Abstract
Purpose
To investigate the associations between baseline and post-treatment circulating tumor cell (CTC) gene expression and outcome of patients enrolled in four NCCTG metastatic breast cancer (MBC) trials where specimens were shipped (at 4°C) from community-based sites to a reference laboratory (Mayo Clinic-Rochester, MN).
Experimental Design
Blood was collected at treating sites from MBC patients before (baseline), during, and at end of treatment with erlotinib+gemcitabine (N0234), sorafenib (N0336), irinotecan+cetuximab (N0436), or paclitaxel-poliglumex+capecitabine (N0437). CTCs were enriched from 10 mls of EDTA blood using CD45-depletion, 24-30 hours post-blood collection. Reverse transcription/quantitative PCR was used to determine cytokeratin-19 (CK19) and mammaglobin (MGB1) mRNA levels in CTCs from up to 13 (N0234), 16 (N0336), 18 (N0436), and 39 (N0437) patients. The gene expressions were normalized to β2-microglobulin and calibrated to healthy blood using the 2−ΔΔCq algorithm; positivity was defined as ≥2.
Results
CK19+mRNA cells were detected in 56%–75% and MGB1+mRNA cells in 23%–38% of 86 patients at baseline. CK19+mRNA cells were detected in 30%–67% and MGB1+mRNA cells in 14%–64% of 110 post-baseline serial samples. The presence of baseline CK19+mRNA cells (p=0.01) but not MGB1+mRNA cells (p=0.14) was significantly associated with shorter overall survival. A decrease in MGB1+mRNA levels (baseline-week 8) appeared to be associated with clinical response (p=0.05).
Conclusions
CTC gene expression analysis performed by a reference laboratory is feasible when blood is collected from treating sites and processed 24-30 hours post-collection. The presence of baseline CK19+mRNA CTCs was associated with poor prognosis; a decrease in MGB1+mRNA CTCs may help predict response to therapy of MBC patients.
Keywords: CTCs, breast cancer, molecular detection, community-based clinics
INTRODUCTION
Breast tumors shed malignant epithelial cells into the circulation, which can cause disease relapse. As metastasis is the main cause of death in patients with solid tumors, accurately detecting and characterizing these circulating tumor cells (CTCs) can positively impact the management of breast cancer patients with metastatic disease (1, 2). The clinical utility of CTCs in breast cancer has shown promise in predicting risk, tailoring treatment, response monitoring, and developing novel therapies (3–5). The CellSearch™ Assay (Veridex, Warren NJ) is an immunofluorescent-based cell imaging system for enumerating CTCs (6, 7) and is FDA-approved for metastatic breast cancer (MBC) patient prognosis and treatment monitoring. The presence and persistence of ≥five CTCs/7.5ml blood in MBC patients before and after treatment predicted for poor clinical outcome and treatment failure (6–9), has been shown to be a superior surrogate endpoint than current radiology imaging (10), and is, perhaps, superior to standard measurements of tumor burden (11).
In addition to CTC enumeration, gene expression analysis of CTCs is becoming increasingly important in determining breast cancer patient prognosis and treatment response and understanding metastatic biology. The high multiplexing capabilities associated with reverse transcription/real-time, quantitative PCR (RT-qPCR) allows for CTCs to be characterized both molecularly and biologically. Specific profiles of CTCs may result in a more effective evaluation of the clinical significance of CTCs in risk prediction and treatment monitoring (12). The AdnaTest BreastCancerSelect (AdnaGen AG, Langenhagen, Germany) is a multiplex PCR test that detects human epidermal growth factor receptor-2 (HER2), mucin 1 (MUC1) and epithelial glycoprotein (GA733-2) transcripts (13). This test has shown clinical significance of CTCs in breast cancer patients (14), but is not FDA-approved and was not available in the United States at the time of our analyses. Other multiple marker assays that include cytokeratin-19 (CK19), mammaglobin (MGB1), and HER2 are being developed to effectively detect CTCs with promising clinical significance (i.e., predicts outcome and treatment response of patients) in early and late stage disease (15–20). Thus, characterizing CTCs in breast cancer patients may result in an important clinical tool to aid in staging, predicting prognosis, and in designing more personalized therapeutic regimens for these patients (12).
Characterization of CTCs is attractive because blood collection is a minimally invasive procedure and blood can be shipped from the community laboratory to the research center. However, the timing between blood collection and processing may be critical for certain analytes. Special blood collection tubes with proprietary preservatives or special phlebotomy/handling requirements are necessary for the CellSearch Assay and the Adna tests. For gene expression analyses of CTCs, EDTA tubes are typically used and in most reported studies, the blood has been processed and CTCs isolated within 2-4 hours after collection at the same institution of blood collection. This makes it difficult to translate a gene expression-based CTC test to local/treating sites, many of which do not have access to the required instrumentation for RT-qPCR. As such, the blood needs to be shipped overnight to a reference laboratory. Therefore, we examined whether gene expression in CTCs can still predict outcome and treatment response of breast cancer patients when CTCs are isolated from EDTA blood 24-30 hours post-collection at a reference laboratory. We determined the mRNA levels of CK19 and MGB1 in CTCs from MBC patients enrolled in community-based, North Central Cancer Treatment Group (NCCTG) clinical trials and investigated associations between baseline and post-treatment gene expression and patient outcome.
METHODS
Patients and Specimens
Patients registered to four phase II NCCTG MBC treatment trials, N0234 (N=59 eligible patients), N0336 (N=20 eligible patients), N0436 (N=19 eligible patients), and N0437 (N=48 eligible patients) (http:ClinicalTrials.gov) (Supplemental Table 1 and Supplemental Figure 1) were eligible for this CTC study. The CTC correlative studies associated with the clinical trials were embedded in the clinical trial protocols, which were approved by participating Institutional Review Boards. The clinical trials accrued patients between 2003 and 2007, and the blood samples were collected prospectively for CTC analyses, which were performed blinded to the clinical trials endpoints.
Usable blood samples from up to 13 (N0234), 16 (N0336), 18 (N0436), and 39 (N0437) patients were obtained at baseline (day 1 of cycle 1, before start of therapy), during (day 1 of cycle 3; eight weeks), and at the treatment end, typically at disease progression. Although we did not pre-specify the number of patients for this CTC correlative study, blood samples for CTC analysis were not collected on all patients enrolled in the clinical trials. Compliance by the local treating hospitals and clinics of blood sample collection for CTC analysis was low (22%) for the first study (N0234) in which CTC analysis was implemented in 2002. Appropriate site training improved compliance to 80% for N0336 initiated in 2003 and 85% for N0436 and N0437 initiated in 2004. A patient flow diagram describing the number of patients and blood samples (baseline and serial) for CTC analysis is provided in Supplemental Figure 1. Comparisons of the clinicopathological characteristics between cohort and non-cohort patients with baseline and week 8 samples are provided in Supplemental Tables 2 and 3. More non-cohort patients appeared to have ≥3 metastatic sites compared to cohort patients (p = 0.01; Supplemental Table 2).
Blood samples (~10 mls) for CTC gene expression analysis were collected in EDTA tubes and were drawn in conjunction but after routine clinical/hematological blood draws to minimize skin epithelial cell contamination. Blood samples were collected at the local treating hospitals and clinics, stored at 4°C, and were shipped, on ice packs (4°C), overnight to the NCCTG research base, Mayo Clinic, Rochester, MN.
Between 24 and 30 hours post-blood collection, CTCs were isolated from blood samples using the RosetteSep Human CD45 Depletion Cocktail followed by density centrifugation over Ficoll-Paque according to the manufacturers’ instructions (StemCell Technologies Inc, Vancouver, BC). Density centrifugation over Ficoll-Paque is necessary for the appropriate separation of rosetted red and white blood cells from the mononuclear cells of interest. The enriched peripheral blood mononuclear cell (PBMNC) layer was isolated and washed thrice with phosphate-buffered saline/2% fetal-bovine serum (PBS/2%FBS). The PBMNC pellet was resuspended with lysis binding buffer and the cell homogenate was stored frozen at −80°C until processed for mRNA isolation.
mRNA Isolation, Reverse Transcription, and Second-strand Synthesis
The procedures for mRNA isolation and cDNA synthesis have been previously described (15). In brief, mRNA was isolated from the cell lysate using the mRNA Direct kit and Dynabeads Oligo (dT)25 according to the manufacturers’ instructions (Dynal, Invitrogen, Carlsbad, CA). A solid phase cDNA library was generated using AMV reverse transcriptase and oligo (dt)25 priming and stored at 4°C. Gene-specific cDNA second strand synthesis was performed using gene-specific forward primers as previously described (15).
Quantitative Real-time PCR (qPCR)
CK-19, MGB1, and β2-microglobulin (B2M) mRNA levels were quantified using qPCR and hydrolysis probes chemistries using a BioRad iCycler IQ (Hercules, CA). The primers and probes for CK-19 (NM_002276.3) were designed using Universal Probe Library from Roche Applied Science (Indianapolis, IN). The forward (5′-GTCATGGCCGAGCAGAAC) and reverse (3′- CCGGTTCAATTCTTCAGTCC) primers were obtained from Integrated DNA Technologies (Coralville, Iowa). The probe (GGATGCTG) was obtained from Roche Applied Science. The primers and probe for MGB1 (HSU33147) were designed using Primer Express from Applied Biosystems Inc. (Palo Alto, CA). The forward (5′- AGAACTGCAGGGTATGGTGAGAA) and reverse (3′- ACATGTATAGCAGGTTTCAACAATTGT) primers and the probe- ([6-FAM]CCAACTACGGATTGCTGCAAACCACA[BHQ1a-6FAM) were obtained from Integrated DNA Technologies. The primers and probe for B2M (NM_004048) were designed using Beacon Designer from PREMIER Biosoft International (Palo Alto, CA). The forward (5′-CATTCCTGAAGCTGACAGCATTC) and reverse (3′-CAGAAAGAGAGAGTAGCGCGAG) primers and the probe ([6-FAM] TGTCTCGCT CCGTGGCCTTAGCTG [BHQ1a-6FAM]) were obtained from Integrated DNA Technologies. Specific oligonucleotides representing the respective amplicons were obtained from Integrated DNA Technologies and were used to construct standard curves for the analytes. The oligonucleotides were serially diluted in Tris-EDTA (TE) buffer and eight to nine working concentrations (in multiples of 10) between 10 transcript copies/5μl and 108 transcript copies/5μl for CK19 and MGB1 and between 10 transcript copies/5μl and 109 transcript copies/5μl for B2M were assayed in duplicate. The average ± standard deviation intra-run and inter-run (13 runs total for the 4 clinical trials) percent coefficient of variation (%CV) for CK19 across the oligonucleotide transcript levels were 1.58 ± 0.40% and 10.5 ± 4.64%, respectively. The average intra-run and inter-run %CV for MGB1 across the oligonucleotide transcript levels were 1.28 ± 0.96% and 8.21 ± 3.59%, respectively. The average (± standard deviation) intra-run and inter-run %CV for B2M across the oligonucleotide transcript levels were 0.82 ± 0.38% and 11.3 ± 6.61%, respectively. These %CVs are within the Food and Drug Administration (FDA) guidelines that recommend %CV values of less than 15% for analytical assay precision and at least two-thirds of the oligonucleotide transcript levels (standards) have %CV less than 15% (21).
Appropriate negative (e.g., water and known amount of mRNA from the MDA MB-361 cell line that contains the marker of interest reverse transcribed without AMV reverse transcriptase) and positive (e.g., specific amplicon oligonucleotides and known amount of mRNA from the MDA MB-361 cell line reverse transcribed with AMV reverse transcriptase) controls were included in each qPCR assay. In accordance to the Minimum Information for Publication of Quantitative PCR Experiments (MIQE) guidelines (22), the specific oligonucleotide standard curves were used to optimize amplification efficiency and to ensure that reaction efficiencies are comparable between genes of interest and the reference gene. In addition, blood samples obtained from healthy individuals were identically processed and were analyzed in each RT-qPCR run. The analytic detection limit of our model assay system is 1 in 106 cells (15), and using oligonucleotide standard curves, we can routinely detect at least 3 transcript copies per reaction. All patient samples were run in triplicate, and a sample had to have at least two Cq values < 40 to be evaluable for the respective transcript. Samples with at least two Cq values of 40 were considered non-detectable. The average Cq value was used as the quantitative value, and the average %CVs (± standard deviation) of the detectable samples were 1.66 ± 0.12, 0.97 ± 0.07, and 0.48 ± 0.02 for CK19, MGB1, and B2M, respectively, across the four studies.
The target messages in the CTC samples were determined using a relative quantification method (22–24). The relative quantification, 2−ΔΔCq algorithm, is the fold expression relative to a calibrator (i.e., blood from healthy individuals; “normal blood”) and normalized to a reference gene (B2M). For the CK19 calibrator, of 62 blood samples tested from healthy individuals, CK19 Cq<35 was observed in 11% of samples. For the MGB1 calibrator, of 56 samples tested from healthy individuals, MGB1 Cq<35 was observed in less than 2% of samples. The average calibrator ΔCq value was obtained from these samples and individual ΔCqs of the calibrator were within 2 standard deviations of the average calibrator ΔCq value. Samples were classified as positive for a particular gene if the 2−ΔΔCq was ≥ 2.0 (i.e., 100% or greater than what is found in healthy blood). Samples with non-detectable target messages (Cq = 40) were assigned a 2−ΔΔCq value of 0.
Statistical Analysis
The primary endpoints of this analysis were to evaluate the impact of baseline and follow-up changes of CK19mRNA+ and MGB1mRNA+ CTCs on progression free survival (PFS) and overall survival (OS) of MBC patients enrolled in four NCCTG phase II treatment trials. This study followed REMARK guidelines (25). Wilcoxon Rank Sum tests were used to assess changes in CK19 and MGB1 gene expression values between baseline and week 8 and end of treatment between responders and non-responders. Wilcoxon Signed Rank tests were used to assess changes in CK19 and MGB1 gene expression levels between baseline and week 8 and treatment end. Chi-Square tests were used to determine whether CK19 and MGB1 gene expression values differed between studies. Non-stratified Cox Regression models and Kaplan-Meier curves were used to determine associations between PFS and OS and CK19mRNA and MGB1mRNA positivity. Multivariate modeling methods included backward elimination, stepwise and assessing all potential subset models based on score criterion. The Kaplan-Meier method was used to estimate overall (OS) and progression-free survival (PFS). Median follow-up was 2.3 years. For each clinical trial, clinical response rates as defined by RECIST v1.0 criteria (26) among patients included in this study are listed in Supplemental Table 1.
RESULTS
Distribution of CK-19 and MGB1 relative gene expression levels
Table 1 shows that 38% and 67% of patients had CK19+mRNA and MGB1mRNA levels <2.0, respectively, at baseline. Of the 53 patients with CK19 2−ΔΔCq ≥2 at baseline, the relative gene expression level ranged between 2.1 and16,286 (mean: 569 median: 54) (Supplemental Table 4). Of the 28 patients with MGB1 2−ΔΔCq ≥2 at baseline, the relative gene expression level ranged between 2.3 and 5806 (mean: 354 median: 14) (Supplemental Table 5).
Table 1.
Distribution of CK19 and MGB1 Relative Gene Expression Levels
| CK-19 (2−ΔΔCq) | Baseline (N=86) N (%) |
Week 8 (N=63) N (%) |
Treatment End (N=47) N (%) |
|---|---|---|---|
|
| |||
| Negative | 33 (38) | 31 (49) | 22 (47) |
| 0 | 25 (29) | 24 (38) | 19 (40) |
| 0.01-1.0 | 3 (3) | 4 (6) | 1 (2) |
| 1.01-1.99 | 5 (6) | 3 (5) | 2 (4) |
|
| |||
| Positive | 53 (62) | 32 (51) | 25 (53) |
| 2.0-9.99 | 15 (17) | 10 (16) | 9 (19) |
| >10.0 | 38 (44) | 22 (35) | 16 (34) |
|
| |||
| MGB1 (2−ΔΔCq) | |||
|
| |||
| Negative | 58 (67) | 42 (67) | 32 (68) |
| 0 | 51 (59) | 36 (57) | 29 (62) |
| 0.01-1.0 | 5 (6) | 5 (8) | 3 (6) |
| 1.01-1.99 | 2 (2) | 1 (2) | 0 (0) |
|
| |||
| Positive | 28 (33) | 21 (33) | 15 (32) |
| 2.0-9.99 | 8 (9) | 8 (13) | 5 (11) |
| >10.0 | 20 (23) | 13 (21) | 10 (21) |
Incidence of CK-19mRNA+ and MGB1mRNA+ CTCs
CK-19mRNA+ CTCs (at 2−ΔΔCq ≥ 2) were detected in 55-75%, 44-60%, and 30-67% patients at baseline, during, and end treatment/progression, respectively (Table 1, Supplemental Figure 2A). MGB1mRNA+ CTCs were detected in 23-38%, 22-64%, and 14-60% patients at baseline, during, and treatment end, respectively (Table 1, Supplemental Figure 2B). No significant differences were observed between the timepoints across the four studies. At baseline, the incidence of CK19mRNA+ CTCs (62%; 95% CI: 51-72%) was higher than the incidence of MGB1mRNA+ CTCs (33%; 95% CI: 23-42%). Of the 28 MGB1+mRNA samples, 23 (82%) were also CK19mRNA+ (Supplemental Table 6), and of the 53 CK19+mRNA samples, 23 (43%) were also MGB1mRNA+. At baseline, CK19+mRNA and MGB1+mRNA incidence was significantly correlated (Supplemental Table 6; p=0.007) and the relative gene expression levels between CK-19 and MGB1 were weakly correlated (Spearman r2= 0.23; p=0.03).
Correlation between CTC positivity and clinico-pathological characteristics
The presence of CK-19mRNA+ CTCs (2−ΔΔCq ≥ 2) at baseline correlated with hormone receptor positivity, >3 metastatic sites, and non-ductal histology but was not correlated with nodal status (Table 2). The presence of MGB1mRNA+ CTCs (2−ΔΔCq ≥ 2) at baseline correlated with hormone receptor positivity (Table 2).
Table 2.
Correlation between CTC Baseline Positivity and Clinicopathological Characteristics.
| Characteristic | CK19 Baseline (N=86) |
MGB1 Baseline (N=86) |
||
|---|---|---|---|---|
| N (%) ≥ 2 | p-value* | N (%) ≥ 2 | p-value | |
|
| ||||
| Nodal Status | 0.82 | 0.65 | ||
| Node Negative | 27/43 (63) | 13/43 (30) | ||
| Node Positive | 26/43 (61) | 15/43 (35) | ||
|
| ||||
| Histologic Tumor Grade (Elston/SBR) | (N=69) | 0.71 | (N=69) | 0.48 |
| Well/Intermediate | 12/20 (60) | 7/20 (35) | ||
| Poor | 27/49 (55) | 13/49 (27) | ||
|
| ||||
| Surgery | (N=70) | 0.19 | (N=70) | 0.12 |
| Breast Conserving | 16/22 (73) | 5/22 (23) | ||
| Mastectomy | 27/48 (56) | 20/48 (42) | ||
|
| ||||
| ER/PR Status: | 0.03 | 0.009 | ||
| ER or PgR Positive | 32/44 (73) | 20/44 (46) | ||
| Other | 21/42 (50) | 8/42 (19) | ||
|
| ||||
| # of Metastatic Sites | 0.009 | 0.90 | ||
| < 3 | 25/50 (50) | 16/50 (32) | ||
| ≥ 3 | 28/36 (78) | 12/36 (33) | ||
|
| ||||
| Histology | (N=57) | 0.004 | (N=57) | 0.12 |
| Ductal | 23/48 (48) | 14/48 (29) | ||
| Other | 9/9 (100) | 5/9 (56) | ||
|
| ||||
| Performance Status | 0.15 | 0.66 | ||
| 0 | 27/49 (55) | 15/49 (31) | ||
| 1,2 | 26/37 (70) | 13/37 (35) | ||
|
| ||||
| Menopausal Status | 0.38 | 0.35 | ||
| Pre or <50 yrs | 13/24 (54) | 6/24 (25) | ||
| Post or ≥ 50 yrs | 40/62 (65) | 22/62 (36) | ||
|
| ||||
| Age Group | 0.61** | 0.72** | ||
| < 40 | 6/10 (60) | 4/10 (40) | ||
| 40-49 | 10/17 (59) | 5/17 (29) | ||
| 50-59 | 19/31 (61) | 11/31 (36) | ||
| 60-60 | 14/23 (61) | 6/23 (26) | ||
| 70+ | 4/5 (80) | 2/5 (40) | ||
: χ2;
: Mantel-Haesnzel; SBR:Scarff-Bloom-Richardson; ER: estrogen receptor; PgR: progesterone receptor
Association between baseline CTC positivity and patient survival
Univariate analysis showed that histologic grade and performance status (PS) were independent significant predictors of PFS (Table 3). The hazard ratio (HR) for PFS was 1.37 (95%CI: 0.87-2.18; p=0.18) for patients with CK19mRNA+ CTCs (2−ΔΔCq ≥ 2) at baseline (Table 3, Figure 1A). Progression-free survival for patients with and without MGB1mRNA+ CTCs were similar, and there was no significant association between baseline MGB1mRNA+ positivity and PFS (Figure 1B). A comparison across four subgroups based on baseline CTC gene expression positivity (e.g., CK19mRNA−/MGB1mRNA−; CK19mRNA−/MGB1mRNA+; CK19mRNA+/MGB1mRNA−; CK19mRNA+/MGB1mRNA+) demonstrated no significant differences in PFS (Supplemental Table 8).
Table 3.
Univariate Analysis for DFS and OS
| Covariate | HR | 95% CI | p-value |
|---|---|---|---|
| DFS | |||
| Age | 1.00 | 0.98-1.02 | 1.0 |
| ER (neg vs pos) | 1.48 | 0.94-2.32 | 0.09 |
| PgR (neg vs pos) | 1.27 | 0.80-2.02 | 0.32 |
| Histologic Tumor Grade Elston/SBR (poor vs well/intermediate) | 1.89 | 1.05-3.40 | 0.03 |
| Node (pos vs neg) | 0.89 | 0.57-1.38 | 0.59 |
| Menopausal Stat (<50 vs ≥50 yrs) | 1.12 | 0.68-1.85 | 0.65 |
| Histology (other vs ductal) | 0.86 | 0.40-1.83 | 0.69 |
| Performance Status (1,2 vs 0) | 1.61 | 1.03-2.51 | 0.04 |
| #Metastatic Sits (≥3 vs <3) | 1.15 | 0.73-1.81 | 0.54 |
| CK19 (≥ 2.0 vs < 2.0) | 1.37 | 0.87-2.18 | 0.18 |
| MGB1 (≥ 2.0 vs <2.0) | 1.05 | 0.65-1.69 | 0.84 |
| OS | |||
| Age | 1.00 | 0.98-1.03 | 0.87 |
| ER (neg vs pos) | 1.52 | 0.90-2.55 | 0.11 |
| PgR (neg vs pos) | 1.52 | 0.89-2.61 | 0.12 |
| Histologic Tumor Grade Elston/SBR (poor vs well/intermediate) | 2.28 | 1.08-4.85 | 0.03 |
| Node (pos vs neg) | 1.12 | 0.67-1.88 | 0.66 |
| Menopausal Stat (<50 vs ≥50 yrs) | 1.23 | 0.67-2.17 | 0.47 |
| Histology (other vs ductal) | 1.48 | 0.61-3.61 | 0.38 |
| Performance Status (1,2 vs 0) | 1.13 | 0.67-1.91 | 0.64 |
| #Metastatic Sits (≥3 vs <3) | 2.00 | 1.18-3.41 | 0.01 |
| CK19mRNA(≥ 2.0 vs < 2.0) | 2.05 | 1.16-3.62 | 0.01 |
| MGB1mRNA (≥ 2.0 vs <2.0) | 1.49 | 0.88-2.52 | 0.14 |
SBR: Scarff-Bloom-Richardson; ER: estrogen receptor; PgR: progesterone receptor
Figure 1.
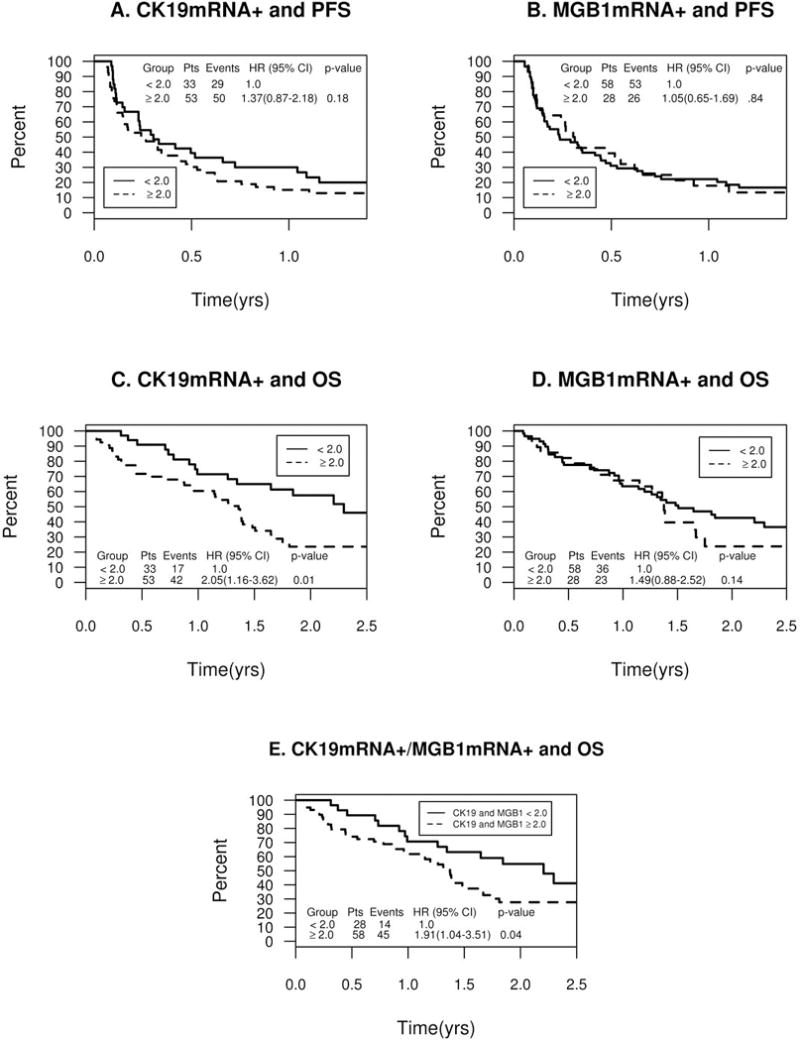
Baseline CTC Positivity and Patient Survival. A. CK19mRNA+ and Progression-free Survival. B. MGB1mRNA+ and Progression-free Survival C. CK19mRNA+ and Overall Survival. D. MGB1mRNA+ and Overall Survival. E. CK19mRNA+ and MGB1mRNA+ and Overall Survival.
Univariate analyses demonstrated that histologic grade, number of metastatic sites, and CK19mRNA+ CTCs were independent predictors for OS (Table 3). Patients with CK19mRNA+ CTCs had a significantly worse OS than patients without CK19mRNA+ CTCs at baseline (Figure 1C). The HR for patients with CK19+mRNA CTCs was 2.05 (95%CI:1.16–3.62) (Figure 1D). The HRs for patients with CK19+mRNA and MGB1mRNA+ CTCs and CK19+mRNA or MGB1mRNA+ CTCs were 1.85 (95%CI:1.07-3.20; p=0.03) and 1.91 (95%CI:1.04-3.51; p=0.04), respectively (Figure 1E). In a multivariate analysis, the histologic grade and number of metastatic sites were not significant covariates in the prognostic utility of CK19mRNA+ CTCs on OS. CK19mRNA+ (p=0.001) and estrogen receptor negativity (p=0.007) were the only predictors of worse OS in the final multivariate model (Supplemental Table 7).
An analysis assessing the association of baseline CTC gene expression positivity with overall survival demonstrated that patients with CK19mRNA+/MGB1mRNA- CTCs had a non-significant HR of 1.77 (p=0.10) and patients with CK19mRNA+/MGB1mRNA+ had a significant HR of 2.44 (p=0.01) when compared to patients with CK19mRNA-/MGB1mRNA- (Supplemental Table 9).
Association between baseline and week 8 CTC positivity change and patient OS
Patients who remained positive (2−ΔΔCq ≥ 2) between baseline and week 8 for CK19mRNA+ (HR: 2.67, p=0.05) or MGB1mRNA+ (HR: 2.13, p=0.09) CTCs appeared to have a worse OS compared to patients who remained negative (2−ΔΔCq < 2) for CK19mRNA or MGB1mRNA CTCs, respectively (Table 4). Similarly, the 35 patients who were positive for CK19mRNA CTCs at baseline irrespective of status at 8 weeks had a worse OS outcome than patients who were negative at baseline irrespective of status at 8 weeks (HR=2.7; p=0.01). The patients who were positive for MGB1mRNA CTCs at baseline irrespective of status at 8 weeks tend to have a worse OS outcome than patients who were negative at baseline irrespective of status at 8 weeks (HR=1.91; p=0.07).
Table 4.
Overall Survival (OS) by Change in CTC Positivity Between Baseline and Week 8.
| CTC Change | N=59 | #Events | HR | 95% CI | p-value | OS | |
|---|---|---|---|---|---|---|---|
| 1 yr | 2 yr | ||||||
|
| |||||||
| CK19 | |||||||
| < 2.0 to < 2.0 | 14 | 6 | 1.0 | — | — | 85.7 | 61.9* |
| < 2.0 to ≥ 2.0 | 10 | 3 | 0.63 | 0.15-2.54 | 0.51 | 87.5* | 87.5* |
| ≥ 2.0 to < 2.0 | 16 | 12 | 1.93 | 0.71-5.22 | 0.20 | 75.0 | 35.7* |
| ≥ 2.0 to ≥ 2.0 | 19 | 13 | 2.67 | 0.99-7.15 | 0.05 | 73.7 | 21.6* |
|
| |||||||
| MGB1 | |||||||
| < 2.0 to < 2.0 | 30 | 15 | 1.0 | — | — | 80.0 | 53.8 |
| < 2.0 to ≥ 2.0 | 9 | 4 | 0.92 | 0.30-2.79 | 0.88 | 76.2* | 63.5* |
| ≥ 2.0 to < 2.0 | 11 | 7 | 1.63 | 0.65-4.08 | 0.29 | 80.8* | 25.3* |
| ≥ 2.0 to ≥ 2.0 | 9 | 8 | 2.13 | 0.90-5.06 | 0.09 | 77.8* | 25.9* |
< 10 at risk
Association between CTC Positivity and Patient Response
No significant correlations were observed between treatment response and CK19mRNA+ CTCs at any timepoint (data not shown). At week 8, 41% (17/42) of patients without MGB1mRNA+ CTCs had a response compared to 19% (4/21) of patients with MGB1mRNA+ CTCs (p=0.09). At treatment end, 38% (12/32) of patients without MGB1mRNA+ CTCs had a response compared to 7% (1/15) of patients with MGB1mRNA+ CTCs (p=0.03). Table 5 shows that the decrease in MGB1 mRNA levels appeared to be larger for patients who had a treatment response compared to those who did not have treatment response at week 8 (p=0.05). However, it appears that MGB1 mRNA levels increased between baseline and treatment end in patients with a treatment response (p=0.09). There were no significant associations between changes in CK19+mRNA levels and treatment response (data not shown).
Table 5.
Gene Expression Level Change and Treatment Response
| Response Group | Change in CTC Levels from Baseline to Week 8 | ||||
|---|---|---|---|---|---|
| Mean | Median | Range | Quartiles | p-value | |
|
| |||||
| CK19 | |||||
| Yes (N=21) | − 350 | 0 | −4063 - 865 | 25%: −1 75%: −204 |
0.25 |
| No (N=38) | −75 | −2 | −16,272-11,906 | 25%: 2 75%: −13 |
|
|
| |||||
| Mammoglobin | |||||
| Yes (N=21) | − 67 | 0 | −738 - 36.0 | 25%: 0 75%: −11 |
0.05 |
| No (N=38) | − 32 | 0 | −970 - 313 | 25%: 7 75%: − 0.1 |
|
|
| |||||
| Response Group | Change in CTC Levels from Baseline to End of Treatment | ||||
| Mean | Median | Range | Quartiles | p-value | |
|
| |||||
| CK19 | |||||
| Yes (N=13) | 2732 | 0 | −3392 - 39,385 | 25%: 2 75% −141 |
0.34 |
| No (N=31) | − 479 | 0 | −15,332 - 999 | 25%: 10 75%: − 8 |
|
|
| |||||
| Mammoglobin | |||||
| Yes (N=13) | 1176 | 0 | −192 -15,506 | 25%: 0 75%: −0.6 |
0.09 |
| No (N=31) | −0.06 | 0 | −970 -1459 | 25%: 9 75% −0.1 |
|
DISCUSSION
The detection of CTCs has been shown to predict for poor clinical outcome and treatment failure in MBC patients (6, 7). The biologic characteristics (e.g., protein and mRNA expression) of CTCs may further improve predicting risk, monitoring response, and tailoring treatment strategies for individual patients with breast cancer (4, 5, 12). As CTC molecular characterization is becoming more relevant in personalized disease management of breast cancer patients, it would be beneficial to develop a gene expression-based test in which a commonly used blood collection tube (e.g., EDTA) can be easily utilized by local sites and the blood easily shipped to a reference laboratory and reliably tested for molecular analysis. This would potentially increase the accessibility of these types of tests to many patients, particularly in the United States. Therefore, we determined the feasibility of analyzing CK19 and MGB1 gene expression in CTCs when isolated from EDTA blood at a central laboratory 24-30 hours post-collection in context of outcome of patients enrolled in NCCTG community-based treatment trials.
We observed that CK19+mRNA was detected overall in 62% of MBC patients at baseline, a slightly higher incidence than the ~50% incidence of patients with ≥5 CTCs typically observed using the FDA-approved CellSearch Assay (6) and 52% incidence using the AdnaTest BreastCancer (27). In two individual studies (N0436 and N0437) with several patients treated in the first-line setting, we did observe similar incidences of baseline CTC positivity (55-56%) compared to literature findings. MGB1+mRNA has been detected in 33-39% of patients (28, 29), which is within the 20%–40% range of baseline incidence we observed using our immunodepletion/RT-qPCR method. A recent comparison analysis of the CellSearch Assay, Adna Test Breast Cancer Select/Detect, and CK-19/MGB1 RT-qPCR assay detected CTCs in 36%, 22%, and 26% (CK19)/54% (MGB1), respectively, in MBC patients (30). Our increased sensitivity of CK-19+ CTCs compared to previous results may be due to different patient populations (first versus second/third/fourth line), cell enrichment and RNA extraction techniques, oligonucleotide primers, and PCR approaches (standard versus real-time) used throughout the various studies. These technical differences for gene expression-based tests emphasize the importance of implementing standardize approaches to isolate and characterize CTCs, similar to what has been proposed to analyze disseminated tumor cells in the bone marrow (31).
The MGB1+mRNA incidence was lower than the incidence of CK19mRNA+ detection. Although the majority (87%) of the MGB1mRNA+ samples were also CK19mRNA+, 43% of CK19mRNA+ samples were MGB1mRNA+ and CK19 and MGB1 transcript levels were only weakly correlated. We previously observed a significant correlation between CK19 and MGB1 expression in patients with primary breast cancer (15). The presence of CK19mRNA+ CTCs at baseline was correlated with hormone receptor positivity, non-ductal histology, >3 metastatic sites, but not with nodal status, and the presence of baseline MGB1mRNA+ CTCs was strongly associated with hormone receptor positivity. It is interesting that significant correlations between CTC gene expression (particularly for MGB1) and hormone receptor positivity were observed. Elevated MGB1 expression has been associated with clinical and biological features defining a less aggressive tumor phenotype in breast tumors (32) including steroid-receptor positive breast tumors (33). This may help explain why baseline MGB1 gene expression did not predict for reduced PFS or OS in our study. Our results are consistent with previous findings that demonstrated a positive association between the presence of CTCs as detected by laser scanning cytometry and RT-qPCR and distant metastases (34–36). Although earlier results suggest that the incidence of CTCs is higher for node-positive than node-negative patients (34, 36, 37), the lack of correlation between CTCs and nodal status has been previously observed and indicates that two distinct routes exist for breast cancer metastases (38–40). Our incidence results and clinico-pathologic correlations are consistent with previous findings indicating that our immunodepletion coupled with RT-qPCR approach produces reliable gene expression analysis of CTCs.
Importantly, we observed that the presence of CK19mRNA+ CTCs is associated with shorter OS and thus, correlates with poor prognosis. We also observed a potential trend towards a predictive significance of MGB1mRNA+ CTCs in that the decrease in MGB1+ transcript levels between baseline and 8 weeks appeared to correlate with patient response to treatment. Similarly, MGB1 transcripts have been recently shown to reflect the effect of therapy on adjuvant breast cancer patients (41). As MGB1 is more highly breast specific compared to CK19, which is used as a general epithelial marker, MGB1 may be a more appropriate measure of treatment response (e.g., tumor shrinkage) for breast cancer compared to CK19 (CK19 gene expression also could be confounded by its loss through epithelial-mesenchymal transition). However, increased response rates may not always result in improved PFS or OS (42). In our study, the baseline or the change in MGB1 gene expression did not significantly predict for patient PFS or OS. Additionally, the detection rate of CK19+ or MGB1+ transcripts in blood samples was not shown to be associated with tumor progression in MBC patients (30). It is also interesting that a trend (p=0.09) was observed between increased MGB1 mRNA levels (between baseline and treatment end) and treatment response. Our initial hypothesis was that a decrease in CTC gene expression would predict that a patient would respond to treatment. We did observe this for MGB1 at the first follow-up at 8 weeks but potentially the opposite was observed at treatment end. A possible explanation could be that initially, the treatment is rapidly clearing the tumor cells from the circulation and by treatment end the tumor is shrinking, releasing cells into the circulation at a greater rate than the cells being cleared from the circulation giving the appearance that an increase in CTC MGB1 gene expression at treatment end may be indicative of treatment response. However, it is important to note that the change in MGB1 gene expression between baseline and treatment end was non-significant and could be due to the limited sample size and various treatment regimens used between the different clinical trials.
Our findings, however, are consistent with previous results that demonstrated an association between the presence of CK19+ CTCs and poor prognosis (16, 27, 43, 44) and demonstrate the validity of the immunodepletion/RT-qPCR method used in this study. In addition, it appears that patients who tested negative for CK19+CTCs at baseline, irrespective of changing to positive at follow-up timepoints, tend to have a better outcome than patients who tested positive for CK19+CTCs at baseline irrespective of changing to negative at follow-up timepoints. This is in slight contrast to the CellSearch data that demonstrated that patients who changed from negative (< 5 CTCs/7.5ml blood) to positive (≥ 5 CTCs/7.5ml blood) had worse outcome than those patients who changed from positive to negative and the outcome of those patients who turned positive was similar to those patients who remained positive for CTCs (6).
Concern does exist regarding the decreased gene expression levels observed in room temperature EDTA blood samples six hours post-collection (45). The blood samples in our studies were stored at 4°C and shipped on ice packs. This cold temperature may slow RNA degradation as our developmental work using blood from healthy individuals spiked with 1,000 cells/ml blood of MDA-MB-361 cells and stored at 4°C suggests that mRNA levels are decreased to a lesser extent when blood was maintained at 4°C (average Cq increase of 1.63 and 1.21 for CK19 and MGB1, respectively, as observed in our hands) versus a Cq increase of 2 to 4 when EDTA blood was kept at room temperature for 24 hours (45). A Cq increase of 6.29 and 5.14, for CK19 and MGB1, respectively, was observed when blood was collected in a CellSave tube maintained at room temperature (Supplemental Figure 3). Although RNA degradation occurs, small transcripts are still detectable by real-time PCR, a technique that is tailored to amplify small amplicon sizes. In addition, large and small tumor cell fragments and tumor microparticles have been observed in blood from colon cancer patients using the CellSearch Assay, and these small fragments were still associated with poor clinical outcome of the patients (46). In addition, EpCam antigenicity was still present three days after blood collection (47). Thus, it appears that RNA may be coming from tumor cell fragments that are still clinically significant and select cells remain viable for several days after collection. Lastly, our quantification method couples immunodepletion cell enrichment with RT-qPCR for which the gene expression results are calibrated to the gene expression observed in healthy blood that is identically processed (i.e., processed 24 hours post-collection) as the samples obtained from patients enrolled in NCCTG clinical trials. The negative selection has been shown to reduce background signals of HER2 and CK19 in hematopoietic cell populations to improve the specificity of RT-qPCR (48). We used normal blood as a calibrator in our quantification method to further decrease and normalize the influence of marker gene expression potentially found in hematopoietic cells as we did observe relatively elevated CK19 gene expression (e.g., Cq < 35) in 11% of normal blood samples. Molecular characterization of a single CTC by a multiplex real-time PCR also has been demonstrated in large quantities of contaminating leukocytes (49).
As the comparison of established assay findings with clinical outcome will define the clinically relevant threshold, clinical assays are only useful when the results can be accurately correlated with clinical outcome of cancer patients and reflect the nature of the disease (12). In our study, we demonstrated for the first time the prognostic significance of CTC gene expression analysis when blood has been processed 24-30 hours post-collection in context of NCCTG MBC clinical trials, suggesting that our findings are very promising for establishing gene expression-based CTC assays for clinical utility as prognostic/diagnostic tests in a community clinic setting. This will enable a broader application of CTC gene expression analyses/studies in clinical trials and different practice settings. As this study examined a limited number of patients with different treatment regimens and lines of therapy, our results should be interpreted as hypothesis-generating.
Additional CTC gene expression analyses are ongoing in other NCCTG treatment trials including early (neo-adjuvant and adjuvant settings) and metastatic breast cancer and advanced lung cancer trials (50) to validate the feasibility of CTC gene expression testing in a community-based setting.
Supplementary Material
Statement of Translational Relevance.
Previous findings using circulating tumor cells (CTCs) that are isolated at the same location of collection (and typically within four hours of collection) have demonstrated that the presence of CK19+mRNA cells is a poor prognostic indicator for advanced breast cancer patients. However, it is difficult to translate a gene expression-based CTC test to local/community-based treating hospitals and clinics because many do not have the required instrumentation for reverse transcription/quantitative PCR (RT-qPCR) analysis. As such, the blood needs to be shipped overnight to a reference laboratory. Therefore, we examined whether gene expression in CTCs can still predict for outcome and treatment response for patients with metastatic breast cancer (MBC) when blood is collected from local/treating sites (stored at 4°C) and CTCs are isolated from EDTA blood 24-30 hours post-collection at a reference laboratory. We observed that the presence of baseline CK19+mRNA CTCs was associated with traditional poor prognostic indicators (≥ 3 metastatic sites and higher grade), a trend towards shorter progression-free survival, and significantly shorter overall survival, independent of number of metastatic sites and grade. We also observed that a decrease in MGB1+mRNA CTCs may help predict response to therapy of MBC patients. Our data confirms literature findings and indicate for the first time that CK19 gene expression analysis in CTCs performed by a reference laboratory still predicts for outcome of advanced breast cancer patients when blood is collected from local sites and processed 24-30 hours post-collection in the context of the North Central Cancer Treatment Group (NCCTG) treatment trials. This will allow for a wider application of CTC gene expression monitoring in clinical trials and different practice settings. Validation studies are ongoing to confirm these findings.
Acknowledgments
This work was supported by National Institutes of Health grants CA25224-31 and CA114740 (PI: JC Buckner) for the North Central Cancer Treatment Group and associated Biospecimen Resource, respectively, and the Breast Cancer Research Foundation (PI: EA Perez).
References
- 1.Andreopoulou E, Cristofanilli M. Circulating tumor cells as prognostic marker in metastatic breast cancer. Expert Rev Anticancer Ther. 2010;10:171–7. doi: 10.1586/era.09.105. [DOI] [PubMed] [Google Scholar]
- 2.Mostert B, Sleijfer S, Foekens JA, Gratama JW. Circulating tumor cells (CTCs): detection methods and their clinical relevance in breast cancer. Cancer Treat Rev. 2009;35:463–74. doi: 10.1016/j.ctrv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Ring A, Smith IE, Dowsett M. Circulating tumour cells in breast cancer. Lancet Oncol. 2004;5:79–88. doi: 10.1016/S1470-2045(04)01381-6. [DOI] [PubMed] [Google Scholar]
- 4.Hayes DF, Smerage JB. Circulating tumor cells. Prog Mol Biol Transl Sci. 2010;95:95–112. doi: 10.1016/B978-0-12-385071-3.00005-8. [DOI] [PubMed] [Google Scholar]
- 5.Ignatiadis M, Georgoulias V, Mavroudis D. Micrometastatic disease in breast cancer: clinical implications. Eur J Cancer. 2008;44:2726–36. doi: 10.1016/j.ejca.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 6.Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 7.Cristofanilli M, Hayes DF, Budd GT, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–30. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 8.Hayes DF, Cristofanilli M, Budd GT, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res. 2006;12:4218–24. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 9.Riethdorf S, Fritsche H, Muller V, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13:920–8. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 10.Budd GT, Cristofanilli M, Ellis MJ, et al. Circulating tumor cells versus imaging–predicting overall survival in metastatic breast cancer. Clin Cancer Res. 2006;12:6403–9. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 11.Cristofanilli M, Broglio KR, Guarneri V, et al. Circulating tumor cells in metastatic breast cancer: biologic staging beyond tumor burden. Clin Breast Cancer. 2007;7:471–9. [PubMed] [Google Scholar]
- 12.Riethdorf S, Pantel K. Advancing personalized cancer therapy by detection and characterization of circulating carcinoma cells. Ann N Y Acad Sci. 2010;1210:66–77. doi: 10.1111/j.1749-6632.2010.05779.x. [DOI] [PubMed] [Google Scholar]
- 13.Demel U, Tilz GP, Foeldes-Papp Z, Gutierrez B, Albert WH, Bocher O. Detection of tumour cells in the peripheral blood of patients with breast cancer. Development of a new sensitive and specific immunomolecular assay. J Exp Clin Cancer Res. 2004;23:465–8. [PubMed] [Google Scholar]
- 14.Hauch S, Zimmermann S, Lankiewicz S, Zieglschmid V, Bocher O, Albert WH. The clinical significance of circulating tumour cells in breast cancer and colorectal cancer patients. Anticancer Res. 2007;27:1337–41. [PubMed] [Google Scholar]
- 15.Reinholz MM, Nibbe A, Jonart LM, et al. Evaluation of a panel of tumor markers for molecular detection of circulating cancer cells in women with suspected breast cancer. Clin Cancer Res. 2005;11:3722–32. doi: 10.1158/1078-0432.CCR-04-1483. [DOI] [PubMed] [Google Scholar]
- 16.Bosma AJ, Weigelt B, Lambrechts AC, et al. Detection of circulating breast tumor cells by differential expression of marker genes. Clin Cancer Res. 2002;8:1871–7. [PubMed] [Google Scholar]
- 17.Apostolaki S, Perraki M, Kallergi G, et al. Detection of occult HER2 mRNA-positive tumor cells in the peripheral blood of patients with operable breast cancer: evaluation of their prognostic relevance. Breast Cancer Res Treat. 2009;117:525–34. doi: 10.1007/s10549-008-0239-3. [DOI] [PubMed] [Google Scholar]
- 18.Ignatiadis M, Xenidis N, Perraki M, et al. Different prognostic value of cytokeratin-19 mRNA positive circulating tumor cells according to estrogen receptor and HER2 status in early-stage breast cancer. J Clin Oncol. 2007;25:5194–202. doi: 10.1200/JCO.2007.11.7762. [DOI] [PubMed] [Google Scholar]
- 19.Ignatiadis M, Kallergi G, Ntoulia M, et al. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clin Cancer Res. 2008;14:2593–600. doi: 10.1158/1078-0432.CCR-07-4758. [DOI] [PubMed] [Google Scholar]
- 20.Xenidis N, Perraki M, Kafousi M, et al. Predictive and prognostic value of peripheral blood cytokeratin-19 mRNA-positive cells detected by real-time polymerase chain reaction in node-negative breast cancer patients. J Clin Oncol. 2006;24:3756–62. doi: 10.1200/JCO.2005.04.5948. [DOI] [PubMed] [Google Scholar]
- 21.Administration USDoHaHSFaD, (CDER) CfDeaR, (CVM) CfVM. Guidance for Industry. Bioanalytical Method Validation. 2001 [Google Scholar]
- 22.Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 23.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–93. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 24.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–91. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–16. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 27.Tewes M, Aktas B, Welt A, et al. Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat. 2009;115:581–90. doi: 10.1007/s10549-008-0143-x. [DOI] [PubMed] [Google Scholar]
- 28.Obermayr E, Sanchez-Cabo F, Tea MK, et al. Assessment of a six gene panel for the molecular detection of circulating tumor cells in the blood of female cancer patients. BMC Cancer. 2010;10:666. doi: 10.1186/1471-2407-10-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen C, Hu L, Xia L, Li Y. The detection of circulating tumor cells of breast cancer patients by using multimarker (Survivin, hTERT and hMAM) quantitative real-time PCR. Clinical Biochemistry. 2009;42:194–200. doi: 10.1016/j.clinbiochem.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Van der Auwera I, Peeters D, Benoy IH, et al. Circulating tumour cell detection: a direct comparison between the CellSearch System, the AdnaTest and CK-19/mammaglobin RT-PCR in patients with metastatic breast cancer. Br J Cancer. 2010;102:276–84. doi: 10.1038/sj.bjc.6605472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fehm T, Braun S, Muller V, et al. A concept for the standardized detection of disseminated tumor cells in bone marrow from patients with primary breast cancer and its clinical implementation. Cancer. 2006;107:885–92. doi: 10.1002/cncr.22076. [DOI] [PubMed] [Google Scholar]
- 32.Nunez-Villar MJ, Martinez-Arribas F, Pollan M, et al. Elevated mammaglobin (h-MAM) expression in breast cancer is associated with clinical and biological features defining a less aggressive tumour phenotype. Breast Cancer Res. 2003;5:R65–70. doi: 10.1186/bcr587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Span PN, Waanders E, Manders P, et al. Mammaglobin is associated with low-grade, steroid receptor-positive breast tumors from postmenopausal patients, and has independent prognostic value for relapse-free survival time. J Clin Oncol. 2004;22:691–8. doi: 10.1200/JCO.2004.01.072. [DOI] [PubMed] [Google Scholar]
- 34.Pachmann K, Clement JH, Schneider CP, et al. Standardized quantification of circulating peripheral tumor cells from lung and breast cancer. Clin Chem Lab Med. 2005;43:617–27. doi: 10.1515/CCLM.2005.107. [DOI] [PubMed] [Google Scholar]
- 35.Giribaldi G, Procida S, Ulliers D, et al. Specific detection of cytokeratin 20-positive cells in blood of colorectal and breast cancer patients by a high sensitivity real-time reverse transcriptase-polymerase chain reaction method. J Mol Diagn. 2006;8:105–12. doi: 10.2353/jmoldx.2006.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahn HJ, Presta A, Yang LY, et al. Enumeration of circulating tumor cells in the blood of breast cancer patients after filtration enrichment: correlation with disease stage. Breast Cancer Res Treat. 2004;86:237–47. doi: 10.1023/B:BREA.0000036897.92513.72. [DOI] [PubMed] [Google Scholar]
- 37.Taubert H, Blumke K, Bilkenroth U, et al. Detection of disseminated tumor cells in peripheral blood of patients with breast cancer: correlation to nodal status and occurrence of metastases. Gynecol Oncol. 2004;92:256–61. doi: 10.1016/j.ygyno.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 38.Lang JE, Mosalpuria K, Cristofanilli M, et al. HER2 status predicts the presence of circulating tumor cells in patients with operable breast cancer. Breast Cancer Res Treat. 2009;113:501–7. doi: 10.1007/s10549-008-9951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jatana KR, Balasubramanian P, Lang JC, et al. Significance of circulating tumor cells in patients with squamous cell carcinoma of the head and neck: initial results. Arch Otolaryngol Head Neck Surg. 2010;136:1274–9. doi: 10.1001/archoto.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin YC, Chen SC, Hsueh S, et al. Lack of correlation between expression of human mammaglobin mRNA in peripheral blood and known prognostic factors for breast cancer patients. Cancer Sci. 2003;94:99–102. doi: 10.1111/j.1349-7006.2003.tb01359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bitisik O, Saip P, Saglam S, Derin D, Dalay N. Mammaglobin and maspin transcripts in blood may reflect disease progression and the effect of therapy in breast cancer. Genet Mol Res. 2010;9:97–106. doi: 10.4238/vol9-1gmr649. [DOI] [PubMed] [Google Scholar]
- 42.Miller KD, Chap LI, Holmes FA, et al. Randomized phase III trial of capecitabine compared with bevacizumab plus capecitabine in patients with previously treated metastatic breast cancer. J Clin Oncol. 2005;23:792–9. doi: 10.1200/JCO.2005.05.098. [DOI] [PubMed] [Google Scholar]
- 43.Wulfing P, Borchard J, Buerger H, et al. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clin Cancer Res. 2006;12:1715–20. doi: 10.1158/1078-0432.CCR-05-2087. [DOI] [PubMed] [Google Scholar]
- 44.Weigelt B, Bosma AJ, Hart AA, Rodenhuis S, van ’t Veer LJ. Marker genes for circulating tumour cells predict survival in metastasized breast cancer patients. Br J Cancer. 2003;88:1091–4. doi: 10.1038/sj.bjc.6600868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benoy IH, Elst H, Van Dam P, et al. Detection of circulating tumour cells in blood by quantitative real-time RT-PCR: effect of pre-analytical time. Clin Chem Lab Med. 2006;44:1082–7. doi: 10.1515/CCLM.2006.210. [DOI] [PubMed] [Google Scholar]
- 46.Coumans FA, Doggen CJ, Attard G, de Bono JS, Terstappen LW. All circulating EpCAM+CK+CD45- objects predict overall survival in castration-resistant prostate cancer. Ann Oncol. 2010;21:1851–7. doi: 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- 47.Hekimian K, Stein E-L, Pachmann K, Pachman K. Detection of tumor stem cells among circulating epithelial tumor cells. 7th International Symposium on Minimal Residual Cancer. 2009 abstract 012:76. [Google Scholar]
- 48.You F, Roberts LA, Kang SP, et al. Low-level expression of HER2 and CK19 in normal peripheral blood mononuclear cells: relevance for detection of circulating tumor cells. J Hematol Oncol. 2008;1:2. doi: 10.1186/1756-8722-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sieuwerts A, Kraan J, Bolt-de Vries J, et al. Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Research and Treatment. 2009;118:455–68. doi: 10.1007/s10549-008-0290-0. [DOI] [PubMed] [Google Scholar]
- 50.Reinholz M, Mandrekar S, Foster N, et al. Circulating tumor cell cytokeratin-19 gene expression as a prognostic factor in lung cancer: Analysis based on North Central Cancer Treatment Group (NCCTG) clinical trials (N0423/426) International Association for the Study of Lung Cancer (IASLC) Annual Meeting 14th World Conference on Lung Cancer. 2011 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


