Abstract
The bacterial cell wall separates the cell from its surrounding and protects it from environmental stressors. Its integrity is maintained by a highly regulated process of cell wall biosynthesis. The membrane-located lipid II cycle provides cell wall building blocks that are assembled inside the cytoplasm to the outside for incorporation. Its carrier molecule, undecaprenyl phosphate (UP), is then recycled by dephosphorylation from undecaprenyl pyrophosphate (UPP). In Bacillus subtilis, this indispensable reaction is catalyzed by the UPP phosphatases BcrC and UppP. Here, we study the physiological function of both phosphatases with respect to morphology, cell wall homeostasis and the resulting cell envelope stress response (CESR). We demonstrate that uppP and bcrC represent a synthetic lethal gene pair, which encodes an essential physiological function. Accordingly, cell growth and morphology were severely impaired during exponential growth if the overall UPP phosphatase level was limiting. UppP, but not BcrC, was crucial for normal sporulation. Expression of bcrC, but not uppP, was upregulated in the presence of cell envelope stress conditions caused by bacitracin if UPP phosphatase levels were limited. This homeostatic feedback renders BcrC more important during growth than UppP, particularly in defense against cell envelope stress.
Keywords: lipid II, bactoprenol, undecaprenyl pyrophosphate, undecaprenol, undecaprenyl phosphate, bacitracin, cell wall biosynthesis
Introduction
The bacterial cell wall is an essential structure that gives the cell its shape and counteracts the turgor pressure. The sacculus is one large macromolecule made up of peptidoglycan that has amazing properties: It is rigid, yet flexible and is constantly expanded and recycled during growth and cell division in a highly regulated manner, both spatially and temporally (Laddomada et al., 2016). Due to its essentiality, it is a prime antibiotic target at virtually any of the numerous steps leading to cell wall assembly.
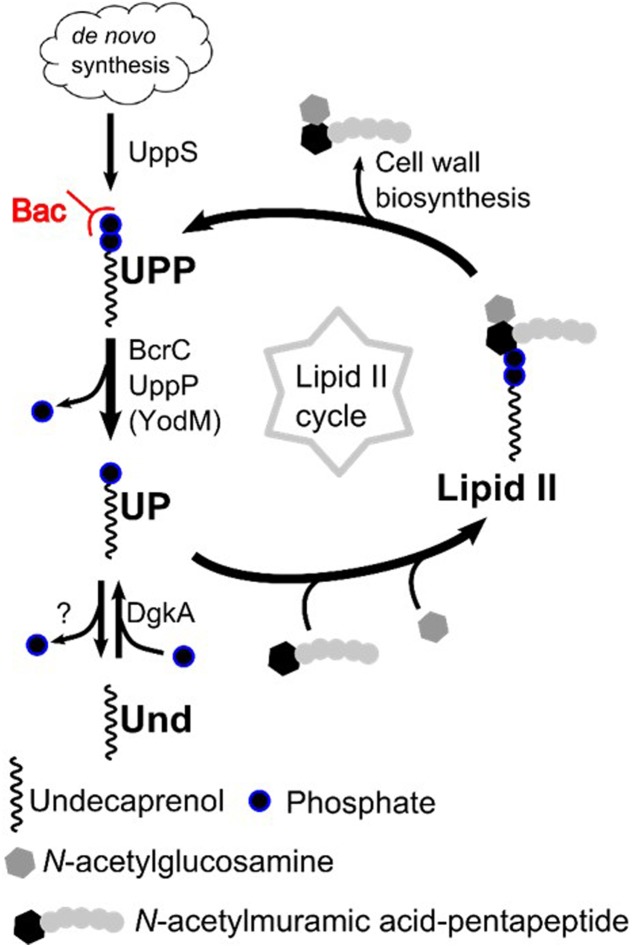
The lipid II cycle describes the membrane-associated steps of this process (Figure 1). Briefly, N-acetylmuramic acid (MurNAc)-pentapeptide building blocks are assembled in the cytosol and linked to the lipid carrier, a C55-phosphate called bactoprenol or undecaprenyl phosphate (UP), thereby forming lipid I. An N-acetylglucosamine (GlcNAc) molecule is added, resulting in lipid II. This cell wall building block is subsequently shuttled across the membrane by the flippases Amj and MurJ (Meeske et al., 2015; Laddomada et al., 2016). On the outside, the GlcNac-MurNAc-pentapeptide building block is incorporated into the existing cell wall by transgylcosylation and transpeptidation reactions, thereby releasing the lipid carrier in its pyrophosphate form (undecaprenyl pyrophosphate, UPP). For its recycling, UPP is then dephosphorylated to UP by specialized UPP phosphatases (Bernard et al., 2005; Manat et al., 2014) and flipped back to the cytosolic leaflet of the membrane, where it can be reloaded to enter the Lipid II cycle again.
FIGURE 1.
Simplified scheme of the Lipid II-cycle. UPP is dephosphorylated to UP by the UPP phosphatases BcrC, UppP and YodM. The peptide antibiotic bacitracin specifically binds to UPP, thereby inhibiting its dephosphorylation. The carrier UP is loaded with a cell wall precursor, resulting in lipid II. After incorporation of the cell wall precursor into the existing cell wall, UPP is released and recycled by the UPP phosphatases. De novo synthesis of UPP occurs from isoprenoids via the enzyme UppS. Undecaprenol can also serve as an unphosphorylated carrier, which is phosphorylated by the kinase DgkA to UP. Bac, Bacitracin; Und, Undecaprenol; UP, Undecaprenyl phosphate; UPP, Undecaprenyl pyrophosphate.
Apart from this recycling, the cellular UP pool can also be replenished by de novo synthesis of UPP via the UPP synthetase UppS (Guo et al., 2005). The subsequent dephosphorylation to UP is likely performed by the same UPP phosphatases that are required for recycling UPP (Manat et al., 2014). In Gram-positive bacteria, UP can also originate from phosphorylating undecaprenol, e.g., by the kinase DgkA in Bacillus subtilis (Higashi et al., 1970; Jerga et al., 2007).
UP is the carrier for both peptidoglycan and wall teichoic acids (WTA) building blocks and its availability represents the central bottleneck for the synthesis of lipid II both in vitro and in vivo (Breukink et al., 2003; Breukink and de Kruijff, 2006; Egan et al., 2015). Only ∼ 2∗105 UP molecules (0.5–1% of all phospholipids) are present per cell (Kramer et al., 2004) and it has been estimated that each of the carriers shuttles one to three cell wall building blocks per seconds during exponential growth (McCloskey and Troy, 1980). The amount of WTA and peptidoglycan synthesis is reduced under UP-limitation, especially if conditions favor the competing pathway (Anderson et al., 1972). Antibiotics that target the lipid II cycle benefit from this bottleneck, because blocking any step will lead to accumulation of intermediates, shortage of free carrier molecules and impaired cell wall biosynthesis that depends on UP.
Maintaining envelope integrity is absolutely essential for the survival of any bacterial cell, as are the metabolic processes that ensure it. Bacteria have therefore evolved appropriate countermeasures to detect and remove threats or damages to cell envelope homeostasis before they become lethal. These responses are collectively termed cell envelope stress response (CESR) (Jordan et al., 2008). Bacillus subtilis is one of the main model organisms for studying the Gram-positive cell wall and member of the Firmicutes phylum (low G + C Gram-positives). In this organism, the CESR is orchestrated by two-component systems and extracytoplasmic sigma factors (ECFs) (Radeck et al., 2016a). While many antibiotics can trigger the CESR, the molecular nature behind these stimuli has only been identified for very few cases. The antibiotic itself seems rarely to be detected directly. Instead, downstream effects of antibiotic threat, such as envelope damage or – more importantly – the accumulation of certain intermediates, are suspected to be the actual triggers of CESR (Meeske et al., 2015; Helmann, 2016). A similar effect to such an antibiotic-mediated blockade can also be achieved by reducing the availability of the corresponding enzyme. Consequently, the lipid II cycle, cell wall homeostasis and cell envelope stress (CES) are interconnected processes that can hardly be studied independently. In fact, a B. subtilis mutant with reduced UppS activity (and therefore reduced de novo synthesis of UPP) had altered antibiotic resistance properties and elevated σM-activity (Lee and Helmann, 2013). Here, we will focus on the CESR caused by limitations of the crucial UPP phosphatase activity, provided, e.g., by BcrC.
The expression of bcrC is controlled by multiple stress-inducible alternative sigma factors, including σM, σI, σX, σV, and potentially also σW (Cao and Helmann, 2002; Tseng and Shaw, 2008; Guariglia-Oropeza and Helmann, 2011; Zweers et al., 2012). σM controls approximately 60 genes involved in cell wall synthesis, shape determination, detoxification and DNA damage response (Eiamphungporn and Helmann, 2008). It is activated by multiple triggers, including antibiotics, high salt, heat stress, and acidic pH (Thackray and Moir, 2003). While all of these inducers affect cell envelope synthesis or integrity, the molecular cue for the activation of this and other ECFs is yet to be identified (as reviewed in Helmann, 2016).
Induction of PbcrC can be triggered, e.g., by the addition of the antibiotic bacitracin (Cao and Helmann, 2002; Radeck et al., 2016b). Bacitracin is a cyclic antimicrobial peptide produced by some strains of Bacillus licheniformis and B. subtilis (Azevedo et al., 1993; Ishihara et al., 2002). It was shown that bacitracin tightly binds UPP, thereby blocking the dephosphorylation reaction mediated by UPP phosphatases and consequently slowing down the lipid II cycle (Siewert and Strominger, 1967; Storm and Strominger, 1973; Economou et al., 2013). The deletion of bcrC may have similar consequences, since the loss of one UPP phosphatase might reduce the rate of UPP dephosphorylation to UP.
A very sensitive indicator of CES is the LiaR-controlled liaI promoter (PliaI) (Mascher et al., 2004). The cognate three-component system, LiaFSR reacts to a broad range of cell envelope stressors, including alkaline shock, oxidative stress, or bacitracin addition (Jordan et al., 2006; Wolf et al., 2010). In turn, it regulates a phage-shock protein-like response that provides a secondary layer of protection against CES (Radeck et al., 2016b). The low basal activity and strong, highly dynamic induction of PliaI made this promoter an ideal candidate for the development of a highly sensitive CESR-inducible whole cell biosensor (Mascher et al., 2004; Wolf and Mascher, 2016; Kobras et al., 2017). Recently, we demonstrated that PliaI activity in response to bacitracin is elevated in a bcrC deletion mutant and decreased in a bcrC overexpression strain. These findings indicate that the CES caused by bacitracin is relieved in the presence of the UPP phosphatase BcrC (Radeck et al., 2016b).
In the same study, we observed that PbcrC activities were increased in a bcrC null mutant. Together, this lead to the hypothesis that changes in UP and UPP levels can be sufficient to create CES (Radeck et al., 2016b). Due to their crucial role in the lipid II cycle, we hypothesize that impaired UPP phosphatase activity leads to a limitation in cell wall synthesis, which in turn should increase the CESR. Here, we aimed at challenging this hypothesis by studying the effects of enzymes potentially involved in UP turnover on B. subtilis physiology and stress responses in detail.
The genome of B. subtilis encodes three UPP phosphatases, BcrC, UppP and YodM. YodM and BcrC belong to the large group of type II phosphatidic acid phosphatases (PAP2s) that share their catalytic mechanism while pursuing a wide range of functions from signaling to export. Both proteins are homologues to the crystalized UPP phosphatase PgpB of Escherichia coli (El Ghachi et al., 2005; Fan et al., 2014; Kelley et al., 2015). While YodM seems to be dysfunctional due to insufficient expression (Zhao et al., 2016), BcrC has been studied to some extent. It seems to be the major B. subtilis UPP phosphatase (Bernard et al., 2005; Inaoka and Ochi, 2012) and is highly expressed at most culture conditions, as judged by a comprehensive tiling array study (Supplementary Figure S1) (Nicolas et al., 2012). The monocistronic gene bcrC (Supplementary Figure S1) is regulated by the CES-inducible ECFs σM and σX (Cao and Helmann, 2002). The latter responds to CES that might be caused by changing UP levels or other intermediates of the lipid II cycle (Helmann, 2016).
The minor UPP phosphatase UppP (Inaoka and Ochi, 2012) is homologous to BacA from E. coli. The latter accounts for about 75% of the UPP phosphatase activity in this organism (El Ghachi et al., 2004). uppP is the second gene of the yubA-uppP operon and its PyubA-dependent expression is not induced by bacitracin (Cao and Helmann, 2002). YubA is predicted to be a membrane protein belonging to the autoinducer-2 exporter (ai-2e) family and might be associated with cell wall synthesis (Fenton et al., 2016; The UniProt Consortium, 2017).
To investigate the correlation between UPP phosphatases and cell envelope homeostasis, we analyzed strains depleted for (combinations of) both the UPP phosphatases BcrC/UppP and the undecaprenol kinase DgkA on cell physiology and morphology. First, we demonstrate the synthetic lethality of BcrC and UppP and a severe morphological defect in UPP phosphatase depleted strains. Next, UppP is shown to be indispensable for efficient sporulation. Unexpectedly, uppP or bcrC deletion and complementation mutants did not activate a classical CESR, as judged by the lack of PliaI induction. Instead, the resulting limitation in UPP phosphatase levels is perceived by the broader ECF-dependent signaling network. As a result, PbcrC activity was increased in those mutants, thereby providing a homeostatic feedback mechanism by which the cell can autoregulate its UPP phosphatase level according to needs. Furthermore, we provide the first evidence that DgkA is indeed involved in UPP homeostasis: While a lack of this predicted undecaprenol kinase did not result in an observable deficiency, a (most likely minor) role in UP turnover is indicated by an increased activity of PbcrC in a dgkA mutant in stationary phase. Taken together, our data provides the first insight into the fine-tuning of UP homeostasis that adjusts the Lipid II cycle, and hence cell wall biosynthesis, in response to growth rates and envelope stress levels.
Results
High Level Expression of the UPP Phosphatase Encoding Genes bcrC and uppP in B. subtilis
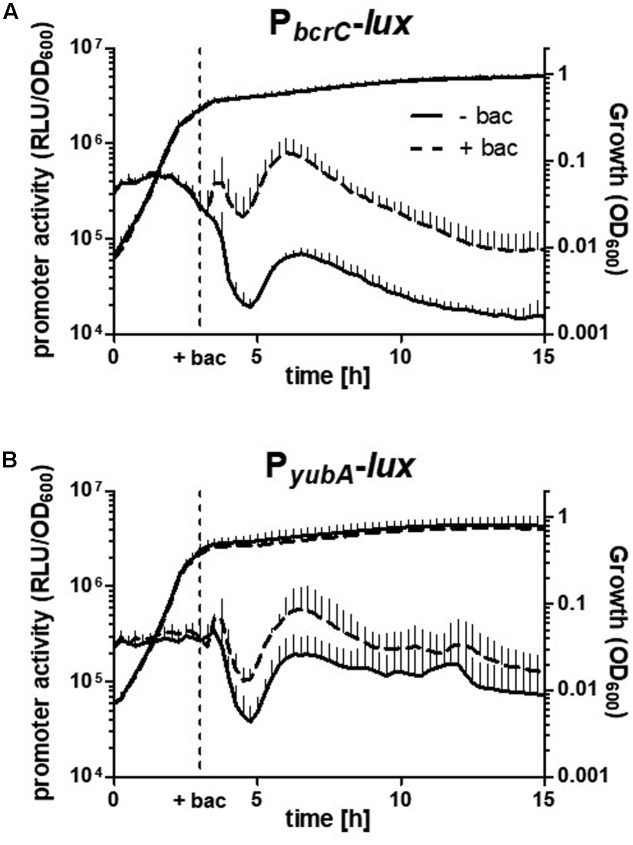
We first wanted to analyze the expression of the two UPP phosphatase-encoding genes bcrC and uppP by monitoring the activity of strains harboring the corresponding promoter-lux fusions, PbcrC and PyubA, respectively. Under our experimental conditions, the activity of PbcrC remained at high levels (Figure 2A) from early exponential to late stationary phase - with exception of the known decrease during transition state, which is frequently observed for online promoter activity measurements (Radeck et al., 2013). As discovered previously, PbcrC activity was increased by the addition of bacitracin (30 μg ml-1; Figure 2A) (Mascher et al., 2003; Radeck et al., 2016b).
FIGURE 2.
Expression of UPP phosphatase genes. Growth (OD600) and activity levels (RLU/OD600) of PbcrC (A) and PyubA (B) in B. subtilis W168 (TMB1620 and TMB3688) from early exponential to late stationary phase in absence or presence of 30 μg ml-1 bacitracin (+ bac; dashed lines), respectively. PbcrC activity was significantly increased upon bacitracin addition (p = 0.021, 2-way ANOVA), but not PyubA (p = 0.29, 2-way ANOVA). Measurements were obtained in a microtiter plate reader at 37°C in MCSEC medium. Data is shown for three independent biological replicates (mean and SD).
The activity of PyubA is comparable to PbcrC during exponential growth, but about three-fold higher during stationary phase (Figure 2B). In contrast to PbcrC, and in agreement with a previous study, PyubA was not significantly induced by bacitracin (Figure 2B, Cao and Helmann, 2002). Our data based on the promoter-lux fusions agrees well with the tiling array data on mRNA levels of uppP and bcrC (Supplementary Figure S1; Nicolas et al., 2012). Under most conditions, bcrC is expressed at a slightly higher level than uppP (yubB), with the exception of sporulation, during which bcrC expression drops at early sporulation and uppP only at late sporulation (Supplementary Figure S1).
For the third UPP phosphatase, the tiling array data shows that there is almost no transcription of yodM, but instead high levels of counter-transcription (Supplementary Figure S1). This finding has recently been verified (Zhao et al., 2016). Due to these observations, YodM and its promoter, PyodM, were not considered further for our analysis.
Hence, there are two well-transcribed UPP phosphatase genes in B. subtilis cells, bcrC and uppP. We therefore decided to study the role of their gene products in cell wall homeostasis and CES in B. subtilis. Toward that goal, we investigated single and combined deletion and complementation strains for their effect on cell morphology, sporulation, CESR and antibiotic resistance.
uppP and bcrC Are a Synthetic Lethal Gene Pair
Initially, we aimed at replacing all three UPP phosphatase genes with resistance cassettes (bcrC::tet, uppP::MLS and yodM::spc) in single and double mutants. For simplicity reasons, all allelic replacements are noted as deletions throughout the manuscript and figure legends. All single mutants and double mutants with ΔyodM were readily obtained. Since the lack of any observable phenotype during the initial characterization of all ΔyodM strains can readily be explained by the lack of yodM expression (Figure 2A and Zhao et al., 2016), these strains were not considered further.
In contrast to the single mutants, multiple attempts to construct a ΔuppP ΔbcrC double mutant failed, indicative of synthetic lethality of bcrC and uppP. To support this assumption, we constructed complementation strains, in which uppP or bcrC were ectopically integrated into the thrC locus under control of the xylose-inducible promoter PxylA. In strains carrying a complementation copy of either bcrC or uppP, the deletion of both native genes was possible in the presence of xylose. These strains (ΔuppP ΔbcrC PxylA-bcrC and ΔuppP ΔbcrC PxylA-uppP) will be referred to as depletion strains to distinguish them from the complementation strains ΔbcrC PxylA-bcrC and ΔuppP PxylA-uppP. Our findings are in agreement with a recent study from the Helmann laboratory, which independently demonstrated the synthetic lethality of the bcrC/uppP gene pair using a CRISPR-dCas9 knockdown approach (Zhao et al., 2016).
Cell Morphology Is Impaired in UPP Phosphatase Mutants during Exponential Growth
Depletion of essential envelope-associated proteins often leads to bulging, filamentation or lysis of cells (Peters et al., 2016). Since uppP and bcrC are synthetic lethal and the lipid II-cycle and cell wall synthesis depend on the recycling of UP by UPP phosphatases, we hypothesized that a depletion of UPP phosphatases in fast-growing cells leads to a morphological phenotype similar to that observed for other essential cell envelope functions.
Single uppP or bcrC deletions, the respective complementation mutants, and the wild type showed no or less than 0.1% misshaped cells (data not shown). In contrast, both UPP phosphatase depletion strains TMB3739 (ΔuppP ΔbcrC PxylA-bcrC) and TMB3740 (ΔuppP ΔbcrC PxylA-uppP) showed a severe phenotype during exponential growth phase (Figure 3). In the absence of xylose, about 20–30% (TMB3739) or 80% (TMB3740) of the cells were bulging and sometimes bending (Figure 3A). This phenotype could be completely suppressed by the addition of xylose, resulting in high expression levels of the complemented UPP phosphatase (Figure 3B). This phenotype could not be observed at slow growth, e.g., in stationary phase (Supplementary Figure S2) even though some cells look swollen compared to wild type cells (e.g., the ΔuppP mutant TMB3408).
FIGURE 3.
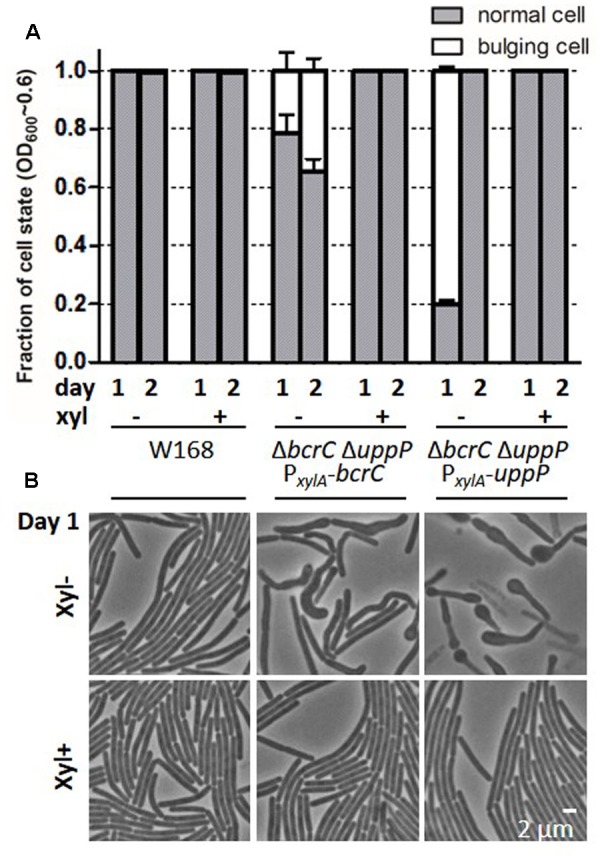
Cell morphology during exponential growth in bcrC and uppP complementation mutants. Strains W168, TMB3739, TMB3740 were inoculated from fresh overnight cultures (day 1) or 24 h-cultures (on day 2), grown in MCSEC at 37°C, 220 rpm without xylose (xyl-) to deplete the respective UPP phosphatase or with 0.2% xylose (xyl+) to fully induce the production in complementation mutants. Overnight cultures were always supplemented with xylose, whereas the inoculum for day 2 was taken from samples either with (xyl+) or without (xyl-) xylose added. Phase contrast pictures were taken in late exponential phase (OD600∼0.6, ∼6 h post-inoculation). (A) Fraction of cells with normal (gray) or bulging morphology (white). At least 1000 cells were counted for each of the three independent biological replicates. (B) Representative pictures of cells with normal or bulging morphology. Samples were taken from day 1. The scale bar represents 2 μm.
In summary, we could show that a very low expression of only uppP or bcrC leads to severe morphological changes, e.g., bulging cells during exponential growth – concomitant with depleted peptidoglycan or WTA synthesis (Muchova et al., 2011; Botella et al., 2014) caused by a lack of UP. This phenotype was most severe for the uppP depletion strain.
A uppP Mutant Is Impaired in Efficient Sporulation
During the morphology studies, we observed altered sporulation rates between the wild type and UPP phosphatase mutants, especially in ΔuppP. We therefore quantified the sporulation efficiency in our strains by determining the fractions of vegetative versus sporulating cells and endospores in a culture 24 h after inoculation (summarized in Figure 4, see Supplementary Figure S3 for the complete dataset). Under our experimental conditions, about 30% of the wild type cells (Figure 4B, i) were in the process of sporulation or had already sporulated. Mutants with a native copy of uppP (ii–iv), and mutants with wild type copy of bcrC in combination with an ectopic inducible copy of uppP (viii, ix) had similar sporulation rates. Sporulation was impaired (<7%), if the native copy of uppP was lost and no ectopic copy was introduced (v, vi), or if uppP was depleted in the phosphatase double mutant (x). The sporulation deficiency of the latter could partially be restored by the addition of xylose to induce uppP expression (xi).
FIGURE 4.
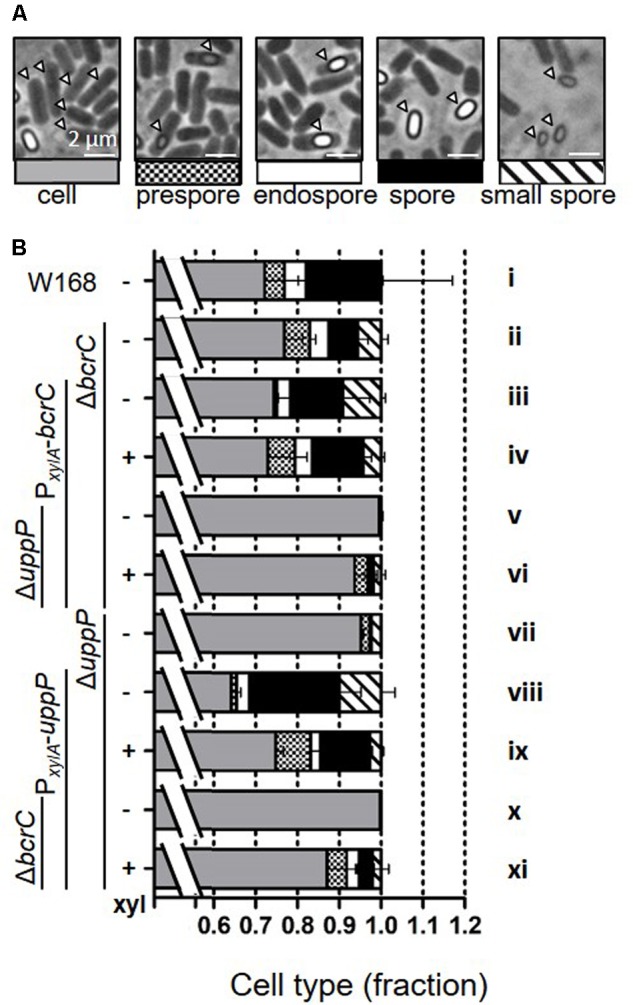
Sporulation efficiencies of bcrC and uppP deletion and complementation mutants. Strains (W168, TMB0297, TMB3694, TMB3739, TMB3408, TMB3695, and TMB3740) were grown as described in Figure 3 and phase contrast microscopy pictures were taken 24 h post-inoculation. (A) Representative pictures for normal cells (gray), prespores without fully established phase-bright endospore (small checkered), completed endospores (white), free spores (black) and small free spores (striped). (B) Cell type fractions are shown as stacked bar graphs. Data is shown for at least 1000 cells per measurement and the error bars represent the standard deviation between independent biological triplicates. The full data set is shown in Supplementary Figure S3.
The reduced sporulation frequencies in ΔuppP mutants did not originate from delayed sporulation, since a similar reduction in sporulation rates (2–5% compared to >30% in the wild type) was also observed after 48 h (data not shown). However, using a spore-crust marker (GFP-CotZ), we detected that some of the phase-gray particles in a ΔuppP mutant were spores instead of cells (Supplementary Figure S4). This phenotype is indicative of alterations in stage IV or V of sporulation, where mutants have thinner or no germ cell wall or cortex (Coote, 1972; Piggot and Coote, 1976). Both spore layers consist of peptidoglycan, a defect in their synthesis therefore points toward UppP being the responsible UPP phosphatase for the lipid II cycle during sporulation. This observation is in agreement with a recent screen for sporulation mutants, in which a reduced sporulation efficiency and phase-gray spores were also detected in a uppP mutant (Meeske et al., 2016).
Here we could show that the rates of normal, phase bright spores drastically decreased in absence of UppP. The physiological relevance of this UPP phosphatase for efficient sporulation is underscored by the dramatic loss in the number of heat resistant spores, which was determined to be only 0,04% of the wild type (Meeske et al., 2016). In the presence of BcrC, low levels of UppP still allow a normal sporulation (Figure 4, vii, viii), while this residual UppP amount is not sufficient in the absence of BcrC (x). Either the native copy of uppP or the combination of native bcrC and an ectopic version of uppP is required for efficient sporulation.
The combined results from our sporulation counts (Figure 4) and the cell morphology study (Figure 3) indicate that limited amounts of either UPP phosphatase alone (TMB3739, TMB3740 without xylose) are not sufficient to retain normal cell shape during fast growth or ensure efficient sporulation. While each native phosphatase is sufficient to keep normal cell shape in exponential growth, BcrC cannot compensate for the lack of UppP during sporulation. Both phenotypes point toward defects in cell wall synthesis. This provoked the question if under such circumstances this bottleneck in cell wall synthesis leads to a CESR, which is normally triggered by the external addition of cell wall antibiotics, such as bacitracin (Radeck et al., 2016a). To address this question, two well established reporters for CESR [the PliaI and PbcrC promoters fused to the lux reporter cassette (Radeck et al., 2016b)] were combined with the mutant collection and probed for their activity under UPP phosphatase-limiting conditions.
Limitations in UPP Phosphatases Are Perceived As Envelope Stress by the PbcrC Reporter
PliaI is a very sensitive reporter of cell envelope damage due to its wide inducer spectrum and high dynamic activity range (Mascher et al., 2004; Rietkötter et al., 2008). But we did not observe any UPP phosphatase-dependent induction of the PliaI-controlled CESR, even if we additionally challenged the cells with bacitracin (data not shown).
In contrast to the damage-sensing PliaI reporter, the PbcrC-derived reporter is postulated to respond to alterations/limitations in cell wall homeostasis (Minnig et al., 2003) and could therefore be more suitable to detect stress caused by changes in the UPP phosphatase levels. In light of this study, PbcrC is particularly relevant since it controls the expression of one of the two UPP phosphatases, BcrC. It therefore provides a direct read-out for the cells ability to respond to limitations in UPP phosphatases by upregulating bcrC expression. Toward this end, we measured PbcrC-activity in the wild type as well as bcrC and uppP deletion, complementation and depletion strains. Promotor activity as relative luminescence units normalized to cell density (RLU/OD600) and growth (OD600) were measured in a microtiter plate reader for 15 h (Figure 5).
FIGURE 5.
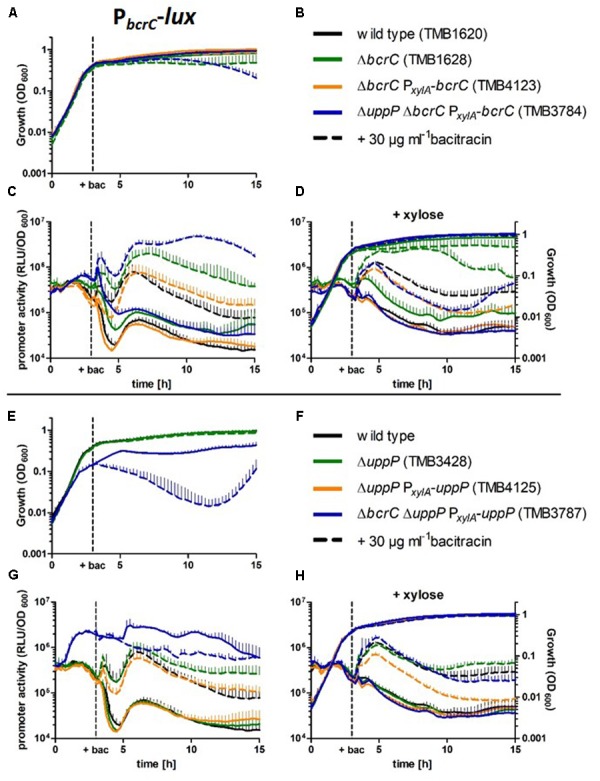
Growth and PbcrC promoter activities in the wild type and bcrC and uppP complementation mutants. Strains were grown in MCSEC at 37°C in 96-well plates in a microtiter plate reader. OD600 and luminescence was measured every 15 min for 15 h. (A–D) Growth and PbcrC-activity in bcrC deletion, complementation and depletion strains. (E–H) Growth and PbcrC-activity in uppP deletion, complementation and depletion strains. The strains are defined by the color, while solid or dashed lines indicate the absence or presence of 30 μg ml-1 bacitracin. (D,H) Samples were grown with 0.2% xylose to fully induce PxylA-driven gene expression. In these cultures, promoter activity steadily decreased from the transition phase onward. This phenomenon was observed for all strains either harboring PbcrC or the constitutive reference promoter PlepA (data not shown). We therefore postulate that the change in promoter dynamics is caused by the presence of an additional C-source (xylose). Thin lines represent the standard deviation of at least three biological replicates.
For the wild type reporter strain (TMB1620), PbcrC activity of 3–5∗105 RLU/OD600 was observed during exponential growth and the transition phase (Figures 5A,C; black lines, 0–3 h). The activity decreased about 10-fold during early stationary phase (4–6 h), briefly increased (6–8 h) and then steadily declined during late stationary phase. Upon bacitracin addition (30 μg ml-1), the promotor activity was increased about 10-fold, while no change in growth behavior was detected.
In the ΔbcrC mutant (TMB1628, green) and the bcrC depletion strain (TMB3784, blue), the PbcrC activity increased without and especially with bacitracin addition and a slightly reduced optical density was observed during stationary phase compared to the wild type (Figures 5A–D). These effects were revoked by the addition of xylose (TMB3784) or the introduction of a complementing copy of bcrC (TMB4123, orange), even without xylose.
Deletion of uppP (TMB3428, green) only had a minor effect on PbcrC activity (approximately three-fold elevation during late stationary phase upon bacitracin addition, Figures 5E,G). However, in the uppP depletion strain (TMB3787, blue) impaired growth – especially in the presence of bacitracin – and strongly increased PbcrC activity was observed throughout growth, even without bacitracin addition. A subsequent in-depth analysis supported these findings: The uppP depletion strain (TMB3740), but not the bcrC depletion strain (TMB3739) showed a clear growth defect in absence of xylose. All phenotypes of complementation mutants reverted to wild type levels in presence of xylose (see Supplementary Figure S5 for details).
Taken together, the single bcrC deletion, as well as two phosphatase depletion strains (ΔbcrC ΔuppP PxylA-bcrC and ΔbcrC ΔuppP PxylA-uppP) had the strongest effect on PbcrC activity, especially in the presence of bacitracin.
The Undecaprenol Kinase DgkA Contributes to the Cellular UP Pool
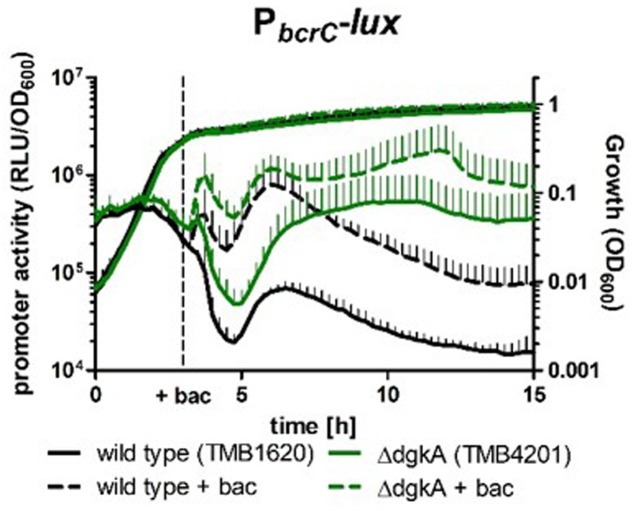
The results described in the previous section demonstrate that the cell is indeed capable of perceiving limitations in UPP phosphatase levels, most likely at the level of the resulting UP shortage. A second enzymatic activity potentially contributing to the cellular UP pool is the undecaprenol kinase DgkA that phosphorylates undecaprenol to UP (Jerga et al., 2007). Based on the results of the previous section, the activity of the PbcrC reporter might provide an ideal read-out to probe if DgkA indeed provides a measurable contribution to the UP pool, particularly if the cellular amount of UPP phosphatases is severely limited. We therefore deleted dgkA in the wild type and all phosphatase deletion, complementation and depletion strains and then measured the PbcrC activity throughout the growth cycle.
Surprisingly, a strong DgkA-dependent effect was already observed in the wild type reporter strain: the PbcrC activity was elevated ∼10-fold in the dgkA mutant during late stationary phase relative to the wild type, both in the presence or absence of bacitracin (Figure 6). A similar effect was observed for all phosphatase mutants (Supplementary Figure S6). This result indicates that a DgkA-dependent phosphorylation of undecaprenol indeed detectably contributes to the cellular UP pool, even though a dgkA mutant did not show any (additional) morphological phenotype during fast growth (data not shown). It has previously been shown that a B. subtilis dgkA mutant produces less and cortex-deficient endospores – a peptidoglycan structure that depends on UP for its synthesis (Amiteye et al., 2003; Supplementary Figure S3). This suggests that the role of DgkA to contribute to the UP pool is rather during sporulation.
FIGURE 6.
PbcrC promoter activity depends on DgkA. Strains were grown as described in Figure 5. Black, wild type; green, dgkA mutant. Samples induced with bacitracin are shown as dashed lines. Thin lines represent the standard deviation of three biological replicates.
Deletion and Depletion of UPP Phosphatases Increases Sensitivity toward the UPP-Binding Antibiotic Bacitracin
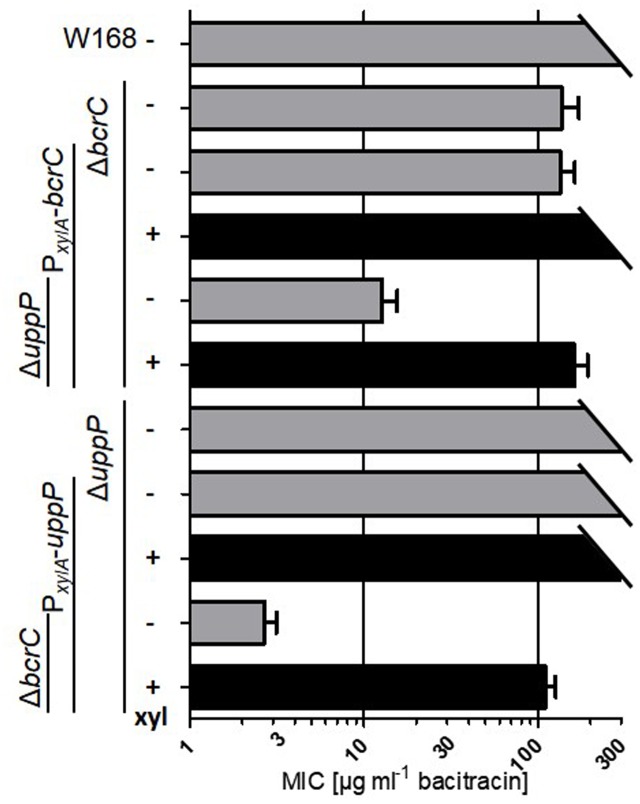
Bacillus subtilis wild type cells are highly resistant against the UPP-binding antibiotic bacitracin (minimal inhibitory concentration, MIC, >256 μg ml-1). The primary resistance determinant is the bacitracin-specific ABC-transporter BceAB (Mascher et al., 2003; Ohki et al., 2003; Rietkötter et al., 2008). But BcrC provides a (secondary) layer of bacitracin resistance, most likely by competing with the antibiotic for the same target molecule, UPP (Bernard et al., 2005; Radeck et al., 2016a,b). The inhibitory effect of bacitracin is based on depleting the UP pool by formation of a UPP-bacitracin complex, finally leading to an arrest of the lipid II cycle (Figure 1). It stands to reason to postulate that deletions in genes encoding UPP phosphatases or undecaprenol kinases might also contribute to the sensitivity of the cells toward bacitracin. We therefore measured the MIC for bacitracin in UPP phosphatase deletion and depletion mutants, using Etest® strips (Figure 7 and Supplementary Figure S7).
FIGURE 7.
Minimal inhibitory bacitracin concentration of bcrC and uppP deletion and complementation mutants. Strains (W168, TMB297, TMB3694, TMB3739, TMB3408, TMB3695, and TMB3740) were inoculated from fresh overnight cultures (xyl+) in MCSEC at 37°C with or without 0.2% xylose. During exponential growth, cells were embedded in soft agar and plated as an overlay on MCSEC agar. One Etest® strip (bacitracin 0.016–256 μg ml-1) was placed on the soft agar (see Material and Methods). The MIC was determined after 24 h of incubation at 37°C. Data is shown for at least three independent biological replicates (two replicates, if MIC > 256 μg ml-1) with mean and standard deviation. The full data set is depicted in Supplementary Figure S7.
While the individual deletion of uppP had no measurable effect on bacitracin MIC, the deletion of bcrC lead to the known reduction of the MIC to ∼ 120 μg ml-1. This phenotype could be complemented by the addition of xylose, thereby inducing the ectopically integrated PxylA-bcrC. Without xylose, the MIC of this strain is comparable to the bcrC deletion mutant, indicating very little background activity of PxylA under non-inducing conditions. If uppP is deleted in this genetic background, the MIC was even further decreased to ∼15 μg ml-1. In this depletion strain, the PxylA-mediated expression of bcrC can no longer fully compensate for the loss of both UPP phosphatases (MIC of ∼150 compared to >256 μg ml-1 in ΔuppP). The uppP depletion mutant (TMB3739, ΔbcrC ΔuppP PxylA-uppP) exhibited the lowest MIC (∼3 μg ml-1). Upon addition of xylose, a 35-fold increase in bacitracin MIC could be observed (Figure 7).
Taken together, the resistance toward the UPP-binding bacitracin is indeed severely reduced in mutants limited for UPP phosphatases. This phenotype can be (almost fully) compensated for by induction of ectopically integrated UPP phosphatase genes under control of PxylA. Again, the phenotype of the uppP-depletion strain (TMB3740) is more severe than that of the bcrC-depletion (TMB3739), in line with the morphological defects observed above (Figure 3). The additional deletion of dgkA or yodM had no effect on the observed MIC in any of the mutants tested, again indicating a very minor contribution of these two gene products. Removing the main bacitracin resistance determinant, bceAB, lead to overall lower basal MICs, but had no additional influence on the behavior described above (Supplementary Figure S7).
Discussion
Together, bcrC and uppP Encode the Essential UPP Phosphatase Function of B. subtilis
In our study, we demonstrated that uppP and bcrC constitute a synthetic lethal gene pair – a result that is in perfect agreement with an independent study performed in parallel using CRISPR-dCas9 knock-downs to study the effect of UPP phosphatase levels in B. subtilis (Zhao et al., 2016). These observations thereby correct two previous studies, which independently reported the successful construction of a uppP/bcrC double deletion mutant that showed the same phenotype as a single bcrC deletion mutant (Bernard et al., 2005; Inaoka and Ochi, 2012). Both groups used deletion constructs based on the vector pMUTIN, which disrupted either uppP or bcrC by integrating via single homologous recombination (Vagner et al., 1998; Kobayashi et al., 2003; Bernard et al., 2005; Inaoka and Ochi, 2012). Based on their data, it must be postulated that these deletion constructs generated a gene fragment up- or downstream the integration site, which was still large enough to maintain (residual) UPP phosphatase activity. In contrast, both recent studies only used complete allelic replacement mutants based on double homologous recombination of a resistance cassette (this study and Zhao et al., 2016).
One important difference between the latter two studies is the occurrence of suppressor mutants in zone-of-inhibition assays, as reported by Zhao et al. (2016). In the timescale of our experiments, we did not observe such suppressors, most likely due to the low but detectable leakiness of the xylose-dependent expression system. While the xylose system is tight enough to allow observing morphological defects and envelope stress in our study, it is not as tight as the CRISPR-dCas9 system that was used in the parallel study (Zhao et al., 2016). This approach obviously generated a much higher selective pressure that ultimately lead to the occurence of suppressors. According to our results, the basal expression of bcrC driven by PxylA is still sufficient for growth at normal doubling times. This is in agreement with the parallel study, which initially failed to generate uppP or bcrC depletion strains due to the high basal activity of Pspac(hy) which was used for complementation: The strains still grew in the absence of IPTG, despite the knock-out of the native uppP and bcrC genes (Zhao et al., 2016).
In contrast to BcrC and UppP, the third UPP phosphatase, YodM, did not provide a measurable contribution to the cellular UP pool. While the Zhao et al. (2016) study demonstrated that the gene product of yodM indeed has UPP phosphatase activity, this was only sufficient to support growth if expression was artificially improved, in line with our own observations. This is not surprising, considering the expression profiles from a comprehensive transcriptome study, which demonstrates a lack of yodM expression, but instead a strong counter-transcription (Supplementary Figurexs S1). Together, the data provided in this study and the recent report from the Helmann group unequivocally demonstrates that the UPP phosphatase activity in B. subtilis is primarily – if not exclusively – provided by BcrC and UppP. While both can functionally complement each other, our study indicates that the two phosphatases have slightly different functions in wild type cells.
BcrC Is More Relevant during Vegetative Growth, While UppP Is Important for Efficient Sporulation
Table 1 summarizes the main findings of our study with regard to bacitracin sensitivity, PbcrC activity, cell morphology and growth rates. While our data demonstrates that either phosphatase is sufficient to support growth, the respective mutants do show significant differences in their overall behavior. The ΔbcrC single mutant had a decreased MIC for bacitracin and an elevated PbcrC activity, in contrast to the ΔuppP strain. If the UPP phosphatase levels are further reduced or if the cultures are additionally challenged with bacitracin, the phenotypes are overall less severe if BcrC is complemented compared to UppP under similar conditions. Hence, the uppP depletion strain (that completely lacks BcrC) shows a severe growth defect and strong CESR in the absence of xylose. While an elevated PbcrC activity is also measured for the bcrC depletion strain, this effect is rather weak in the absence of bacitracin and only a mild growth defect is observed. Our data therefore not only supports previous findings that BcrC is the major UPP phosphatase during vegetative growth in B. subtilis (Bernard et al., 2005; Inaoka and Ochi, 2012), but also demonstrates that an ectopically complementing copy of bcrC is more efficient in providing the UPP phosphatase activity than a similar construct for uppP.
Table 1.
Summary of phenotypes associated with UPP phosphatase mutants.
| Strains |
Phenotypes1 |
||||
|---|---|---|---|---|---|
| Genotype (condition) | Phosphatase presence | Morphology (bulging) | CESR (PbcrC)3 | Sensitivity (bacitracin) | Sporulation defect5 |
| Wild type | BcrC, UppP | - | - | - | - |
| ΔuppP | BcrC | - | - | - | + |
| ΔbcrC ΔuppP PxylA-bcrC (xyl+) | BcrC | - | - | o | + |
| ΔbcrC ΔuppP PxylA-bcrC (xyl-) | (BcrC)2 | + | + | + | ++ |
| ΔbcrC | UppP | - | oo | o | - |
| ΔbcrC ΔuppP PxylA-uppP (xyl+) | UppP | - | - | o | oo |
| ΔbcrC ΔuppP PxylA-uppP (xyl-) | (UppP)2 | ++ | ++4 | ++ | ++ |
1Severity of phenotype (for details see corresponding results section): -, none; o, weak; oo, medium; +, strong; ++, very strong. 2Residual amounts of these phosphatases are based on the slight leakiness of PxylA. 3CESR expressed as increased PbcrC activity in stationary phase. 4In this strain, the PbcrC activity is even increased in exponential growth phase. 5Decreased amount of mature spores after 24 and 48 h growth in MCSEC medium.
In contrast, UppP seems to play the more prominent role with regard to sporulation. At strongly reduced levels of UppP, BcrC can support but never fully compensate the function of UppP. Its role in the formation of mature spores was recently also observed in a screen for sporulation mutants (Meeske et al., 2016).
When triggering a CESR, as monitored by an increased σM-dependent PbcrC activity (this study, Cao and Helmann, 2002; Radeck et al., 2016b), we recently observed that a deletion of bcrC further increases the CESR (Radeck et al., 2016a,b). Here, we could demonstrate that this effect holds true for UPP phosphatases in general: While a uppP deletion alone does not trigger the CESR, very low levels of UPP phosphatase activity (especially in the uppP depletion strain) cause a stronger CESR than the bcrC single mutant (Figure 5). This finding perfectly fits to the working model that low levels of UP (or downstream effects thereof) are the stimulus for σM activation, rather than the protein levels of BcrC (Lee and Helmann, 2013; Zhao et al., 2016). It is also supported by the finding that PbcrC is induced in presence of bacitracin, which blocks the dephosphorylation to UP by binding to UPP (Cao and Helmann, 2002).
Outlook and Open Questions
Our study clearly demonstrates the essential role of UPP phosphatases for the lipid II cycle, in perfect agreement with results from an independent study performed in parallel (Zhao et al., 2016). In addition, we could demonstrate that these phosphatases also provide a direct link in connecting cell envelope homeostasis with CESR. Nevertheless, some questions are still open and need to be addressed in subsequent studies.
Quite surprisingly, PbcrC activity is unchanged in the uppP depletion strain treated with xylose, while the native uppP copy present in ΔbcrC is not sufficient to prevent CESR. This phenomenon could not be observed with regard to the bacitracin sensitivity and provokes the question if and how uppP is regulated during growth and CES. For further investigations, protein and/or activity levels of UPP phosphatases and the abundance of UPP and UP in challenged and non-challenged cells will help to better understand the stoichiometry of UPP dephosphorylation, a crucial step of the lipid II cycle. The lack of PliaI induction in UPP phosphatase-depleted strains – that was also observed by Zhao et al. (2016) – seems to be even more puzzling. Such a bottleneck in cell wall biosynthesis should result in perturbations affecting envelope integrity and hence activate PliaI. While we do not have an explanation for this behavior, it is reminiscent to the lack of response to bacitracin of PliaI in cell wall-less L-forms of B. subtilis (Wolf et al., 2012). Here, the lack of a PliaI response in the presence of bacitracin and other cell wall antibiotics was attributed to the absence of an intact cell wall biosynthesis machinery. If this would be true, it is tempting to postulate that the severe depletion of UPP phosphatases – as done in the present study – somehow also affects the integrity of the cell wall biosynthesis machinery, thereby removing the (still unknown) source of PliaI induction in the presence of peptide antibiotics that interfere with the lipid II cycle. But such a farfetched (and far reaching) speculation will require follow-up experiments.
Another aspect that needs to be taken into account is the substantial contribution of UPP de novo synthesis to the lipid II cycle: Reducing the UppS protein levels by 50% significantly altered cell wall antibiotic sensitivities (Lee and Helmann, 2013). But so far, very little is known about the stoichiometry between UPP recycling and de novo synthesis, which are both essential and depend on UPP phosphatases.
Two reactions are known to generate UP independent of UPP phosphatases: (i) recycling from WTA-shuttling, which depends on UP and is therefore not self-sustaining (Brown et al., 2013), and (ii) phosphorylation of undecaprenol, e.g., via DgkA (Jerga et al., 2007; Van Horn and Sanders, 2012). The cellular abundance and dynamics of undecaprenol has so far not been studied for B. subtilis, but data from other species indicates that this molecule is present in the membrane of Gram-positive bacteria and absent in Gram-negative bacteria (Higashi et al., 1970; Barreteau et al., 2009). In Staphylococcus aureus, a UP phosphatase activity was detected, but could not be assigned to a certain protein (Willoughby et al., 1972). Future studies – particularly for B. subtilis – will hopefully address the source of undecaprenol and its role as a possible resource for the lipid II cycle.
The localization and cellular dynamics of UPP phosphatases throughout the growth cycle and into sporulation might provide further insights into their activity pattern and hence their cellular roles. Such studies would also allow studying their proximity to active cell wall biosynthesis clusters (peptidoglycan and WTA), which could be a relevant proxy for efficient carrier supply (Kawai et al., 2011; Typas et al., 2012). Unfortunately, our initial attempts to generate functional translational GFP-fusions to the N- or C-terminus of UppP or BcrC were not successful. Some fusion constructs did not provide (sufficient) UPP phosphatase activity to complement the synthetic lethal gene pair in a uppP and bcrC deletion background. And those constructs that maintained the phosphatase activity lacked a fluorescent signal, potentially due to the fluorophore localizing to the extracellular side of the membrane (data not shown). Future studies, that employ protein linkers or fluorophors that mature in the periplasm (such as mCherry or superfolder GFP, Dammeyer and Tinnefeld, 2012) will hopefully circumvent these obstacles.
The data provided by our and other recent studies (Meeske et al., 2016; Zhao et al., 2016) are an important first step in gaining a mechanistic understanding on UPP phosphatases. But despite the insights gained during these studies, there is still a lot to be learned about the dynamics of the UP pool and how the functions that make and break this essential intermediate of cell envelope biosynthesis contribute to cell growth, differentiation and cellular stress responses.
Experimental Procedures
Bacterial Strains and Growth Conditions
Escherichia coli strains were routinely grown in lysogeny broth (LB) and B. subtilis in MOPS-based chemically defined medium with succinate and glutamate (MCSE) (Radeck et al., 2013), supplemented with casamino acids (1%, CAA) and L-threonine (50 μg ml-1) (MCSEC) at 37°C with agitation (220 rpm). Addition of CAA was necessary to prevent background activity of Phom, which is located upstream of the integration site of the uppP and bcrC complementation constructs (Radeck et al., 2013). Transformations of B. subtilis were carried out as described previously (Harwood and Cutting, 1990). All B. subtilis strains used in this study are derivatives of the laboratory wild type strain W168 and are listed in Supplementary Table S1. All allelic replacements are shown as gene deletions in the main text and figure captions for better readability. Selective media for E. coli contained ampicillin (100 μg ml-1) or chloramphenicol (35 μg ml-1). Selective media for B. subtilis contained chloramphenicol (5 μg ml-1), kanamycin (10 μg ml-1), spectinomycin (200 μg ml-1), tetracycline (12.5 μg ml-1) and/or a combination of erythromycin (1 μg ml-1) and lincomycin (25 μg ml-1) for macrolide-lincosamide-streptogramin B (MLS) resistance. Solid media additionally contained 1.5% (w/v) agar. For complementation studies, full induction of the promoter PxylA was achieved by adding xylose to a final concentration of 0.2% (w/v). Overnight cultures contained xylose per default to ensure normal growth of depletion strains.
DNA Manipulation
Plasmids were generated by using standard cloning techniques (Sambrook and Russell, 2001) with enzymes and buffers from New England Biolabs® (NEB) according to the manufacturer’s protocols. Phusion® polymerase was used for polymerase chain reaction (PCR) amplification for cloning purposes, otherwise OneTaq® was used. PCR purification was performed with HiYield PCR Gel Extraction/PCR Clean-up Kit (Süd-Laborbedarf Gauting, SLG®). For complementation studies, uppP or bcrC were placed under control of the xylose-inducible promoter PxylA inserted into the thrC-integration vector pBS4S. For measurements of promoter activity, promoter fragments spanning about 400 bp upstream of the Shine-Dalgarno sequence of the respective gene were cloned into pAH328, which carried the luxABCDE operon as an online luminescence reporter (Schmalisch et al., 2010). All plasmids generated during this study and a brief description of the construction are provided in Supplementary Table S2.
Allelic replacement mutations of bcrC and uppP were generated via long flanking homology PCRs, as described previously (Mascher et al., 2003). The integration of plasmids or DNA fragments into the B. subtilis genome via double recombination was verified with threonine auxotrophy (thrC) or colony PCR (sacA, uppP, bcrC, yodM, dgkA). All primer sequences are listed in Supplementary Table S3.
Luciferase Assay
Luciferase activities of B. subtilis strains harboring promoter-lux fusions were assayed using a SynergyTM NEOALPHAB multi-mode microplate reader from BioTek® (Winooski, VT, United States). The reader was controlled using the software Gen5TM (version 2.06). Hundred microliter culture volume were used per well in 96-well plates (black wall, clear bottom, clear lid, Greiner Bio-One). Incubation in the reader occurred at 37°C with linear agitation (567 cpm) and luminescence and OD600 were measured every 5 min. Strains were grown in MCSEC medium. Overnight cultures contained 0.2% xylose, to ensure protein production in complementation strains. (i) Day cultures (containing 0.2% xylose) were inoculated 1:5,000 from fresh overnight cultures, and strains were grown until exponential phase (OD600 = 0.1–0.4) (ii) Cells were harvested by centrifugation, washed twice in MCSEC, resuspended in MCSEC and the optical density was adjusted to OD600 = 0.025. (iii) 0.2% xylose was added if indicated and incubation in the reader occurred for 3 h. (iv) 30 μg ml-1 of bacitracin was added, if applicable, and the incubation and measurement continued for 17 h. Specific luminescence activity is given by the raw luminescence output (RLU) normalized by cell density (RLU/OD) (Radeck et al., 2013). For Supplementary Figure S5, cultures were handled as described, but the resuspended cultures (ii) were set to OD600 = 0.1 and 1:2, 1:4, 1:8, 1:16, and 1:100 dilutions thereof.
Microscopy
Cell morphologies and sporulation frequencies were studied with an Olympus Microscope (AX70, 100x oil objective, camera XC10) and the accompanying software (Olympus cellSens Dimension 1.14). Phase contrast and GFP fluorescence channels (filter cube: U-MNIB, FF blue longpass, Ex. 470-490 nm, Em. > 515 nm) were used. The exposure time for the GFP-channel was 100 ms. Strains were grown as described above (see “Luciferase assay” (i)) and incubated for up to 48 h in flasks. The day cultures were only supplemented with 0.2% xylose if indicated in figure legends. Samples were taken at late exponential phase (OD600 ∼0.6–0.8, typically after 5–6 h), late/very late stationary phase (24 h/48 h post inoculation). Phase contrast pictures were adjusted in brightness and contrast to improve cell shape detection. All GFP-channel pictures were adjusted in brightness and contrast with the identical settings.
Determination of Minimal Inhibitory Concentration
Bacitracin resistance in B. subtilis strains was determined using Etest® strips on bacterial lawns (bioMérieux, Marcy l’Etoile, France), as described previously (Radeck et al., 2016b), with the following changes: (i) MCSEC medium was used instead of MH, (ii) overnight cultures contained 0.2% xylose, and (iii) day cultures, soft agar and agar plates contained 0.2% xylose, if applicable (see figure legends).
Author Contributions
JR and TM conceptualized the study. NL and JR designed the experiments and generated the B. subtilis strains. JR performed MIC assays and coordinated the experimental work, NL performed all remaining experiments. JR and TM wrote the manuscript. All authors approved the manuscript and agree to be accountable for the content of the work.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Vanessa Gilly for obtaining some of the micrographs shown in Supplementary Figures S2, S4 and Philipp Popp for the construction of TMB4201-4205.
Footnotes
Funding. This project was funded by the Deutsche Forschungsgemeinschaft (DFG) priority program SPP1617 ‘Phenotypic Heterogeneity and Sociobiology of Bacterial Populations’ (grant MA 2837/3-2 to TM).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02403/full#supplementary-material
References
- Amiteye S., Kobayashi K., Imamura D., Hosoya S., Ogasawara N., Sato T. (2003). Bacillus subtilis diacylglycerol kinase (DgkA) enhances efficient sporulation. J. Bacteriol. 185 5306–5309. 10.1128/jb.185.17.5306-5309.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Hussey H., Baddiley J. (1972). The mechanism of wall synthesis in bacteria. The organization of enzymes and isoprenoid phosphates in the membrane. Biochem. J. 127 11–25. 10.1042/bj1270011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo E. C., Rios E. M., Fukushima K., Campos-Takaki G. M. (1993). Bacitracin production by a new strain of Bacillus subtilis. Extraction, purification, and characterization. Appl. Biochem. Biotechnol. 42 1–7. 10.1007/BF02788897 [DOI] [PubMed] [Google Scholar]
- Barreteau H., Magnet S., El Ghachi M., Touze T., Arthur M., Mengin-Lecreulx D., et al. (2009). Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877 213–220. 10.1016/j.jchromb.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Bernard R., El Ghachi M., Mengin-Lecreulx D., Chippaux M., Denizot F. (2005). BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J. Biol. Chem. 280 28852–28857. 10.1074/jbc.M413750200 [DOI] [PubMed] [Google Scholar]
- Botella E., Devine S. K., Hubner S., Salzberg L. I., Gale R. T., Brown E. D., et al. (2014). PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol. Microbiol. 94 1242–1259. 10.1111/mmi.12833 [DOI] [PubMed] [Google Scholar]
- Breukink E., de Kruijff B. (2006). Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5 321–332. 10.1038/nrd2004 [DOI] [PubMed] [Google Scholar]
- Breukink E., van Heusden H. E., Vollmerhaus P. J., Swiezewska E., Brunner L., Walker S., et al. (2003). Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 278 19898–19903. 10.1074/jbc.M301463200 [DOI] [PubMed] [Google Scholar]
- Brown S., Santa Maria J. P., Jr., Walker S. (2013). Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67 313–336. 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Helmann J. D. (2002). Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function σ factors. J. Bacteriol. 184 6123–6129. 10.1128/JB.184.22.6123-6129.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote J. G. (1972). Sporulation in Bacillus subtilis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J. Gen. Microbiol. 71 1–15. 10.1099/00221287-71-1-1 [DOI] [PubMed] [Google Scholar]
- Dammeyer T., Tinnefeld P. (2012). Engineered fluorescent proteins illuminate the bacterial periplasm. Comput. Struct. Biotechnol. J. 3:e201210013. 10.5936/csbj.201210013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou N. J., Cocklin S., Loll P. J. (2013). High-resolution crystal structure reveals molecular details of target recognition by bacitracin. Proc. Natl. Acad. Sci. U.S.A. 110 14207–14212. 10.1073/pnas.1308268110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan A. J., Biboy J., van’t Veer I., Breukink E., Vollmer W. (2015). Activities and regulation of peptidoglycan synthases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20150031. 10.1098/rstb.2015.0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W., Helmann J. D. (2008). The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67 830–848. 10.1111/j.1365-2958.2007.06090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghachi M., Bouhss A., Blanot D., Mengin-Lecreulx D. (2004). The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J. Biol. Chem. 279 30106–30113. 10.1074/jbc.M401701200 [DOI] [PubMed] [Google Scholar]
- El Ghachi M., Derbise A., Bouhss A., Mengin-Lecreulx D. (2005). Identification of multiple genes encoding membrane proteins with undecaprenyl pyrophosphate phosphatase (UppP) activity in Escherichia coli. J. Biol. Chem. 280 18689–18695. 10.1074/jbc.M412277200 [DOI] [PubMed] [Google Scholar]
- Fan J., Jiang D., Zhao Y., Liu J., Zhang X. C. (2014). Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B. Proc. Natl. Acad. Sci. U.S.A. 111 7636–7640. 10.1073/pnas.1403097111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton A. K., El Mortaji L., Lau D. T. C., Rudner D. Z., Bernhardt T. G. (2016). CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat Microbiol. 2:16237. 10.1038/nmicrobiol.2016.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guariglia-Oropeza V., Helmann J. D. (2011). Bacillus subtilis σV confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J. Bacteriol. 193 6223–6232. 10.1128/JB.06023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R. T., Ko T. P., Chen A. P., Kuo C. J., Wang A. H., Liang P. H. (2005). Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. J. Biol. Chem. 280 20762–20774. 10.1074/jbc.M502121200 [DOI] [PubMed] [Google Scholar]
- Harwood C. R., Cutting S. M. (1990). Molecular Biological Methods for Bacillus. Chichester: John Wiley & Sons. [Google Scholar]
- Helmann J. D. (2016). Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr. Opin. Microbiol. 30 122–132. 10.1016/j.mib.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y., Strominger J. L., Sweeley C. C. (1970). Biosynthesis of the peptidoglycan of bacterial cell walls. XXI. Isolation of free C55-isoprenoid alcohol and of lipid intermediates in peptidoglycan synthesis from Staphylococcus aureus. J. Biol. Chem. 245 3697–3702. [PubMed] [Google Scholar]
- Inaoka T., Ochi K. (2012). Undecaprenyl pyrophosphate involvement in susceptibility of Bacillus subtilis to rare earth elements. J. Bacteriol. 194 5632–5637. 10.1128/JB.01147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Takoh M., Nishibayashi R., Sato A. (2002). Distribution and variation of bacitracin synthetase gene sequences in laboratory stock strains of Bacillus licheniformis. Curr. Microbiol. 45 18–23. 10.1007/s00284-001-0041-5 [DOI] [PubMed] [Google Scholar]
- Jerga A., Lu Y. J., Schujman G. E., de Mendoza D., Rock C. O. (2007). Identification of a soluble diacylglycerol kinase required for lipoteichoic acid production in Bacillus subtilis. J. Biol. Chem. 282 21738–21745. 10.1074/jbc.M703536200 [DOI] [PubMed] [Google Scholar]
- Jordan S., Hutchings M. I., Mascher T. (2008). Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32 107–146. 10.1111/j.1574-6976.2007.00091.x [DOI] [PubMed] [Google Scholar]
- Jordan S., Junker A., Helmann J. D., Mascher T. (2006). Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188 5153–5166. 10.1128/JB.00310-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., et al. (2011). A widespread family of bacterial cell wall assembly proteins. EMBO J. 30 4931–4941. 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., et al. (2003). Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100 4678–4683. 10.1073/pnas.0730515100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobras C. M., Mascher T., Gebhard S. (2017). “Application of a Bacillus subtilis whole-cell biosensor (PliaI-lux) for the identification of cell wall active antibacterial compounds,” in Antibiotics: Methods and Protocols, ed. Sass P. (New York, NY: Springer; ), 121–131. [DOI] [PubMed] [Google Scholar]
- Kramer N. E., Smid E. J., Kok J., De Kruijff B., Kuipers O. P., Breukink E. (2004). Resistance of Gram-positive bacteria to nisin is not determined by Lipid II levels. FEMS Microbiol. Lett. 239 157–161. 10.1016/j.femsle.2004.08.033 [DOI] [PubMed] [Google Scholar]
- Laddomada F., Miyachiro M. M., Dessen A. (2016). Structural insights into protein-protein interactions involved in bacterial cell wall biogenesis. Antibiotics 5:E14. 10.3390/antibiotics5020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Helmann J. D. (2013). Reducing the level of undecaprenyl pyrophosphate synthase has complex effects on susceptibility to cell wall antibiotics. Antimicrob. Agents Chemother. 57 4267–4275. 10.1128/AAC.00794-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manat G., Roure S., Auger R., Bouhss A., Barreteau H., Mengin-Lecreulx D., et al. (2014). Deciphering the metabolism of undecaprenyl-phosphate: the bacterial cell-wall unit carrier at the membrane frontier. Microb. Drug Resist. 20 199–214. 10.1089/mdr.2014.0035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T., Margulis N. G., Wang T., Ye R. W., Helmann J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50 1591–1604. 10.1046/j.1365-2958.2003.03786.x [DOI] [PubMed] [Google Scholar]
- Mascher T., Zimmer S. L., Smith T. A., Helmann J. D. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48 2888–2896. 10.1128/AAC.48.8.2888-2896.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey M. A., Troy F. A. (1980). Paramagnetic isoprenoid carrier lipids. 2. Dispersion and dynamics in lipid membranes. Biochemistry 19 2061–2066. 10.1021/bi00551a009 [DOI] [PubMed] [Google Scholar]
- Meeske A. J., Rodrigues C. D., Brady J., Lim H. C., Bernhardt T. G., Rudner D. Z. (2016). High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLOS Biol. 14:e1002341. 10.1371/journal.pbio.1002341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske A. J., Sham L. T., Kimsey H., Koo B. M., Gross C. A., Bernhardt T. G., et al. (2015). MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 112 6437–6442. 10.1073/pnas.1504967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnig K., Barblan J. L., Kehl S., Moller S. B., Mauel C. (2003). In Bacillus subtilis W23, the duet σX σM, two sigma factors of the extracytoplasmic function subfamily, are required for septum and wall synthesis under batch culture conditions. Mol. Microbiol. 49 1435–1447. 10.1046/j.1365-2958.2003.03652.x [DOI] [PubMed] [Google Scholar]
- Muchova K., Wilkinson A. J., Barak I. (2011). Changes of lipid domains in Bacillus subtilis cells with disrupted cell wall peptidoglycan. FEMS Microbiol. Lett. 325 92–98. 10.1111/j.1574-6968.2011.02417.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335 1103–1106. 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- Ohki R., Giyanto, Tateno K., Masuyama W., Moriya S., Kobayashi K.,et al. (2003). The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49 1135–1144. 10.1046/j.1365-2958.2003.03653.x [DOI] [PubMed] [Google Scholar]
- Peters J. M., Colavin A., Shi H., Czarny T. L., Larson M. H., Wong S., et al. (2016). A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165 1493–1506. 10.1016/j.cell.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Coote J. G. (1976). Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40 908–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeck J., Fritz G., Mascher T. (2016a). The cell envelope stress response of Bacillus subtilis: from static signaling devices to dynamic regulatory network. Curr. Genet. 63 79–90. 10.1007/s00294-016-0624-0 [DOI] [PubMed] [Google Scholar]
- Radeck J., Gebhard S., Orchard P. S., Kirchner M., Bauer S., Mascher T., et al. (2016b). Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol. Microbiol. 100 607–620. 10.1111/mmi.13336 [DOI] [PubMed] [Google Scholar]
- Radeck J., Kraft K., Bartels J., Cikovic T., Dürr F., Emenegger J., et al. (2013). The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J. Biol. Eng. 7:29. 10.1186/1754-1611-7-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietkötter E., Hoyer D., Mascher T. (2008). Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68 768–785. 10.1111/j.1365-2958.2008.06194.x [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning - A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schmalisch M., Maiques E., Nikolov L., Camp A. H., Chevreux B., Muffler A., et al. (2010). Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol. 192 5402–5412. 10.1128/JB.00534-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert G., Strominger J. L. (1967). Bacitracin: an inhibitor of the dephosphorylation of lipid pyrophosphate, an intermediate in the biosynthesis of the peptidoglycan of bacterial cell walls. Proc. Natl. Acad. Sci. U.S.A. 57 767–773. 10.1073/pnas.57.3.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm D. R., Strominger J. L. (1973). Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248 3940–3945. [PubMed] [Google Scholar]
- Thackray P. D., Moir A. (2003). SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185 3491–3498. 10.1128/JB.185.12.3491-3498.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45 D158–D169. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C. L., Shaw G. C. (2008). Genetic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis. J. Bacteriol. 190 1561–1567. 10.1128/JB.01497-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A., Banzhaf M., Gross C. A., Vollmer W. (2012). From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10 123–136. 10.1038/nrmicro2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V., Dervyn E., Ehrlich S. D. (1998). A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144(Pt 11), 3097–3104. 10.1099/00221287-144-11-3097 [DOI] [PubMed] [Google Scholar]
- Van Horn W. D., Sanders C. R. (2012). Prokaryotic diacylglycerol kinase and undecaprenol kinase. Annu. Rev. Biophys. 41 81–101. 10.1146/annurev-biophys-050511-102330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby E., Higashi Y., Strominger J. L. (1972). Enzymatic dephosphorylation of C55-isoprenylphosphate. J. Biol. Chem. 247 5113–5115. [PubMed] [Google Scholar]
- Wolf D., Dominguez-Cuevas P., Daniel R. A., Mascher T. (2012). Cell envelope stress response in cell wall-deficient L-forms of Bacillus subtilis. Antimicrob. Agents Chemother. 56 5907–5915. 10.1128/AAC.00770-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D., Kalamorz F., Wecke T., Juszczak A., Mäder U., Homuth G., et al. (2010). In-depth profiling of the LiaR response of Bacillus subtilis. J. Bacteriol. 192 4680–4693. 10.1128/JB.00543-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D., Mascher T. (2016). The applied side of antimicrobial peptide-inducible promoters from Firmicutes bacteria: expression systems and whole-cell biosensors. Appl. Microbiol. Biotechnol. 100 4817–4829. 10.1007/s00253-016-7519-3 [DOI] [PubMed] [Google Scholar]
- Zhao H., Sun Y., Peters J. M., Gross C. A., Garner E. C., Helmann J. D. (2016). Depletion of undecaprenyl pyrophosphate phosphatases disrupts cell envelope biogenesis in Bacillus subtilis. J. Bacteriol. 198 2925–2935. 10.1128/JB.00507-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweers J. C., Nicolas P., Wiegert T., van Dijl J. M., Denham E. L. (2012). Definition of the σW regulon of Bacillus subtilis in the absence of stress. PLOS ONE 7:e48471. 10.1371/journal.pone.0048471 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.