content and viscosity properties in rice (Oryza sativa L.) Journal of Cereal Science. 2012;56:706–712. doi: 10.1016/j.jcs.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Umemoto T, Yano M, Satoh H, Shomura A, Nakamura Y. Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theoretical and Applied Genetics. 2002;104:1–8. doi: 10.1007/s001220200000. [DOI] [PubMed] [Google Scholar]
- 41.Kharabian-Masouleh A, Waters DL, Reinke RF, Ward R, Henry RJ. SNP in starch biosynthesis genes associated with nutritional and functional properties of rice. Scientific reports. 2012;2:557. doi: 10.1038/srep00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo J, Jobling SA, Millar A, Morell MK, Li Z. Allelic effects on starch structure and properties of six starch biosynthetic genes in a rice recombinant inbred line population. Rice. 2015;8:15. doi: 10.1186/s12284-015-0046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebadi AA, Farshadfar E, Rabiei B. Mapping QTLs controlling cooking and eating quality indicators of Iranian rice using RILs across three years. Australian Journal of Crop Science. 2013;7:1494. [Google Scholar]
- 44.Wan XY, et al. QTL detection for eating quality of cooked rice in a population of chromosome segment substitution lines. Theoretical and Applied Genetics. 2004;110:71–79. doi: 10.1007/s00122-004-1744-3. [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Wan J, Xia J, Yano M. Mapping of quantitative trait loci controlling physico-chemical properties of rice grains (Oryza sativa L.) Breeding Science. 2003;53:209–215. doi: 10.1270/jsbbs.53.209. [DOI] [Google Scholar]
- 46.Leng Y, et al. Mapping of QTLs for eating and cooking quality-related traits in rice (Oryza sativa L.) Euphytica. 2014;197:99–108. doi: 10.1007/s10681-013-1055-3. [DOI] [Google Scholar]
- 47.Isshiki M, et al. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. The Plant Journal. 1998;15:133–138. doi: 10.1046/j.1365-313X.1998.00189.x. [DOI] [PubMed] [Google Scholar]
- 48.Hirano HY, Eiguchi M, Sano Y. A single base change altered the regulation of the Waxy gene at the posttranscriptional level during the domestication of rice. Molecular biology and evolution. 1998;15:978–987. doi: 10.1093/oxfordjournals.molbev.a026013. [DOI] [PubMed] [Google Scholar]
- 49.Cai XL, Wang ZY, Xing YY, Zhang JL, Hong MM. Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. The Plant Journal. 1998;14:459–465. doi: 10.1046/j.1365-313X.1998.00126.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.