Abstract
The Myogenic Regulatory Factors (MRFs) Myf5, MyoD, myogenin and MRF4 are members of the basic helix-loop-helix family of transcription factors that control the determination and differentiation of skeletal muscle cells during embryogenesis and postnatal myogenesis. The dynamics of their temporal and spatial expression as well as their biochemical properties have allowed the identification of a precise and hierarchical relationship between the four MRFs. This relationship establishes the myogenic lineage as well as the maintenance of the terminal myogenic phenotype. The application of genome-wide technologies has provided important new information as to how the MRFs function to activate muscle gene expression. Application of combined functional genomics technologies along with single cell lineage tracing strategies will allow a deeper understanding of the mechanisms mediating myogenic determination, cell differentiation and muscle regeneration.
Keywords: Myogenic Regulatory Factors, MyoD, Skeletal Muscle, Myogenesis
I. Introduction
The myogenic regulatory factors (MRFs) form a select family of transcription factors whose function and activity represent a paradigm where a series of molecular switches determine the fate of an entire cell lineage. The MRFs are a group of four muscle-specific proteins including MyoD, Myf5, Myogenin and Myogenic Regulatory Factor4 (MRF4) that act at multiple points in the muscle lineage to cooperatively establish the skeletal muscle phenotype through regulation of proliferation, irreversible cell cycle arrest of precursor cells, followed by a regulated activation of sarcomeric and muscle specific genes to facilitate differentiation and sarcomere assembly (1).
The discovery of MyoD was a landmark in the understanding of the processes of muscle differentiation. MyoD was the first of the MRF family identified and was cloned by screening a library containing a 1:1 mixture of myoblasts and differentiated myotubes in expression vectors for cDNAs that would convert cultured fibroblasts into skeletal myocytes. The three remaining MRFs were discovered shortly after this seminal finding through cloning by similar functional screens and by recovering cDNAs with homology to MyoD (2,3). Further investigations led to the notion that MRFs would be essential for muscle development, based on observations that over-expression of MyoD was sufficient to convert non-muscle cells into myoblast-like cells. Thus, MRFs form the core of the transcriptional network that leads to skeletal muscle development through direct binding to DNA. MyoD, Myf5, myogenin and MRF4 contain a basic helix–loop–helix (bHLH) domain that confers them with the ability to recognize the E-box sequence (CANNTG) in the regulatory sequences of target genes upon heterodimerization with a member of the ubiquitously expressed E-protein family of bHLH proteins (4).
Although MRF genes are expressed exclusively in myogenic cells during embryonic myogenesis, there are intrinsic differences at the protein domain level and in the timing and stages of myogenesis where they are expressed, reflecting underlying specificities in the roles of the RFs in muscle cell commitment and differentiation. These differences highlight the specific response of each of the MRFs genes to signaling cues and their interaction partners at the protein level, that to date remain poorly explored.
II. Embryonic development of skeletal muscle
Distinct transcriptional gene regulatory networks hierarchically control myogenic differentiation, each under the precise control of a master regulator present at specific temporal and spatial developmental stages (1). Each of the MRFs can act as a master regulator of myogenesis, whose ectopic expression can circumvent the natural gene regulatory program of a non-muscle cell towards a myogenic-like cell;however, the location, timing and expression levels of the MRFs during development are finely modulated to ensure the accurate progression of the developmental process.
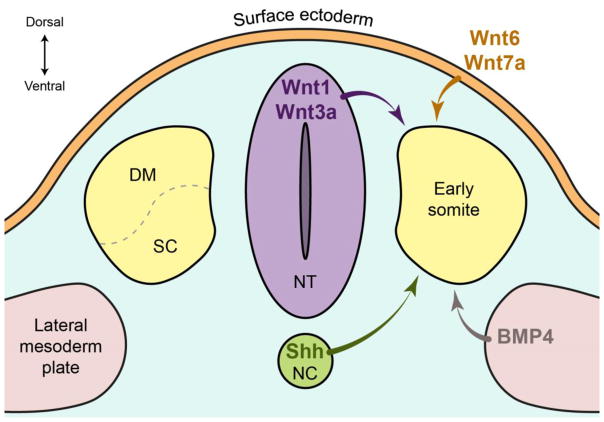
The basic structure of skeletal muscle is patterned during embryogenesis (1,5,6). Paraxial mesoderm is formed bilaterally adjacent to the neural tube in the mouse embryo and becomes progressively segmented along the rostral-to-caudal axis to generate the somites. Through the influence of signaling molecules—such as Sonic Hedgehog (Shh), Wnt, and BMP emanating from the notochord, neural tube, surface ectoderm, and lateral plate mesoderm—the somites undergo a transition, partitioning into an underlying mesenchymal sclerotome and an overlying dermomyotome, where around embryonic day E8.0 expression of MRFs is first detected (Figure 1) (7). The lateral lips of the dermomyotome migrate to its ventral surface with the cells undergoing an epithelial-to mesenchymal transition to form a distinct myotome beneath the dermotome. The myotome eventually forms the skeletal muscles of the body and limbs, as a result of Shh signaling from the notochord and floor plate, which induces Myf5 expression by the mab-related transcription factor 2 (Dmrt2) (8,9). Then the dorsomedial part of the myotome forms the epaxial myotome and is responsible for building the deep back muscles. The ventrolateral region or hypaxial myotome control the formation of the muscle of the body wall, where MyoD expression is induced at E10.5 by Wnt signaling from the dorsal endoderm and by BMP4 from the lateral mesoderm (10). MyoD activation also depends on the paired-like homeodomain factor 2 (Pitx2), which lies genetically upstream of Pax3 (11). Sine oculis homeobox transcription factors 1/4 (Six1/4), with their coactivators Eya1/2, are also important players in MyoD expression (2). Together with Pax3, they respond to enhancer elements responsible for hypaxial somite and limb expression of Myf5 and MyoD. Finally, hypaxial myoblasts adjacent to the nascent limb buds are induced to delaminate and migrate into the limbs to form the limb muscles.
Figure 1. Schematic representation of transverse sections through the embryo at the early stage of somitogenesis.
Several signaling molecules are secreted from different domains in the embryo in order to specify the early somite to give rise to the sclerotome (SC) and the dermomyotome (DM). Wnt proteins are secreted from the dorsal neural tube (NT) and the surface ectoderm while Shh is secreted from the notochord (NC) and BMP4 from the lateral mesoderm plate. Altogether, these signaling molecules regulate the early myogenesis.
Transcripts for myogenin and MRF4 are first detected at E8.5 and E9.0, respectively, and their expression is evenly distributed throughout the myotome (10,12). Consistent with a dual role in both determination and differentiation of the muscle lineage, MRF4 transcripts show biphasic expression, whereby they are downregulated by E11.5, but reappear at E16.0 in differentiated muscle fibers. Differences between the epaxial and hypaxial pool of myoblasts are evidenced at the molecular level by differing dependencies upon Myf5 and MyoD (13,14). Myogenesis then proceeds through several waves of differentiation, beginning with an initial group of embryonic myoblasts, which form the primary muscle fibers. These fibers serve as a scaffold for the secondary myofibers formed by the fusion of fetal myoblasts around E14.0 (15). Finally, a third wave of myogenic progenitors residing adjacent to existing fibers, called satellite cells, appear at the end of postnatal development and will allow for the hypertrophic growth of skeletal muscle after birth. At the end of postnatal development, some satellite cells enter quiescence but remain primed for activation and are responsible for skeletal muscle regeneration during the extent of the animal's lifetime (5).
Embryonic myogenic progenitors originated from the central region of the somitic dermomyotome are characterized by expression of the paired box transcription factors Pax3 and Pax7 (16,17). Myogenesis during this stage of embryogenesis is controlled by the precise hierarchical induction of Myf5 and MyoD, followed by myogenin and MRF4. It is hypothesized that a subpopulation of myogenic precursor cells that do not express MRFs but retain Pax3 and Pax7 expression, are precursors of the adult satellite cells.
III. Post-natal skeletal muscle differentiation
At the postnatal satellite cell stage, Pax3 and Pax7 mark the presence of these muscle progenitors located underneath the basal lamina of adult myofibers (18). Although Pax7 is expressed in all satellite cells in the postnatal myofiber, not all satellite cells express Pax3. Gene expression analyses coupled with ChIP-seq studies of Pax7 and Pax3 in primary myoblasts have shown that while both transcription factors recognize the same DNA motifs, Pax7 has a higher affinity than Pax3 to homeodomain-binding motifs. While Pax3 binds a subset of Pax7 target genes that are mainly involved in the regulation of embryonic functions and maintenance of an undifferentiated phenotype, Pax7 specifically activates genes involved in the maintenance of adult satellite cell phenotype from regulation of proliferation to inhibition of differentiation (19).
How Myf5 and MyoD expression is controlled in satellite cells and consequently their commitment towards the myogenic lineage is a topic of intense research. Recent reports have shown that although adult satellite cells do not express MyoD in resting conditions, the use of a MyoD-iCre mouse strain with a lineage-tracing reporter allele, suggests that irrespective of their anatomical location and embryological origin, all the progenitors derived from satellite cells transcribe MyoD prenatally (20). Notably, distinct populations of Myf5-positive and Myf5-negative satellite cells are present in adult muscles, as observed in Myf5-nlacZ reporter mice and by the direct detection of Myf5 protein levels (21,22).
To determine whether the Myf5-negative satellite cells represent a distinct population that has never expressed Myf5 during development, our laboratory generated a Myf5 Cre/R26R-YFP mouse model, in which cells expressing Myf5 and their progeny are permanently labelled with yellow fluorescent protein (YFP). We determined that a subpopulation of 10% of total satellite cells has never expressed Myf5 during development (21). Importantly, satellite cells that do not transcribe Myf5 (YFP-) display long-term self-renewing capacity, whereas satellite cells that do transcribe Myf5 (YFP+) appear to behave more like committed progenitors.
Induction of Myf5 expression requires arginine methylation of Pax7 by the arginine methyltransferase Carm1 (23), which triggers the recruitment of the histone methyltransferase complex Wdr5-Ash2l-Mll2 (Kmt2) to the Myf5 locus, resulting in permissive chromatin modifications that stimulate transcriptional activation of Myf5 through asymmetric muscle stem cell divisions (24). In addition, it was found that satellite cells indeed transcribe the Myf5 gene, but the transcript remains untranslated in order to maintain these cells in a quiescent state through sequestration of the Myf5 transcripts in mRNP granules in a mechanism orchestrated by miR-31. Upon satellite cell activation, mRNP granule dissociation results in the release of sequestered transcripts and rapid translation of the Myf5 mRNAs (25).
In proliferating myoblasts, MyoD expression is induced by the transcription factors FoxO3, Six1/4 and Pax3 and Pax7 (26,27). An interesting aspect not largely addressed in the field is the relationship between nuclear localizationof loci containing genes expressed during myogenesis with their transcriptional status. For example, low-level MyoD transcription is induced through TFIID, correlating with MyoD locus spatial localization near the periphery of the nucleus. As differentiation proceeds towards myotube formation, the MyoD locus is relocated to the lumen of the nucleus where the factors TAF3/TRF3 promote MyoD expression (28). Under these conditions, MyoD induces Myogenin expression, which results in downregulation of Myf5 expression (29). This switch in expression from Myf5 to myogenin coincides with cell cycle exit and a commitment to differentiate (30). The combined activity of MyoD and Myogenin leads to the expression of the MRF-4 gene and other late muscle differentiation genes to permit the formation of multinucleated fibers. In mature muscle fibers, expression of MyoD and Myogenin is then downregulated, whereas MRF4 continues to be expressed at high levels to act as the predominant MRF in mature differentiated muscle (12).
IV. Regulation of MRFs by signaling molecules
During embryonic development, vertebrate myogenesis is tightly regulated by a combination of signaling molecules secreted from the neural tube and the surrounding structures that synergistically induce the direct expression of MRFs to specify myogenic progenitors in somites and allow their differentiation (Figure 1) (31). These molecules are capable to induce myogenic specification, which include Wnts, Sonic hedgehog (Shh), the Notch receptor and bone morphogenetic proteins (BMPs) (5,32). Wnt proteins constitute a large family of glycoproteins with several members shown to be required for early myogenesis in somites (33). The canonical Wnt signaling pathway requires Wnts binding to the Frizzled receptors to activate the β-catenin/TCF transcriptional complex (34). In the presence of Wnt proteins, β-catenin translocates into the nucleus and binds members of the Tcf and Lef family of transcription factors, acting as a transcriptional coactivator to induce expression of target genes. Wnts binding to the Frizzled receptors can also activate non-canonical pathways, independently of the β-catenin/Tcf transcriptional complex (34).
During somitogenesis for example, Wnt1 and Wnt3a are secreted from the dorsal neural tube whereas Wnt6 and Wnt7a are secreted from the surface ectoderm (35), allowing for the specification of the dorsal part of the somites that will give rise to the dermomyotome. Explant cultures of paraxial mesoderm showed that Wnt1 binds to Fzd receptors 1 and 6 to induce myogenesis through direct activation of Myf5 expression via the canonical β-catenin pathway (36,37). Wnt7a however, preferentially activates MyoD through a PKC-dependent β–catenin-independent non-canonical pathway (38).
Along with the Wnt proteins, Shh, which is secreted from the notochord and dorsal neural tube, also acts in somitic tissue to induce myogenesis in vitro (39). Mammalian Hedgehog proteins-binding to the Patched1 receptor leads to de-repression of Smoothened activity, allowing for regulation of gene expression through nuclear translocation of Gli transcription factors. Interestingly, Shh knockout mice fail to express MyoD and Myf5 and to form epaxial myotome (8,40,41). During hypaxial muscle development, Shh signaling maintains Myf5 and MyoD expression in mouse limb buds and Shh−/− mice display severe deficiency of hypaxial limb muscles (42). Mechanistically, Shh directly induces
Myf5 expression through essential Gli-binding sites located in the Myf5 enhancer leading to the specification of myogenic progenitor cells (43). Using P19 cells, Voronova et al. showed that Gli2 transcription factor binds MyoD gene elements to regulate its transcription but also binds MyoD itself regulating its ability to induce skeletal myogenesis (44).
Altogether, these findings demonstrate that Wnts and Shh act synergistically for myogenic determination of unspecified cells. Supporting this idea, both Gli binding sites and Tcf/Lef sequences were identified in the early epaxial Myf5 enhancer explaining how both Hedgehog and Wnt signaling pathways respectively, can act together to activate Myf5 (37).
In contrast to the Wnt and Shh proteins positively regulating the specification of myogenic progenitors, BMPs and Notch receptor inhibit the expression of the MRFs (45–47). They maintain the cells in an undifferentiated state, thus promoting the expansion of the progenitor cells rather than their differentiation. Belonging to the TGF-β superfamily, BMPs act through serine-threonine kinase receptors to activate the SMAD proteins and allow their translocation to the nucleus leading to the activation or repression of the target genes (48). In Xenopus, it was shown that MyoD and Myf5 expression requires a specific level of BMP signaling regulated by BMP4 from the ventral parts of the embryo and BMP antagonists such as Noggin from the dorsal regions of the embryo (49). In the chicken embryo, BMP4 is secreted from the plate mesoderm and maintains the lateral somitic cells as undifferentiated muscle progenitors by repressing Myf5 and MyoD expression (50). As observed for BMP signaling, active Notch signaling represses MyoD expression through the DNA-binding protein RBP-J and the transcriptional repressor Hes1 (51). Correlating with these findings, mutation of the Notch ligand Delta1 impairs Notch signaling, leading to an excessive myogenic differentiation and a loss of myogenic progenitors (47).
V. Mouse models to understand the contribution of MRFs on myogenesis
Despite this canonical perspective, MRFs are under strict regulatory mechanisms to ensure their expression during development and muscle regeneration. Moreover, regardless of the differences in their origin during embryogenesis, all myogenic progenitor cells share the same core components of the myogenic pathway. The generation of knockout mouse models for these genes has been instrumental to identify their role in myogenesis and to understand how they regulate each other’s expression.
Interestingly, mice lacking a functional MyoD gene were found to be viable and fertile and did not exhibit any apparent deficits in skeletal muscles (52). However, the amount of Myf5 mRNA was increased in homozygous mutant mice, suggesting that Myf5 was able to substitute for the absence of MyoD in the development of skeletal muscle. Similar to MyoD mutant mice, Myf5 deficient animals did not develop any abnormalities in skeletal muscle. However, these mice died perinatally due to severe rib abnormalities that prevented normal respiratory function of the lung (53–55). Remarkably in Myf5 mutant mice, the onset of MyoD expression occurs normally, which is followed by the expression of Myogenin. These results suggested that the activation of MyoD occurs independently of Myf5 and that MyoD can substitute for the absence of Myf5 in the development of skeletal muscle (55).
Most of the myogenic program is severely affected only when both Myf5 and MyoD are absent in a double knockout model. Newborn mice deficient for the two MRFs were totally devoid of skeletal myoblasts and muscle. Mutant pups were born alive, but were immobile and died within minutes (55). Mesenchymal-like cells were present at the cavities normally populated by muscle, where no desmin-positive cells were found. Furthermore, except for truncated ribs and the lack of muscle tendon insertion points, the skeleton appeared normal. Early observations that Myf5 and MyoD are expressed in different subdomains of the dermomyotome, suggests that the two compartments give rise to different muscle lineages (56). Indeed, the presence of one functional Myf5 allele in homozygous MyoD-mutant mice resulted in the formation of normal ribs as well as skeletal muscle, but these mice still died soon after birth. These mice contained considerably fewer muscle fibers, suggesting that perinatal death is a consequence of impairment in muscle function. In contrast, one functional MyoD allele in homozygous Myf5-mutant mice resulted in normal skeletal muscle development (55).
Mice homozygous for targeted mutations in the myogenin gene are born immobile and decease immediately after birth (57,58). Myogenin knockout has a remarkable phenotype with perinatal death; while myoblasts are formed, there is a complete absence of functional skeletal muscle, supporting the idea that Myogenin regulates the later stages of myogenic differentiation, whilst Myf5 and MyoD are involved in the process of determination (58). In addition, normal numbers of myoblasts were present and these were organized in groups similar to wild-type skeletal muscle, showing that myogenin is not involved in skeletal muscle specification. However these animals displayed a marked reduction in fiber density with mononucleated cells, replacing most of the mature muscle cells with overall severe reduction of muscle mass. Therefore, most of these mononucleated cells were observed to express virtually normal levels of MyoD, but lack myosin heavy chain and actin, both markers for differentiated muscle cells. This clearly indicates an essential role for myogenin as a differentiation factor
Generation of three MRF4 knockout mice models showed different phenotypes despite their similar design. These three knockouts vary in the severity of defects, which range from deficiencies in myotomal myogenesis and deep back muscle formation along with rib stubs with lethality at birth in the strongest allele (59), to the presence of a few misshapen bifurcated ribs and full viability in the weakest allele (60). The third allele falls between the first two on a rib phenotype and produces rare homozygous viable animals (61). Interestingly, these rib abnormalities resemble those described for Myf5 knockout. Indeed, mRNA expression of Myf5 is reduced in a direct correlation with the phenotype observed on each MRF4 allele. Since Myf5 and MRF4 are adjacent to each other on the same chromosome, it was later proposed that this phenotype might have been the result of cis effects of the MRF4 null on the linked Myf5 locus (59,60).
As mentioned before, double knockouts of MyoD and Myf5 were originally reported to lack muscle (55). Later work showed that MRF4 expression is also compromised in cis in the Myf5 allele. Indeed, in MyoD/Myf5 null animals which retain functional MRF4, this gene is able to compensate and initiate myogenesis, indicating that MRF4 has both differentiation and determination activities (62).
Interestingly, MRF4/MyoD double null mice died at birth, showing a phenotype highly similar to that of the myogenin null mutant mice (63). In these mice, myogenin was expressed, but was insufficient to support normal myogenesis in vivo. This suggests that MRF4 and MyoD play a redundant role in mediating skeletal muscle differentiation during development.
All these together, show that MyoD and Myf5 function as determination factors, whereas myogenin and MRF4 act to permit terminal differentiation (Figure 2).
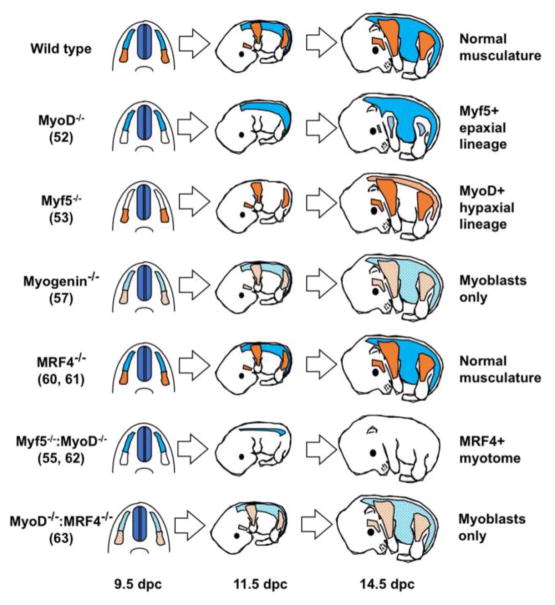
Figure 2. Effects of MRF null mutations on skeletal muscle development.
Wild type embryos with normal development of epaxial (blue) and hypaxial (orange) muscles. MyoD null embryos have a 2-day delay in differentiation of all hypaxial musculature (shown in light blue), and normal epaxial musculature (shown in blue). Myf5 null embryos have a 2-day delay in translocation of for expaxial musculature (light orange) and normal development of hypaxial musculature (shown in orange). Mice lacking myogenin contain myoblasts that fail to differentiate efficiently into myotubes (light orange and light blue). Newborn mice lacking both Myf5 and MyoD display a complete absence of skeletal myoblasts and myofibers, however MRF4 is sufficient to specify myotomal musculature earlier in development (shown in blue). Myogenin absence results in failure of myoblast fusion and an absence of differentiated myofibers. MRF4 deficient mice have normal musculature, whereas compound MyoD/MRF4 mutant embryos resemble the myogenin phenotype with an absence of myofibers. Numbers after each genotype denote the reference to the original publication. Embryonic days of development are indicated below the diagram as days post coitus (dpc).
VI. Structural divergence between MRFs
Early observations that each of the MRFs is able to initiate myogenesis when artificially expressed in a wide variety of non-muscle cells such as primary fibroblasts, nerve, fat and liver, suggested a functional overlap among them in establishing muscle commitment. Although these experiments suggested redundancy within the myogenic bHLH family of proteins, genetic studies demonstrated that MyoD and Myf5 act to establish the myogenic lineage, whereas myogenin and MRF4 mediate the expression of the terminal phenotype (64). Specifically, newborn mice lacking both MyoD and Myf5 are totally devoid of myoblasts and myofibers (55). In contrast, mice lacking myogenin generate myoblasts, but show incomplete skeletal muscle differentiation, with fewer and smaller myotubes (57,58) . The differences between the myogenic factors that confer these specific attributes and their fundamental functional differences are a combination of elements. For example, the dynamics of recruitment to DNA binding motifs at genomic regulatory elements and functional divergence between their proteins sequences.
Phylogenetic analysis of amino acid sequences of the mouse MRFs suggests the following relationships among them . Myf5 and MyoD are more similar to one another (53%) than either are to myogenin (40% and 38%, respectively) or to MRF4 (40% and 43%, respectively). Similarly, myogenin and MRF4 are more related to one another (53%) than to either Myf5 or MyoD (Figure 3) (65). Therefore, these relationships suggest an evolutionary scheme by which the family arose. Myf5 appears to represent the ancestral gene of the family. MRF4 arose from Myf5 by gene duplication at the same locus, while myogenin arose from MRF4 after a gene-duplication event to a second chromosome, and finally, MyoD arose from Myf5 after a gene-duplication event to a third chromosome. Further phylogenetic analysis indicates that the four vertebrate MRF genes have evolved from a single ancestral MRF gene as a result of gene duplication events and subsequent divergent mutations (66). Indeed, many invertebrates, such as Caenorhabditis elegans, Drosophila, sea urchins, and acidians, continue to develop their musculature in the presence of a single MRF gene (67–69). However, the more complex musculature of vertebrates has forced the evolution of four MRFs to regulate the complex gene expression program in myogenesis.
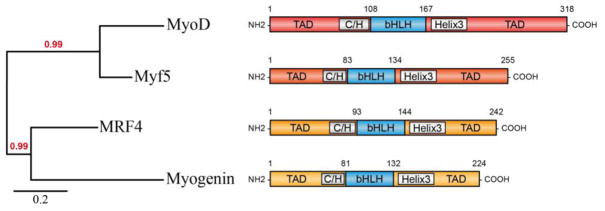
Figure 3. Protein structure of the myogenic regulatory factors.
Mouse amino-acid sequences of each myogenic regulatory factor were obtained from Uniprot and the phylogenetic tree was generated using Phylogeny analyzer (www.phylogeny.fr/index.cg). The phylogenetic tree reveals the close relationship between Myf5 and Myod, and myogenin and MRF4, correlating with their function during myogenesis, specification and differentiation respectively. All MRFs share highly similar and conserved bHLH domain, essential for their myogenic function. The bHLH domain is flanked by less conserved N-terminal (NH2) region containing a C/H domain and C-terminal (COOH) region containing a helix 3 domain, mediating transcriptional regulation.
Analysis of their protein sequences shows that MRFs are highly similar, in that they possess a conserved bHLH domain for DNA binding that is flanked by less conserved N-terminal and C-terminal domains that mediate transcriptional regulation. A series of 65 amino acids embedded in the bHLH domain is highly conserved among MRFs. In MyoD, only 68 amino acids inside the bHLH domain are necessary and sufficient for myogenic conversion of stably transfected 10T1/2 fibroblasts (70). The helix-loop-helix region permits dimerization with bHLH E-proteins such as El2, E47, or HEB, which on their own are non-myogenic. The basic domain of MRFs recognizes E-box sites of the consensus sequence CANNTG present at regulatory elements of target genes and show additional preferences for internal and flanking sequences. Activation of the muscle transcriptional program by MyoD depends on two amino acids in the basic domain, an alanine and threonine (AT), referred to as the myogenic code (71,72). These essential determinants of myogenic specificity are conserved in all muscle bHLH proteins from worms to humans and are absent from all non-muscle bHLH proteins (73). The myogenic code is absolutely required for dominant induction of myogenesis by the MRFs, since replacement of these residues with the asparagines found in the corresponding positions in the E-protein E-12, renders MyoD non-myogenic potential (73). Furthermore, when the AT along with a lysine in the junctional region of the first helix of MyoD are substituted for the corresponding amino acids in E12, these myogenic code amino acids are sufficient to induce myogenesis (70,74).
Reintroduction of MyoD and Myf5 into fibroblasts isolated from the MyoD−/−;Myf5−/− mouse, allowed the dissection of functional differences between these MRFs. MyoD is strikingly more effective than Myf5 at inducing differentiation-phase target genes than Myf5, whereas both MyoD and Myf5 control relatively the same type of genes related to cell proliferation (75,76). This difference is explained by the fact that both the N-terminal, containing a C/H domain and the C-terminal, containing a helix III domain in MyoD, act in a synergistic manner to induce differentiation. In fact unlike Myf5, the MyoD C-terminal domain is necessary to interact with bHLH domain and to promote chromatin-remodeling activity and to further provide access to silenced locus for the N-terminal activation domain (75). Furthermore, swapping of the C/H domain and the helix III domain of MyoD into myogenin allows myogenin to efficiently activate the expression of silent muscle genes (77).
Interestingly, neither the C/H nor 3 helix domains of myogenin are able to induce chromatin accessibility at muscle regulatory elements (78) but are greatly dependent on MyoD to mark an accessible chromatin environment for myogenin target genes. Indeed, promoters of inactive MyoD target genes are constitutively bound by Pbx/Meis factors, with which MyoD interacts through helix III (79). This interaction recruits histone acetyl-transferase complexes to acetylate surrounding histones and MyoD itself (80). Acetylation leads to engagement of SWI/SNF chromatin remodeling complexes that provide access to DNA binding sites for the MRFs and other transcription factors such as Mef2 and recruitment of RNA polymerase II for transcription (81), in a mechanism where presumably Pol ll is kept in a poised state for later activation. Then, myogenin interacts with other factors such as Mef2d to recruit chromatin-remodeling machinery at target genes (81). These mechanistic observations explain the fundamental differences related to the ability of MyoD and Myf5 to establish myogenic lineage by opening chromatin, whereas myogenin is a strong activator of transcription through recruitment of chromatin remodeling complexes.
Functional differences between MyoD and MRF4 have been also determined through swapping strategies (82). The MRF4 activation domain and the related amino-terminal MyoD activation domain are capable of substituting for one another in converting fibroblasts to a myogenic phenotype, however this capacity is achieved at the expense of target specificity, since the type of muscular reporter genes induced by the wild type version of MRF4 are different when a MyoD-MRF4 chimera is used. It was reported that MRF4 could act as either a transcriptional activator or as a repressor, depending on the promoter context. Among the target genes repressed by MRF4 is the cardiac α-actin gene, which is activated by MyoD expression in vitro (82). Direct competition studies on the cardiac α-actin promoter showed that the repressive property of MRF4 predominates over MyoD-mediated transactivation, suggesting that the relative levels of different MRFs may modulate the transcriptional output of specific muscle genes (82). Interestingly, MRF4 activity is modulated by the p38 mitogen-activated protein kinase (MAPK) signaling pathway during the course of myoblast differentiation (83). Phosphorylation of MRF4 by p38 MAPK at Ser31 and Ser42 within the N-terminal transactivation domain inhibits its function, permitting activation of the cardiac α-actin gene while blocking creatine kinase M-type (CKM) gene expression (83). Thus, the N-terminal domain of MRF4 cooperates with p38 MAPK to selectively modulate the expression of the late myogenic transcriptional program. Recent reports showed that transcriptional activity of Mef2d is reduced by its interaction with MRF4, to control hypertrophy in adult skeletal muscle and the expression of Mef2 target genes. Taken together, these results suggest that divergent transactivation domains within the MRF family play a role in modulating their targeting to specific genomic loci.
VII. Regulation of gene expression by MRFs
Given the functional differences among the MRFs related to their ability to induce different gene expression programs, it is of the utmost interest to interrogate the dynamics of binding to the genome during muscle development and regeneration. Binding of MyoD has been the most thoroughly explored in cultured cell systems with high-throughput technologies such as ChIP-sequencing (75,84–86).
Using C2C12 myoblasts and myotubes, Cao et al., (87) identified the sites to which MyoD is bound during in vitro muscle differentiation. Apart from the expected regulatory elements of genes that are known to be downregulated or upregulated during differentiation, MyoD was found to be at a surprisingly considerable large number of E-box sites where no obvious genes involved in myogenesis are present. They reported 23000 MyoD binding sites in myoblasts and 26000 sites in myotubes. More intriguingly, they found that MyoD binding is stable after induction of differentiation as most of the sites are the same in the two cell differentiation stages. Interestingly, MyoD binding correlates with regional rather than local histone acetylation. Importantly, the functionality and biological significance of most of these sites remains to be elucidated, as most of them are inactive when tested for enhancer functionality in vitro.
Increased number of MyoD sites in myotubes with respect to myoblasts (39700 and 18142, respectively) were defined by Mousavi et al. (88). Differences between these analyses may arise based on peak discovery methodology, genomic compartmentalization regarding boundaries considered and other experimental variability. In this study, the authors also used RNA-seq to show that the majority of 35000 binding sites shared by MyoD and myogenin are also bound by Pol II, are marked by H3K4me1 and H3K27Ac, are also transcribed in both senses in myotubes. Expression of these elements, termed eRNAs, was reduced upon siRNA knockdown of MyoD but not of myogenin and interestingly, blocking the expression of the eRNA transcribed from the core enhancer element of MyoD, caused the reduction of MyoD mRNA levels.
One interesting aspect of these series of genomic studies is the confirmation that nucleotide composition around the canonical E-box affects the output of the MRF binding. For instance, Soleimani et al., (86) used retroviral transduction to introduce TAP-tagged derivatives of MyoD, Myf5 and Snail1 into primary myoblasts isolated from hind limb in order to recover the protein and its bound DNA. Using this method, they identified 1428 MyoD sites in myoblasts and 9300 MyoD sites in myotubes whereas Myf5 was identified at 1053 genomic regions in myoblasts. Interestingly there was an overlap of 30% between MyoD and Myf5 binding sites in myoblasts, supporting the notion that these factors share a role in defining myoblasts identity. In an effort to identify a negative regulator of MyoD binding in myoblasts that could explain the dramatic differences in the number of MyoD peaks between myoblasts and myotubes, the study focused on transcriptional repressors that share the same DNA binding motif as helix-loop-helix factors such as MRFs. They show that Snail1 binds to E-boxes that have a G/C-rich central dinucleotide and that such sites are associated with genes that are expressed almost exclusively in myotubes. By contrast, Snail does not bind to E-boxes with A/T-rich central dinucleotides, which are associated with genes expressed in myoblasts. Thus at the onset of differentiation Snail must be removed in order to allow MyoD access to the myotube genes. Finally, evidence is presented showing that MRFs induce expression of miRNA-30 and miRNA-206, which inhibit the expression of Snail1 and Snail2, respectively in myotubes, thus as Snails are downregulated MyoD can access to its binding sites in myotubes to induce differentiation-specific genes (86).
Examination of the sites co-bound by MyoD and myogenin identified genes involved in muscle development (85). Interestingly, microarray studies examining the role of myogenin in differentiation recently identified genes involved in cell cycle progression as key transcriptional targets that are downregulated by myogenin during differentiation. This suggests that myogenin is an important modulator of cell cycle exit during differentiation. Thus, in contrast to MyoD, which promotes proliferation in growing myoblasts, myogenin attenuates the expression of genes that mediate cell cycle progression (85).
More recently, analysis of Myf5 and MyoD binding was performed in transduced double knock-out MyoD−/−/Myf5−/− mouse embryonic fibroblasts (76). Upon expression of both MRFs at comparable levels of that detected in C2C12 cells, ChIP-Seq experiments demonstrated that the difference between MyoD and Myf5 relies on transcriptional activity rather than a preference for binding sites throughout the genome, as genome-wide binding profiles of Myf5 and MyoD were identical. However only MyoD was able to recruit RNA Pol II and activate transcription of target genes whereas Myf5 modifies chromatin by inducing histone H4 acetylation but fails to recruit RNA Pol II. This fundamental difference in the number of shared MyoD/Myf5 binding sites with respect to previous reports (87), where only 30% of sites are shared may be related to the peak calling method or to the effect of the C-terminal TAP-tag. It has been previously reported that MyoD is able to direct histone acetylation, probably by recruiting p300 and PCAF (89). One interesting finding of this work is the potential activity of Myf5 as a chromatin remodeler to specify muscle progenitors, despite its inability to induce transcription, which can also indicate that Myf5 might functionally precede MyoD during lineage specification by reorganizing chromatin prior to robust transcription of differentiation specific genes (76). In fact, myogenin also binds the same sites as MyoD but its capacity to access native chromatin is limited (85).
All these studies show the properties of MRFs during myogenic differentiation and identify each of them in the hierarchical sequence of molecular events in the initiation, activation and maintenance of muscle gene transcription.
VIII. Conclusions and future directions
Despite the great progress that functional genomic approaches have generated towards our understanding of the regulatory networks mediated by some of the MRFs during myogenesis, these genome-wide technologies have used cultured cells. The next level of advances will necessary focus on identifying the basis of different muscle-lineages present during embryonic development as well as in adult skeletal muscle stem cells. Single-cell approaches coupled with lineage tracing experiments are currently under the scope of developmental biology, giving an exciting opportunity to reveal new mechanistic knowledge towards upstream regulatory networks in the embryo and link it to our current biochemical understanding of muscle differentiation. Finally, this would provide a satisfying and integrating understanding to establish new therapeutic strategies to treat affections of skeletal muscle, such as muscular dystrophies and age-related regenerative issues.
Acknowledgments
These studies from the laboratory of M.A.R. were supported by grants from the the Canadian Institutes of Health Research [FDN-148387], US National Institutes for Health [R01AR044031], E-Rare-2: Canadian Institutes of Health Research/Muscular Dystrophy Canada [ERA-132935], the Muscular Dystrophy Association, and the Stem Cell Network.
Abbreviations
- MRFs
Myogenic Regulatory Factors
- bHLH
basic Helix-loop-Helix, Embryonic myogenesis, Regenerative Myogenesis
- ChIP
chromatin immunoprecipitation
- miRNA
micro-RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
X. References
- 1.Buckingham M, Rigby PWJ. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014 Feb 10;28(3):225–38. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 2.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986 Dec 5;47(5):649–56. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 3.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, et al. The myod gene family: Nodal point during specification of the muscle cell lineage. Science. 1991 Feb 15;251(4995):761–6. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 4.Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, et al. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/e47-like proteins in vivo. Cell. 1991 Jul 26;66(2):305–15. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 5.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: Molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012 Feb 1;4(2) doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005 Jun;132(12):2685–95. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 7.Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991 Apr;111(4):1097–107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- 8.Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP. Sonic hedgehog controls epaxial muscle determination through myf5 activation. Development. 1999 Sep;126(18):4053–63. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- 9.Sato T, Rocancourt D, Marques L, Thorsteinsdóttir S, Buckingham M. A pax3/dmrt2/myf5 regulatory cascade functions at the onset of myogenesis. PLoS Genet. 2010 Apr 1;6(4):e1000897. doi: 10.1371/journal.pgen.1000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sassoon D, Lyons G, Wright WE, Lin V, Lassar A, Weintraub H, Buckingham M. Expression of two myogenic regulatory factors myogenin and myod1 during mouse embryogenesis. Nature. 1989 Sep 28;341(6240):303–7. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- 11.L'honoré A, Ouimette J-F, Lavertu-Jolin M, Drouin J. Pitx2 defines alternate pathways acting through myod during limb and somitic myogenesis. Development. 2010 Nov;137(22):3847–56. doi: 10.1242/dev.053421. [DOI] [PubMed] [Google Scholar]
- 12.Hinterberger TJ, Sassoon DA, Rhodes SJ, Konieczny SF. Expression of the muscle regulatory factor MRF4 during somite and skeletal myofiber development. Dev Biol. 1991 Sep;147(1):144–56. doi: 10.1016/s0012-1606(05)80014-4. [DOI] [PubMed] [Google Scholar]
- 13.Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997 Dec;124(23):4729–38. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- 14.Kablar B, Krastel K, Ying C, Tapscott SJ, Goldhamer DJ, Rudnicki MA. Myogenic determination occurs independently in somites and limb buds. Dev Biol. 1999 Feb 15;206(2):219–31. doi: 10.1006/dbio.1998.9126. [DOI] [PubMed] [Google Scholar]
- 15.Hollway GE, Currie PD. Myotome meanderings. Cellular morphogenesis and the making of muscle. EMBO Rep. 2003 Sep;4(9):855–60. doi: 10.1038/sj.embor.embor920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Relaix F, Rocancourt D, Mansouri A, Buckingham M. Divergent functions of murine pax3 and pax7 in limb muscle development. Genes Dev. 2004 May 1;18(9):1088–105. doi: 10.1101/gad.301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000 Sep 15;102(6):777–86. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 18.Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomès D, Tajbakhsh S. Pax3/pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005 Jun 15;19(12):1426–31. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soleimani VD, Punch VG, Kawabe Y-I, Jones AE, Palidwor GA, Porter CJ, et al. Transcriptional dominance of pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev Cell. 2012 Jun 12;22(6):1208–20. doi: 10.1016/j.devcel.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanisicak O, Mendez JJ, Yamamoto S, Yamamoto M, Goldhamer DJ. Progenitors of skeletal muscle satellite cells express the muscle determination gene, myod. Dev Biol. 2009 Aug 1;332(1):131–41. doi: 10.1016/j.ydbio.2009.05.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007 Jun 1;129(5):999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beauchamp JR, Heslop L, Yu DS, Tajbakhsh S, Kelly RG, Wernig A, et al. Expression of CD34 and myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J Cell Biol. 2000 Dec 11;151(6):1221–34. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawabe Y-I, Wang YX, McKinnell IW, Bedford MT, Rudnicki MA. Carm1 regulates pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012 Sep 7;11(3):333–45. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinnell IW, Ishibashi J, Le Grand F, Punch VGJ, Addicks GC, Greenblatt JF, et al. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat Cell Biol. 2008 Jan;10(1):77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of myf5 mrna targeted by microrna-31 in mrnp granules. Cell Stem Cell. 2012 Jul 6;11(1):118–26. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, et al. Six1 and six4 homeoproteins are required for pax3 and mrf expression during myogenesis in the mouse embryo. Development. 2005 May;132(9):2235–49. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 27.Hu P, Geles KG, Paik J-H, DePinho RA, Tjian R. Codependent activators direct myoblast-specific myod transcription. Dev Cell. 2008 Oct;15(4):534–46. doi: 10.1016/j.devcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao J, Fetter RD, Hu P, Betzig E, Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011 Mar 15;25(6):569–80. doi: 10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deato MDE, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol Cell. 2008 Oct 10;32(1):96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q-C, Zha X-H, Faralli H, Yin H, Louis-Jeune C, Perdiguero E, et al. Comparative expression profiling identifies differential roles for myogenin and p38α MAPK signaling in myogenesis. J Mol Cell Biol. 2012 Dec;4(6):386–97. doi: 10.1093/jmcb/mjs045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nat Rev Genet. 2008 Aug;9(8):632–46. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- 32.Marcelle C, Stark MR, Bronner-Fraser M. Coordinate actions of bmps, wnts, shh and noggin mediate patterning of the dorsal somite. Development. 1997 Oct;124(20):3955–63. doi: 10.1242/dev.124.20.3955. [DOI] [PubMed] [Google Scholar]
- 33.Rudnicki MA, Williams BO. Wnt signaling in bone and muscle. Bone. 2015 Nov;80:60–6. doi: 10.1016/j.bone.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. Wnt signaling in myogenesis. Trends Cell Biol. 2012 Nov;22(11):602–9. doi: 10.1016/j.tcb.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cossu G, Borello U. Wnt signaling and the activation of myogenesis in mammals. EMBO J. 1999 Dec 15;18(24):6867–72. doi: 10.1093/emboj/18.24.6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tajbakhsh S, Borello U, Vivarelli E, Kelly R, Papkoff J, Duprez D, et al. Differential activation of myf5 and myod by different wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of myf5. Development. 1998 Nov;125(21):4155–62. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- 37.Borello U, Berarducci B, Murphy P, Bajard L, Buffa V, Piccolo S, et al. The wnt/beta-catenin pathway regulates gli-mediated myf5 expression during somitogenesis. Development. 2006 Sep;133(18):3723–32. doi: 10.1242/dev.02517. [DOI] [PubMed] [Google Scholar]
- 38.Brunelli S, Relaix F, Baesso S, Buckingham M, Cossu G. Beta catenin-independent activation of myod in presomitic mesoderm requires PKC and depends on pax3 transcriptional activity. Dev Biol. 2007 Apr 15;304(2):604–14. doi: 10.1016/j.ydbio.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Münsterberg AE, Kitajewski J, Bumcrot DA, McMahon AP, Lassar AB. Combinatorial signaling by sonic hedgehog and wnt family members induces myogenic bhlh gene expression in the somite. Genes Dev. 1995 Dec 1;9(23):2911–22. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- 40.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking sonic hedgehog gene function. Nature. 1996 Oct 3;383(6599):407–13. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 41.McDermott A, Gustafsson M, Elsam T, Hui C-C, Emerson CP, Borycki AG. Gli2 and gli3 have redundant and context-dependent function in skeletal muscle formation. Development. 2005 Jan;132(2):345–57. doi: 10.1242/dev.01537. [DOI] [PubMed] [Google Scholar]
- 42.Krüger M, Mennerich D, Fees S, Schäfer R, Mundlos S, Braun T. Sonic hedgehog is a survival factor for hypaxial muscles during mouse development. Development. 2001 Mar;128(5):743–52. doi: 10.1242/dev.128.5.743. [DOI] [PubMed] [Google Scholar]
- 43.Anderson C, Williams VC, Moyon B, Daubas P, Tajbakhsh S, Buckingham ME, et al. Sonic hedgehog acts cell-autonomously on muscle precursor cells to generate limb muscle diversity. Genes Dev. 2012 Sep 15;26(18):2103–17. doi: 10.1101/gad.187807.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voronova A, Coyne E, Al Madhoun A, Fair JV, Bosiljcic N, St-Louis C, et al. Hedgehog signaling regulates myod expression and activity. J Biol Chem. 2013 Feb 8;288(6):4389–404. doi: 10.1074/jbc.M112.400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirsinger E, Malapert P, Dubrulle J, Delfini MC, Duprez D, Henrique D, et al. Notch signalling acts in postmitotic avian myogenic cells to control myod activation. Development. 2001 Jan;128(1):107–16. doi: 10.1242/dev.128.1.107. [DOI] [PubMed] [Google Scholar]
- 46.Hirsinger E, Duprez D, Jouve C, Malapert P, Cooke J, Pourquié O. Noggin acts downstream of wnt and sonic hedgehog to antagonize BMP4 in avian somite patterning. Development. 1997 Nov;124(22):4605–14. doi: 10.1242/dev.124.22.4605. [DOI] [PubMed] [Google Scholar]
- 47.Schuster-Gossler K, Cordes R, Gossler A. Premature myogenic differentiation and depletion of progenitor cells cause severe muscle hypotrophy in delta1 mutants. Proc Natl Acad Sci U S A. 2007 Jan 9;104(2):537–42. doi: 10.1073/pnas.0608281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hinck AP. Structural studies of the tgf-βs and their receptors - insights into evolution of the tgf-β superfamily. FEBS Lett. 2012 Jul 4;586(14):1860–70. doi: 10.1016/j.febslet.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 49.Re'em-Kalma Y, Lamb T, Frank D. Competition between noggin and bone morphogenetic protein 4 activities may regulate dorsalization during xenopus development. Proc Natl Acad Sci U S A. 1995 Dec 19;92(26):12141–5. doi: 10.1073/pnas.92.26.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pourquié O, Fan CM, Coltey M, Hirsinger E, Watanabe Y, Bréant C, et al. Lateral and axial signals involved in avian somite patterning: A role for BMP4. Cell. 1996 Feb 9;84(3):461–71. doi: 10.1016/s0092-8674(00)81291-x. [DOI] [PubMed] [Google Scholar]
- 51.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced notch signaling mediated by RBP-J inhibits myod expression and myogenesis. J Biol Chem. 1999 Mar 12;274(11):7238–44. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 52.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of myod in mice leads to up-regulation of the myogenic HLH gene myf-5 and results in apparently normal muscle development. Cell. 1992 Oct 30;71(3):383–90. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 53.Braun T, Rudnicki MA, Arnold HH, Jaenisch R. Targeted inactivation of the muscle regulatory gene myf-5 results in abnormal rib development and perinatal death. Cell. 1992 Oct 30;71(3):369–82. doi: 10.1016/0092-8674(92)90507-9. [DOI] [PubMed] [Google Scholar]
- 54.Braun T, Bober E, Rudnicki MA, Jaenisch R, Arnold HH. MyoD expression marks the onset of skeletal myogenesis in myf-5 mutant mice. Development. 1994 Nov;120(11):3083–92. doi: 10.1242/dev.120.11.3083. [DOI] [PubMed] [Google Scholar]
- 55.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or myf-5 is required for the formation of skeletal muscle. Cell. 1993 Dec 31;75(7):1351–9. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 56.Smith TH, Kachinsky AM, Miller JB. Somite subdomains, muscle cell origins, and the four muscle regulatory factor proteins. J Cell Biol. 1994 Oct;127(1):95–105. doi: 10.1083/jcb.127.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993 Aug 5;364(6437):501–6. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 58.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993 Aug 5;364(6437):532–5. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 59.Braun T, Arnold HH. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development. The EMBO Journal. 1995;14(6):1176–1186. doi: 10.1002/j.1460-2075.1995.tb07101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W, Behringer RR, Olson EN. Inactivation of the myogenic bhlh gene MRF4 results in up-regulation of myogenin and rib anomalies. Genes Dev. 1995 Jun 1;9(11):1388–99. doi: 10.1101/gad.9.11.1388. [DOI] [PubMed] [Google Scholar]
- 61.Patapoutian A, Yoon JK, Miner JH, Wang S, Stark K, Wold B. Disruption of the mouse MRF4 gene identifies multiple waves of myogenesis in the myotome. Development. 1995 Oct;121(10):3347–3358. doi: 10.1242/dev.121.10.3347. [DOI] [PubMed] [Google Scholar]
- 62.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, et al. MRF4 determines skeletal muscle identity in Myf5:Myod double-mutant. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 63.Rawls A, Valdez MR, Zhang W, Richardson J, Klein WH, Olson EN. Overlapping functions of the myogenic bhlh genes MRF4 and myod revealed in double mutant mice. Development. 1998 Jul;125(13):2349–58. doi: 10.1242/dev.125.13.2349. [DOI] [PubMed] [Google Scholar]
- 64.Rudnicki MA, Jaenisch R. The myod family of transcription factors and skeletal myogenesis. Bioessays. 1995 Mar;17(3):203–9. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 65.Megeney LA, Rudnicki MA. Determination versus differentiation and the myod family of transcription factors. Biochem Cell Biol. 1995;73(9–10):723–32. doi: 10.1139/o95-080. [DOI] [PubMed] [Google Scholar]
- 66.Atchley WR, Fitch WM, Bronner-Fraser M. Molecular evolution of the myod family of transcription factors. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11522–6. doi: 10.1073/pnas.91.24.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michelson AM, Abmayr SM, Bate M, Arias AM, Maniatis T. Expression of a myod family member prefigures muscle pattern in drosophila embryos. Genes Dev. 1990 Dec;4(12A):2086–97. doi: 10.1101/gad.4.12a.2086. [DOI] [PubMed] [Google Scholar]
- 68.Venuti JM, Goldberg L, Chakraborty T, Olson EN, Klein WH. A myogenic factor from sea urchin embryos capable of programming muscle differentiation in mammalian cells. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6219–23. doi: 10.1073/pnas.88.14.6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krause M, Fire A, Harrison SW, Priess J, Weintraub H. CeMyoD accumulation defines the body wall muscle cell fate during C. Elegans embryogenesis. Cell. 1990 Nov 30;63(5):907–19. doi: 10.1016/0092-8674(90)90494-y. [DOI] [PubMed] [Google Scholar]
- 70.Davis RL, Weintraub H. Acquisition of myogenic specificity by replacement of three amino acid residues from myod into E12. Science. 1992 May 15;256(5059):1027–30. doi: 10.1126/science.1317057. [DOI] [PubMed] [Google Scholar]
- 71.Black BL, Molkentin JD, Olson EN. Multiple roles for the myod basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol Cell Biol. 1998 Jan;18(1):69–77. doi: 10.1128/mcb.18.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis RL, Cheng PF, Lassar AB, Weintraub H. The myod DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990 Mar 9;60(5):733–46. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 73.Olson EN, Klein WH. BHLH factors in muscle development: Dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994 Jan;8(1):1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 74.Weintraub H, Dwarki VJ, Verma I, Davis R, Hollenberg S, Snider L, et al. Muscle-specific transcriptional activation by myod. Genes Dev. 1991 Aug;5(8):1377–86. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- 75.Ishibashi J, Perry RL, Asakura A, Rudnicki MA. MyoD induces myogenic differentiation through cooperation of its NH2- and cooh-terminal regions. J Cell Biol. 2005 Nov 7;171(3):471–82. doi: 10.1083/jcb.200502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conerly ML, Yao Z, Zhong JW, Groudine M, Tapscott SJ. Distinct activities of myf5 and myod indicate separate roles in skeletal muscle lineage specification and differentiation. Dev Cell. 2016 Feb 22;36(4):375–85. doi: 10.1016/j.devcel.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol Cell Biol. 2001 Apr;21(7):2404–12. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of myod mediate transcriptional activation of genes in repressive chromatin: A mechanism for lineage determination in myogenesis. Genes Dev. 1997 Feb 15;11(4):436–50. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 79.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by myod indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004 May 21;14(4):465–77. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 80.Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ. In vitro transcription system delineates the distinct roles of the coactivators pcaf and p300 during myod/e47-dependent transactivation. Proc Natl Acad Sci U S A. 2004 Aug 10;101(32):11593–8. doi: 10.1073/pnas.0404192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable dna-bound complex. Mol Cell Biol. 2005 May;25(10):3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moss JB, Olson EN, Schwartz RJ. The myogenic regulatory factor MRF4 represses the cardiac alpha-actin promoter through a negative-acting n-terminal protein domain. J Biol Chem. 1996 Dec 6;271(49):31688–94. doi: 10.1074/jbc.271.49.31688. [DOI] [PubMed] [Google Scholar]
- 83.Suelves M, Lluís F, Ruiz V, Nebreda AR, Muñoz-Cánoves P. Phosphorylation of MRF4 transactivation domain by p38 mediates repression of specific myogenic genes. EMBO J. 2004 Jan 28;23(2):365–75. doi: 10.1038/sj.emboj.7600056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005 Mar 1;19(5):553–69. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, et al. Global and gene-specific analyses show distinct roles for myod and myog at a common set of promoters. EMBO J. 2006 Feb 8;25(3):502–11. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CEM, van Ijcken WFJ, et al. Snail regulates myod binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell. 2012 Aug 10;47(3):457–68. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, et al. Genome-wide myod binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev Cell. 2010 Apr 20;18(4):662–74. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mousavi K, Zare H, Dell'orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, et al. ERNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013 Sep 12;51(5):606–17. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997 Dec;1(1):35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]