Abstract
The germ cell lineage is specified early in embryogenesis and undergoes complex developmental programs to generate gametes. Here, we conducted genetic studies to investigate the role of Utf1 (Undifferentiated embryonic cell transcription factor 1) in mouse germ cell development. Utf1 is expressed in pluripotent embryonic stem (ES) cells and regulates ES cell differentiation. In a proteomics screen, we identified UTF1 among 38 proteins including DNMT3L and DND1 that associate with chromatin in embryonic testes. We find that UTF1 is expressed in embryonic and newborn gonocytes and in a subset of early spermatogonia. Ubiquitous inactivation of Utf1 causes embryonic lethality in mice with a hybrid genetic background. Male mice with a germline-specific deletion of Utf1 resulting from Prdm1-Cre mediated recombination are born with significantly fewer gonocytes and exhibit defective spermatogenesis and reduced sperm count as young adults. These defects are ameliorated in older animals. These results demonstrate that UTF1 is required for embryonic development and regulates male germ cell development.
Introduction
In sexually reproducing organisms, germ cells pass genetic information to the next generation. The germ cell lineage arises in early embryogenesis. Toward the end of gastrulation, murine primordial germ cells (PGCs) arise from the epiblast and, as a small cluster of cells, adopt a germ cell fate outside the embryo proper1,2. Subsequently, PGCs proliferate and migrate to the bipotential genital ridges, which develop into either testes or ovaries. Following sex determination at embryonic day 12.5 (E12.5), male germ cells are enclosed in seminiferous cords to become gonocytes. Gonocytes continue to proliferate and enter mitotic arrest at about E163. Gonocytes undergo genome-wide epigenetic reprogramming and only resume proliferation shortly after birth3. In contrast, female germ cells enter meiosis, progress through early stages of meiosis I, and become arrested at the dictyate stage of meiosis I at birth. The number of oocytes is finite in females at birth. In contrast, males produce sperm continuously throughout adult life due to the presence of spermatogonial stem cells in the testis3,4. Spermatogonial stem cells, a subset of spermatogonia, are capable of self-renewal and differentiation. At puberty, spermatogonia enter meiosis to become spermatocytes, which give rise to post-meiotic haploid spermatids and, finally, to mature spermatozoa.
Embryonic stem (ES) cells are derived from the inner cell mass of the blastocyst. ES cells are pluripotent and capable of self-renewal and differentiation into different cell types. Undifferentiated embryonic cell transcription factor 1 (Utf1) is expressed in pluripotent ES cells and embryonal carcinoma cells, but is rapidly downregulated upon differentiation5. UTF1 is a transcription repressor protein that associates with chromatin in mouse ES cells in a dynamic manner similar to core histone proteins6. UTF1 prevents ES cell chromatin decondensation7. Utf1 is a transcriptional target of OCT4 and SOX28,9. Mechanistically, UTF1 modulates the epigenetic state of bivalent genes and prevents aberrant gene expression through mRNA decapping in ES cells7,10. These UTF1 functions are critical for ES cell pluripotency and differentiation. Indeed, knockdown or inactivation of Utf1 causes a delay in ES cell differentiation6,10.
In mouse embryos, Utf1 is expressed in the inner cell mass, the primitive ectoderm and the extra-embryonic tissue, but not in the mesoderm5. Downregulation of Utf1 expression coincides with differentiation of the primitive ectoderm into mesoderm. Utf1 is not expressed in most adult tissues except in testis and ovary. In rats, Utf1 expression is restricted to a subpopulation of early spermatogonia in the adult testis11. In humans, Utf1 is also expressed in spermatogonia12. The phenotype of Utf1-null mouse mutants depends on the strain background. In mice of C57BL/6J strain background, inactivation of Utf1 causes embryonic developmental delay, resulting in lethality within 2 days after birth, whereas Utf1-null mice on a C57BL/6J × ICR mixed genetic background are viable and fertile13.
Here we report a role of UTF1 in embryogenesis and male germ cell development. In a proteomics screen, we identified UTF1 as one of 38 chromatin-associated proteins enriched in developing male embryonic germ cells. We find that UTF1 is present in the nucleus of embryonic male germ cells. Inactivation of Utf1 results in embryonic lethality on the C57BL/6 × 129 hybrid genetic background. Furthermore, inactivation of Utf1 leads to a reduction in the number of gonocytes at birth and defective spermatogenesis in adult males.
Results
UTF1 is a chromatin-associated protein in male embryonic germ cells
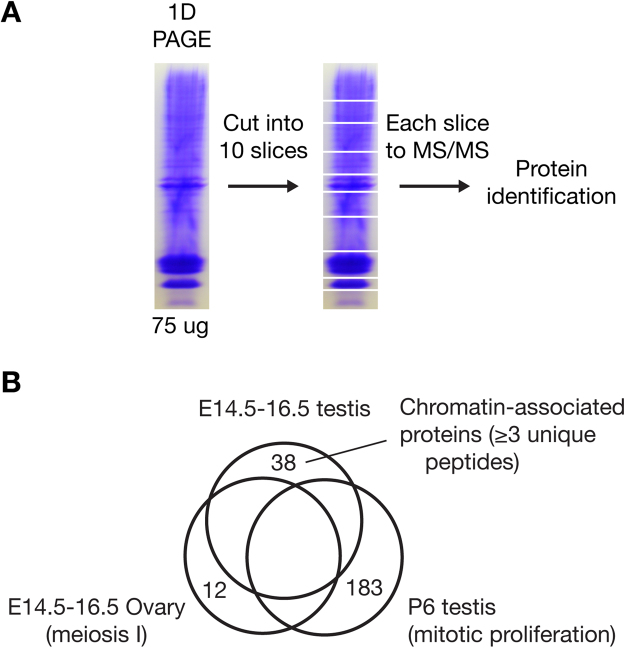
We designed a proteomic screen to identify proteins that are associated with the chromatin of embryonic gonocytes in the developing mouse testis. Adapting our previously described methodology14, we performed a mass spectrometry analysis of chromatin-associated proteins from embryonic (E14.5-16.5) testes (Fig. 1A) and ovaries, respectively. At this stage of development, male germ cells (called gonocytes) still proliferate but undergo genome-wide epigenetic reprogramming, whereas female germ cells enter the prophase of meiosis I3 , 15. In the post-natal day 6 (P6) testis, germ cells (now called spermatogonia) proliferate by mitosis. To identify proteins specific to or enriched in male embryonic germ cells, we subtracted all proteins also found in E14.5-16.5 ovary and/or P6 testis. With this approach, we excluded any chromatin-associated proteins that are common to meiotic germ cells of the embryonic ovary and spermatogonia of the P6 testis and also eliminated proteins from somatic cell types of the testis, which are present in both embryonic and post-natal testes. Using a cutoff of at least three unique peptides, we identified 38 chromatin-associated proteins in embryonic male germ cells (Fig. 1B and Table 1).
Figure 1.
Proteomic identification of chromatin-associated proteins from E14.5-E16.5 testes. (A) Experimental approach. Identification of chromatin-associated proteins from E14.5-16.5 embryonic mouse testes was used here. (B) Systematic identification of chromatin-associated proteins enriched in E14.5-E16.5 testes by subtractive analysis of three proteomic datasets from E14.5-E16.5 testes (771 proteins, each with at least three unique peptides), E14.5-E16.5 ovaries (534 proteins), and postnatal day 6 (P6) testes (1011 proteins). The number of proteins with at least 3 unique peptides in one dataset but none in the other two datasets are shown.
Table 1.
Chromatin-associated proteins identified in male embryonic germ cells.
| Protein | Gene Accession | Protein Accession | No. Unique Peptides | Percent Coverage | Length (aa) | Official Full Name |
|---|---|---|---|---|---|---|
| UTF1 | NM_009482 | NP_033508 | 6 | 32% | 340 | Undifferentiated embryonic cell transcription factor 1 |
| DND1 | NM_173383 | NP_775559 | 6 | 23% | 353 | DND microRNA-mediated repression inhibitor 1 |
| DNMT3L | NM_001081695 | NP_001075164 | 5 | 16% | 422 | DNA (cytosine-5)-methyltransferase 3-like |
| WDR43 | NM_175639 | NP_783570 | 5 | 11% | 678 | WD repeat domain 43 |
| ZFP326 | NM_018759 | NP_061229 | 4 | 8% | 581 | Zinc finger protein 326 |
| INTS7 | NM_178632 | NP_001293132 | 4 | 6% | 967 | Integrator complex subunit 7 |
| UTP11L | NM_026031 | NP_080307 | 4 | 15% | 254 | UTP11-like, U3 small nucleolar ribonucleoprotein |
| KDM3A | NM_001038695 | NP_001033784 | 4 | 4% | 1324 | Lysine demethylase 3A |
| EXOSC10 | NM_016699 | NP_057908 | 4 | 9% | 888 | Exosome component 10 |
| PHF10 | NM_024250 | NP_077212 | 4 | 12% | 498 | PHD finger protein 10 |
| PPP2R5C | NM_001135001 | NP_001128473 | 4 | 10% | 510 | Protein phosphatase 2, regulatory subunit B′, gamma |
| THOC2 | NM_001033422 | NP_001028594 | 3 | 2% | 1595 | THO complex 2 |
| PPP2CB | NM_017374 | NP_059070 | 3 | 13% | 310 | Protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform |
| WTAP | NM_175394 | NP_780603 | 3 | 31% | 152 | Wilms tumour 1-associating protein |
| YLPM1 | NM_178363 | NP_848140 | 3 | 3% | 2140 | YLP motif containing 1 |
| NOC2L | NM_021303 | NP_067278 | 3 | 5% | 751 | NOC2 like nucleolar associated transcriptional repressor |
| SUPT6 | NM_009297 | NP_033323 | 3 | 3% | 1727 | Suppressor of Ty 6 |
| CTNNB1 | NM_001165902 | NP_001159374 | 3 | 6% | 782 | Catenin (cadherin associated protein), beta 1 |
| NFYC | NM_001048168 | NP_001041633 | 3 | 10% | 336 | Nuclear transcription factor-Y gamma |
| KAT7 | NM_177619 | NP_808287 | 3 | 8% | 523 | K(lysine) acetyltransferase 7 |
| LDB1 | NM_001113408 | NP_001106879 | 3 | 11% | 412 | LIM domain binding 1 |
| ACTR2 | NM_146243 | NP_666355 | 3 | 13% | 395 | ARP2 actin-related protein 2 |
| DDB1 | NM_015735 | NP_056550 | 3 | 4% | 1141 | Damage specific DNA binding protein 1 |
| MRPS9 | NM_023514 | NP_076003 | 3 | 11% | 391 | Mitochondrial ribosomal protein S9 |
| SCFD2 | NM_001114660 | NP_001108132 | 3 | 8% | 685 | Sec1 family domain containing 2 |
| MLLT4 | NM_010806 | NP_034936 | 3 | 3% | 1821 | Myeloid/lymphoid or mixed-lineage leukemia; translocated to, 4 |
| KDM2A | NM_001001984 | NP_001001984 | 3 | 3% | 1162 | Lysine (K)-specific demethylase 2A |
| GFM2 | NM_001146043 | NP_001139515 | 3 | 6% | 742 | G elongation factor, mitochondrial 2 |
| ATM | NM_007499 | NP_031525 | 3 | 2% | 3067 | Ataxia telangiectasia mutated |
| ANAPC5 | NM_001042491 | NP_001035956 | 3 | 6% | 728 | Anaphase-promoting complex subunit 5 |
| BZW2 | NM_025840 | NP_080116 | 3 | 12% | 420 | Basic leucine zipper and W2 domains 2 |
| TCF20 | NM_013836 | NP_038864 | 3 | 3% | 1966 | Transcription factor 20 |
| ACOT7 | NM_133348 | NP_579926 | 3 | 10% | 380 | Acyl-CoA thioesterase 7 |
| CUL4B | NM_028288 | NP_082564 | 3 | 5% | 971 | Cullin 4B |
| ARID4B | NM_194262 | NP_919238 | 3 | 4% | 1315 | AT rich interactive domain 4B (RBP1-like) |
| SAP30BP | NM_020483 | NP_065229 | 3 | 12% | 309 | SAP30-binding protein |
| NEK1 | NM_175089 | NP_001280568 | 3 | 4% | 1204 | NIMA (never in mitosis gene a)-related expressed kinase 1 |
| CWC27 | NM_026072 | NP_080348 | 3 | 10% | 470 | CWC27 spliceosome-associated protein |
Several proteins identified in our screen have a well-characterized role in germline development, including DND1 and DNMT3L (Table 1). DND microRNA-mediated repression inhibitor 1 (DND1) is essential for primordial germ cell migration and survival in zebrafish, and has a strong influence on differentiation and cell cycle control in the mammalian germ line16,17. A mutation in Dnd1 causes germ cell loss and testicular germ cell tumors in mice18. DNA methyltransferase 3-like (DNMT3L) establishes maternal genomic imprints and maintains silencing of LINE1/IAP retrotransposons in the male germ line19–21. Deletion of Dnmt3l causes loss of embryos in knockout females and sterility in males. The identification of known proteins in this proteomics screen confirmed the validity of our approach and suggests that other proteins identified may also play an important role in the development of male embryonic germ cells.
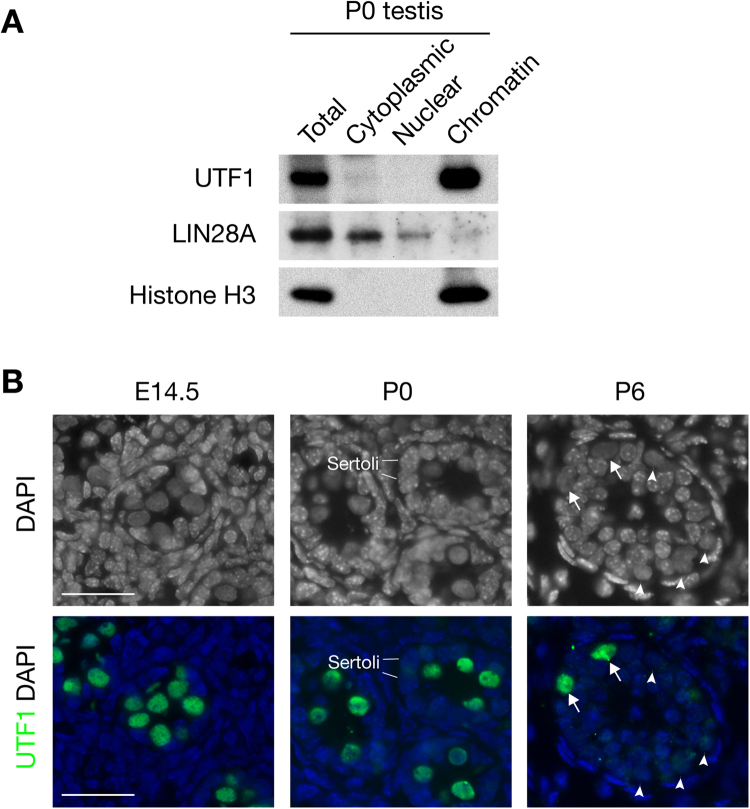
In our proteomics screen, UTF1 was the protein with the highest number of unique peptides (Table 1). Consistent with proteomic data, UTF1 was detected in the chromatin fraction of protein lysates from neonatal (P0) testes, but not in the cytoplasmic and soluble nuclear extracts (Fig. 2A). To assess the developmental and cell-type specific expression and intracellular localization of UTF1, we performed immunostaining of testis sections. UTF1 was present in the nuclei of all gonocytes in embryonic (E14.5) and P0 testes, but only in a subpopulation of spermatogonia in postnatal day 6 testis (Fig. 2B). UTF1 was not detected in spermatocytes or spermatids in postnatal testis. These results suggest that UTF1 may play a role in early male germ cell development.
Figure 2.
UTF1 is associated with chromatin in early male germ cells. (A) Western blot analysis of UTF1 in total and subcellular fractions of neonatal (P0) testis. LIN28A serves as a cytoplasmic protein control35. Histone H3 serves as a chromatin protein control. Images were cropped to show the relevant bands. The full-length blots are presented in Supplementary Figure S1. (B) Immunolocalization of UTF1 in sections from wild-type E14.5, neonatal (P0), and postnatal day 6 (P6) testes. DAPI staining of DNA shows the nuclear appearance of germ cells and supporting cells. Gonocytes (E14.5 and P0) and early spermatogonia (P6) lack heterochromatin and exhibit a uniform dark appearance, whereas somatic cells such as Sertoli cells are rich in heterochromatin visualized as punctate bright dots. In P6 testes, UTF1-positive and UTF1-negative spermatogonia are indicated by arrows and arrowheads respectively. Scale bars, 25 µm.
Ubiquitous Utf1 deletion is embryonic lethal
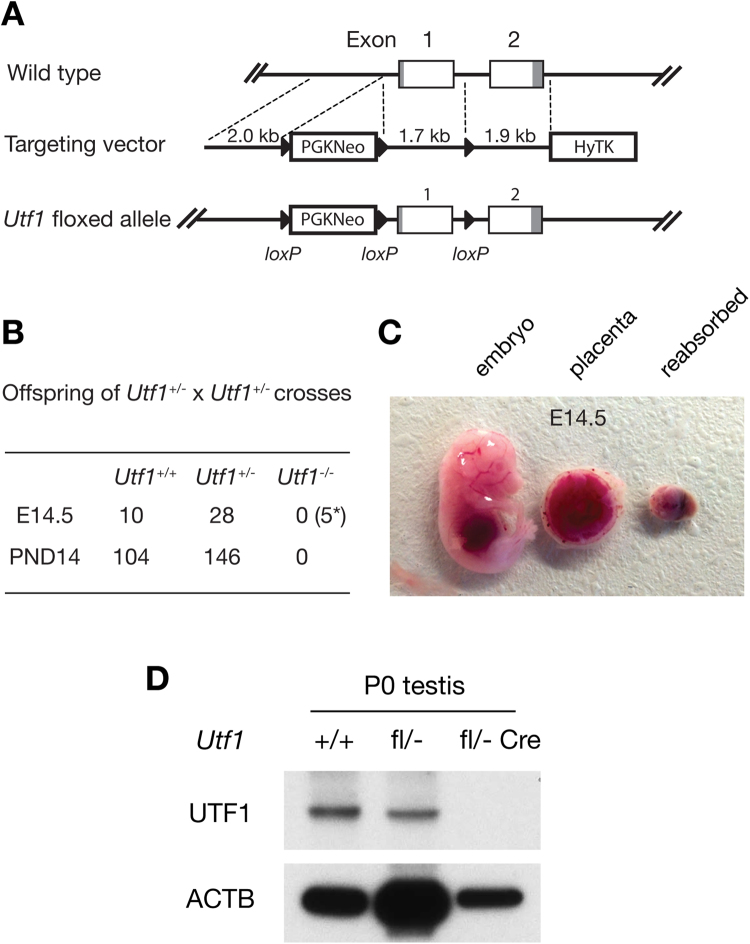
To ascertain the tissue-specific requirement for Utf1 in the gonad, we generated a conditional knockout (floxed) allele by homologous recombination in mouse ES cells. The Utf1 gene is comprised of two exons located on mouse chromosome 7 and encodes a protein of 339 amino acids. In the conditional knockout allele, the first exon was flanked by loxP sites (Utf1 fl) (Fig. 3A), such that Cre recombinase-mediated deletion of exon 1 produces a null allele that lacks the potential promoter region and the coding sequence for the N-terminal 188 amino acids including the start codon. Utf1 fl/fl mice were viable and fertile with no gross abnormalities.
Figure 3.
UTF1 is required for embryonic development. (A) Schematic diagram of the Utf1 wild-type allele, targeting construct, and conditional knockout allele. Mouse Utf1 gene maps to chromosome 7 and consists of two exons. (B) Embryos and pups from Utf1 +/− × Utf1 +/− matings. The number in the bracket labelled by * indicates the number of resorptions found. (C) E14.5 mouse embryos from Utf1 +/− × Utf1 +/− matings. Left and center: wild type embryo and placenta. Right: reabsorbed embryo of presumptive Utf1 −/− genotype. (D) Absence of UTF1 protein in neonatal (P0) Utf1 fl/− Prdm1-Cre testes. Note the reduction in UTF1 abundance in Utf1 fl/− testes. ACTB serves as a loading control. The full-length blots are presented in Supplementary Figure S1.
From the intercross of Utf1 fl/fl mice with Actb-Cre mice, in which Cre recombinase is expressed ubiquitously22, we obtained Utf1 +/− mice. The Utf1 +/− mice were on a mixed genetic background (FVB/N, C57BL/6, and 129). Interbreeding of Utf1 heterozygous mice failed to yield any Utf1 −/− offspring, and the genotype frequency of live-born pups was skewed towards the wild type (Utf1 +/+ = 104, Utf1 +/− = 146) [χ2(1, N = 250) = 34.54, p < 4 × 10−9] (Fig. 3B). To determine the time of embryonic death, we performed timed heterozygote matings and isolated embryos from the uteri of Utf1 +/− females at various embryonic stages. Homozygous Utf1 −/− embryos were absent at E10.5 (N = 38, Utf1 +/+ = 10, Utf1 +/− = 28, reabsorbed = 5) [χ2 = 4.29, p < 0.04] (Fig. 3B), however, residual placental tissues from reabsorbed embryos were detectable (Fig. 3C). This result demonstrates that Utf1 is essential for early embryo viability.
Conditional inactivation of Utf1 causes a reduction in the number of gonocytes
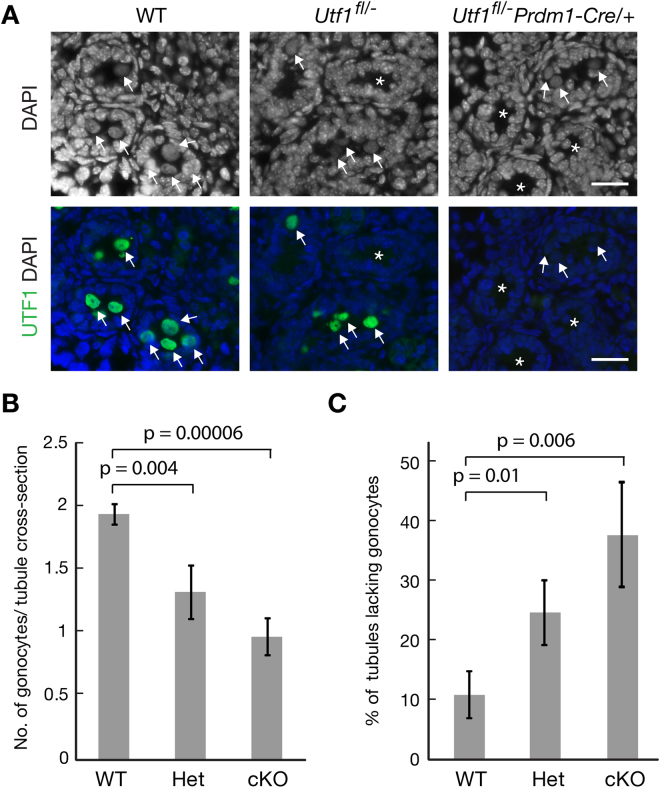
To elucidate the role of Utf1 in germ cells, we inactivated the conditional allele using a mouse strain that expresses Cre recombinase under the control of the mouse Prdm1 promoter (Prdm1-Cre). Prdm1 expression is induced by BMP4 early in the post-implantation embryo23. PRDM1 is a marker of the founding population of primordial germ cells (PGCs), and Prdm1-Cre expression in PGCs begins at E6.2523. Utf1 fl/fl mice were bred with Prdm1-Cre mice to produce Utf1 fl/+ Prdm1-Cre tg/+ mice, which were then crossed with Utf1 fl/fl females to produce Utf1 fl/− Prdm1-Cre tg/+ mice (referred to as Utf1 cKO). The Utf1 cKO mice were on a mixed genetic background (C57BL/6 × 129). Western blot analyses showed that UTF1 protein was absent in Utf1 cKO testes and reduced in abundance in Utf1 fl/− testes (Fig. 3D). Immunostaining of testis sections from newborn mice confirmed that UTF1 was present in the nucleus of both wild-type and Utf1 fl/− gonocytes, but absent in Utf1 cKO gonocytes (Fig. 4A), indicating that Cre-mediated deletion was complete at birth. Gonocytes were present in the testes of Utf1 cKO neonate males after early embryonic deletion of Utf1. However, in both heterozygous and cKO testes, a subset of tubules were apparently devoid of gonocytes (Fig. 4A). Quantitative analysis revealed that Utf1 fl/− and cKO testes contained significantly fewer gonocytes compared with wild-type testes, with a relatively larger reduction of gonocyte number in cKO testes (Fig. 4B). Consistent with this observation, the percentage of tubules lacking gonocytes was significantly higher in Utf1 fl/− and Utf1 cKO testes than in the wild type, again with a relatively larger increase in Utf1 cKO testes (Fig. 4C).
Figure 4.
Reduced number of gonocytes in Utf1 cKO neonatal testis. (A) Detection of gonocytes in seminiferous tubule cross-sections from newborn (P0) wild type (WT), Utf1 fl/− (het), and Utf1 fl/− Prdm1-Cre/+ (cKO) testes. Sections were immunostained with anti-UTF1 antibody and counterstained with DAPI. Gonocytes are indicated by arrows. Tubule cross-sections lacking gonocytes are designated by asterisks. Scale bars, 25 µm. (B,C) Quantitative analysis of gonocytes in seminiferous tubules from the testes of newborn WT (n = 4 animals), het (n = 5), and cKO (n = 6). For each animal, at least 150 tubule cross sections were counted. (B) Number of gonocytes per seminiferous tubule cross-section. (C) Percentage of tubules lacking gonocytes.
Conditional inactivation of Utf1 causes spermatogenic defects in adult mice
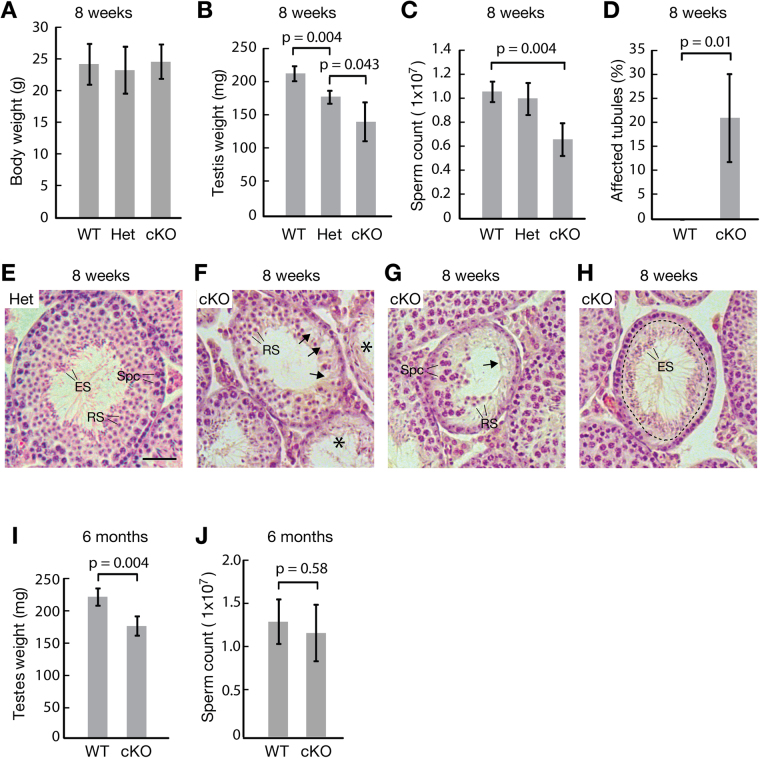
We examined the effect of Utf1 deletion on spermatogenesis in 8-week-old males. The body weight was similar between different genotypes (Fig. 5A). Testis weight was significantly reduced in both heterozygous and cKO animals compared to wild type (Fig. 5B). Sperm count of cKO males was decreased by approximately 40% compared to wild type, but sperm count of Utf1 heterozygous males was not significantly reduced (Fig. 5C). Histological analysis revealed that approximately 20% of seminiferous tubules from 8-week-old cKO males exhibited severe defects in spermatogenesis, while the remaining tubules appeared to be normal (Fig. 5D). Seminiferous tubules from Utf1 +/− testes contained a full spectrum of spermatogenic cells and lacked apparent histological defects (Fig. 5E). However, seminiferous tubules from Utf1 cKO testes exhibited a variety of defects. Some mutant tubules displayed a general paucity of germ cells (the middle tubule in Fig. 5F) and other tubules (indicated by asterisks) lacked almost all germ cells (Fig. 5F). Some mutant tubules showed a localized loss of all germ cells, including spermatogonia, spermatocytes, and round spermatids (Fig. 5G). In the remaining mutant tubules, loss of an entire middle layer of germ cells - spermatocytes - was observed (Fig. 5H). These testicular defects are heterogeneous and suggest that UTF1 plays an important role in spermatogenesis.
Figure 5.
Impaired spermatogenesis in adult Utf1 cKO males. (A–D) Phenotype of adult (8-week-old) wild-type (n = 4), heterozygous (n = 3), and Utf1 fl/− Prdm1-Cre (cKO) males (n = 5). (A) Body weight. (B) Testis weight. (C) Sperm count. (D) Percentage of tubules with defective spermatogenesis. Only cross-sections were counted. Histological analysis of testes from 8-week-old Utf1 f/− (E) and cKO (F–H) males. Arrows indicate areas of germ cell loss (F,G), asterisks tubules depleted of nearly all germ cells (F), and a dashed circle layers of missing germ cells (H). Abbreviations: Spc, spermatocytes; RS, round spermatids; ES, elongating spermatids. (I,J) Phenotype of 6-month-old wild-type (n = 3) and cKO (n = 4) males. (I) Testis weight. (J) Sperm count.
Mating tests showed that Utf1 cKO males exhibited significantly reduced fertility (Table 2). Wild type males sired seven pups per litter on average, in contrast, Utf1 cKO males only produced two offspring per litter on average. The dramatic reduction in the fertility of Utf1 cKO males is consistent with the reduced sperm count.
Table 2.
Reduced fertility in Utf1 cKO malesa.
| Genotype | No. of litters | No. of pups | litter sizeb |
|---|---|---|---|
| WT (n = 3 males) | 8 | 55 | 6.9 ± 1.1 |
| Utf1 cKO (n = 3 males) | 12 | 23 | 1.9 ± 1.2c |
aEach male was housed with two females at the age of 8-weeks for 2–3 months. Pups born were checked and recorded daily.
bLitter size is shown as average ± s.d.
cThe p vale is 0.0001 by Student’s t-Test.
Given that not all tubules in relatively young adult cKO mice exhibited abnormalities, we evaluated whether spermatogenic defects would become more severe with age. The testes from 6-month-old cKO testes remained smaller than wild type on average (Fig. 5I), although the relative difference was reduced compared to the 8-week-old group. The sperm count of cKO animals was indistinguishable from that of wild-type males (Fig. 5J). Histological analysis of testes revealed that tubules with abnormal morphology were rare ( < 1%) in both wild-type or cKO 6-month-old adult males. These results suggest that spermatogenesis in Utf1 cKO testes may recover over time due to the activity of spermatogonial stem cells in the intact regions of the seminiferous epithelium.
Discussion
Here we report that UTF1 plays important roles during two stages of mouse male germ cell development: embryonic gonocyte development and postnatal spermatogenesis. Inactivation of Utf1 results in a significant reduction in the number of gonocytes at birth. Two potential mechanisms may explain the observed gonocyte depletion in Utf1 cKO mice: reduction of the founder population of PGCs or a defect in PGC proliferation. Because Prdm1 is required for PGC specification, Prdm1-Cre-triggered deletion of Utf1 is expected to occur concomitantly with PGC specification. Therefore, the reduction in gonocytes could result from a defect in the specification of PGCs from epiblast cells, leading to a smaller founding population of PGCs. Utf1 cKO female mice are fertile and display no apparent histological defects in ovaries (data not shown). A mild reduction in the number of PGCs prior to sex determination might not have an observable effect on oogenesis, as fetal oocytes are present in excess, and two thirds of fetal oocytes become eliminated before birth24. On the other hand, the fertility of Utf1 cKO female mice argues against an exclusive role for UTF1 in PGC specification. Alternatively, loss of UTF1 may affect PGC proliferation. PGCs proliferate during migration to the genital ridges and are enclosed in seminiferous cords in males to become gonocytes, which continue to proliferate and enter mitotic arrest at E16 in males3. Studies in ES cells and iPS cells have revealed a role for Utf1 in cell proliferation10,25. Therefore, paucity of gonocytes in Utf1 cKO testes may result from reduced proliferation by mitosis prior to late-gestational mitotic arrest of male germ cells.
Adult Utf1 cKO males exhibit defects in spermatogenesis in a subset of seminiferous tubules. Shortly after birth, mouse gonocytes resume mitotic divisions and differentiate, with some becoming spermatogonial stem cells and the remainder developing into spermatocytes3. The paucity of germ cells (Fig. 5F) or localized loss of all stages of germ cells (Fig. 5G) in Utf1 mutant tubules could be due to the reduction or loss of gonocytes at birth. As a more likely scenario, loss of spermatocytes and the presence of elongated spermatids from the previous round of spermatogenesis in the same tubules suggest that gonocytes might fail to produce spermatogonial stem cells capable of differentiation (Fig. 5H). Recovery of spermatogenesis in older Utf1 cKO mice suggests that unaffected spermatogonial stem cells have the capacity to repopulate adjacent germ cell-depleted regions of seminiferous tubules. We tried to distinguish these two possibilities (defects in proliferation of gonocytes vs spermatogonial stem cells) by using the Ddx4-Cre, which begins to express at E1526. However, breeding of Ddx4-Cre male and Utf1 fl/fl female mice failed to produce the desired Utf1 fl/+ Ddx4-Cre offspring but instead resulted in Utf1 +/− Ddx4-Cre (genotype of tail genomic DNA) offspring, demonstrating global deletion of the Utf1 floxed allele. The global deletion of the Utf1 fl allele was likely due to the close proximity of the two loxP sites, the increased chromatin access of the Utf1 locus, and leaky Ddx4-Cre expression in somatic tissues.
The phenotype of Utf1-deficient mice depends on the genetic background. On a complete C57BL/6J background, Utf1-deficient mice are significantly smaller than wild-type littermates and all die within two days after birth13. Utf1 is expressed in extraembryonic ectodermal cells and its deletion causes a developmental delay of the placenta5,13. Therefore, the neonatal lethality phenotype most likely attributes to placental insufficiency. However, on a hybrid genetic background (75% ICR and 25% C57BL/6J), Utf1-deficient mice are viable and fertile albeit still smaller than wild-type mice13. In this study, we find that, on a mixed background (FVB/N, C57BL/6, and 129), inactivation of Utf1 causes a much more severe phenotype - embryonic lethality by E10.5. Genetic backgrounds can potentially influence the phenotypic expression of specific genes27,28. Indeed, the phenotypes of an increasing number of mouse mutants are strongly influenced by genetic background29–32. It is known that testicular germ cell tumors arise preferentially on the 129 strain background18. DND1 was also identified as a chromatin-associated protein in our proteomics screen (Table 1) and its mutation is responsible for the formation of testicular germ cell tumors on the 129 background18. Intriguingly, the role of DND1 in the regulation of mitotic arrest in male embryonic germ cells is strongly modulated by the genetic background17. The variable phenotypes of Utf1-deficient mutants on different genetic backgrounds is therefore likely due to the presence of different alleles of genes involved in the UTF1-mediated transcriptional and epigenetic pathways.
Methods
Ethics statement
All experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. All the methods were carried out in accordance with the approved guidelines.
Chromatin purification and proteomic screen
Chromatin was isolated from 100 mg of E14.5-E16.5 testes or ovaries as described previously14. 75 μg of chromatin-associated proteins from testes or ovaries were separated on 10% SDS-PAGE gels and stained with Coomassie blue. Each gel lane was cut into 10 slices, digested with trypsin and subjected to HPLC followed by MS/MS at the University of Pennsylvania Proteomic Core Facility. All spectra from 10 MS/MS runs per sample were pooled and analyzed using Scaffold 4 software (Proteome Software, Portland, Oregon), producing E14.5-E16.5 testis and E14.5-E16.5 ovary datasets. Mass spec data for chromatin-associated proteins from P6 testes were generated previously14.
Generation of Utf1fl mice
For Utf1 targeting construct generation, three Utf1 genomic DNA fragments were amplified from a BAC clone (RP23-182C11) template by high-fidelity PCR (Fig. 3A). A floxed neomycin-resistance cassette was subcloned 5′ of the Utf1 gene, and a loxP site was added between exons 1 and 2 by PCR. After validation of the gene targeting plasmid (pUtf1Ex1Flox) by sequencing, the KpnI-linearized construct was used to electroporate V6.5 (C57BL/6 × 129S4/SvJae) mouse ES cells33. ES cells were cultured in the presence of G418 (for positive selection) and ganciclovir (for negative selection). Among 96 G418-resistant ES clones, 18 had undergone homologous recombination at both homology arms. Two Utf1 fl/+ ES clones (Utf1B9 and Utf1C12) were injected into blastocysts, which were transferred to pseudopregnant female mice. Matings of male chimeras with C57BL/6J females confirmed germline transmission of the Utf1 fl allele. Genotyping for the floxed allele (181 bp) was performed by PCR with primers 5′-GTGGAGCAAGGTAGGAGCA-3′ and 5′-CAGGTTCGTCATTTTCCGCA-3′. Following Cre-mediated recombination, the Utf1 knockout allele (465 bp) was assayed by PCR with primers 5′-CAGAGTGTCGGTGCTCGTAA-3′ and 5′-GTTCAAGCCCCTAGTCACAAATC-3′. Prdm1-Cre and Actb-Cre transgenic mice were purchased from Jackson Laboratory (Stock numbers: 008827 and 003376, respectively)22,23. The Prdm1-Cre strain had been backcrossed to C57BL/6 four times by the donating investigator, and Actb-Cre transgenic mice were on a FVB/N background.
UTF1 antibody production
The cDNA sequence encoding the N-terminal 200 amino acids of mouse UTF1 was subcloned into the pQE-30 expression construct (Qiagen, Valencia, CA), and the resulting 6xHis-UTF1 fusion protein was expressed in M15 bacteria carrying the pREP4 lac repressor plasmid and purified using Ni-NTA agarose. Two rabbits were inoculated with the recombinant protein, resulting in antisera UP2390 and UP2391 (Cocalico Biologicals, Reamstown, PA). Antisera were affinity-purified against the UTF1 fusion protein prior to use. Other antibodies used for western blotting: Anti-histone H3 monoclonal antibody (1:10,000, Cat No. 05-499, clone 6.6.2, Millipore), ACTB monoclonal antibody (1:50,000, Cat No. A5441, clone AC-15, Sigma), and goat anti-LIN28A (1:3,000, Cat No. AF3757, R&D systems). Immuno-detection was performed using horseradish peroxidase-conjugated secondary antibodies and ECL Select detection reagents (Amersham).
Histology and immunostaining
For histological analysis, testes were fixed in Bouin’s fixative, dehydrated, and embedded in paraffin. 8-micron sections were collected and stained with hematoxylin and eosin before mounting with Permount (Fisher Scientific, Pittsburgh, PA). Slides were imaged on a Leica DM5500B microscope with a DFC450 camera (Leica Microsystems, Buffalo Grove, IL).
For immunofluorescence, testes were fixed in 4% paraformaldehyde at 4 °C for 3–4 hours and cut into 8-micron sections in a cryostat. Slides were blocked with 10% goat serum in Tris buffered saline containing 0.1% Tween-20. The slides were incubated with a 1:200 dilution of the purified anti-UTF1 antibody at 37 °C overnight followed by one-hour incubation with a 1:200 dilution of FITC-conjugated goat anti-rabbit IgG secondary antibody. Slides were mounted in VectaShield with DAPI (Vector Laboratories, Burlingame, CA) and imaged with an ORCA-Flash4.0 digital camera (Hamamatsu Photonics, Hamamatsu City, Japan) on a Leica DM5500B microscope.
Gonocyte quantification and sperm count
Testes from individual animals were cut into 8-micron sections and placed on slides in groups of 5 sections with each approximately 80 microns apart. After staining with DAPI, each section was imaged, and the images stitched to create full mosaics of each section. Gonocytes were counted in all seminiferous tubule cross-sections. Per genotype, testis sections from several animals were quantified. Student’s t-test was used to calculate p-values. Sperm count of cauda epididymides was performed as previously described34.
Statistical methods
For each genotype, at least three animals were analyzed. All variables were presented as mean ± SD. P-values were calculated by Student’s t-Test and p values of less than 0.05 were considered statistically significant.
Electronic supplementary material
Acknowledgements
We thank Chao-Xing Yuan at the UPenn Proteomics Core facility for help with proteomic analysis and Sigrid Eckardt for help with manuscript preparation. National Institutes of Health/National Institute of Child Health and Human Development and National Institute of General Medical Sciences (R01HD069592 and R35GM118052 to P.J.W.) provided financial support.
Author Contributions
S.D.K. and M.L. designed the experiments and performed the experiments. J.M. and N.A.L. performed the experiments. S.D.K. and P.J.W. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-17482-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Leitch HG, Tang WW, Surani MA. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr. Top. Dev. Biol. 2013;104:149–187. doi: 10.1016/B978-0-12-416027-9.00005-X. [DOI] [PubMed] [Google Scholar]
- 2.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 3.de Rooij DG. Stem cells in the testis. Int. J. Exp. Pathol. 1998;79:67–80. doi: 10.1046/j.1365-2613.1998.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol. 2006;419:259–282. doi: 10.1016/S0076-6879(06)19011-4. [DOI] [PubMed] [Google Scholar]
- 5.Okuda A, et al. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 1998;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Boom V, et al. UTF1 is a chromatin-associated protein involved in ES cell differentiation. J. Cell Biol. 2007;178:913–924. doi: 10.1083/jcb.200702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kooistra SM, et al. Undifferentiated embryonic cell transcription factor 1 regulates ESC chromatin organization and gene expression. Stem Cells. 2010;28:1703–1714. doi: 10.1002/stem.497. [DOI] [PubMed] [Google Scholar]
- 8.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 1999;19:5453–5465. doi: 10.1128/MCB.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimoto M, et al. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Mol. Cell. Biol. 2005;25:5084–5094. doi: 10.1128/MCB.25.12.5084-5094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia J, et al. Regulation of pluripotency and self- renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell. 2012;151:576–589. doi: 10.1016/j.cell.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Bragt MP, et al. Expression of the pluripotency marker UTF1 is restricted to a subpopulation of early A spermatogonia in rat testis. Reproduction. 2008;136:33–40. doi: 10.1530/REP-07-0536. [DOI] [PubMed] [Google Scholar]
- 12.Kristensen DM, et al. Presumed pluripotency markers UTF-1 and REX-1 are expressed in human adult testes and germ cell neoplasms. Hum. Reprod. 2008;23:775–782. doi: 10.1093/humrep/den010. [DOI] [PubMed] [Google Scholar]
- 13.Nishimoto M, et al. In vivo function and evolution of the eutherian-specific pluripotency marker UTF1. PLoS One. 2013;8:e68119. doi: 10.1371/journal.pone.0068119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo M, et al. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat. Commun. 2013;4:2788. doi: 10.1038/ncomms3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarrey, J. R. In Cell and Molecular Biology of the Testis (eds Desjardins, C. & Ewing, L.) 58–89 (Oxford University Press, New York, 1993).
- 16.Weidinger G, et al. Dead End, a Novel Vertebrate Germ Plasm Component, is Required for Zebrafish Primordial Germ Cell Migration and Survival. Curr. Biol. 2003;13:1429–1434. doi: 10.1016/S0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- 17.Cook MS, Munger SC, Nadeau JH, Capel B. Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background. Development. 2011;138:23–32. doi: 10.1242/dev.057000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youngren KK, et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 20.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 21.Webster KE, et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl. Acad. Sci. USA. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:159–168. doi: 10.1101/SQB.1997.062.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 24.Malki S, van der Heijden GW, O’Donnell KA, Martin SL, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev. Cell. 2014;29:521–533. doi: 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell. Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linder CC. Genetic variables that influence phenotype. ILAR J. 2006;47:132–140. doi: 10.1093/ilar.47.2.132. [DOI] [PubMed] [Google Scholar]
- 28.Yoshiki A, Moriwaki K. Mouse phenome research: implications of genetic background. ILAR J. 2006;47:94–102. doi: 10.1093/ilar.47.2.94. [DOI] [PubMed] [Google Scholar]
- 29.Takada Y, et al. Mammalian Polycomb Scmh1 mediates exclusion of Polycomb complexes from the XY body in the pachytene spermatocytes. Development. 2007;134:579–590. doi: 10.1242/dev.02747. [DOI] [PubMed] [Google Scholar]
- 30.Pan J, et al. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev. Biol. 2009;330:167–174. doi: 10.1016/j.ydbio.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saunders PT, et al. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction. 2003;126:589–597. doi: 10.1530/rep.0.1260589. [DOI] [PubMed] [Google Scholar]
- 32.Lin Y, Page DC. Dazl deficiency leads to embryonic arrest of germ cell development in XY C57BL/6 mice. Dev. Biol. 2005;288:309–316. doi: 10.1016/j.ydbio.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 33.Eggan K, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, et al. Abnormal sperm in mice lacking the Taf7l gene. Mol. Cell. Biol. 2007;27:2582–2589. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng K, Wu X, Kaestner KH, Wang PJ. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev. Biol. 2009;9:38. doi: 10.1186/1471-213X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.