Abstract
In 1987, Robert Davis, Hal Weintraub and I reported the identification of MyoD, a transcription factor that could reprogram fibroblasts into skeletal muscle cells. In this recollection, I both summarize the prior work of Helen Blau, Woody Wright, Peter Jones and Charlie Emerson that inspired my entry into this field, and the subsequent events that led to finding MyoD. Lastly, I highlight some of the principles in developmental biology that have emerged during the past 30 years, which are particularly relevant to skeletal muscle biology.
Recollection
Inspiration comes from Helen, Woody, Peter, and Charlie
When I was finishing up my graduate work in 1983, one of the mysteries that fascinated me was how a cell becomes specified to a particular cell fate. At the time, precious little was known about the molecules that regulated either the determination of a cell to a particular lineage or the differentiation of that cell, once committed, to a particular cell fate. Although it was clear that transcription factors would likely play a role in this process, the tools to identify such putative regulators of cell type specification during vertebrate development were fairly primitive. However, just as I was finishing up my graduate work in Bob Roeder’s lab, several papers were published that suggested the path to identify one such regulator that would control skeletal muscle formation.
In 1983 and 1984, papers were published by both Helen Blau’s group at Stanford and Woody Wright at the University of Texas, that pointed to the existence of a factor(s) present in skeletal muscle cells which could activate the expression of muscle genes in non-muscle cell-types. Both of these groups had made heterokaryons between differentiated skeletal muscle cells and either amniocytes [1] or neural cells [2], and both had found that the non-muscle cell nucleus in these heterokaryons had been reprogrammed to express muscle-specific gene products. These observations got me quite excited because they both suggested that there must be a trans-acting factor(s) present in muscle cells that could activate expression of muscle-specific genes in a normally non-myogenic cell type. However, although these elegant cell fusion experiments had convincingly argued for the existence of a trans-acting factor in muscle cells that could activate expression of the muscle differentiation program, it was not clear if activation of the myogenic program was induced by a single factor or by an army of muscle regulatory molecules working in a combinatorial fashion.
A logical path to identify such myogenic regulators would have been to study the transcription factors that bind to the regulatory regions of muscle structural genes. However, given the technology of the early '80s, this pursuit would have entailed fractionating nuclear extracts of muscle cells in search of DNA binding activities that recognized these regulatory regions. During my graduate career, I realized that I had an aversion to cold-room work and I knew that this approach entailed months if not years of column chromatography at 4°C. In addition, I was a bit tired of the biochemistry of my graduate school days and wondered if there was another approach I could take to identifying muscle regulatory molecules. The key to unraveling this problem came from my recollection of two papers published by Peter Jones and Shirley Taylor in '79 and '82 [3, 4]. In these studies, Jones and Taylor had made the striking finding that a brief treatment of a line of embryonic mouse fibroblasts, C3H10T1/2 (10T1/2) cells, with the DNA demethylating agent, 5-azacytidine, could stably convert these fibroblasts into either adipogenic, chondrogenic, or myogenic cell lines. At the time, it was already known that incorporation of 5-azacytidine into cellular DNA would block the activity of cellular DNA methyltransferases and thereby induce the demethylation of CpG residues [5]. Because there was evidence that methylated DNA directed its own assembly into inactive chromatin [6–8], it seemed plausible that a brief treatment with 5-azacytidine was converting 10T1/2 cells into different cellular phenotypes by inducing demethylation of transcriptionally silent regulatory genes. The consequent reactivation of these transcriptional regulators would in turn induce the differing cellular phenotypes. Subsequent work by Konieczny and Emerson firmly established that the frequency of myogenic conversion of 5-azacytidine-treated 10T1/2 cells was quite high and was consistent with the notion that demethylation of only one or a few loci was sufficient to convert 10T1/2 cells into the myogenic lineage [9].
These observations suggested to me a genetic approach to identify the myogenic regulator whose expression was induced by 5-azacytidine treatment. I reasoned that this locus would be demethylated, and therefore expressed in myogenic cell lines; whereas in 10T1/2 cells it would be methylated and transcriptionally silent. If myogenic conversion of 10T1/2 cells was due to the demethylation of a single regulatory locus, I reasoned that transfection of this unmethylated locus into 10T1/2 cells would convert these cells into myocytes in the absence of any 5-azacytidine treatment. This argument relied upon the prior observations that methylated genes tend not to be expressed [6–8], and that transfected DNA can propagate its methylation pattern during subsequent rounds of host cell division [10, 11]. I thought that this approach could eventually identify the myogenic regulatory locus if the donor unmethylated DNA was from either human muscle DNA (and linked to Alu sequences) or from an unmethylated murine cosmid library.
Breakfast at the “The Surrogate Hostess”
I remember being pretty excited about this approach, as it could both keep me out of the cold room and potentially identify a regulator of cell fate. I had already arranged to do my postdoc with Hal Weintraub, whose lab was at the Fred Hutchinson Cancer Research Center in Seattle. Before finishing up my graduate work in October '83, I sent Hal a postcard asking him if he would be interested in identifying the myogenic regulators whose existence Helen Blau had recently documented in her heterokaryon experiments [1]. Upon arriving in Seattle the following month, I outlined my scheme to Hal at "The Surrogate Hostess", a restaurant with great cinnamon rolls, which served as Hal's early morning office. Although Hal initially wanted me to work on transcription factors that regulated globin gene expression, he was extremely enthusiastic about any new idea to identify regulators of cell fate and immediately gave me the go ahead to work on the 10T1/2 project in his lab.
One of the first indications that treatment of 10T1/2 cells with 5-azacytidine converted these cells into skeletal muscle by demethylating a regulatory locus rather than by demethylating muscle structural genes was a result of a collaboration between myself and Bruce Paterson at the NIH. Bruce had recently demonstrated that a cloned chicken muscle actin gene was expressed only in transfected myogenic cell lines [12]. I reasoned that if 5-azacytidine treatment of 10T1/2 cells activated the expression of a myogenic regulator(s), a transfected chicken muscle actin gene, which both lacked DNA-methylation and contained its muscle-specific transcriptional regulatory regions, would be expressed in 5-azacytidine derived myogenic cell lines, but transcriptionally inert in the parental 10T1/2 cell line. On the other hand, if 5-azacytidine induced muscle gene expression by demethylating the muscle structural genes themselves, then the transfected chicken muscle actin gene (which lacked DNA-methylation) would be expressed in both cellular contexts. We found that the transfected chicken muscle actin gene was only expressed in the 5-azacytidine derived myogenic cell lines, indicating that these cells expressed a transcriptional regulator(s) that was absent from the parental 10T1/2 cell [13]. Similar experiments by Charlie Emerson's lab had led to the same conclusion [14].
I spent my first three years in Hal's lab deriving myogenic cell lines from 10T1/2 cells and transfecting the genomic DNA from such myogenic cell lines back into the parental 10T1/2 cells. These transfection experiments were somewhat massive, entailing 50–100 15cm tissue culture dishes at a shot. I was continually battling mold contamination in these tissue culture dishes, and it was usually a race to analyze the thousands of 10T1/2 cell colonies for muscle cells before the mold would do them in. Fortunately for me a veteran muscle biologist, Steve Hauschka, was in the neighborhood at the University of Washington. I told Steve of my ambitions and he very generously showed me how to perform an immunodetection technique to identify myosin heavy chain expressing colonies. After several lack luster transfection experiments, which failed to display any discernable muscle conversion, I finally had one experiment that yielded a bona-fide myogenic colony complete with myosin expressing cells in multinucleate myotubes. I celebrated my first muscle colony by jetting off to New York to be with my girlfriend over the Thanksgiving holiday.
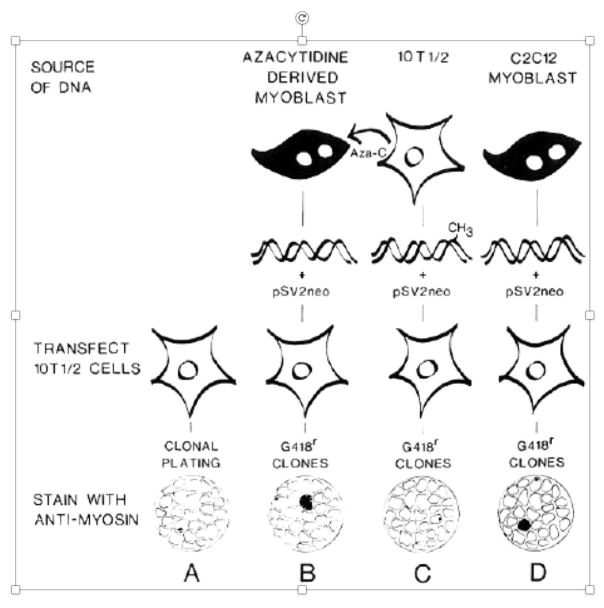
After returning to Seattle, my elation quickly turned to concern that perhaps my precious muscle colony was not due to introduction of a demethylated regulatory locus into 10T1/2 cells, but instead reflected a spontaneous conversion of 10T1/2 cells into myocytes. Thus began a year of control experiments, during which I analyzed the frequency of skeletal muscle colonies in either non-transfected 10T1/2 cells or in 10T1/2 cells transfected with DNA derived from parental 10T1/2 DNA, 5-azacytidine derived myoblasts, or myogenic cell lines isolated from muscle tissue (C2C12 cells). The bottom line from these transfection experiments was that 10T1/2 cells can spontaneously convert into muscle cells at a frequency of about 2.5 X 10−8; however when transfected with genomic DNA derived from either 5-azacytidine derived myoblasts or C2C12 cells this frequency jumps to about 8 X 10−5 [13]. In contrast, transformation of 10T1/2 cells with their own DNA did not increase the frequency of myogenic conversion over the low spontaneous conversion rate. The conversion rate of 10T1/2 cells into myogenic colonies following transfection with muscle derived DNA (about 1 in 104 transfected colonies) was consistent with the notion that introduction of a single regulatory locus had activated the muscle cell program in these cells (schematic of these results is displayed in Figure 1). I soon learned at a Keystone conference on Muscle Biology that Steve Konieczny, Al Baldwin, and Charlie Emerson had been performing similar experiments at the University of Virginia, and had arrived at the same conclusion [15].
Figure 1. The scheme employed to investigate whether transfection of 10T1/2 cells with genomic DNA derived from either parental 10T1/2 DNA, 5-azacytidine derived myoblasts, or a myogenic cell line (C2C12 cells) could convert fibroblasts into myoblasts.
Black cells represent myoblasts; white cells represent 10T1/2 cells. Both macroscopic and microscopic muscle colonies are indicated by blackening. (Figure taken from [13].)
As an initial approach towards identifying the muscle regulatory locus, I investigated if 10T1/2 cells could be converted into muscle cells following transfection with genomic cosmid libraries. The rationale behind using cosmid libraries was that any locus would be linked to the cosmid DNA, which contained a selectable marker encoding G418 resistance, and therefore could potentially be identified by this linkage. The logic was to firstly isolate G418 resistant myogenic colonies from 10T1/2 cells transfected with a genomic cosmid library. If the muscle regulatory locus could fit into a cosmid (about 40 kilobases), then the frequency of myogenic conversion of 10T1/2 cells transfected with cosmid DNA would be expected to be similar to that following transfection with muscle genomic DNA. Genomic DNA from such primary myogenic clones was then used to transfect a new round of 10T1/2 cells. If the muscle regulatory locus remained linked to the G418 resistance gene, then the frequency of myogenic, G418 resistant colonies in this second round of transfection would be expected to significantly increase. In my hands, this technique was a complete flop. While I found that transfection of 10T1/2 cells with cosmid DNA could induce myogenic conversion, I was unable to establish that the myogenic regulatory locus was linked to the G418 resistance marker. In contrast, Charlie Emerson's group, taking a similar approach, demonstrated myogenic conversion of 10T1/2 cells following transfection with a cosmid library and noted an increase in the frequency of myogenic, G418 resistant colonies in the secondary transfection [16]. The increase in myogenic frequency observed in this secondary transfection suggested that Charlie and his colleagues had established linkage between a myogenic regulatory locus, which they termed myd, and the G418 resistance marker [16].
A new approach finally yields success
Faced with my own failure to establish linkage of the G418 resistance marker with the myogenic regulatory locus, I began to think of other schemes that might enable me to identify this regulator. I was present at a Gordon Conference listening to Dan Littman describe how he had identified CD4 when the idea came to me. Dan had transfected NIH 3T3 cells with human genomic DNA and isolated stable cell lines that expressed human CD4. Dan and colleagues had ultimately cloned human CD4 by performing the recently developed subtractive hybridization technique to identify transcripts that were present in the CD4 expressing NIH3T3 cells and absent from the parental NIH3T3 cells [17]. While listening to Dan, I realized that the myogenic cell lines derived from 10T1/2 cells by 5-azacytidine treatment would similarly contain RNA transcripts that were absent from the parental 10T1/2 cells, and that one of these transcripts would be the myogenic regulator that I sought. The problem however was that the myogenic cell lines expressed not only the putative myogenic regulator but also expressed muscle structural genes, such as muscle myosins and actins which were activated during the muscle differentiation program. The problem was to somehow identify the RNA encoding the muscle regulatory factor in a sea of transcripts encoding more prosaic muscle structural proteins. I realized that the biology of muscle differentiation might provide me with the means to discriminate between a putative myogenic regulator and the multitude of muscle structural proteins, whose expression was in turn activated by this myogenic regulator. It had been appreciated for a number of years that myogenic cell lines only activate their differentiation program upon mitogen withdrawal [18, 19]. However, the myogenic cell lines that I had derived following 5-azacytidine treatment of 10T1/2 cells had undergone an epigenetic change that was stably maintained in proliferating myoblasts prior to their differentiation. Thus, I conjectured that 5-azacytidine may have activated the expression of a myogenic regulator in proliferating cells and that expression of this regulator allowed these cells to activate the muscle differentiation program upon mitogen withdrawal. Indeed, Charlie Emerson's group had already established that proliferating 5-azacytidine derived myoblasts expressed a new spot on a 2D protein gel in comparison to the parental 10T1/2 cell line [9], increasing my confidence that there might be muscle-specific transcripts encoding lineage determinators which were expressed in proliferating myoblasts, prior to execution of the differentiation program.
At this point, I was very fortunate to be joined in this project by Robert Davis, a “laboratory dynamo”, who had recently joined Hal’s lab for his graduate work. Armed with the hypothesis that proliferating myoblasts express a “determination gene” that maintained their cellular phenotype, Robert and I screened a skeletal muscle cDNA library with subtracted cDNA probe made from proliferating myoblasts, from which we had removed transcripts common to proliferating 10T1/2 cells. As soon as I saw that this myoblast–specific probe was hybridizing with only 1% of the cDNAs in the skeletal muscle cDNA library, I felt a great relief, as I knew that hidden amongst these spots must be the gene we were after. The problem then became identifying this candidate regulator out of the hundred or so cDNAs showing a hybridization signal. Robert and I methodically slogged though the myoblast cDNAs, characterizing them into hybridization families and analyzing their expression pattern. While working with the azacytidine-derived myogenic cell lines, I noticed that if I was not diligent to passage them prior to their reaching confluency, these cells would differentiate, leaving non-differentiating variants to proliferate. Consequently, it was a fairly simple task to isolate non-differentiating skeletal muscle cell variants. I made clonal lines out of these non-differentiating muscle cell variants, with the hope that they would have lost the expression of the putative muscle determination gene. Indeed, by identifying the myoblast-specific cDNAs whose expression was specifically lost in the non-differentiating variants, Robert and I were able to focus our attention on 3 out of the 26 families of “myoblast-specific” genes we had found.
Because genomic muscle DNA could convert 10T1/2 cells into skeletal muscle, Robert and I decided to explore whether transfection of these cells with expression vectors programmed to encode one of the three myoblast-specific cDNAs might also exhibit this capacity. Because sequencing DNA at that time was fairly time consuming, this functional assay was performed prior to even sequencing the three candidate cDNAs! The experiment was to co-transfect an expression vector encoding each cDNA individually into 10T1/2 cells along with a selectable marker. I remember carefully monitoring this transfection on a daily basis for a couple of weeks to allow the drug resistant colonies to develop, and seeing to my great astonishment that nearly half of the resulting colonies transfected with one of the cDNAs were fusing into myotubes. When greeted with this news, Hal was of course elated, but we both realized that this transfection experiment needed to be repeated over and over to make sure that we were not being misled by some terrible artifact. Happily, this cDNA maintained its myogenic potency in both 10T1/2 cells and in other fibroblast and adipoblast cells lines, and thus was named myogenic determination gene number 1 (a.k.a. MyoD) [20]. In subsequent studies, both Hal and Harold Holtzer’s group demonstrated that MyoD could elicit the skeletal muscle differentiationprogram not only in fibroblasts but also in other differentiated cell types [21, 22].
Since these early studies, it is now appreciated that MyoD is part of a larger group of myogenic basic-helix-loop-helix (bHLH) proteins (including Myf5, MRF4, and myogenin) which work together to both initiate and execute the myogenic differentiation program in differing regions of the developing mouse embryo (reviewed in [23, 24]). Activation of either MyoD, Myf5, or MRF4, is necessary for determination of skeletal muscle progenitor cells [25, 26]. Myogenin expression is induced by these upstream muscle regulators, and in turn drives terminal differentiation of myoblasts [27–29]. MyoD works in the context of both bHLH E protein dimerization partners [30] and the homeobox protein Pbx [31] to bind to relevant targets in both proliferating myoblasts and in differentiated myotubes. In addition, MyoD requires interaction with the BAF60c subunit of the Brg1-based SWI/SNF chromatin-remodeling complex to induce expression of target genes [32]. It is now clear that MyoD activates the expression of other muscle regulators (i.e., MEF2, myogenin, lncRNAs, and microRNAs) that work in combination to induce the skeletal muscle differentiation program (reviewed [23]). While expression of the myogenic bHLH proteins are induced by differing regulatory pathways in cranial, trunk and limb musculature (reviewed in [23, 24]) their induction signals a nodal point for commitment to the skeletal muscle lineage.
Cell reprogramming since MyoD, and some of the lessons I’fve learned along the way
A mechanistic understanding of how cell-type specification is regulated was quite elusive when I was in graduate school during the late ’70s-early ‘80s. Two years prior to the discovery of MyoD, Walter Gehring and colleagues had found that forced expression of the transcription factor, Eyeless (i.e., Pax6) in various imaginal disc primordia of Drosophila induced ectopic eye structures on either the wings, legs, or antennae [33]. Since this early work on MyoD and Eyeless, a number of outstanding discoveries have led to our current sophisticated understanding of how cell-type is regulated. Interestingly, the ability of a single transcription factor to reprogram cell fate is clearly not a universal theme in cell-lineage specification. During the past 30 years cellular reprogramming has exploded to include both several additional somatic cell lineages [34–41] and most notably pluripotent stem cells [42]. In striking contrast to the MyoD result, reprogramming fibroblast-like cells into differentiated cell types (other than skeletal muscle) frequently requires a combination of transcription factors, and speaks to the combinatorial logic of specifying cellular identity. The stunning finding by Takahashi and Yamanaka that a combination of 4 transcription factors can reprogram somatic cells into induced pluripotent stem (iPS) cells [42] sparked the current revolution in cellular reprogramming. Indeed the derivation of human iPS cells that can be coaxed to differentiate into various somatic cell lineages is now providing substrates for either disease modeling in vitro or cell therapy in vivo (reviewed in [43]). The history of these advances in reprogramming cell fate, following ectopic expression of transcription factors, non-coding RNAs, and even small molecules, has recently been reviewed by others [44–47]. In addition to the identification of key transcriptional regulators that control cell identity, a number of important principles have emerged during the past 30 years regarding the regulatory logic that controls cell type specification—many of which are relevant to the formation of both skeletal muscle and other cell types. Below I discuss some of these principles, and highlight their relevance to skeletal muscle biology.
Extrinsic signals that induce cell fate activate intrinsic auto-regulatory loops that maintain cell identity
Expression of the transcription factors that regulate cell-type specification are frequently induced by transient signaling cues in the embryo, and then subsequently maintained by positive feedback loops that maintain the expression of these key regulators. Work by Tom Jessell, James Briscoe, Andy McMahon and their colleagues established that signals (such as Shh, BMP, and Wnt) which pattern cell identity in the neural tube are transiently expressed, but induce the stable expression of transcription factors that in turn maintain neuronal cell identity (reviewed in [48]). Similarly, Wnt signals (secreted by the dorsal neural tube) can work in combination with Shh (secreted by the notochord and floor plate of the neural tube) to promote the stable expression of the myogenic bHLH genes, MyoD and Myf5, in the somite [49]. While the signals that induce MyoD and Myf5 are transient, because the myogenic bHLH proteins can activate their own expression [50], transient signaling leads to stable cell type specification. In the case of MyoD, a feed-forward circuit is activated which induces the expression of myogenin [50], MEF2C [51], microRNAs [52–55], and lncRNAs [56, 57], which work in combination with MyoD to both maintain MyoD expression [50] and coordinate the skeletal muscle differentiation program.
Gene duplication allows new functions to evolve in regulatory factors
Another principle that is amply represented in the circuitry that regulates skeletal muscle specification is that of redundancy. It became evident soon after the identification of MyoD, that it was a member of a larger family of myogenic bHLH proteins including Myf5, MRF4, and myogenin. In the case of MyoD, Myf5, and MRF4, these proteins display overlapping function to regulate determination of skeletal muscle progenitors [25, 26] In contrast, myogenin is necessary to promote the terminal differentiation of myoblasts [27–29]. Overlapping roles for members of a transcription factor family seems to be a fairly common occurrence in vertebrates, and speaks to the path of evolution of new transcription factors, following duplication and diversification of the ancestral gene(s). This process allows the evolution of new transcription factors to be expressed in either new temporal or spatial patterns, while maintaining functional overlap with the ancestral family member. In the case of Myf5, distinct upstream regulatory elements have been found to drive expression of this gene at various times of development or in various muscle groups [58–61]; consistent with the finding that different transcription factors drive Myf5 expression in differing muscle groups (reviewed in [23]). The seemingly piecemeal acquisition of new upstream regulatory sequences, which drive Myf5 expression in diverse regions of the embryo, no doubt speaks to the ability of evolution to “tinker” (to borrow the words of François Jacob; [62]) and evolve new enhancer sequences, which enable new spatial and temporal expression patterns for Myf5 gene expression.
Pioneer transcription factors and their followers coordinate cell-type specification and signaling pathways
Stephen Tapscott and colleagues made the important observation that MyoD can remodel chromatin at binding sites in muscle gene enhancers and activate transcription at previously silent loci [63]. Since these early studies, it has been established that MyoD recruits both histone acetyl-transferases [64] and chromatin remodeling complexes [65–67] to make regions of the genome (proximal to its binding sites) accessible to both other transcription factors and to the basic transcriptional machinery. Work by Ken Zaret and colleagues has established that not all transcription factors are equal. Some, which have been termed “pioneer” transcription factors, are able to access their binding sites in transcriptionally inert chromatin, while other transcription factors can only access their binding sites in already transcriptionally active regions of the genome (reviewed in [68, 69]). It seems likely that transcription factors that are able to reprogram cellular identity, such as MyoD, are able to achieve this function by acting as pioneer transcription factors. Indeed, recent findings by Marius Wernig and colleagues, have shown that Ascl1 (which like MyoD is a bHLH protein) works with other transcription factors, Brn2 and Myt1l, to reprogram fibroblasts into a neuronal identity by acting as a pioneer transcription factor [70]. In this case, Ascl1 can occupy most cognate genomic sites in fibroblasts. In contrast, Brn2 and Myt1l do not access fibroblast chromatin on their own; but require Ascl1 to designate their binding sites [70]. The feature that likely distinguishes pioneer transcription factors from “follower” transcription factors is the ability of pioneer transcription factors to target partial DNA binding motifs displayed on the nucleosome surface [71]. Interestingly, MyoD also needs guidance to properly access some of its key transcriptional targets, as Stephen Tapscott and colleagues noted that interaction of MyoD with the homeobox protein Pbx1 was critical for initial interaction of MyoD with the myogenin promoter [31].
Importantly, transcription factors that regulate cell fate such as MyoD, Oct, Pu.1, C/EBPα, and GATA1 recruit transcription factors that carry signaling information from the cell surface, such as R-Smads (which transduce TGFβ/BMP signals) and TCF (which transduces Wnt signals), to different transcriptional targets in a cell context-dependent manner [72, 73]. It seems likely that these cell lineage regulators direct differential targeting of R-Smads and TCF to cell-type specific targets, by both direct protein-protein interaction [72] and by initiating the formation of super-enhancers [74], which in turn establish differential accessibility of enhancer elements in different cell types. External signals from the cell surface can also modulate the interaction of lineage determining transcription factors with essential co-activators. In the case of the skeletal muscle differentiation program, MyoD provides a platform for p38-kinase induced recruitment of SWI/SNF chromatin remodeling complexes [32, 75]. In this manner, chromatin remodeling at MyoD binding sites is held in check until external signals activate p38-kinase, which then enables SWI/SNF complexes to properly assemble on MyoD-bound enhancers, and finally cue the initiation of skeletal muscle differentiation.
Mutually antagonistic regulatory pathways are a key aspect of cell lineage specification
The decision of a multi-potent progenitor cell to choose one cell fate over another often involves a mutually antagonistic regulatory circuit, that precludes the simultaneous expression of both daughter fates. This sort of logic has been seen in patterning of the neural tube, in which a gradient of Shh works as a morphogen to establish expression domains for differing homeobox transcriptional repressors. Subsequently, the overlapping expression of these transcription factors is refined by cross-repressive inhibition to generate sharp mutually exclusive expression domains for these transcriptional repressors [76]. In this manner, the ventral neural tube progenitor cell is forced to express only one of a pair of mutually repressive transcription factors, and thus initiate the process of cell fate selection. The somite also displays mutually exclusive domains of gene expression driven by mutually repressive transcription factors. In the somite, Pax3 and Nkx3.2 are expressed in the dermomyotome and sclerotome, respectively. These transcription factors can mutually repress one another’s expression, and thus promote the exclusive development of either the dermomyotome/myotome (Pax3) or sclerotome (Nkx3.2) [77]. Furthermore, mutual repression between Pax3 and Foxc2 in dermomyotomal cells serves to drive cells towards either a skeletal muscle or an endothelial cell fate, respectively [78].
An aspect of skeletal muscle biology that certainly was not anticipated by the initial MyoD experiments, was the realization that protein accumulation of this key regulator was limited to activated satellite cells (i.e., transiently amplifying myoblasts) and differentiated myotubes, but was absent from the quiescent satellite stem cell. Studies from Margaret Buckingham’s group, my own lab, and Michael Rudnicki’s group established that Pax3 and Pax7 act upstream of MyoD [79, 80], and are required for the generation of either skeletal muscle precursors (in the embryo) [79] or satellite cells (in the adult) [81]. Interestingly, production of MyoD and Myf5 proteins in quiescent satellite cells is held in check via either mRNA degradation [82] or microRNA-mediated sequestration into non-translated RNPs [83], respectively. Thus, evolution has selected for regulatory mechanisms in quiescent skeletal muscle stem cells that both repress the stability or translation of mRNAs encoding MyoD or Myf5, and in addition block the activity of residual levels of these proteins, via Pax7-induced expression of the negative HLH regulators Id2 and Id3 [84]. I suspect that this is testimony to the distinct patterns of gene expression necessary to maintain quiescent satellite cells (which express Pax7) versus that necessary to sustain their more “differentiated” transient amplify progeny (which express MyoD and Myf5) prior to terminal differentiation. A regulatory circuitry has evolved, such that gene expression in satellite cells can switch from that directed by Pax7 to a program directed by MyoD/Myf5 (discussed in [85]). Regulatory mechanisms in quiescent satellite cells both block the stability and/or translation of MyoD/Myf5 mRNAs [82, 83] and dimerization of their encoded proteins with E protein partners [84]. Conversely in differentiating skeletal muscle cells, microRNAs, which are induced by MyoD, block the expression of Pax7 [53]. Thus, mutually antagonistic regulatory circuits seem to control the balance between satellite cell quiescence, growth and differentiation. In addition, this delicate balance is also regulated by exogenous signals, such as BMP [86] and Notch signaling [87], which play a critical role in both driving the expansion of Pax3/7 expressing cells while blocking the premature expression of MyoD/Myf5.
Acknowledgments
This recollection is dedicated to the memory of Hal Weintraub, an inspirational mentor and a great friend.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blau HM, Chiu CP, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32(4):1171–80. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- 2.Wright WE. Induction of muscle genes in neural cells. J Cell Biol. 1984;98(2):427–35. doi: 10.1083/jcb.98.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17(4):771–9. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 4.Taylor SM, Jones PA. Changes in phenotypic expression in embryonic and adult cells treated with 5-azacytidine. J Cell Physiol. 1982;111(2):187–94. doi: 10.1002/jcp.1041110210. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 6.Keshet I, Yisraeli J, Cedar H. Effect of regional DNA methylation on gene expression. Proc Natl Acad Sci U S A. 1985;82(9):2560–4. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busslinger M, Hurst J, Flavell RA. DNA methylation and the regulation of globin gene expression. Cell. 1983;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 8.Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci U S A. 1982;79(11):3418–22. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konieczny SF, Emerson CP., Jr 5-Azacytidine induction of stable mesodermal stem cell lineages from 10T1/2 cells: evidence for regulatory genes controlling determination. Cell. 1984;38(3):791–800. doi: 10.1016/0092-8674(84)90274-5. [DOI] [PubMed] [Google Scholar]
- 10.Stein R, Gruenbaum Y, Pollack Y, Razin A, Cedar H. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proc Natl Acad Sci U S A. 1982;79(1):61–5. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- 12.Seiler-Tuyns A, Eldridge JD, Paterson BM. Expression and regulation of chicken actin genes introduced into mouse myogenic and nonmyogenic cells. Proc Natl Acad Sci U S A. 1984;81(10):2980–4. doi: 10.1073/pnas.81.10.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lassar AB, Paterson BM, Weintraub H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell. 1986;47(5):649–56. doi: 10.1016/0092-8674(86)90507-6. [DOI] [PubMed] [Google Scholar]
- 14.Konieczny SF, Emerson CP., Jr Differentiation, not determination, regulates muscle gene activation: transfection of troponin I genes into multipotential and muscle lineages of 10T1/2 cells. Mol Cell Biol. 1985;5(9):2423–32. doi: 10.1128/mcb.5.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konieczny SF, Baldwin AS, Emerson CP., Jr Myogenic determination and differentiation of 10T1/2 cell lineages: evidence for a simple genetic regulatory system. In: Emerson CP Jr, Fischman DA, Nadal-Ginard B, Siddiqui MAQ, editors. Molecular Biology of Muscle Development, UCLA Symposium on Molecular and Cellular Biology; New York: Alan R. Liss, Inc; 1985. pp. 21–34. [Google Scholar]
- 16.Pinney DF, Pearson-White SH, Konieczny SF, Latham KE, Emerson CP., Jr Myogenic lineage determination and differentiation: evidence for a regulatory gene pathway. Cell. 1988;53(5):781–93. doi: 10.1016/0092-8674(88)90095-5. [DOI] [PubMed] [Google Scholar]
- 17.Maddon PJ, Littman DR, Godfrey M, Maddon DE, Chess L, Axel R. The isolation and nucleotide sequence of a cDNA encoding the T cell surface protein T4: a new member of the immunoglobulin gene family. Cell. 1985;42(1):93–104. doi: 10.1016/s0092-8674(85)80105-7. [DOI] [PubMed] [Google Scholar]
- 18.Linkhart TA, Clegg CH, Hauschka SD. Control of mouse myoblast commitment to terminal differentiation by mitogens. J Supramol Struct. 1980;14(4):483–98. doi: 10.1002/jss.400140407. [DOI] [PubMed] [Google Scholar]
- 19.Clegg CH, Linkhart TA, Olwin BB, Hauschka SD. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J Cell Biol. 1987;105(2):949–56. doi: 10.1083/jcb.105.2.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 21.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86(14):5434–8. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci U S A. 1990;87(20):7988–92. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28(3):225–38. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 24.Comai G, Tajbakhsh S. Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol. 2014;110:1–73. doi: 10.1016/B978-0-12-405943-6.00001-4. [DOI] [PubMed] [Google Scholar]
- 25.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75(7):1351–9. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 26.Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431(7007):466–71. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 27.Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364(6437):501–6. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 28.Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364(6437):532–5. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 29.Venuti JM, Morris JH, Vivian JL, Olson EN, Klein WH. Myogenin is required for late but not early aspects of myogenesis during mouse development. J Cell Biol. 1995;128(4):563–76. doi: 10.1083/jcb.128.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66(2):305–15. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- 31.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell. 2004;14(4):465–77. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 32.Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R, Wang K, Wu Z, Baranovskaya S, Miller A, Dilworth FJ, Puri PL. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 2012;31(2):301–16. doi: 10.1038/emboj.2011.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267(5205):1788–92. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 34.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–76. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455(7213):627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long JE, Meyers KA, Aaronson SA, Miki T. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet. 1996;14(1):50–4. doi: 10.1038/ng0996-50. [DOI] [PubMed] [Google Scholar]
- 37.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–86. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468(7323):521–6. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 40.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–3. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 41.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–9. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat Rev Mol Cell Biol. 2016;17(3):183–93. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 44.Mall M, Wernig M. The novel tool of cell reprogramming for applications in molecular medicine. J Mol Med (Berl) 2017 doi: 10.1007/s00109-017-1550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adlakha YK, Seth P. The expanding horizon of MicroRNAs in cellular reprogramming. Prog Neurobiol. 2017;148:21–39. doi: 10.1016/j.pneurobio.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Hochedlinger K, Jaenisch R. Induced Pluripotency and Epigenetic Reprogramming. Cold Spring Harb Perspect Biol. 2015;7(12) doi: 10.1101/cshperspect.a019448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graf T. Historical origins of transdifferentiation and reprogramming. Cell Stem Cell. 2011;9(6):504–16. doi: 10.1016/j.stem.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 48.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135(15):2489–503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 49.Munsterberg AE, Kitajewski J, Bumcrot DA, McMahon AP, Lassar AB. Combinatorial signaling by Sonic hedgehog and Wnt family members induces myogenic bHLH gene expression in the somite. Genes Dev. 1995;9(23):2911–22. doi: 10.1101/gad.9.23.2911. [DOI] [PubMed] [Google Scholar]
- 50.Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989;58(2):241–8. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- 51.Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128(22):4623–33. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- 52.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103(23):8721–6. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31(1):203–14. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175(1):77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Munsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol. 2008;321(2):491–9. doi: 10.1016/j.ydbio.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 56.Mueller AC, Cichewicz MA, Dey BK, Layer R, Reon BJ, Gagan JR, Dutta A. MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol Cell Biol. 2015;35(3):498–513. doi: 10.1128/MCB.01079-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, Glass DJ. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell. 2015;34(2):181–91. doi: 10.1016/j.devcel.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 58.Summerbell D, Ashby PR, Coutelle O, Cox D, Yee S, Rigby PW. The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development. 2000;127(17):3745–57. doi: 10.1242/dev.127.17.3745. [DOI] [PubMed] [Google Scholar]
- 59.Teboul L, Hadchouel J, Daubas P, Summerbell D, Buckingham M, Rigby PW. The early epaxial enhancer is essential for the initial expression of the skeletal muscle determination gene Myf5 but not for subsequent, multiple phases of somitic myogenesis. Development. 2002;129(19):4571–80. doi: 10.1242/dev.129.19.4571. [DOI] [PubMed] [Google Scholar]
- 60.Hadchouel J, Carvajal JJ, Daubas P, Bajard L, Chang T, Rocancourt D, Cox D, Summerbell D, Tajbakhsh S, Rigby PW, Buckingham M. Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development. 2003;130(15):3415–26. doi: 10.1242/dev.00552. [DOI] [PubMed] [Google Scholar]
- 61.Buchberger A, Nomokonova N, Arnold HH. Myf5 expression in somites and limb buds of mouse embryos is controlled by two distinct distal enhancer activities. Development. 2003;130(14):3297–307. doi: 10.1242/dev.00557. [DOI] [PubMed] [Google Scholar]
- 62.Jacob F. Evolution and tinkering. Science. 1977;196(4295):1161–6. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 63.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11(4):436–50. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 64.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol Cell. 1997;1(1):35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 65.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27(2):187–90. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 66.de la Serna IL, Ohkawa Y, Berkes CA, Bergstrom DA, Dacwag CS, Tapscott SJ, Imbalzano AN. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol Cell Biol. 2005;25(10):3997–4009. doi: 10.1128/MCB.25.10.3997-4009.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. Embo J. 2006;25(3):490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwafuchi-Doi M, Zaret KS. Cell fate control by pioneer transcription factors. Development. 2016;143(11):1833–7. doi: 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, Drechsel D, Martynoga B, Castro DS, Webb AE, Sudhof TC, Brunet A, Guillemot F, Chang HY, Wernig M. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155(3):621–35. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, Zaret KS. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161(3):555–68. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147(3):565–76. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, Li P, Durand EM, Mosimann C, Heffner GC, Daley GQ, Paulson RF, Young RA, Zon LI. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147(3):577–89. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat Genet. 2004;36(7):738–43. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 76.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101(4):435–45. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 77.Cairns DM, Sato ME, Lee PG, Lassar AB, Zeng L. A gradient of Shh establishes mutually repressing somitic cell fates induced by Nkx3.2 and Pax3. Dev Biol. 2008;323(2):152–65. doi: 10.1016/j.ydbio.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lagha M, Brunelli S, Messina G, Cumano A, Kume T, Relaix F, Buckingham ME. Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev Cell. 2009;17(6):892–9. doi: 10.1016/j.devcel.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 79.Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89(1):127–38. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 80.Maroto M, Reshef R, Munsterberg AE, Koester S, Goulding M, Lassar AB. Ectopic Pax-3 activates MyoD and Myf-5 expression in embryonic mesoderm and neural tissue. Cell. 1997;89(1):139–48. doi: 10.1016/s0092-8674(00)80190-7. [DOI] [PubMed] [Google Scholar]
- 81.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA. Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–86. doi: 10.1016/s0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 82.Hausburg MA, Doles JD, Clement SL, Cadwallader AB, Hall MN, Blackshear PJ, Lykke-Andersen J, Olwin BB. Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. Elife. 2015;4:e03390. doi: 10.7554/eLife.03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11(1):118–26. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 84.Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 Is a Direct Transcriptional Target of Pax7 in Quiescent Satellite Cells. Mol Biol Cell. 2009 doi: 10.1091/mbc.E08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol. 2007;177(5):769–79. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Noulet F, Edom-Vovard F, Le Grand F, Duprez D. Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Dev Cell. 2010;18(4):643–54. doi: 10.1016/j.devcel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 87.Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, Tajbakhsh S. A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells. 2012;30(2):243–52. doi: 10.1002/stem.775. [DOI] [PubMed] [Google Scholar]