Abstract
Introduction:
Colorectal cancer (CRC) is one of the most common malignancies and cause of cancer mortality worldwide. Given the importance of predicting the survival of CRC patients and the growing use of data mining methods, this study aims to compare the performance of models for predicting 5-year survival of CRC patients using variety of basic and ensemble data mining methods.
Methods:
The CRC dataset from The Shahid Beheshti University of Medical Sciences Research Center for Gastroenterology and Liver Diseases were used for prediction and comparative study of the base and ensemble data mining techniques. Feature selection methods were used to select predictor attributes for classification. The WEKA toolkit and MedCalc software were respectively utilized for creating and comparing the models.
Results:
The obtained results showed that the predictive performance of developed models was altogether high (all greater than 90%). Overall, the performance of ensemble models was higher than that of basic classifiers and the best result achieved by ensemble voting model in terms of area under the ROC curve (AUC= 0.96).
Conclusion:
AUC Comparison of models showed that the ensemble voting method significantly outperformed all models except for two methods of Random Forest (RF) and Bayesian Network (BN) considered the overlapping 95% confidence intervals. This result may indicate high predictive power of these two methods along with ensemble voting for predicting 5-year survival of CRC patients.
Keywords: colorectal cancer, survival, data mining, machine learning, AUC
1. INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer morality in the world. Predicting survival in patients with CRC is essential to determine their eligibility to participate in clinical trials, development of treatment planning and follow-up programs (1, 2).
Given the quite strong correlation between the stage and the prognosis of cancer, the staging systems developed by medical experts by using just 3 predictive factors including tumor extent, regional lymph node metastasis and distant metastasis, have been commonly used to predict survival of CRC patients (2, 3). These systems, while are convenient, and easy to understand, still have their own faire share of limitations. To begin with, survival time in patients with the same stage of cancer varies, depending on the individual case. Moreover, the survival prediction for advanced cancer patients is even less accurate (4). That is because the outcome of CRC patients relies heavily on not only the anatomical extent of the disease but also many factors related to the patient and the tumor (1).
The complex procedures of predicting survival rate are not easy when considering the dozens or even hundreds predictive factors that the physicians has to evaluate. In these cases, physicians’ experience and conventional techniques do not generally work. Instead it seems necessary to rely on unconventional, intensively computational approaches such as data mining (5). Data mining is a set of methods based on machine learning in order to develop accurate prediction models. In other words, data mining is a process that extracts knowledge from a set of data using intelligent techniques (6, 7). A variety of data mining methods which have been widely utilized in cancer prediction and prognosis are Decision Trees (DTs), Artificial Neural Networks (ANNs), Bayesian Networks (BNs) and Support Vector Machines (SVMs) (8). The research conducted by Snow et al is one of the first studies that predicted 5-year survival of CRC patients using data mining methods (9). While the studies of (10-15) were used one data mining method to develop a predictive model, the majority of the works makes use of several data mining techniques (5,16-18). More recently, the ensemble methods have received increasing attention in which multiple learning algorithms used to obtain better predictive performance (19-21).
2. AIM
Given the significance of accurate prediction of survival rate and the growing reliance on a variety of data mining methods, this study aims at comparing the efficiency of prediction models based on multiple basic and ensemble data mining methods for 5-year survival of CRC patients. Applying data mining methods in the prediction of survival of CRC patients can assist physicians, researchers and healthcare centers to better predict patient’s survival and consequently make better treatment planning, follow-up programs and prioritize healthcare resources. In addition, the obtained comparison results may also be taken into consideration by data mining and medical informatics specialist when selecting proper classifiers for decision support systems.
3. PROGNOSTIC FACTORS OF CRC
According to our survey, several prognostic factors have been proposed for CRC in the literature. Compton et al reviewed more than 200 studies on the prognostic factors for CRC and classified these factors into five categories. The first category is the factors that are proven by statistical certainty and are practically applied in patient’s treatment decisions which are tumor extend, regional lymph node metastasis, lymphovascular invasion, carcinoembryonic antigen (CEA) and residual tumor following surgery. The second category of factors are extensively studied clinically and biologically and included in pathological reports and validated in statistical studies. These factors include tumor grade and residual tumor following palliative resection. The third category is the factors that lack sufficient data for inclusion in the first and second categories, but have been introduced as prognostic factors in many studies. These factors include histological type, high level of MSI (MSI-H) and tumor border configuration. The fourth category is factors such as DNA content and all other molecular markers that have not adequately examined and finally, the fifth category is the factors which have no prognostic value according to the results of conducted studies (22).
Further investigation into CRC prognostic indicators could result in the introduction of totally new factors or even may alter the significance of already existing and known factors. For instance, BMI have been recently found to serve as powerful prognostic (predictive) indicator. These studies have shown that although there is almost direct relationship between being overweight and different types of cancer, after the onset of the disease, the prognosis of patients with slightly overweight is better than those with normal BMI (23, 24). Overall, the prognostic factors of CRC which have been mainly presented in the literature and need to be considered in the modeling of survival prediction are: tumor extension, regional lymph node metastasis, distant metastasis, stage (cancer stage), tumor grade, CEA, lymphovascular invasion, BMI, residual tumor following surgery, residual tumor following palliative resection, inherited or acquired type of cancer, histological type, bowel obstruction or perforation, intestinal inflammatory disease (IBD), hypertension, treatment methods, diabetes, tumor location, age, gender, smoking, education level, MSI-H, tumor border configuration, DNA content (22-31).
4. MATERIALS AND METHODS
4.1. Dataset
This retrospective study uses data from Cancer Registry Center of Research Center of Gastroenterology and Liver Disease, Shahid Beheshti University of Medical Sciences, Tehran, Iran. Originally, the dataset contains 1127 records and 36 raw attributes of CRC patients who registered during January 2002 to 2007. The death of the patient was confirmed through contact with family and relatives. The cause of death in all patients was CRC and survival time was calculated in term of month. The data attributes can be broadly classified as demographic attributes (such as, age, gender, occupation, marital status), diagnosis attributes (such as primary site), tumor characteristics attributes (such as histology, tumor grade, tumor size, stage) treatment and outcome attributes (such as survival time, cause of death) which gathered using interview and pathology reports stored in cancer registry forms.
4.2. Preprocessing
Raw data is rarely suitable for data mining and need to be processed before final analysis. This phase that is commonly called as data preprocessing is known that it is very often time consuming and compute intensive (32, 33). In the data preprocessing, any of data cleaning operation (such as removing or replacing missing values, identifying and eliminating outliers), data transformation (such as integration, normalization and construction of new features), data balancing and features selection might be done when necessary (34).
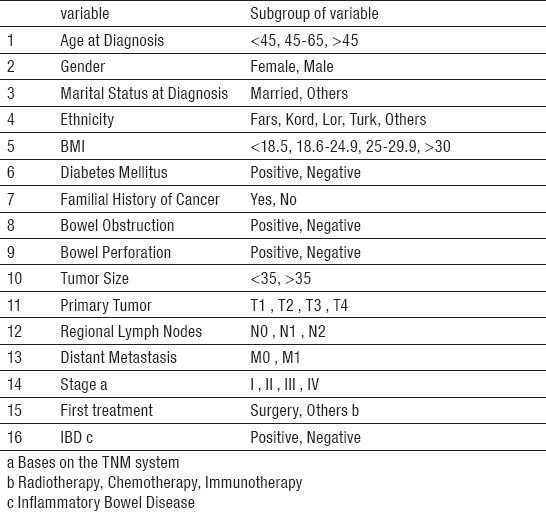
For the sake of preprocessing, variables that were not identified as prognostic factors based on previous studies or were not included either in calculation or creation of new variables were excluded. After removing these irrelevant attributes, we were left with 21 attributes including Age at Diagnosis (Dx), Gender, Marital Status at Dx, Ethnicity, BMI, Hypertension, Diabetes Mellitus, Familial History of Cancer, Personal History of Cancer, Bowel Obstruction, Bowel Perforation, Site (tumor location), Histological Type, Tumor Size, Tumor Grade, Tumor Extension, Regional Lymph Node Metastasis, Distant Metastasis, Tumor Stage, IBD, Treatment Methods. Furthermore, the records in which 5-year survival could not be determined due to being lost follow-up were excluded as well as the records with missing values of key variables including tumor extension, regional lymph node metastasis, distant metastasis and tumor stage. Missing values of other variables were less than 10% which imputed using the K-Nearest Neighbors (KNN) algorithm (35). Subsequently, the instances were classified by response variable into two groups as survival (patients who survived five years after the diagnosis date) and non-survival (patients who did not survived five years after the diagnosis date).
Following this, 261 records were left in which the number of survival (26%) and non-survival (74%) patients was a significant imbalance. Since this imbalance in data can potentially affect the performance of the developed model (36, 37), Synthetic Minority Oversampling Technique (SMOTE) (38) was employed in WEKA to address this problem. Using this technique the dataset were approximately balanced and the resulting total number of records increased to 395 in which 201 instances (51%) related to survival and 194 instances (49%) related to none-survival patients.
Finally, since feature selection techniques can improve the predictive performance of models by selecting the most informative subset of variables (32, 39), we applied both filter and wrapper methods to select the predictor variables from the 21 existing variables in the original dataset. Compared with the feature sets selected by filter method, the 16 features selected by the wrapper method provided better classification results. These features shown in Table 1, were used to construct models. Thus, the research dataset left with 17 variables and 395 records in which 16 columns indicate the features and one column indicates the response variable.
Table 1.
Selected data set attributes
5. MODEL DEVELOPMENT
Herein several different types of supervised classification methods in WEKA toolkit were employed to predict survival of CRC patients, at the end of 5 years of diagnosis. The basic classifier methods encompasses C4.5 (using Weka’s J48), SVM (using Weka’s SMO), Naive Bayes (NB, using Weka’s WAODE), BN, ADTree, Radial Basis Function (RBF, using Weka’s RBFNetwork), REPTree, KNN (using Weka’s KStar) and RF were used to generate classier models along with ensemble classifiers of bagging and voting. Generally speaking, ensemble classifiers are a type of meta model that use a set of base classifiers as input to a combination function (32). In this study, ensembles methods are of two types, namely bagging and voting (using Weka’s vote). Bagging as the acronym of bootstrap aggregating, is a homogenous ensemble method which constructs component classifiers of a same type on different bootstrap replicates of the dataset and combines prediction by a simple majority voting across (40), whereas ensemble voting is a heterogeneous method which uses different classifier over the same dataset and able to combine prediction generated by each classifier in different ways like average of probabilities, majority voting and median (32). Herein the average of probabilities was chosen since all the ways gave similar results.
It is worth noting that the RF algorithm is very much like the bagging algorithm, but specifically designed for decision trees (40, 41). In other words RF is hard wired to RandomTree and cannot use other base classifiers as underlying learner. That is why it was regarded as a base classifier in this study. A group of basic classifiers were selected to be used in theses ensembles methods. The bagging models used C4.5, REPTree, NB, ADTree, RBF, SVM, BN, KNN classifiers as base learners. The voting model used SVM, C4.5, RF, BN and NB as base learners. Finally 9 individual basic, 8 bagging models and a voting ensemble classifier were used to generate 18 models for survival prediction of CRC patients, at the end of 5 years of diagnosis.
For evaluation purposes, stratified 10 fold Cross-validation (CV) method was employed in order to avoid over-fitting problem. In this approach, the dataset is split into 10 stratified segments, and this operation is performed 10 times, each time all folds but one are used for training and the remaining single fold is used for testing. Therefore, the overall result is the average of the 10 sub results (32,42, 43). Further, the performance of the prediction models were measured by the AUC, since AUC is considered as the most widely used metric to measure the ability of the model to discriminate between the different class values (44). In addition, the method of DeLong et al was used to compare the difference between two ROC curves in MedCalc software. The significant level was defined at 0.05.
6. RESULTS
A total of 18 models have been developed to predict 5-year survival of CRC patients. These models include 9 basic individual classifier, 8 ensemble bagging models together with an ensemble voting model of five basic classifiers including SVM, C4.5, RF, BN and NB. From the foregoing, the research database on which the models were built, composed of 17 variables (table 1) and 395 records in which 16 columns indicate the features and one column indicates the response variable.
Figure 1 indicates the corresponding performance of developed models in terms of AUC for results of the 5-year survivability.
Figure 1.
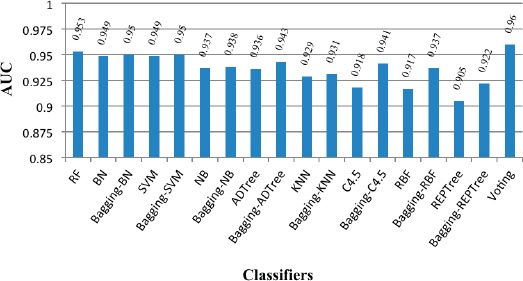
Prediction performance of developed models in terms of AUC
As it can be seen in Figure 1, AUC statistics for all developed models is above 0.90. In other words all models perform over 90%. This statistics is generally higher in ensemble models than basic individual models, among which the highest efficiency is reported in voting method (AUC = 0.96). The differences in predictive performances between basic and ensemble models are studied by using statistical analysis in MedCalc software in which the equality of the surfaces area under the ROC curves was tested by defining the significant level at 0.05. The results are shown in Table 2.
Table 2.
Comparison of the AUC for Developed Models with CIa =95%
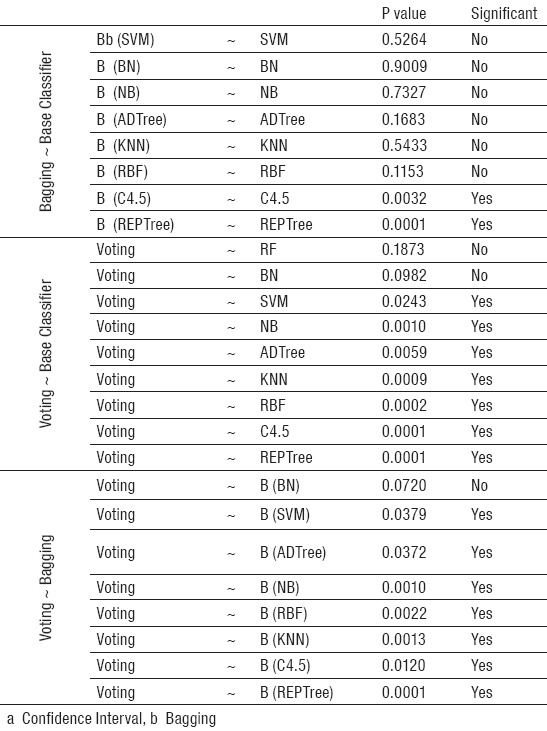
According to Table 2, the significant performance difference exist between ensemble bagging methods and individual basic classifiers include C4.5 and REPTree, but this difference was not observed in other basic classifiers. In other words, ensemble bagging did not improve the predictive performance of SVM, BN, NB, KNN, RBF and ADTree basic classifiers (P>0.05). Additionally from Figure 1 it is clear that ensemble voting method yielded the best prediction performance in terms of AUC, although it was found not to be significantly better than the RF and BN, at 5% significance level.
7. DISCUSSION AND CONCLUSION
The present study predicted the 5-year survivability of CRC patients by conducting a comparative study of basic (C4.5, SVM, NB, ADTree, RBF, REPTRee, KNN, BN and RF) and ensemble (bagging and voting) classifier methods. The wrapper feature selection method was used to select 16 relevant variables, while the SMOTE technique was applied to resolve imbalanced data problem. Finally, the differences in predictive performances between the models were measured by comparing the AUCs using MedCalc software, while the significant level was defined at 0.05.
The obtained results showed that all built models have achieved high classification performance. Overall, the ensembles performed better than the individual base classifiers in terms of AUC. Similarly, the ensemble voting was found to result in the best prediction performance and showed the highest AUC of 0.96 which is consistent with the previous studies (19-21). However ensemble voting could not significantly improve predictive performance of RF and BN classifiers. Even though recent results in solving classification problems indicate that the use of ensembles often leads to improved performance over using single classier models (45), it is difficult to see any advantages of using ensemble voting method over the RF and BN classifiers, based on the findings of this study.
Similar to the finding of (18,19, 21), this study proposed the RF method as a robust and powerful machine learning technique to predict survival in patients with CRC. In addition, in our experiments, we found the BN method not only could accurately estimate survivability of CRC patients but also could easily be understood by those who need to use it. This is critically important in medicine area since the studies have shown that the clinicians are reluctant to accept black-box models. Domain experts may select the final model based on its performance and ability to explain (46).
Obviously, accurate prediction of survival in patients with cancer could support clinical decisions and improve institutional performance in cancer management, which may be achieved by utilizing correct data mining algorithms in making decision support systems. One way to motivate why particular data mining techniques were suitable for a particular learning task is through comparative studies (47). Looking at the findings of this comparative study, we can conclude that the RF, BN works as good as ensemble voting method which may make them appealing techniques when selecting suitable data mining models for decision support systems.
This study has some limitations. Firstly, the research database lacks prognostic factors such as lymphovascular invasion, CEA and residual tumor after surgery which evidence suggests that may have an impact on survival prediction in CRC patients. Secondly, this study is a retrospective single center experience. Finally, due to the relatively small database size, the study may not have been powered enough to assess the generality of the models. Therefore, it will be of great interest to see how it performs in different settings in order to integrate the patient’s dataset and increase reliability of results for future researches.
Acknowledgments
All valuable contribution of the Research Institute of Gastroenterology and Liver Disease, Shahid Beheshti University of Medical Sciences, Tehran are kindly appreciated for providing data in this study.
Footnotes
v• Author Contribution: All authors made substantial contribution to conception and design, analysis and interpretation of data, critically revising the article for important intellectual content, and final approval of the version to be published. Plus Pourhoseingholi M.A & Kheirian S. participated in drafting the article and also Pourhoseingholi M.A & Zali M.R made substantial contribution to acquisition of data
• Conflict of interest: none declared.
REFERENCES
- 1.Stewart B, Wild C. World Cancer Report 2014. Lyon: International Agency for Research on Cancer/World Health Organization; 2014. [Google Scholar]
- 2.Weiser MR, Gonen M, Chou JF, Kattan MW, Schrag D. Predicting survival after curative colectomy for cancer: individualizing colon cancer staging. Journal of Clinical Oncology. 2011;29(36):4796–802. doi: 10.1200/JCO.2011.36.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Annals of surgical oncology. 2010;17(6):1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 4.Roncucci L, Fante R, Losi L, Di Gregorio C, Micheli A, Benatti P, et al. Survival for colon and rectal cancer in a population-based cancer registry. European Journal of Cancer. 1996;32(2):295–302. doi: 10.1016/0959-8049(95)00532-3. [DOI] [PubMed] [Google Scholar]
- 5.Gao P, Zhou X, Wang ZN, Song YX, Tong LL, Xu YY, et al. Which is a more accurate predictor in colorectal survival analysis?. Nine data mining algorithms vs. the TNM staging system. PLoS One. 2012;7(7):e42015. doi: 10.1371/journal.pone.0042015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kantardzic M. Data mining: concepts, models, methods, and algorithms. John Wiley & Sons; 2011. [Google Scholar]
- 7.Coenen F. Data mining: past, present and future. The Knowledge Engineering Review. 2011;26(1):25–9. [Google Scholar]
- 8.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Computational and structural biotechnology journal. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snow PB, Kerr DJ, Brandt JM, Rodvold DM. Neural network and regression predictions of 5-year survival after colon carcinoma treatment. Cancer. 2001;91(S8):1673–8. doi: 10.1002/1097-0142(20010415)91:8+<1673::aid-cncr1182>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 10.Eschrich S, Yang I, Bloom G, Kwong KY, Boulware D, Cantor A, et al. Molecular staging for survival prediction of colorectal cancer patients. Journal of Clinical Oncology. 2005;23(15):3526–35. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 11.Fathy SK. A predication survival model for colorectal cancer. Proceedings of the 2011 American conference on applied mathematics and the 5th WSEAS international conference on Computer engineering and applications. 2011:36–42. [Google Scholar]
- 12.Stojadinovic A, Nissan A, Eberhardt J, Chua TC, Pelz JO, Esquivel J. Development of a Bayesian Belief Network Model for personalized prognostic risk assessment in colon carcinomatosis. The American Surgeon. 2011;77(2):221–30. [PubMed] [Google Scholar]
- 13.Liu Z, Chen D, Tian G, Tang ML, Tan M, Sheng L. Efficient support vector machine method for survival prediction with SEER data. In Advances in Computational Biology. 2010:11–18. doi: 10.1007/978-1-4419-5913-3_2. [DOI] [PubMed] [Google Scholar]
- 14.Dolgobrodov SG, Moore P, Marshall R, Bittern R, Steele RJ, Cuschieri A. Artificial neural network: predicted vs. observed survival in patients with colonic cancer. Diseases of the colon & rectum. 2007;50(2):184–91. doi: 10.1007/s10350-006-0779-8. [DOI] [PubMed] [Google Scholar]
- 15.Spelt L, Nilsson J, Andersson R, Andersson B. Artificial neural networks –A method for prediction of survival following liver resection for colorectal cancer metastases. European Journal of Surgical Oncology (EJSO) 2013;39(6):648–54. doi: 10.1016/j.ejso.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 16.Van Stiphout RG, Postma EO, Valentini V, Lambin P. The contribution of machine learning to predicting cancer outcome. Artificial Intelligence. 2010;350:400. [Google Scholar]
- 17.Anderson B, Hardin JM, Alexander DD, Meleth S, Grizzle WE, Manne U. Comparison of the predictive qualities of three prognostic models of colorectal cancer. Frontiers in bioscience (Elite edition) 2010;2:849. doi: 10.2741/e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sailer F, Pobiruchin M, Bochum S, Martens UM, Schramm W. Prediction of 5-Year Survival with Data Mining Algorithms. ICIMTH. 2015:75–8. [PubMed] [Google Scholar]
- 19.Al-Bahrani R, Agrawal A, Choudhary A. Colon cancer survival prediction using ensemble data mining on SEER data. InBig Data, 2013 IEEE International Conference. 2013:9–16. [Google Scholar]
- 20.Hosseini N. Predicting Colorectal Cancer Survival: A Data Mining Approach. Gastroenterology. 2014;146(5):S–688. [Google Scholar]
- 21.Silva A, Oliveira T, Neves J, Novais P. Treating Colon Cancer Survivability Prediction as a Classification Problem. ADCAIJ: Advances in Distributed Computing and Artificial Intelligence Journal. 2016;5(1):37–50. [Google Scholar]
- 22.Compton CC, Fielding LP, Burgart LJ, Conley B, Cooper HS, Hamilton SR, et al. Prognostic factors in colorectal cancer: College of American Pathologists consensus statement 1999. Archives of pathology & laboratory medicine. 2000;124(7):979–94. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 23.Kroenke C, Neugebauer R, Meyerhardt J, Prado C, Weltzien E, Kwan, et al. Analysis of Body Mass Index and Mortality in Patients With Colorectal Cancer Using Causal Diagrams. JAMA oncology. 2016;2(9):1137–45. doi: 10.1001/jamaoncol.2016.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter V, Jansen L, Hoffmeister M, Ulrich A, Roth W, Bläker H, et al. Prognostic relevance of prediagnostic weight loss and overweight at diagnosis in patients with colorectal cancer. The American journal of clinical nutrition. 2016;104(4):1110–20. doi: 10.3945/ajcn.116.136531. [DOI] [PubMed] [Google Scholar]
- 25.Nan KJ, Qin HX, Yang G. Prognostic factors in 165 elderly colorectal cancer patients. World journal of gastroenterology. 2003;9(10):2207. doi: 10.3748/wjg.v9.i10.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills KT, Bellows CF, Hoffman AE, Kelly TN, Gagliardi G. Diabetes and colorectal cancer prognosis: a meta-analysis. Diseases of the colon and rectum. 2013;56(11):1304–19. doi: 10.1097/DCR.0b013e3182a479f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Y, Li MD, Hu HG, Dong CX, Chen JQ, Li XF, et al. Prognostic and survival analysis of 837 Chinese colorectal cancer patients. World Journal of Gastroenterology. 2013;19(17):2650. doi: 10.3748/wjg.v19.i17.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulendran M, Stebbing JF, Marks CG, Rockall TA. Predictive and prognostic factors in colorectal cancer: a personalized approach. Cancers. 2011;3(2):1622–38. doi: 10.3390/cancers3021622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Mauldin P, Ebeling M, Hulsey T, Liu B, Thomas M, et al. Effect of metabolic syndrome and its components on recurrence and survival in colon cancer patients. Cancer. 2012;119(8):1512–20. doi: 10.1002/cncr.27923. [DOI] [PubMed] [Google Scholar]
- 30.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. Journal of the National Cancer Institute. 2004;96(19):1420–5. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 31.Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. International journal of molecular sciences. 2017;18(1):197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witten IH, Frank E, Hall MA, Pal CJ. Data Mining: Practical machine learning tools and techniques. Morgan Kaufmann. 2016 [Google Scholar]
- 33.Pal N. Advanced techniques in knowledge discovery and data mining. Springer Science & Business Media; 2007. [Google Scholar]
- 34.Han J, Pei J, Kamber M. Data mining: concepts and techniques. Elsevier; 2011. [Google Scholar]
- 35.Schmitt P, Mandel J, Guedj M. A comparison of six methods for missing data imputation. Journal of Biometrics & Biostatistics. 2015;6(1):1. [Google Scholar]
- 36.He H, Garcia EA. Learning from imbalanced data. IEEE Transactions on knowledge and data engineering. 2009;21(9):1263–84. [Google Scholar]
- 37.Maciejewski T, Stefanowski J. Local neighbourhood extension of SMOTE for mining imbalanced data. Computational Intelligence and Data Mining (CIDM): IEEE. 2011:104–111. [Google Scholar]
- 38.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research. 2002;16:321–57. [Google Scholar]
- 39.Guyon I, Elisseeff A. An introduction to variable and feature selection. Journal of machine learning research. 2003:1157–82. [Google Scholar]
- 40.Breiman L. Bagging predictors. Machine learning. 1996 Aug 1;24(2):123–40. [Google Scholar]
- 41.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Biometrics. 2002 [Google Scholar]
- 42.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of International Joint Conference on AI. 1995;14(2):1137–45. [Google Scholar]
- 43.Delen D. Analysis of cancer data: a data mining approach. Expert Systems. 2009;26(1):100–12. [Google Scholar]
- 44.Chimieski BF, Fagundes RD. Association and classification data mining algorithms comparison over medical datasets. Journal of health informatics. 2013;5(2) [Google Scholar]
- 45.Banfield RE, Hall LO, Bowyer KW, Kegelmeyer WP. A comparison of decision tree ensemble creation techniques. IEEE transactions on pattern analysis and machine intelligence. 2007;29(1):173–80. doi: 10.1109/tpami.2007.250609. [DOI] [PubMed] [Google Scholar]
- 46.Bellazzi R, Zupan B. Predictive data mining in clinical medicine: current issues and guidelines. International journal of medical informatics. 2008;77(2):81–97. doi: 10.1016/j.ijmedinf.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Joachims T. A statistical learning learning model of text classification for support vector machines. Proceedings of the 24th annual international ACM SIGIR conference on Research and development in information retrieval. 2001:128–36. [Google Scholar]


