Abstract
Platelets are small, anucleate cell fragments that are central to hemostasis, thrombosis and inflammation. They are derived from megakaryocytes from which they inherit their organelles. As platelets can synthesize proteins and contain many of the enzymes of the secretory pathway, one might expect all mature human platelets to contain a stacked Golgi apparatus, the central organelle of the secretory pathway. By thin section electron microscopy, stacked membranes resembling the stacked Golgi compartment in megakaryocytes and other nucleated cells can be detected in both proplatelets and platelets. However, the incidence of such structures is low and whether each and every platelet contains such a structure remains an open question. By single-label, immunofluorescence staining, Golgi glycosyltransferases are found within each platelet and map to scattered structures. Whether these structures are positive for marker proteins from multiple Golgi subcompartments remains unknown.
Here we have applied state-of-the-art techniques to probe the organization state of the Golgi apparatus in resting human platelets. By the whole cell volume technique of serial-block-face scanning electron microscopy (SBF-SEM), we failed to observe stacked, Golgi-like structures in any of the 65 platelets scored. When antibodies directed against Golgi proteins were tested against HeLa cells, labeling was restricted to an elongated juxtanuclear ribbon characteristic of a stacked Golgi apparatus. By multi-label immunofluorescence microscopy, we found that each and every resting human platelet was positive for cis, trans and trans Golgi network (TGN) proteins. However, in each case, the proteins were found in small puncta scattered about the platelet. At the resolution of deconvolved, wide field fluorescence microscopy, these proteins had limited tendency to map adjacent to one another. When the results of 3D structured illumination microscopy (3D SIM), a super resolution technique, were scored quantitatively, the Golgi marker proteins failed to map together indicating at the protein level considerable degeneration of the platelet Golgi apparatus relative to the layered stack as seen in the megakaryocyte.
In conclusion, we suggest these results have important implications for organelle structure/function relationships in the mature platelet and the extent to which Golgi apparatus organization is maintained in platelets. Our results suggest that Golgi proteins in circulating platelets are present within a series of scattered, separated structures. As separate elements, selective sets of Golgi enzymes or sugar nucleotides could be secreted during platelet activation. The establishment of the functional importance, if any, of these scattered structures in sequential protein modification in circulating platelets will require further research.
Keywords: Platelets, Golgi apparatus, Serial-block-face scanning electron microscopy, fluorescence microscopy, 3D SIM
INTRODUCTION
Although anucleate cells, circulating platelets, the primary cell type involved in the initial steps of hemostasis, are capable of de novo protein synthesis [1–5], albeit likely time limited due to mRNA and ribosomal RNA decay [6]. Many of the synthesized proteins are secretory proteins [7]. Circulating platelets possess many of the organelles required to form a secretory pathway including endoplasmic reticulum (ER, known in platelets as the dense tubular system, DTS) and protein storage granules [1]. Yet the organizational status within circulating platelets of the Golgi apparatus, the central organelle, within the secretory pathway remains elusive. In the secretory pathway, the Golgi apparatus receives newly synthesized proteins from the ER, further processes these proteins and supports their sorting to destinations such as storage granules from which the proteins will be released in response to selective agonist [8]. The standard features of a Golgi apparatus might be expected to be a prominent feature of circulating platelets. However, that is not the case; the stacked series of Golgi subcompartments common in most nucleated mammalian cells is rarely seen in platelets [9].
At least five reasons might explain the rarity of stacked Golgi apparatus in healthy human platelets. First, this may reflect the sum of what is biosynthesized by the platelet or inherited from the precursor cell, the megakaryocyte [4,10]. Between the two there may be little capacity for Golgi formation [9]. Second, the rarity of stacked Golgi profiles may reflect the organizational state of microtubules within the platelet [11]. In nucleated mammalian cells, microtubules radiate outward from a juxtanuclear microtubule organizing center (MTOC) [12]. Platelets are anucleate and their microtubules are arranged in a circumferential belt underlying the plasma membrane [9,13]. Third, there may be no reason to have a Golgi apparatus in platelets because of a lack of glycosyltransferases, sugar nucleotide transporters and other proteins that constitute important enzymes/transporters in protein processing within mammalian Golgi apparatus. However, that is not the case as recent work indicates the presence of glycosyltransferases and stored sugar nucleotides that can contribute to remodeling of the platelet cell surface during platelet activation [14]. Fourth, a stacked Golgi apparatus may simply be hard to detect in thin section electron micrographs, an approach that only samples a very small volume of the platelet. Fifth, the Golgi apparatus itself may degenerate in organization as platelets enter the circulation as mRNA and ribosomal RNA have been recently reported to do in circulating platelets [6].
In the present work, we have applied a series of structural techniques including SBF-SEM, fluorescence microscopy and super-resolution 3D SIM light microscopy to determine how the Golgi apparatus is organized in healthy, circulating human platelets. Based on microtubule organization, we hypothesized that the organization might well resemble more that of the yeast, Saccharomyces cerevisiae, in which there is no radiating microtubule network [15]. In this cell type, a functional Golgi apparatus consists of a series of separated Golgi subcompartments that have no close physical association [16]. Our results, both from localizing proteins and characterizing organelle structure in whole cell depths, lead to the conclusion that Golgi proteins in healthy, circulating human platelets are dispersed over a series of scattered, non-stacked subcompartments much like that in yeast [16–17]. Based on distribution of Golgi protein staining across hundreds of platelets, we suggest that this is a general trait of human platelets. We suggest that the dispersion of Golgi enzymes across separated Golgi subcompartments may well be functionally important to the selectivity of platelet cell surface remodeling in response to platelet activation. Whether this organization is functional as a mature Golgi apparatus within platelets or may represent Golgi degeneration with respect to the proplatelet and the megakaryocyte remains an open question.
Methods
Antibodies for Golgi-specific immunofluorescence and immunoblotting
Immunostaining was performed using Golgi-specific antibodies raised against the human antigen. Cis-Golgi was labeled with GM130 mouse monoclonal antibody (BD Transduction Labs.), trans-Golgi was labeled with either SialylT ((α-N-acetyl-neuraminyl-2,3β-galactosyl-1,3)-N-acetylgalactosaminide α2,β-sialyltransferase 4), goat polyclonal antibodies (Santa Cruz Biotechnologies Inc.), or GalT rabbit polyclonal antibodies (β1.4-galactosyltransferase 1, generous gift from Dr. E. Berger), and the trans Golgi network (TGN) was labeled with TGN46, sheep polyclonal antibodies (AbD Serotech). Secondary antibodies were Alexa fluor (AF) 488 donkey anti-rabbit, AF488 donkey anti-sheep, Cy3 donkey anti-goat, AF647 donkey anti-mouse antibodies (Jackson ImmunoResearch), and Hilyte fluor 647 cat anti-rabbit antibodies (AnaSpec, Inc.). All primary antibodies used for immunofluorescence were also tested for reactivity in western blotting against total HeLa cell extracts. Of these only GM130 and GalT gave a positive reaction. Failure of antibodies to react in both immunofluorescence and western blotting is common. Rabbit polyclonal antibodies directed against human TGN46 (Abcam) were used for the respective western blot. For quantification of the blots, secondary antibodies conjugated with IRDye 800CW (LI-COR Biosciences) were used. GAPDH, rabbit monoclonal antibody (Cell Signaling) was used as a loading control for western blot analysis.
Cell culture
Wild type HeLa cells were used as a positive control to show that the Golgi-protein specific antibodies gave the juxtanuclear, overlapping staining pattern expected for the known stacked Golgi structure of a HeLa cell. In brief, HeLa cells were grown on glass coverslips for 24–48 h, fixed and processed for immunofluorescence [18].
Platelet purification
a) For SBF-SEM
Human blood was drawn in citrate anticoagulant tubes and mixed immediately with an equal amount of 2x fixative using a mixture of 0.2 M sodium cacodylate buffer containing 4 mM calcium chloride, 0.2% glutaraldehyde and 6% paraformaldehyde and fixed for 30 min at room temperature (RT). To obtain a platelet rich plasma (PRP), blood was centrifuged (Eppendorf centrifuge 5702 RH) at 300 × g for 15 min at RT. The PRP was then centrifuged at 500 × g for 15 min at RT to obtain platelets and then pelleted using a microcentrifuge (Eppendorf centrifuge 5417C) at 2200 × g for 8 min at RT.
b) For immunofluorescence
Human blood (12 mL) was drawn intravenously following IRB approved informed consent into citrated 5 ml vacutainers and mixed immediately with an equal amount of 8% paraformaldehyde in phosphate buffered saline (PBS, pH 7.4) to give a final concentration of 4% and fixed for 30 min at room temperature (RT). To give a platelet rich plasma (PRP), blood was centrifuged (Eppendorf centrifuge 5702 RH) at 300 × g for 15 min at RT. The PRP was centrifuged at 1200 × g for 12 min at RT to obtain platelets. Platelets were washed with 1 ml PBS (pH 7.4), then pelleted using a microcentrifuge (Eppendorf centrifuge 5417C) at 800 × g for 1 min at RT and finally resuspended in PBS. Platelet count was measured with a Hemavet HV 950FS platelet counter (Drew Scientific Inc., Oxford, CT, USA) and was adjusted to 380,000–400,000 per μl in the final suspension. Platelets were then pelleted and resuspended to the same final volume of 4% paraformaldehyde in PBS and fixed for an additional 30 min at RT.
Serial-Block-Face Scanning Electron Microscopy
a) SBF-SEM sample staining
Platelets were stained as previously described [19–22]. The purified platelet pellet was fixed using 0.1 M cacodylate buffer containing 2.5% glutaraldehyde and 2 mM calcium chloride for 1 h in ice. The pellet was resuspended and wash three times with cold 0.1 M sodium cacodylate buffer containing 2 mM calcium chloride and spin at 600 × g for 5 min. Samples were fixed in 3% potassium ferrocyanide in 0.3 M cacodylate buffer with 4mM calcium chloride combined with an equal volume of 4% aqueous tetroxide for 1 h in ice. Samples were then placed in a 0.22 μm-Millipore-filtered 1% thiocarbohydrazide (TCH) solution in ddH2O for 20 min following five washes with double-distilled water (ddH2O) at RT each for 3 min. Sample were fixed in 2% osmium tetroxide in ddH2O for 30 min at RT following five washes with ddH2O at RT each for 3 min. Samples were then placed in 1% uranyl acetate (aqueous) and left overnight at 4°C. The next day, samples were washed five times with ddH2O at RT each for 3 min and processed for en bloc Walton’s lead aspartate staining [23]. Sample were then placed in the lead aspartate solution at 60°C for 30 min following five washes with ddH2O at RT each for 3 min. Samples were dehydrated and proceed for resin embedding.
b) SBF-SEM imaging
Resin-embedded blocks of stained pelleted platelets were first mounted on empty resin blocks for trimming in a Leica EM UC6 microtome. After the surface of the pellet was exposed, the block was remounted, exposed-side down, on an aluminum specimen pin (Gatan, Pleasanton, CA) using CircuitWorks Conductive Epoxy (CW2400), which grounded the sample electrically to the pin. Each sample was then trimmed again to expose the opposite side, and sputter-coated with a 40 nm gold layer. The trimmed block was imaged at an accelerating voltage of 1.8 kV in a Zeiss SIGMA-VP (variable pressure) scanning electron microscope (SEM) equipped with a Gatan 3View serial block face (SBF) imaging system [21–22, 24]. The SEM was operated in the high vacuum mode with a 30 μm condenser aperture, and images containing 2000 × 2000 pixels were acquired using the Gatan DigitalMicrograph program with a pixel size of 5.5 nm in the x–y plane. The diamond knife of the SBF system was set to cut the block 1.2 mm/s with a slice thickness of 30 nm, thus providing a 30-nm pixel size along the z-axis. Resulting 250 image slices (total thickness of 7.5 μm) were assembled into a volume file and aligned, using the DigitalMicrograph software.
Immunoblotting analysis
Human platelets were purified as previously described [14]. Human platelets and wild type HeLa cells were lysed with hot 2% SDS as described previously [26]. 50 μg and 20 μg of protein was loaded on each lane for platelets and wild type HeLa cells respectively, proteins were separated on a 10% gel by SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare Life Sciences). Membranes were incubated overnight with primary antibodies, followed by incubation with secondary antibodies for 1 h and imaged on an Odyssey Infrared Imaging System (LI-COR Biosciences).
Immunofluorescence staining
Previously published procedures were followed. In brief, 100 μl of fixed platelet suspension was incubated on a glass coverslip under high humidity at 37 °C in water bath for 90 minutes as described previously [27]. HeLa cells were grown on coverslips and washed with 1x PBS to rinse off growth media and fixed with 3.7% formaldehyde for 20 min at RT. Inverted platelet and HeLa cell coverslips were incubated in 1x PBS for 5 min followed by 50 mM ammonium chloride to quench free aldehydes for 5 min. A saponin-gelatin mix in 1x PBS was used for cell permeabilization, 15 min treatment. Cells were then incubated with the corresponding primary and secondary antibodies for 20 min each and washed three times with saponin-gelatin mix in 1x PBS for 5 min each followed by a final wash in deionized water to remove salt. Coverslips were mounted in Mowiol media (Calbiochem, San Diego, CA, USA).
Spinning-disk confocal, widefield and super resolution (3D SIM) fluorescence microscopy
Spinning-disk confocal image stacks were taken with a 63x/1.40 NA using a BD CARVII (BD Bioscience) spinning-disk confocal microscope accessory fitted to the Zeiss 200M inverted microscope as described previously [26]. Widefield images were taken using a 63x/1.40 numerical aperture (NA) objective with an Optovar setting of 1.6x or 2.5x on a Zeiss 200M inverted microscope (Carl Zeiss MicroImaging Inc., Thornwood, NY, USA). Image stacks were deconvolved using 10 iterations, Huygens Professional software (version 09:26:16; Scientific Volume Imaging, Hilversum, the Netherlands). Maximum intensity projections (MIPs) and mean pixel intensity measurements were made with iVision-Mac™ software (version 4.5.5; BioVision Technologies). Super resolution (3D SIM) stacks were taken using a Deltavision OMX™ super resolution imaging microscope (GE Healthcare) with an Olympus 60x/NA 1.42 or 100x/NA 1.49 objective and analyzed using FIJI software (version 2.0.0-rc-34/1.50a) and iVision-Mac™ software.
Quantitative analysis for co-localization of Golgi proteins
To measure the percent co-localization of proteins in a pairwise comparison manner [28], platelets were co-labeled with either two trans markers SialylT and GalT (n=9) or GalT and TGN46 antibodies (n=17). Platelets stacks were imaged using the Deltavision OMX™ super resolution-imaging microscope (GE Healthcare). Co-localization was done in 3D using the object analyzer algorithms in Huygens professional software.
Results
Research indicates that resting human platelets can synthesize proteins de novo, possess much of the elements of a secretory pathway and contain Golgi glycosyltransferases and their sugar nucleotide substrates [2–5,14]. Yet how the proteins of the central organelle of the secretory pathway, the Golgi apparatus, are distributed in circulated platelets and the characteristics of the structures to which these proteins are localized has been substantiated by little systemic experimentation. The organelle has been reported to be organized into stacked cisternae, even though these appear to be rare [9], while at the same time individual Golgi glycosyltransferases appear by immunofluorescence to be present in multiple scattered sites within the platelet.
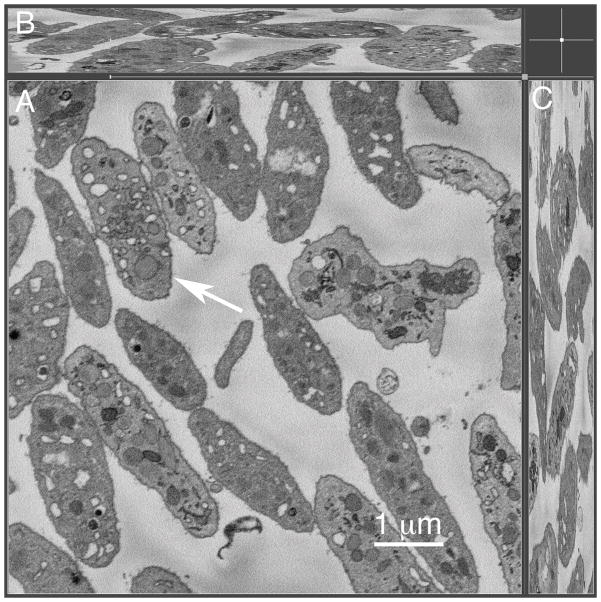
To correlate possible structural features with Golgi protein distribution in a systematic manner in resting human platelets, we first assessed the occurrence of Golgi-like stacked membranes in human platelets using a full platelet cell volume electron microscope approach. We reasoned that previous thin section approaches to the problem might well have under assessed the frequency of stacked platelet Golgi apparatus because the volume sampled is so small, a 50 nm thick section. Platelets may well be non-homogeneous in their subcellular organelle distribution and the organelle might well be missed. We took a SBF-SEM approach, current state-of-the-art technology. In this approach, the block face is repeatedly trimmed and re-imaged to generate a sequential series of images of the block face spanning several microns in depth, in our case 7.5 microns. As can be seen in Figure 1, the technique is sufficient to image many platelets at high resolution in the XY dimension (Figure 1A) and to give full cell volume imaging of 3–4 platelet depths in the Z-dimension (Fig. 1B–C). Our expectation was that if a stacked Golgi apparatus was the dominant feature in which Golgi enzymes were organized in resting human platelets then when imaged over full platelet cell volume, there should be one or more stacked Golgi elements per platelet. Upon careful, full cell depth examination of 65 resting human platelets (see Supplemental Fig. 1 for example), we observed no stacked Golgi-like structures in any of the cells. We conclude that a stacked Golgi apparatus is not a feature of the typical, mature, circulating human platelet.
Figure 1. Serial-block-face scanning electron microscopy of full platelet cell volume indicates that mature platelets do not contain stacked Golgi-like structures.
Resting human platelets (n=65) were analyzed for full platelet cell volume using serial-block-face SEM technique. The representative micrograph showing high-resolution image of the full platelet cell volume on XY dimension (A) and Z dimension (B–C). Arrow in (A) points to the platelet shown in the full cell volume video (Supplemental Figure 1, 2460 nm depth). The overall depth of the SBF-SEM sample imaged was 7.5 μm.
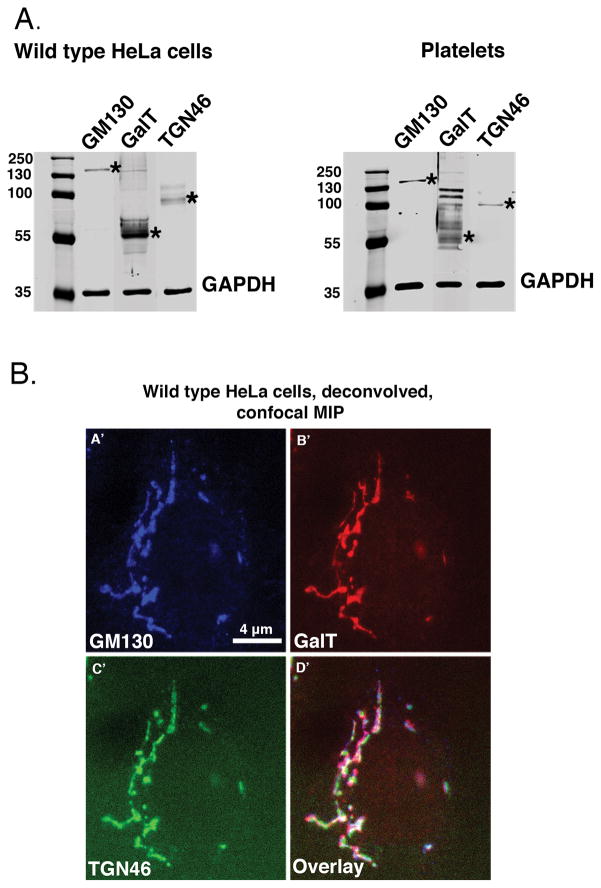
As a second, non-morphological approach, we co-mapped the distribution of protein markers to test how multiple, known Golgi-specific proteins present themselves in human platelets. Antibodies were validated by western blotting and co-labeling of a known stacked Golgi apparatus. As shown in Fig. 2A, antibodies directed against GM130 (cis Golgi, Golgi entry subcompartment) and TGN46 (trans Golgi network, Golgi exit subcompartment) reacted with a single molecular species (asterisks) in both platelets and nucleated HeLa cells. In both cases, GalT (β1.4-galactosyltransferase 1) ran as a diffuse set of bands with a major species at about 55 kDa. As shown in Fig. 2B(A′–D′), these antibodies all triple labeled the same structure, the juxtanuclear stacked Golgi apparatus in HeLa cells by deconvolved, spinning disk confocal microscopy. The three markers (GM130, blue; GalT, red; TGN46, green) often appeared to map laterally adjacent to one another with some overlapping (Fig. 2BA′–D′); the expected distribution for a stacked Golgi apparatus.
Figure 2. Western blot and multilabel immunofluorescence analysis indicates that the marker antibodies recognize distinct protein species.
(A.) Wild type HeLa cell and human platelet lysate was run on a 10% SDS-PAGE gel and blotted with GM130 (M.W. = 130 kDa), GalT (M.W. = ~55 kDa plus possible oligomers) and TGN46 (M.W. = predicted 51 kDa, typically runs as a dimer on SDS-PAGE) antibodies to identify cis, trans and trans Golgi network proteins, respectively. GAPDH is shown as a loading control. Asterisks indicate the position of the Golgi proteins. The blots were imaged with an Odyssey Infrared Imaging System (LI-COR Biosciences). (B.) A representative image of wild type HeLa cells showing a classic” stacked, ribbon-like juxtanuclear Golgi structure in which markers to the Golgi cisternae subcompartments cis, GM130 (blue); Trans, GalT (red); and trans Golgi network, TGN46 (green), overlay image map proximal to each other in deconvolved, spinning-disk confocal micrographs (A′–D′).
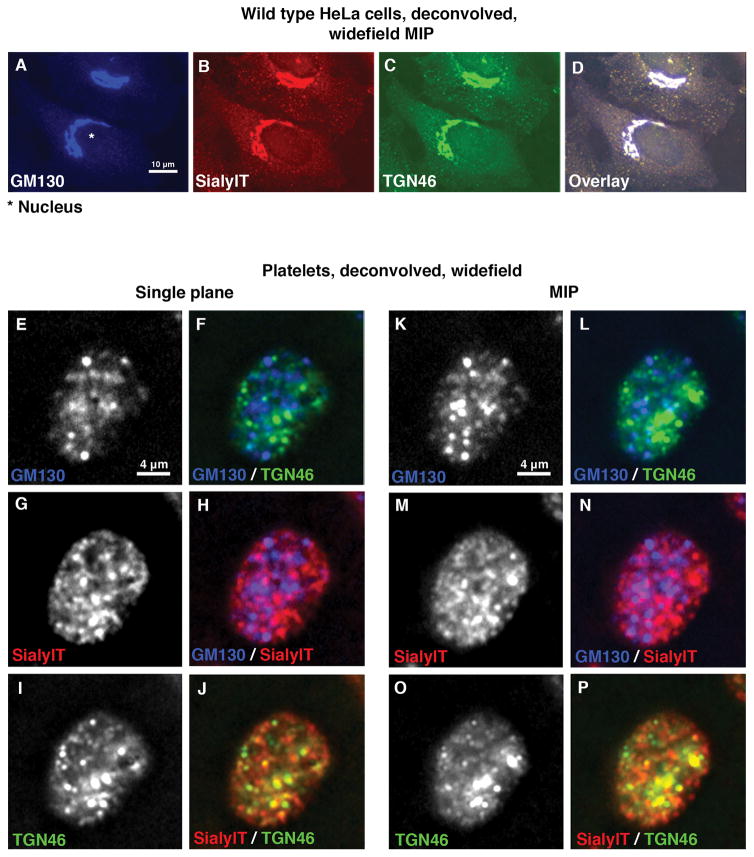
Having validated antibodies as bona fide reagents for the detection of stacked Golgi subcompartments in human cells (see also Fig. 3A–D, SialylT ((α-N-acetyl-neuraminyl-2,3β-galactosyl-1,3)-N-acetylgalactosaminide α2,β-sialyltransferase 4, trans Golgi example), we next prepared resting human platelets and triple labeled them with GM130, SialylT and TGN46 (Fig. 3E–P). Here, we chose to use the technique of deconvolved, widefield microscopy, an approach that produces similar resolution to confocal microscopy. The deconvolved, widefield images are shown in Fig. 3 as either a single plane (2 left columns, Fig. 3E–J) or the maximum intensity projection (MIP, 2 right columns, Fig. 3K–P) of the entire platelet to convey full platelet volume information. In striking contrast, we found in platelets that the three antibodies stained many puncta across the entire cell rather than a centralized Golgi apparatus (Fig. 3E–P). Moreover, when proteins located in distal Golgi subcompartments, cis versus TGN (Fig. 3E,F,K,L) or cis versus trans (Fig. 3G,H,M,N), were compared, we found little to no co-labeling and each marker protein appeared randomly distributed with respect to the other (compare GM130, blue color versus TGN46, green color or SialylT, red color). As an additional test, we compared the distribution of two proteins mapping to adjacent Golgi subcompartments in HeLa cells, SialylT (red, trans Golgi) and TGN46 (green). For platelets, as shown in Fig. 3I,J,O,P limited overlap (yellow color) between the trans and TGN markers was observed. Whether this indicates limited co-compartmentalization is tested further below.
Figure 3. Validated Golgi proteins show a scattered distribution with limited to no overlapping in resting human platelets.
(A–D) Representative deconvolved, widefield MIP fluorescence images of wild type HeLa cells stained with Golgi markers show a “classic” stacked ribbon-like juxtanuclear distribution. This provides validation of the antibodies as markers of Golgi subcompartments. (E–P) Immunofluorescence micrograph of resting human platelets show many puncta when stained for Golgi proteins marking cis, trans or TGN subcompartments using GM130 (blue), SialylT (red) or TGN46 (green) primary antibodies, respectively. Widefield Z-stack images were taken and the deconvolved images are presented as either single plane or maximum intensity projections (MIP).
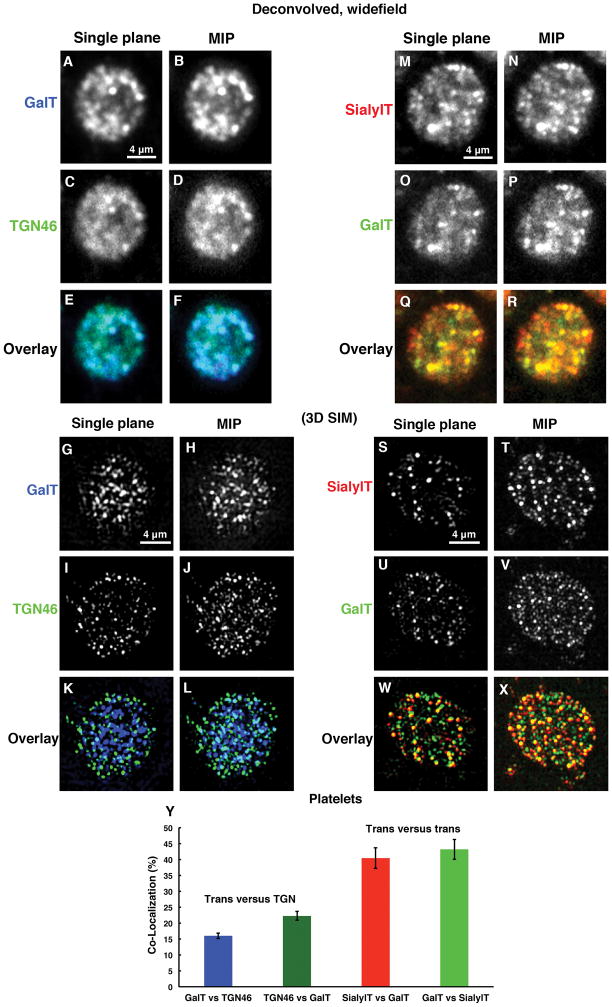
To address this question further, we tested in resting platelets whether there was any overlap in the distribution of a second trans Golgi marker, GalT (blue), with the TGN (green) by deconvolved, widefield immunofluorescence microscopy. As shown in Fig. 4A–F, overlap between the two markers was observed as indicated by the light blue/cyan color in the overlay (Fig. 4E–F). To distinguish whether this apparent overlap was due to the limited resolution of deconvolved, widefield microscopy, we applied the super-resolution approach of 3D SIM, a technique that gives a 2-fold improvement in X, Y and Z resolution, i.e., 8-fold in 3D space [29]. As shown in Fig. 4G–L, the extent of apparent overlap for GalT and TGN46 in platelets was much less by 3D SIM. In particular in single plane images (Fig. 4K), overlap as indicated by a light blue/cyan color was frequently skewed to one side, suggestive of two Golgi subcompartments that are adjacent to one another. Quantitatively, the 3D SIM overlap/Peason co-localization coefficient was ~20% (Fig. 4Y). Both SialylT and GalT are trans Golgi markers and overlap extensively in HeLa cells (data not shown). Taking these two, SialylT (red) and GalT (green), we tested the extent to which two trans markers overlap in distribution in human platelets. As shown in Fig. 4M–R, fairly extensive overlap (yellow color) was indicated by deconvolved widefield microscopy (Fig. 4Q–R). When observed by 3D SIM (Fig. 4S–X), the overlap was still extensive (Fig. 4W–X). Quantitatively, the Peason co-localization coefficient for the 3D SIM images was ~42%, a substantially higher degree of co-localization than observed for the trans/TGN comparison (Fig. 4Y). In sum, these data indicate that two Golgi enzymes normally thought to be part of the same Golgi subcompartment do behave in human platelets as if they share to a significant extent the same compartment. In comparison, markers normally thought to map to adjacent lateral Golgi subcompartments, trans (SialylT and GalT) and TGN (TGN46) show much less tendency to share the compartment in platelets and some low tendency to map adjacent to one another. As a final quantitative assessment of the 3D SIM images, we quantified the XY diameter of the observed puncta and found that these corresponded to resolution-limited values of 110 to 140 nm depending on wavelength of the emitted fluorescent light. The expected resolution limited Z dimension of the structures is a minimum of 220 to 280 nm as the Z-resolution of 3D-SIM is no better than half the XY resolution. Hence, we conclude that the observed Golgi protein rich compartments are small structures of no greater than about 100–140 nm in diameter and at least 220 to 280 nm in depth.
Figure 4. Deconvolved, widefield microscopy and 3D SIM analysis reveals that trans and TGN Golgi markers show little tendency to map in an adjacent manner suggestive of mini-Golgi stacks while the co-localization of two trans markers with one another suggests that the trans Golgi is a distinctive subcompartment in platelets.
(A–F) Trans, GalT (blue) and trans Golgi network, TGN46 (green) proteins showed apparent overlapping (light blue/Cyan color) with each other as seen by deconvolved, widefield microscopy where as (G–L) in three-dimensional structured illumination microscopy (3D SIM) of trans, GalT (blue) and trans Golgi network, TGN46 (green) proteins reveals little to no overlapping with each other. (M–R) Deconvolved, widefield microscopy suggests that two trans proteins SialylT (red) versus GalT (green), share a common compartment and show extensive overlapping (yellow color). (S–X) Furthermore considerable overlapping confirmed by 3D SIM analysis. (Y) Quantification of the limited overlapping. Seventeen individual platelets for trans versus (vs.) trans Golgi network and 9 individual platelets for trans versus trans are quantified in pairings. All pairings were averaged, and bars are presented as the mean ± SEM.
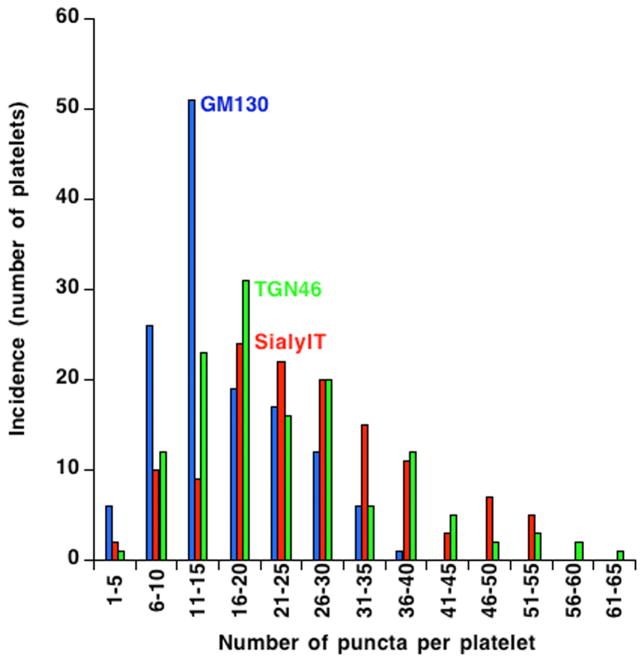
As a concluding step in our light microscope analysis of Golgi protein distribution in human platelets, we analyzed the incidence of fluorescent Golgi protein puncta per platelet over a population of almost 150 platelets. The goal here was to test whether there might be any evidence for discrete platelet subpopulations with respect to the structural arrangement of Golgi-specific proteins. Such might be true if young versus middle-aged versus old platelets had significant differences in Golgi organization. We chose to use the deconvolved, widefield images for analysis as this image set contained a larger population of platelets. As shown in Fig. 5, the incidence of Golgi puncta whether stained for GM130, SialylT or TGN46 was fairly similar for all with the peak incidence for GM130 staining being at about 11–15 puncta per platelet and that for SialylT and TGN46 at 16–20 puncta per platelet. There was little to no evidence from the incidence profiles for distinct populations of platelets with respect to any of the three Golgi markers suggesting that detectable age-dependent variation was not a major factor in determining the organization of Golgi structures in circulating human platelets.
Figure 5. The incidence of Golgi-protein-positive puncta in resting human platelets suggest that circulating platelets behave as a single population with respect to Golgi organization.
The incidence of Golgi-protein-positive puncta were manually calculated for 3 Golgi markers GM130 (blue), SialylT (red) and TGN46 (Green) using deconvolved, widefield images of 135 platelets. The frequency distribution of puncta per platelet is shown as histograms using KaleidaGraph software version 4.5.2. The histogram plots of each of the three markers are smooth and fairly similar suggesting that circulating platelets behave as a single population.
Discussion
The overall aim of the present study was to determine how the distribution of multiple Golgi proteins related to one another and possible stacked Golgi structures in resting human platelet. Our data from the state-of-the-art approach of SBF-SEM in which whole platelet volumes were imaged at electron microscope resolution showed that the incidence of stacked Golgi apparatus in individual, resting platelets was extremely low. In fact, we failed to observe any stacked Golgi apparatus in the 65 full volume platelets scored from SBF-SEM images. Using antibodies validated against reference HeLa cells, we found little to no evidence that these antibodies labeled adjacent Golgi subcompartments in circulating platelets as would be expected for a stacked Golgi apparatus. Instead, we observed scattered, disperse fluorescent puncta. We suggest that the Golgi apparatus in circulating human platelets consists of a series of dispersed Golgi subcompartments as in yeast. Such an arrangement is functional in the yeast, Saccharomyces cerevisiae, and could support protein modifications, for example, glycosylation in platelets [14]. Alternatively, the scattered puncta may be degenerate/decayed Golgi elements of more doubtful function within the circulating platelet.
Disperse Golgi-protein-positive puncta were observed in each of the 100s of circulating human platelets scored by protein-specific immunofluorescence staining using either deconvolved, widefield microscopy or the super resolution approach of 3D SIM. On the other hand, rare examples of apparent stacked Golgi apparatus have been observed in thin section data sets by others [9, 14] and by us [Yadav and Storrie, unpublished]. In White syndrome, the incidence of stacked Golgi structures by thin section electron microscopy increases about 5-fold [9]. What could be the significance of these examples? The rare stacked Golgi-like compartments observed in thin section could be due to the presence of immature platelets in the cell preparation. We suggest that future studies on the distribution of Golgi proteins and Golgi structure in such examples as proplatelets, White syndrome [9], idiopathic thrombocytopenic purpura [IPT, 30] or acute platelet proliferation [31] should reveal important features of organelle biogenesis.
Wandall et al. [14] have shown previously that Golgi enzymes and their substrates are secreted during platelet activation to produce remodeling of the platelet cell surface. We would like to suggest that the Golgi-protein-positive puncta that we observed might well better support platelet surface remodeling than a stacked Golgi apparatus. Dispersed structures place terminal glycosylation enzymes in separate compartments from earlier acting glycosyltransferases. Under this scenario, terminal Golgi compartments could be selectively secreted leading to the selective addition of terminal sugars during platelet cell surface remodeling. In comparison, the existence of a stacked Golgi apparatus is likely to lead to the more complicated problem of secreting the whole Golgi apparatus as a unit.
Finally, two additional questions must be addressed: 1) what is the relationship between the disperse Golgi-protein-positive puncta reported here and stacked Golgi apparatus within the platelet precursor cell, the megakaryocyte and 2) what is the underlying ultrastructural basis of the dispersed puncta. First with respect to the origin of the structures in the mature circulating platelet relative to the megakaryocyte, we would like to suggest that one possibility is that proplatelets are seeded not with stacked Golgi apparatus but with near vesicle size structures positive for selected Golgi enzymes. In other words, the unit of inheritance might well be the individual Golgi subcompartment not the Golgi stack. In mammalian cells during Golgi reorganization in response to drug-induced microtubule breakdown, vesicles carry a select set of Golgi proteins, which then generate Golgi ministacks [12, 32]. Structures of the size of the puncta could be the unit of transfer/inheritance in the proplatelet. Alternatively, the proplatelet may inherit stacked Golgi apparatus and later through Golgi apparatus degeneration/decay come to manifest the scattered puncta. This suggestion is consistent with the recent evidence for time-dependent mRNA and ribosomal RNA decay in circulating platelets [6]. Finally, with respect to the ultrastructural basis of what is a dispersed Golgi-protein-positive puncta, we would like to suggest that the most logical choice is closed elements of what had previously been described as canalicular system. The human canalicular system in contrast to that of other species has been thought to be completely open, i.e., interconnected to the plasma membrane in resting human platelets. Recent work using high volume sampling electron microscopy strongly indicates that in immediately fixed human platelets a significant portion is closed [25]. Many of these closed elements could correspond to the disperse puncta we observed. A future experimental challenge will be to correlate quantitatively structures positive for Golgi proteins in immunogold labeling experiments with the extent of such closed structures in resting human platelets.
In conclusion, we present evidence from both morphology/structure and protein labeling that Golgi proteins in mature human platelets fail to distribute in stacked Golgi structures. Rather the proteins are found in scattered, dispersed puncta by fluorescence microscopy that could be roughly the size of Golgi elements in yeast. Based on these results, we suggest that this organization should optimally support the selective secretion of Golgi proteins during platelet activation. Determining how these structures are inherited at the proplatelet level and their functional importance, if any, in glycoprotein processing in circulating platelets will require further experimentation.
Supplementary Material
Resting platelets were embedded for SBF-SEM, the block mounted and block face imaged sequentially through a depth of 7.5 μm. In the example shown, the center platelet is imaged from top to bottom across 82 sequential block face images (2460 nm depth). No structure resembling a stacked Golgi apparatus was observed in this platelet or the 64 other full-volume imaged platelets scored. Scale bar = 500 nm
Acknowledgments
The authors thank Jeff Kamykowski (Department of Physiology and Biophysics, University of Arkansas for Medical Sciences) for the excellent technical support. The GalT antibody was a generous gift from Dr. E. G. Berger (Zurich, Switzerland).
Footnotes
Declaration of interest
This work was supported in part by NIH grants S10 OD018065, R01 HL119393, and R01 GM092960. Work in the Leapman laboratory was supported by the NIBIB, NIH intramural program. The authors report no other declarations of interest.
References
- 1.King S, Reed GL. Development of platelet secretory granules. Semin Cell Dev Biol. 2002;13:203–302. doi: 10.1016/s1084952102000599. [DOI] [PubMed] [Google Scholar]
- 2.Warshaw AL, Laster L, Shulman NR. Protein Synthesis by Human Platelets. J Biol Chem. 1967;242(9):2094–7. [PubMed] [Google Scholar]
- 3.Weyrich AS, Schwertz H, Kraiss LW, Zimmerman GA. Protein Synthesis by Platelets: Historical and New Perspectives. J Thromb Haemost. 2009;7(2):241–6. doi: 10.1111/j.1538-7836.2008.03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schubert P, Devine DV. De novo protein synthesis in mature platelets: a consideration for transfusion medicine. Vox Sang. 2010;99(2):112–22. doi: 10.1111/j.1423-0410.2010.01333.x. [DOI] [PubMed] [Google Scholar]
- 5.Schwertz H, Rowley JW, Tolley ND, Campbell RA, Weyrich AS. Assessing protein synthesis by platelets. Methods Mol Biol. 2012;788:141–53. doi: 10.1007/978-1-61779-307-3_11. [DOI] [PubMed] [Google Scholar]
- 6.Angénieux C, Maître B, Eckly A, Lanza F, Gachet C, de la Salle H. Time-dependent decay of mRNA and ribosomal RNA during platelet aging and its correlation with translational activity. PLoS One. 2016;11(1):e0148064. doi: 10.1371/journal.pone.0148064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weyrich AS, Lindemann S, Tolley ND, Kraiss LW, Dixon DA, Mahoney TM, Prescott SP, McIntyre TM, Zimmerman GA. Change in protein phenotype without a nucleus: translational control in platelets. Semin Thromb Hemost. 2004;30(4):491–8. doi: 10.1055/s-2004-833484. [DOI] [PubMed] [Google Scholar]
- 8.Jonnalagadda D, Izu LT, Whiteheart SW. Platelet secretion is kinetically heterogeneous in an agonist-responsive manner. Blood. 2012;120:5209–16. doi: 10.1182/blood-2012-07-445080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White JG. Platelet Structure. In: Michelson AD, editor. Platelets. 3. Burlington, MA: Academic Press Elsevier; 2007. pp. 45–73. [Google Scholar]
- 10.Machlus KR, Thon JN, Italiano JE. Interpreting the developmental dance of the megakaryocyte: A review of the cellular and molecular processes mediating platelet formation. British Journal of Haematology. 2014;165(2):227–36. doi: 10.1111/bjh.12758. [DOI] [PubMed] [Google Scholar]
- 11.Yang W, Storrie B. Scattered Golgi elements during microtubule disruption are initially enriched in trans-Golgi proteins. Mol Biol Cell. 1998;9(1):191–207. doi: 10.1091/mbc.9.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storrie B, Yang W. Dynamics of the interphase mammalian Golgi complex as revealed through drugs producing reversible Golgi disassembly. Biochim Biophys Acta. 1998;1404(1–2):127–37. doi: 10.1016/s0167-4889(98)00053-6. [DOI] [PubMed] [Google Scholar]
- 13.Thon JN, Montalvo A, Patel-Hett S, Devine MT, Richardson JL, Ehrlicher A, Larson MK, Hoffmeister K, Hartwig JH, Italiano JE., Jr Cytoskeletal mechanics of proplatelet maturation and platelet release. J Cell Biol. 2010;191(4):861–74. doi: 10.1083/jcb.201006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wandall HH, Rumjantseva V, Sørensen AL, Patel-Hett S, Josefsson EC, Bennett EP, Italiano JE, Jr, Clausen H, Hartwig JH, Hoffmeister KM. The origin and function of platelet glycosyltransferases. Blood. 2012;120(3):626–35. doi: 10.1182/blood-2012-02-409235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rambourg A, Gachet E, Clermont Y, Képès F. Modifications of the Golgi apparatus in Saccharomyces cerevisiae lacking microtubules. Anat Rec. 1996;246(2):162–8. doi: 10.1002/(SICI)1097-0185(199610)246:2<162::AID-AR2>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O’Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang S, Rhee SW, Gleeson PA, Storrie B. Capacity of the Golgi Apparatus for Cargo Transport Prior to Complete Assembly. Mol Biol Cell. 2006;17(9):4105–17. doi: 10.1091/mbc.E05-12-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West JB, Fu Z, Deerinck TJ, Mackey MR, Obayashi JT, Ellisman MH. Structure-function studies of blood and air capillaries in chicken lung using 3D electron microscopy. Respir Physiol Neurobiol. 2010;170:202–209. doi: 10.1016/j.resp.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holcomb PS, Hoffpauir BK, Hoyson MC, Jackson DR, Deerinck TJ, Marrs GS, Dehoff M, Wu J, Ellisman MH, Spirou GA. Synaptic inputs compete during rapid formation of the calyx of Held: a new model system for neural development. J Neurosci. 2013;33:12954–12969. doi: 10.1523/JNEUROSCI.1087-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeifer CR, Shomorony A, Aronova MA, Zhang G, Cai T, Xu H, Notkins AL, Leapman RD. Quantitative analysis of mouse pancreatic islet architecture by serial block-face SEM. J Struct Biol. 2015;189:44–52. doi: 10.1016/j.jsb.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shomorony A, Pfeifer CR, Aronova MA, Zhang G, Cai T, Xu H, Notkins AL, Leapman RD. Combining quantitative 2D and 3D image analysis in the serial block face SEM: application to secretory organelles of pancreatic islet cells. J Microsc. 2015;259(2):155–164. doi: 10.1111/jmi.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walton J. Lead aspartate, an en bloc contrast stain particularly useful for ultrastructural enzymology. J Histochem Cytochem. 1979;27:1337–1342. doi: 10.1177/27.10.512319. [DOI] [PubMed] [Google Scholar]
- 24.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PloS Biol. 2004;2:1900–1909. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokrovskaya ID, Aronova MA, Kamykowski JA, Prince AA, Hoyne JD, Calco GN, Kuo BC, He Q, Leapman RD, Storrie B. STEM tomography reveals that the canalicular system and α-granules remain separate compartments during early secretion stages in blood platelets. J Thromb Haemost. 2016;3:572–84. doi: 10.1111/jth.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majeed W, Liu S, Storrie B. Distinct sets of Rab6 effectors contribute to ZW10--and COG-dependent Golgi homeostasis. Traffic. 2014;15(6):630–47. doi: 10.1111/tra.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sehgal S, Storrie B. Evidence that differential packaging of the major platelet granule proteins von Willebrand factor and fibrinogen can support their differential release. J Thromb Haemost. 2007;5(10):2009–16. doi: 10.1111/j.1538-7836.2007.02698.x. [DOI] [PubMed] [Google Scholar]
- 28.Kamykowski J, Carlton P, Sehgal S, Storrie B. Quantitative immunofluorescence mapping reveals little functional coclustering of proteins within platelet α-granules. Blood. 2011;118(5):1370–3. doi: 10.1182/blood-2011-01-330910. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald L, Baldini G, Storrie B. Does super-resolution fluorescence microscopy obsolete previous microscopic approaches to protein co-localization? Methods Mol Biol. 2015;1270:255–75. doi: 10.1007/978-1-4939-2309-0_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kieffer N, Guichard J, Farcet JP, Vainchenker W, Breton-Gorius J. Biosynthesis of major platelet proteins in human blood platelets. Eur J Biochem. 1987;164(1):189–95. doi: 10.1111/j.1432-1033.1987.tb11010.x. [DOI] [PubMed] [Google Scholar]
- 31.Nishimura S, Nagasaki M, Kunishima S, Sawaguchi A, Sakata A, Sakaguchi H, Ohmori T, Manabe I, Italiano JE, Jr, Ryu T, Takayama N, Komuro I, Kadowaki T, Eto K, Nagai R. IL-1α induces thrombopoiesis through megakaryocyte rupture in response to acute platelet needs. J Cell Biol. 2015;209(3):453–66. doi: 10.1083/jcb.201410052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;4:631–50. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Resting platelets were embedded for SBF-SEM, the block mounted and block face imaged sequentially through a depth of 7.5 μm. In the example shown, the center platelet is imaged from top to bottom across 82 sequential block face images (2460 nm depth). No structure resembling a stacked Golgi apparatus was observed in this platelet or the 64 other full-volume imaged platelets scored. Scale bar = 500 nm