Abstract
We describe experiments that follow species dynamics and gene expression patterns in synthetic bacterial communities including species that compete for the single carbon substrate supplied, methane, and species unable to consume methane, which could only succeed through cooperative interactions. We demonstrate that these communities mostly select for two functional guilds, methanotrophs of the family Methylococcaceae and non-methanotrophic methylotrophs of the family Methylophilaceae, these taxonomic guilds outcompeting all other species included in the synthetic mix. The metatranscriptomics analysis uncovered that in both Methylococcaceae and Methylophilaceae, some of the most highly transcribed genes were the ones encoding methanol dehydrogenases (MDH). Remarkably, expression of alternative MDH genes (mxaFI versus xoxF), previously shown to be subjects to the rare Earth element switch, was found to depend on environmental conditions such as nitrogen source and methane and O2 partial pressures, and also to be species-specific. Along with the xoxF genes, genes encoding divergent cytochromes were highly expressed in both Methylophilaceae and Methylococcaceae, suggesting their function in methanol metabolism, likely encoding proteins serving as electron acceptors from XoxF enzymes. The research presented tested a synthetic community model that is much simplified compared to natural communities consuming methane, but more complex than the previously utilized two-species model. The performance of this model identifies prominent species for future synthetic ecology experiments and highlights both advantages of this approach and the challenges that it presents.
Keywords: synthetic community, methylotrophy, methanotrophs, Methylobacter, Methylosarcina, Methylomonas, Methylophilaceae, Lake Washington
Introduction
Metabolism of methane is an important part of biogeochemical cycling of carbon (Singh et al., 2010). Methane is also a major contributor to climate change (Schuur et al., 2015). A specialized group of microbes, the methanotrophs that consume methane, gaining both energy and carbon from this compound, represent a natural filter preventing an even faster escape of methane into the atmosphere (Malyan et al., 2016). While methanotrophy has been studied for the past 100 years as a metabolic feature of individual pure cultures (Kaserer, 1906; Söhngen, 1906; Trotsenko and Murrell, 2008; Chistoserdova and Lidstrom, 2013), a concept of communal function in methanotrophy has been gaining momentum (Ho et al., 2016; Yu and Chistoserdova, 2017). The mechanistic details are still scarce with regard to how and why the methanotrophs share their carbon with other species, and whether and what they gain in return (Yu and Chistoserdova, 2017). We have previously reported that feeding 13C-labeled methane to natural communities of Lake Washington sediment resulted in label accumulation mainly by the Methylococcaceae and the Methylophilaceae species (Kalyuzhnaya et al., 2008; Beck et al., 2013). Through microcosm manipulation, using methane as the sole source of carbon, followed by metagenomic analysis, we further confirmed that the Methylococcaceae and the Methylophilaceae were active in methane consumption (Hernandez et al., 2015; Oshkin et al., 2015). A more recent study has identified methanol as one metabolic node at which community cross-talk may be taking place (Krause et al., 2017), suggesting that methanol may be a major carbon source released by the methanotrophs, to support satellite communities. In most Gram-negative methylotrophs, methanol oxidation can be carried out by two alternative methanol dehydrogenases (MDH), the classic, MxaFI-type enzyme that has been studied for decades (Anthony, 2004; Williams et al., 2005) and the recently discovered, lanthanide (Ln3+)-dependent, XoxF-type MDH (Fitriyanto et al., 2011; Nakagawa et al., 2012; Pol et al., 2014). Moreover, Ln3+ have been implicated in a regulatory mechanism inversely controlling expression of xoxF and mxaFI genes, this mechanism known as the rare Earth element (REE) switch (Chu et al., 2016). This REE switch has been proposed to be an important factor in community function (Krause et al., 2017). Methanotrophs have also been proposed to excrete multicarbon compounds such as acetate, citrate, lactate, and succinate (Modin et al., 2007; Kalyuzhnaya et al., 2013), thus having a potential of also supporting communities of non-methylotrophic heterotrophs. A mathematical model has been recently implemented to explain carbon flow in microbial consortia consuming methane (Modin, 2017). In accordance with this model, methanotrophs feed methanol to methanol utilizers, and both methanotrophs and methanol utilizers produce organics that feed non-methylotrophic heterotrophs, in conjunction with denitrification. In accordance with the model, methanol should be available to all organisms that are capable of methanol utilization, and other organics should be available to all heterotrophs, as “public goods.” However, experimental evidence is somewhat contradictory to the notion of “public goods,” as specific species cooccurrences, such as cooperative behavior of Methylococcaceae and Methylophilaceae, have been noted not only in manipulated microcosms (Kalyuzhnaya et al., 2008; Beck et al., 2013; Hernandez et al., 2015; Oshkin et al., 2015), but also in natural populations inhabiting methane-rich environments such as permafrosts (Martineau et al., 2010; Crevecoeur et al., 2015) or methane-fueled cave biofilms (Karwautz et al., 2017). These results suggest either that many species detected in natural niches, through DNA profiling, are dormant, that some organisms may be more competitive for “public goods,” or that specific partnerships are taking place. A Black Queen Hypothesis, in accordance with which gene loss plays a role in species coevolution (Morris et al., 2012), has been evoked recently to explain non-random distribution of “public goods” among species involved in a cyanobacterial consortium, and vitamin B12 was proposed as an important metabolite (Lee et al., 2017). Vitamin B12 exchange has been previously implicated in maintaining stable cocultures of methanotrophs and non-methanotrophs (Iguchi et al., 2011).
In this manuscript, we describe experiments addressing the following questions: (1) whether the methanotrophs share carbon with non-methanotrophs as “public goods,” or whether they form specific partnerships, (2) whether all species capable of metabolizing methanol are equally competitive when present in physiologically active state, (3) whether non-methylotrophic heterotrophs also benefit from methane-derived carbon on “public goods” basis, and (4) whether transcription of any key methylotrophy genes changes in response to different environmental conditions. In these experiments, we employed synthetic communities (SCs) built of pure cultures of methanotrophs and non-methanotrophs, all previously isolated from the same ecological niche. A small subsample of these experiments has been previously published, to demonstrate the utility of SCs in basic research (Yu et al., 2016).
Materials and Methods
Bacterial Strains, Growth Conditions, and Experimental Setup
Bacterial strains employed in this study are listed in Table 1 along with their characteristics relevant to methylotrophy. All have been previously isolated from Lake Washington sediment. The cultures were grown on solid media. The methanotrophs were cultivated on the nitrate minimal salts (NMS) medium in the atmosphere of methane (Dedysh and Dunfield, 2014; 25% methane, 75% air V/V). Methylotenera mobilis JLW8, Methylotenera sp. G11, and the Gram-positive methylotrophs were cultivated on the NMS medium supplemented with methylamine (0.2% W/V). The remaining methylotrophs were cultivated on the NMS medium supplemented with methanol (0.2% V/V). For the Rhodocyclaceae, vitamin B12 was added at 0.5 μg L-1 (Smalley et al., 2015). The non-methylotrophic heterotrophs were grown on diluted nutrient broth (BD Difco) medium (1/2 strength). Cultures were allowed to grow at room temperature (approximately 24°C) for 2–3 days, till they formed nice biofilms on the plates, after which the biomass was washed with the NMS medium, and optical density and/or cell counts were quantified, respectively, via spectrophotometry or flow cytometry (see Krause et al., 2017). The two exceptions were the Methylobacter strains, which were grown at 20°C, as they are psychrophilic (Kalyuzhnaya et al., 2015). The 50 strains were mixed at desired proportions (see below), and the master mixtures were incubated at 4°C for 48 h, to mimic starvation. Synthetic community 1 (SC1) master mixture was assembled by mixing the 50 strains in equal proportions, based on optical density (OD600). The SC2 master mixture was assembled of the same strains, as part of a separate experiment, where they were mixed based on cell counts, as determined by flow cytometry. The main difference between SC1 and SC2 was in the proportion of the methanotrophs and the Bacillus strains, whose cells have larger size, thus they were less abundant in SC1 compared to SC2. Incubations were carried out at 18°C in 250 mL vials, with liquid-to-headspace ratio of 1–4, as previously described (Hernandez et al., 2015; Oshkin et al., 2015). Initial inoculum OD600 was 0.1. Gas phase was refreshed daily according to the following scheme. (1) For the high methane high O2 (HH) regimen, the headspace was flushed with air for 60 s (flow rate > 1 L/min), followed by removal of 50 mL of the headspace and replacement with 50 mL of methane; (2) for the high methane low O2 (HL) regimen, the headspace was flushed with N2 for 60 s (flow rate > 1.28 L/min), followed by the removal of 60 mL of headspace and replacement with 50 mL of methane and 10 ml of air; (3) for the low methane high O2 (LH) regimen, the headspace was flushed with air for 60 s, followed by removal of 1 mL of headspace and replacement with 1 ml of methane; and (4) for the low methane low oxygen (LL) regimen, headspace was flushed with N2 for 60 s, followed by removal of 2 mL of headspace and replacement with 1 mL of methane and 1 mL of air. When cultures reached OD600 of approximately 0.5, they were transferred with 10-fold dilution into fresh medium. For the HH regimens, cells were transferred every 2–3 days, for the HL regimens, cells were transferred every 9–20 days, and for the LH and LL regimens, cells were transferred every 19–42 days. For the HH and HL regimens, cells from 11 time points were used for iTag profiling (Illumina sequencing of 16S rRNA gene fragment; Singer et al., 2016), for the LH and LL regimens, cells from five time points were used. For sample map and sampling schedule see Supplementary Table S1. Cells for DNA isolation (3 mL) were collected by centrifugation at 15,000 rpm for 5 min, and pellets were used immediately or stored at -20°C. Cells for RNA isolation (40 mL) were collected by centrifugation at 5,000 rpm for 30 min with the 1:10 volume of stop solution (5% buffer-saturated phenol in ethanol) added to cells prior to harvesting.
Table 1.
Bacterial strains employed in the study.
| Organism/strain | Phylum/class | Reference |
|---|---|---|
| The methanotrophs | ||
| Methylomonas sp. LW13 | Proteobacteria/Gammaproteobacteria | Kalyuzhnaya et al., 2015 |
| Methylomonas sp. MK1 | Kalyuzhnaya et al., 2015 | |
| Methylomonas sp. 11b | Kalyuzhnaya et al., 2015 | |
| Methylobacter tundripaludum 21/22 | Kalyuzhnaya et al., 2015 | |
| Methylobacter tundripaludum 31/32 | Kalyuzhnaya et al., 2015 | |
| Methylosarcina lacus LW14 | Kalyuzhnaya et al., 2015 | |
| Methylosinus sp. PW1 | Proteobacteria/Alphaproteobacteria | Beck et al., 2015 |
| Methylosinus sp. LW3 | Beck et al., 2015 | |
| Methylosinus sp. LW4 | Beck et al., 2015 | |
| Methylosinus sp. LW5 | Beck et al., 2015 | |
| Beck et al., 2015 | ||
| The Methylophilaceae | ||
| Methylotenera mobilis JLW8 | Proteobacteria/Betaproteobacteria | Lapidus et al., 2011 |
| Methylotenera versatilis 301 | Lapidus et al., 2011 | |
| Methylovorus glucosotrophus SIP3-4 | Lapidus et al., 2011 | |
| Methylotenera mobilis 13 | Beck et al., 2014 | |
| Methylophilaceae 73s | Beck et al., 2014 | |
| Methylophilaceae 11 | Beck et al., 2014 | |
| Methylophilus methylotrophus 1 | Beck et al., 2014 | |
| Methylophilus methylotrophus 5 | Beck et al., 2014 | |
| Methylophilus methylotrophus Q8 | McTaggart et al., 2015b | |
| Methylotenera sp. G11 | McTaggart et al., 2015b | |
| Methylophilaceae 7 | McTaggart et al., 2015b | |
| Other methanol utilizers | ||
| Ancylobacter sp. 117 | Proteobacteria/Alphaproteobacteria | Beck et al., 2015 |
| Ancylobacter sp. 202 | Beck et al., 2015 | |
| Ancylobacter sp. 501b | Beck et al., 2015 | |
| Hyphomicrobium sp. 99 | Beck et al., 2015 | |
| Hyphomicrobium sp. 802 | Beck et al., 2015 | |
| Labrys methylaminiphilus JLW10 | Beck et al., 2015 | |
| Methylobacterium sp. 10 | Beck et al., 2015 | |
| Methylobacterium sp. 77 | Beck et al., 2015 | |
| Methylobacterium sp. 88A | Beck et al., 2015 | |
| Methylopila sp. 73B | Beck et al., 2015 | |
| Methylopila sp. 107 | Beck et al., 2015 | |
| Paracoccus sp. N5 | Beck et al., 2015 | |
| Xanthobacter sp. 91 | Beck et al., 2015 | |
| Xanthobacter sp. 126 | Beck et al., 2015 | |
| Methyloversatilis discipulorum FAM1 | Proteobacteria/Betaproteobacteria | Smalley et al., 2015 |
| Methyloversatilis discipulorum RZ18-153 | Smalley et al., 2015 | |
| Methyloversatilis universalis FAM5 | Smalley et al., 2015 | |
| Non-methanol utilizing methylotrophs | ||
| Aminobacter sp. 108 | Proteobacteria/Alphaproteobacteria | Beck et al., 2015 |
| Arthrobacter sp. 31Y | Actinobacteria/Actinobacteria | McTaggart et al., 2015a |
| Arthrobacter sp. 35W | McTaggart et al., 2015a | |
| Arthrobacter sp. MA-N2 | McTaggart et al., 2015a | |
| Mycobacterium sp. 141 | McTaggart et al., 2015a | |
| Mycobacterium sp. 155 | McTaggart et al., 2015a | |
| Bacillus sp. 37MA | Firmicutes/Bacilli | McTaggart et al., 2015a |
| Bacillus sp. 72 | McTaggart et al., 2015a | |
| Non-methylotrophic heterotrophs | ||
| Pseudomonas sp. 11/12A | Proteobacteria/Gammaproteobacteria | McTaggart et al., 2015c |
| Janthinobacterium sp. RA13 | Proteobacteria/Betaproteobacteria | McTaggart et al., 2015d |
| Flavobacterium sp. 83 | Bacteroidetes/Flavobacteria | McTaggart et al., 2015e |
| Flavobacterium sp. Fl | McTaggart et al., 2015e | |
16S rRNA Gene Amplicon and Transcript Sequencing and Analysis
DNA was isolated using the FastDNA SPIN Kit for Soil (MP Biomedicals), according to the manufacturer’s protocol. Total RNA was isolated using the RNeasy minikit (Qiagen) as previously described (Chu et al., 2016). The purified RNA was tested for DNA contamination using 16S rRNA gene fragment PCR. Samples were stored at -80°C for subsequent analyses. iTag sequencing as well as transcript sequencing were performed at the DOE Joint Genome Institute (JGI) using standard JGI pipelines. Briefly, primers for hypervariable rRNA gene regions V4–V5 were used for iTag profiling (Parada et al., 2016). The primer sets contained Illumina linkers; a 12 bp barcode index; a pad region; a 0, 1, 2, or 3 base pair long spacer and the sequence-specific primer. Samples were pooled and run on two 2 by 300 bp Illumina MiSeq runs, essentially as described (Singer et al., 2016). RNAseq sequencing was carried out essentially as described (Beam et al., 2016).
iTag data have been archived with the IMG genome portal under JGI Project IDs 1105725 and 1105727. Transcript sequences have been archived with the IMG Genome Portal IMG genome IDs 3300009726, 3300009727, 3300009729, 3300009731, 3300009733, 3300009734, 3300009736, 3300009737, 3300009740, 3300009741, 3300009746, 3300009749, 3300009751, and 3300009752. Raw reads were aligned to the 50 respective reference genomes (see Table 1 for reference) using the Burrows–Wheeler alignment tool version 0.7.12-r1039 (Li and Durbin, 2009), using default parameters. Raw read counts are shown in Supplementary Table S2. Gene expression for organisms/genes recruiting large numbers of reads were normalized manually, by calculating numbers of reads per million of total reads mapping to a specific genome and normalizing per gene length (kb).
Bioinformatics
The UPARSE method was used for sequence processing and OTU clustering with USEARCH version 9.2.64 (Edgar, 2013). Clustering was performed at 97% sequence identity and chimeras were identified against the ChimeraSlayer reference database in the Broad Microbiome Utilities version r20110519 obtained from the UCHIME distribution (Edgar et al., 2011). Taxonomic assignments were made using the RDP Classifier from the Ribosomal Database Project version 2.12 (Wang et al., 2007). The samples were normalized so that the numbers of reads in each sample were equal.
Phylogenetic Analysis
Protein sequences were aligned using the CLUSTAL W algorithm, and phylogenetic trees were constructed using the maximum-likelihood method, as implemented in the MEGA7 software (Kumar et al., 2016). Statistical support was obtained from 1,000 bootstrap replicates (bootstrap values >50% are reported).
Results
Community Dynamics in Time-Series Experiments Identify Methylococcaceae As the Most Competitive Methanotrophs and Methylophilaceae As Their Most Successful Satellite Species, under a Range of Simulated Environmental Conditions
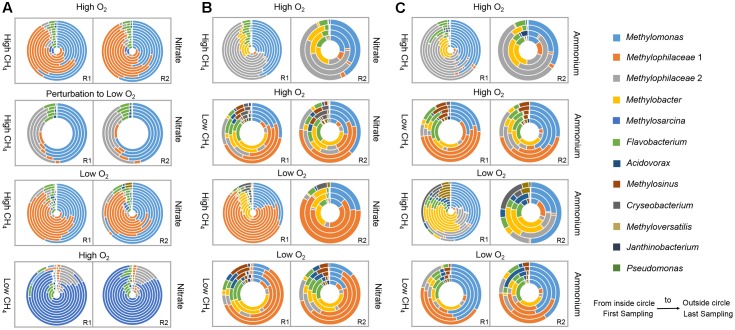
To address the questions posed, we first created SC1, by mixing 50 pure cultures of bacteria representing different functional guilds, all originating from Lake Washington sediment (Table 1). A single master mix was used to inoculate all vials. SC1 was incubated under three simulated environmental conditions: HH, HL, and LH (see section “Materials and Methods” and Supplementary Table S1 for details), in two replicates, nitrate being the nitrogen source in all cases. iTag sequence profiles were generated for 11 time points for each condition, reflecting community dynamics over, respectively, 27, 141, and 212 days (Supplementary Table S1). Under the HH condition, communities were mostly represented by two major OTUs, Methylophilaceae 1 (30–67% of total reads) and Methylomonas (12–62% of total reads), the proportion of Methylomonas increasing with time (Figure 1A). Methylosarcina OTU was detected in early samples (up to 23% of total reads), and the second Methylophilaceae OTU, Methylophilaceae 2 was detected across samples, its relative abundance declining with time (Figure 1A). An OTU representing Flavobacterium was detected in all samples, its relative abundance declining with time. To test for response to perturbation, a switch from HH to HL at day 28 was analyzed additionally. Community response included the switch between the Methylophilaceae OTUs, Methylophilaceae 2 gradually replacing Methylophilaceae 1, with overall increase in relative abundance of Methylophilaceae (Figure 1A).
FIGURE 1.
Species dynamics over time, based on iTag profiling. (A) SC1, incubated under three environmental conditions (two technical replicates, R1 and R2 are shown). The HH communities were perturbed by switching to HL regimen (second panel from top); (B) SC2, incubated under four environmental conditions, with nitrate as a nitrogen source; (C) SC2, incubated under four environmental conditions, with ammonium as a nitrogen source. Each circle represents community composition at a specific sampling point (5–11 time-series samples were analyzed for each microcosm). Refer to Supplementary Table S1 for sampling schedule.
The HL condition also selected for very simple community structure, dominated by Methylophilaceae 1 and Methylomonas, with minor presence of Methylosarcina, Methylophilaceae 2, and Flavobacterium (Figure 1A). An OTU representing Methyloversatilis was detected in some samples at over 2% abundance. Surprisingly, an OTU representing Acidovorax was detected in some samples at up to 5% abundance, even though Acidovorax was not intentionally added to the synthetic mix. Propagation of this organism must be a result of a minor contamination present in one of the frozen culture stocks.
Species composition was dramatically different under the LH condition, with Methylosarcina rapidly outcompeting all other methanotroph species, and with Methylophilaceae 2 outcompeting Methylophilaceae 1 (Figure 1A). An OTU representing Methylobacter was detected in SC1 samples only at very low abundance. Overall, the community dynamics and trajectories were very well replicated under each regimen (Figure 1A).
SC2 was built independently, of the same pure cultures revived from frozen stocks. With SC2, we expanded the range of simulated conditions to include nitrogen source as a variable, substituting ammonium for nitrate, and including an LL condition, a total of eight conditions. The HH and HL regimen communities were sampled at 11 time points, while the LH and LL regimen communities were sampled at five time points (Supplementary Table S1).
Community dynamics under the HH regimen were similar whether nitrate or ammonium were used as nitrogen sources (Figures 1B,C). In both cases, the Methylomonas types gradually outcompeted the Methylobacter types, and Methylophilaceae 2 gradually outcompeted Methylophilaceae 1 (Figures 1B,C). In contrast, community dynamics appeared to be different under the HL regimen, dependent on nitrogen source. While in both conditions, Methylomonas gradually outcompeted Methylobacter, in the nitrate-supplemented medium, Methylophilaceae were represented almost entirely by Methylophilaceae 1, and the sum of Methylococcaceae and Methylophilaceae made up to over 90 and up to 97% of the entire community (Figure 1B). However, in the ammonium-supplemented medium, Methylophilaceae were represented by Methylophilaceae 2, and the sum of Methylococcaceae and Methylophilaceae made up to no more than 80% of the community, other relatively abundant OTUs representing Flavobacterium, Methyloversatilis, and Acidovorax (Figure 1C).
Community dynamics under low methane (LH and LL conditions) were similar to each other, independently of O2 concentration or nitrogen source. Methylomonas again outcompeted Methylobacter. However, in most cases, Methylophilaceae 1 prevailed over Methylophilaceae 2. An exception was one replicate under the LHNH4 regimen, in which abundances of both Methylophilaceae OTUs increased with time. Under low methane, the sum of Methylococcaceae and Methylophilaceae made up approximately 80% of the total community, other abundant OTUs representing Flavobacterium, Methylosinus, and Acidovorax. Under the LHNO3 regimen, we saw a significant population of Chryseobacterium (up to 13%), another organism that must have originated from a minor contamination of one of the frozen stocks. Notably, Methylosarcina was not detected in SC2 samples. Data for technical replicates, again, agreed very well (Figures 1B,C).
To summarize, while alternative major players within Methylococcaceae and Methylophilaceae were involved in community dynamics, especially dramatically contrasting the “high” versus “low” methane conditions, these two guilds were the dominant species in methane consumption, corroborating our previous results (Hernandez et al., 2015; Oshkin et al., 2015). As in our previous experiments, the Methylocystaceae species did not appear competitive under the “high” methane regimens. They were also gradually outcompeted by Methylococcaceae under “low” methane regimens.
Metatranscriptomics Provide Strain-Resolved Data on Community Composition, Identify Major Expressed Pathways, Pinpoint Alternative Methanol Dehydrogenases As Major Subjects to Regulation
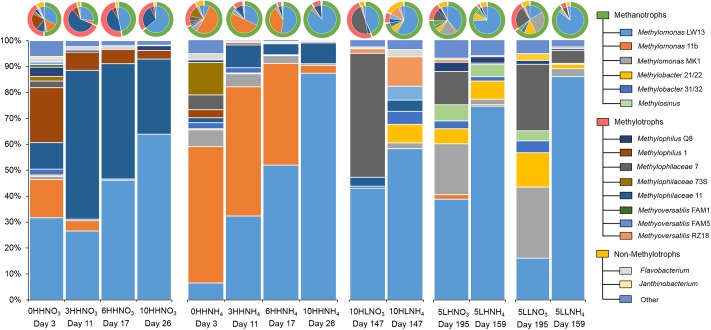
Strain-resolved community compositions of SC2 microcosms were determined through transcript analysis of 14 samples (see Supplementary Table S1). These represented four time points for the two HH regimens (time series), and one time point for each of the remaining six regimens (Figure 2). Dynamics among the Methylococcaceae strains were dominated by Methylomonas strains under the HH regimens, with Methylomonas sp. LW13 gradually outcompeting Methylomonas sp. 11b and Methylomonas sp. MK1. However, the dynamics over time differed dependent on the source of nitrogen (Figure 2). Few reads were mapped to the Methylobacter genomes under these regimens, even though the Methylobacter OTU was relatively abundant at sampling points 1 and 3 in iTag analysis (Figures 1B,C). This suggests that Methylobacter species may have been transcriptionally inactive at the time of sampling. However, Methylobacter transcripts were detected under the HLNH4 and the LH and the LL regimens, in proportions predicted via the iTag analysis. Methylosinus transcripts were relatively abundant under the LH regimens and also under the LLNO3 regimen. Among the Methylophilaceae, Methylophilus species dominated over other Methylophilaceae under the HH regimens, Methylophilus methylotrophus Q8 gradually outcompeting M. methylotrophus 1. Methylophilaceae 7 was the dominant Methylophilaceae type under the HL and LH regimens when nitrate was supplied as the nitrogen source, and under both LL regimens. Methyloversatilis species outcompeted Methylophilaceae under the HLNH4 regimen. Overall, abundance of the methanotroph transcripts as a fraction of total transcripts was higher in samples supplemented with ammonium (75–95%) compared to samples supplemented with nitrate (30–75%), likely due to transcription patterns responsive to the nitrogen source, as discussed below.
FIGURE 2.
Strain-resolved transcript abundances for major expressing species, as fractions of total transcripts. For Methylosinus, transcripts for all four strains were combined. On top, pie charts represent the same data, along with broad-function guild abundances (outside circles). Sample designations (at the bottom) refer to number of transfers, followed by methane and O2 regimens (H or L), followed by the nitrogen source. The day of sampling from the beginning of the experiment is also indicated for each sample. For more information, see Supplementary Table S1.
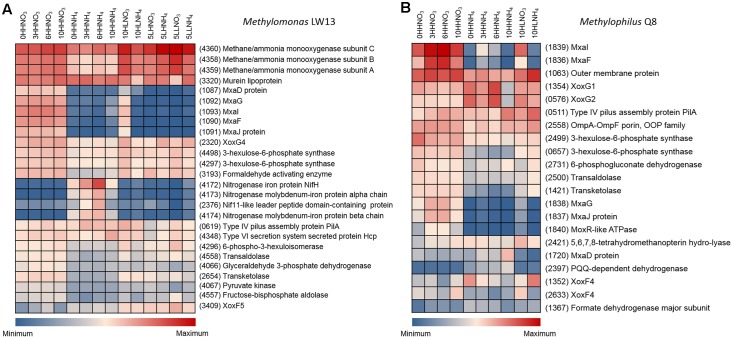
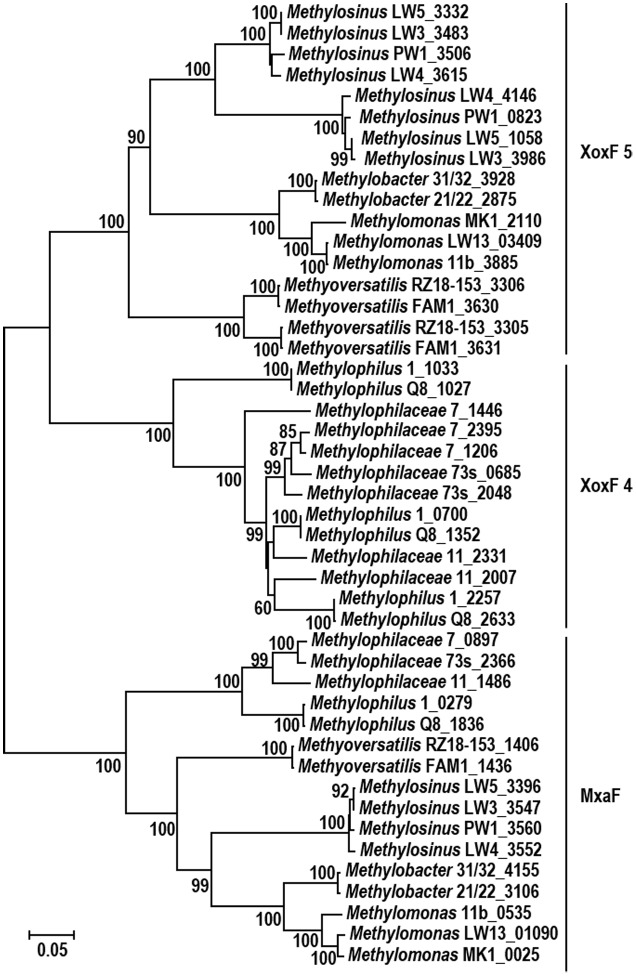
The most highly transcribed genes in the genomes of major species were determined through normalizing the number of transcript reads per gene length as a fraction of each species-specific transcriptome (Figure 3, Supplementary Figures S1–S4 and Table S2). Not surprisingly, genes encoding subunits of the particulate methane monooxygenase (pmoCAB) were most highly transcribed in the methane oxidizing species, both Methylococcaceae and Methylocystaceae, when nitrate served as the nitrogen source (Supplementary Table S2). Other methylotrophy functions were highly expressed in these species, as well as in the Methylophilaceae and the Rhodocyclaceae (Methyloversatilis). Remarkably, in all methylotrophs, some of the genes most responsive to specific experimental conditions were the genes encoding alternative MDH enzymes, MxaFI and XoxF. While the methylotroph genomes contain a single copy of each mxaF and mxaI (encoding the large and the small subunits of the Ca2+-dependent MDH; see Table 1 for genome reference) the number of xoxF genes (encoding the single subunit of the Ln3+-dependent MDH) is variable. The Methylococcaceae possess one (Kalyuzhnaya et al., 2015), the Methylocystaceae and Rhodocyclaceae possess two (Beck et al., 2015; Smalley et al., 2015), and the Methylophilaceae possess up to three (Beck et al., 2014). The phylogenetic relationships of MxaF and XoxF have been reviewed previously (Chistoserdova, 2011; Keltjens et al., 2014). The expression patterns were found very distinct dependent on whether nitrate or ammonium were supplied, or whether “low” or “high” concentrations of O2 and methane were used (Figure 3 and Supplementary Figures S1–S4). Moreover, expression patterns differed among different species and among different MDH enzymes. While the Methylococcaceae, Methylocystaceae, Rhodocyclaceae, and the Methylophilus species expressed mxaFI genes at very high levels when “high” O2, “high” methane, and nitrate were supplied, they expressed xoxF genes at higher levels when ammonium was supplied instead, or when methane was limited. Under the HLNO3 regimen, genes for both enzymes were expressed at approximately equal levels. In contrast, Methylophilaceae other than Methylophilus (Methylophilaceae strains 7, 73s, 11) displayed higher expression of xoxF genes under all regimens. For example, at an early sampling point under the “high” methane, “high” O2, nitrate, while Methylomonas, Methylobacter, Methylophilus, and Methyloversatilis were expressing genes for MxaFI at higher levels, non-Methylophilus Methylophilaceae species were expressing one or two genes encoding XoxF-type MDH enzymes at higher levels (Figure 4, Supplementary Figures S3, S4 and Table S2). Differential mxaF expression in response to the nitrogen source was confirmed on pure cultures of selected species, by real-time quantitative PCR (Supplementary Figure S5). It is noteworthy that the xoxF genes expressed by different community partners belong to two different phylogenetic clades (Chistoserdova, 2011), while Methylococcaceae, Methylocystaceae, and Rhodocyclaceae possess and express xoxF5, the Methylophilaceae possess and express xoxF4, and these genes are so far unique to this latter group (Figure 5).
FIGURE 3.
Heatmaps of some of the most highly/most differentially transcribed genes in (A) Methylomonas sp. LW13 and (B) Methylophilus methylotrophus Q8. Sample designations are as in Figure 2. In parentheses, protein numbers are shown as per genome annotation. See Table 1 for reference. See Supplementary Figures S1–S4 for highly transcribed genes in other major species.
FIGURE 4.
Heatmap depicting expression of xoxF and mxaF genes in major species. On the left, genus and strain names. On the right, gene name followed by gene number as per genome annotation (see Table 1 for reference). Sample designations are as in Figure 2.
FIGURE 5.
Evolutionary relationships between XoxF4, XoxF5, and MxaF proteins present in the species relevant to this study [for XoxF classification, refer to Chistoserdova (2011) and Keltjens et al. (2014)]. Genus names and strain names are shown, followed by protein numbers as per genome annotation (see Table 1 for reference).
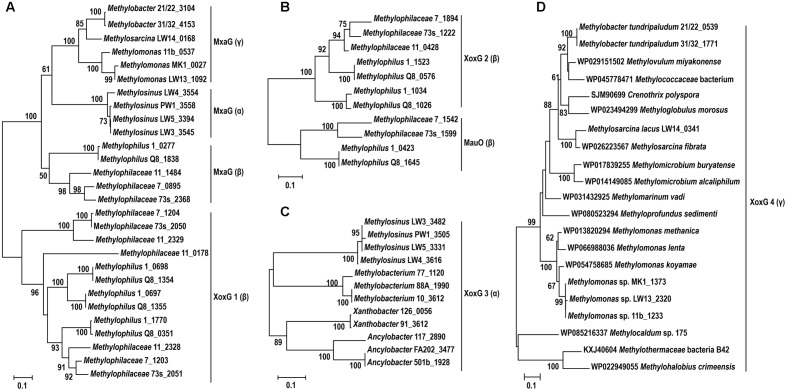
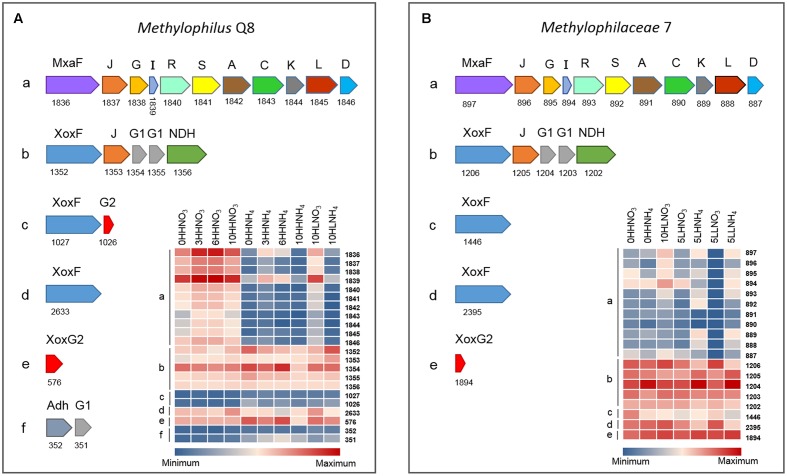
In addition to the multiple genes for the MDH enzymes, genes for multiple cytochromes were highly and differentially expressed. Cytochrome MxaG, whose gene (mxaG) is cotranscribed with mxaFI genes, has been previously established as the electron acceptor from the MxaFI type MDH (Williams et al., 2006). While electron acceptors from XoxF enzymes have not been experimentally demonstrated, genes encoding cytochromes are colocalized on the chromosomes of alphaproteobacterial methylotrophs and Methylophilaceae (Beck et al., 2014, 2015). The arrangement is especially complex in Methylophilaceae, as they encode multiple cytochromes that belong to two phylogenetic lineages, XoxG1 and XoxG2, the former sharing homology with MxaG cytochromes, and the latter sharing homology with the cytochrome with a proposed function in methylamine oxidation (Kalyuzhnaya et al., 2008; Figure 6). In each case, genes for cytochromes representing the two phylogenetic groups were found co-expressed with specific xoxF genes, suggesting that they may be the natural electron acceptors from XoxF enzymes. Note that the coexpressing xoxF and xoxG genes did not always colocalize on the chromosomes (Figure 7).
FIGURE 6.
Evolutionary relationships between different XoxG proteins present in the species relevant to this study. (A–C) Genus names and strain names are shown, followed by protein numbers as per genome annotation (see Table 1 for reference). (D) Full names are shown for organisms used in this study and for organisms whose sequences were retrieved from databases. Note that mxaG genes are found in α-, β-, and γ-proteobacteria (noted in parentheses after the gene name). xoxG1 and xoxG2 are unique to β-proteobacteria, xoxG3 are unique to α-proteobacteria, and xoxG4 are unique to γ-proteobacteria.
FIGURE 7.
Arrangement on the chromosomes and differential expression of different xoxF and xoxG genes in different Methylophilaceae species (A) Methylophilus sp. Q8, (B) Methylophilaceae 7. Protein numbers are used in the heatmaps that correspond to protein numbers in gene clusters. See Table 1 for genome reference.
While no cytochrome genes were present in the vicinity of the xoxF gene on the chromosomes of the Methylococcaceae, in each species we were able to identify a highly expressed gene, tentatively designated xoxG4, predicted to encode a novel cytochrome, with no sequence homology to either alphaproteobacterial xoxG3 genes or the two alternative Methylophilaceae genes, xoxG1 and xoxG2 (Figure 6). While XoxG4 is likely an electron acceptor from XoxF5 in the Methylococcaceae, xoxG4 transcription patterns differed from the ones of xoxF5 genes (Figure 3 and Supplementary Figure S1), suggesting that the role of this cytochrome is complex and may extend beyond methanol oxidation. Investigation of the properties of XoxG4 will be pursued in future studies.
Overall, for the Methylococcaceae, significant differences were noted between the transcriptomes of nitrate-supplemented versus ammonium-supplemented microcosms. In the latter, a series of hypothetical genes were expressed at very high levels, some even higher than the pMMO or the MDH genes. Interestingly, nitrogen fixation functions were expressed at high levels in ammonium-supplemented microcosms, compared to nitrate-supplemented microcosms (Figure 3 and Supplementary Figure S1). This type of regulation was somewhat unexpected. However, it may be potentially explained by faster oxygen consumption in the ammonium-supplemented microcosms, which are characterized by higher abundance of non-methanotroph species, and thus induction of nitrogen fixation genes may be in response to hypoxia. Future research is required to test this hypothesis.
Other highly expressed genes in the Methylococcaceae were the ones for pilin (pilA), proposed to be involved in extracellular electron transfer (Holmes et al., 2017; Lovley, 2017), and for type VI secretion functions, proposed to be involved in warfare among bacterial species (Alcoforado Diniz et al., 2015; Cianfanelli et al., 2016). In the Methylophilaceae, pilA genes were also highly expressed. Multiple porin genes and genes for other outer membrane proteins were also highly expressed (Figure 3 and Supplementary Figures S1–S4), and these may be involved in exchange of metabolites, signaling molecules, or other factors. The roles of these will also be addressed in future studies.
Discussion
In this study, we aimed to make advances toward developing a synthetic model for studying syntrophy in aerobic methane-oxidizing communities. We employed defined SCs of methanotrophs, non-methanotrophic methylotrophs, and non-methylotrophic heterotrophs, to further test whether partnerships in aerobic methanotrophy are specific or non-specific, and also to potentially identify most prominent models for future SC-based experiments. Note that many methylotrophs included into SCs are capable of growth on a variety of organic compounds, and many are capable of denitrification (Beck et al., 2015; McTaggart et al., 2015a,e). These organisms have a potential to consume either single carbon- or multicarbon compounds, and some possess a potential to link carbon consumption to either oxygen respiration or denitrification. Thus, these organisms presented good models for testing the “public goods” hypothesis. The Methyloversatilis strains that require vitamin B12 (Smalley et al., 2015) also served as controls for vitamin B12 exchange potential.
All organisms were supplied into SCs in viable state, and all were presumed to be active, thus the possibility of dormancy was excluded. We assumed that members of each of the three major functional guilds would reveal competitive trends within each respective guild, and that these trends may be determined by fitness at time zero, on the one hand, and by response to specific environmental conditions, on the another hand. Thus, while we expected and indeed observed very good agreement in terms of community dynamics between the technical replicates (Figures 1A–C), good replication between independent experiments was not necessarily expected. In this sense, the SC approach presents some serious methodological challenges. First, as the organisms involved can survive and thrive on their own, as pure cultures, the choice of experimental conditions conducive of cooperative behavior may be important (medium composition, partial pressures of methane and O2, nitrogen sources, etc.). Second, the relative fitness at time zero is hard to control. For example, it is not clear on what substrate the pure cultures should be pregrown, to better reflect their diet in situ. This problem is especially profound when it comes to organisms with broad metabolic capabilities. Assuming that most organisms must be limited by some nutrient in their natural habitat (be it O2, carbon, nitrogen, or an essential metal), we elected to starve the premixed SCs, incubating them in a minimal medium, before supplying them with the carbon source. However, different organisms may respond differently to this mimicked “starvation,” and thus initial fitness of different organisms would depend on their stress response mechanisms and their robustness. Additionally, other factors may be important that may control species ratios in natural communities, including functional guilds not primarily involved in methane utilization or cometabolism, but sporadically influencing community structure, for example, predatory species, species harboring predatory plasmids or phages, or free-living phages.
Despite some differences between the dynamics among the specific strains of Methylococcaceae and Methylophilaceae in SC1 versus SC2, general trends at the level of functional guilds remained the same. In each case, Methylococcaceae and Methylophilaceae became the dominant species, quickly outcompeting Methylocystaceae as well as other methylotrophic and non-methylotrophic heterotrophs. In most cases, the Methylomonas species became dominant. This would be predicted from their growth characteristics in pure cultures, in standard conditions, under which they outperform Methylobacter, Methylosarcina, or Methylosinus (Auman et al., 2000 and unpublished results). However, Methylomonas species have not been detected as dominant in prior experiments that involved natural sediment samples (Kalyuzhnaya et al., 2008; Beck et al., 2013; Hernandez et al., 2015; Oshkin et al., 2015), possibly due to existence of some specific controls (predation, antibiotic regulation, molecular signaling) that were not recreated in SCs in this study.
The trend of the Methylophilaceae outcompeting other methanol utilizers, noted for natural communities (Hernandez et al., 2015; Oshkin et al., 2015), persisted in SCs. While many species included into SCs show robust growth on methanol in laboratory (e.g., Methylobacterium, Paracoccus, Hyphomicrobium, Methylopila; Beck et al., 2015), these species did not seem competitive under most regimens. As an exception, Methyloversatilis species were prominent under the “low” methane regimens in SC2. These observations suggest that the Methylophilacaea, and, under some conditions, Methyloversatilis species have some advantages over alphaproteobacterial methylotrophs in consuming methanol. Success of the latter also suggests that vitamin B12 indeed could be shared among the community members.
The heterotrophs that could potentially consume a variety of organic compounds, i.e., the Gram-positive species and the facultatively methylotrophic alphaproteobacteria, did not persist in the microcosms, in agreement with prior results with “natural” microcosms (Hernandez et al., 2015; Oshkin et al., 2015). In contrast, Pseudomonas and Janthinobacterium were detected in some samples at over 2% abundance, suggesting that they were more competitive. However, Flavobacterium species, and also “uninvited,” contaminants belonging to Chryseobacterium (which are also Bacteroidetes) and Acidovorax (Burkholderiales, closely related to Janthinobacterium) were measured at even higher abundances, suggesting that Bacteroidetes and Acidovorax/other Burkholderiales must have strong competitive advantages over other, methylotrophic or non-methylotrophic heterotrophs. Overall, our data continue to suggest that the carbon released by the methanotrophs is not equally accessible by all organisms present in the community, and thus some mechanisms must exist that make Methylophilaceae and, under certain conditions, Rhodocyclaceae more competitive for methanol, Acidovorax more competitive for acetate and/ other organic acids, while Bacteroidetes appear more competitive in consumption of polymeric substances that are excreted by other species.
Highly covered transcriptomes were obtained for major partner species. Remarkably, among the most highly and most differentially expressed were the genes for alternative MDH enzymes, MxaFI, the classic Ca2+-dependent MDH and XoxF, the recently discovered MDH requiring REEs (Chistoserdova, 2016; Martinez-Gomez et al., 2016). These enzymes have been previously found to be subjects of the so-called REE switch (Chu et al., 2016), a mechanism that inversely regulates transcription from xox and the mxa gene clusters in response to the presence of REEs (Chu and Lidstrom, 2016; Gu et al., 2016; Vu et al., 2016). However, the straightforward nature of the switch has already been questioned. For example, in an alphaproteobacterial methanotroph Methylosinus trichosporium OB3b, copper appears to override the switch (Gu et al., 2016; Gu and Semrau, 2017). The switch also appears to work differently dependent on whether methanotrophs are cultivated as pure cultures or as members of communities (Krause et al., 2017). Thus, further insights are required to better understand the mechanism of the REE switch.
The data we present here indicate that the REE switch must be a subject to much more complex regulation than previously appreciated. (1) First, we demonstrate that the REE switch is responsive to a nitrogen source. As our experimental design did not include added REEs, high expression of the mxa genes was expected. However, this was true only for the HHNO3 regimen, the standard laboratory condition (Chu and Lidstrom, 2016). Even under this regimen, Methylophilaceae other than Methylophilus tended to overexpress xoxF over mxaF. (2) Second, we demonstrate that both O2 and methane partial pressures also have control over the REE switch, high methane selecting for the MxaFI–MDH (under nitrate), “low” methane and “low” O2 selecting for XoxF, and “high” methane “low” O2, nitrate allowing for transcription of both systems at similar levels. (3) Third, we demonstrate that, when multiple copies of xoxF genes are present in a single genome, they are not all following the same regulation pattern. (4) Finally, different organisms operate the RRE switch differently, such as in the samples where Methylophilus types express the mxa genes at a higher level, other Methylophilaceae preferentially express xoxF genes.
Besides multiple xoxF genes, multiple cytochrome-encoding genes (xoxG) were also highly and differentially expressed, adding further complexity to the REE switch. Moreover, while Methylophilaceae tended to coexpress xoxF and xoxG genes, expression of xoxG in Methylococcaceae was not coordinated with expression of xoxF. While the meaning of such complexity for the REE switch remains enigmatic, it further points to the importance of the methanol oxidation step in communal metabolism of methane and warrants further investigations.
Conclusion
We here demonstrated the utility of multispecies SCs in studying complex biogeochemical processes, such as communal metabolism of methane, in a simplified, controllable model. We demonstrate that general trends in SCs mimic the ones in natural communities, selecting for Methylococcaceae and Methylophilaceae under most conditions, thus defying the hypothesis of random distribution of “public goods.” Through metatranscriptome analysis, we uncover the unexpected complexity of transcriptional regulation of methanol oxidation, mediated through the REE switch, which, in turn, is governed by multiple environmental factors. We also demonstrate that the REE switch acts differently in different organisms and on different XoxF/XoxG enzymes. These new data will inform future development of SCs customized toward specific experimental goals, in order to target specific functions that may contribute to cooperative behavior in methane consumption.
Author Contributions
ZY and LC conceived the study, ZY carried out experiments, DB carried out bioinformatics analyses, and ZY and LC analyzed the data and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-SC-0016224. Research was facilitated through the use of advanced computational storage and networking infrastructure provided by the Hyak Supercomputer System supported in part by the University of Washington eScience Institute. The work conducted by the U.S. Department of Energy JGI, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02392/full#supplementary-material
Sample map.
Raw read counts.
References
- Alcoforado Diniz J., Liu Y. C., Coulthurst S. J. (2015). Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell Microbiol 17 1742–1751. 10.1111/cmi.12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C. (2004). The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 428 2–9. 10.1016/j.abb.2004.03.038 [DOI] [PubMed] [Google Scholar]
- Auman A. J., Stolyar S., Costello A. M., Lidstrom M. E. (2000). Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66 5259–5266. 10.1128/AEM.66.12.5259-5266.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam J. P., Jay Z. J., Schmid M. C., Rusch D. B., Romine M. F., Jennings Rde M., et al. (2016). Ecophysiology of an uncultivated lineage of Aigarchaeota from an oxic, hot spring filamentous ‘streamer’ community. ISME J. 10 210–224. 10.1038/ismej.2015.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D. A., Kalyuzhnaya M. G., Malfatti S., Tringe S. G., Glavina del Rio T., Ivanova N., et al. (2013). A metagenomic insight into freshwater methane-utilizing communities and evidence for cooperation between the Methylococcaceae and the Methylophilaceae. PeerJ 1:e23. 10.7717/peerj.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck D. A., McTaggart T. L., Setboonsarng U., Vorobev A., Kalyuzhnaya M. G., Goodwin L., et al. (2015). Multiphyletic origins of methylotrophy in Alphaproteobacteria, exemplified by comparative genomics of Lake Washington isolates. Environ. Microbiol. 17 547–554. 10.1111/1462-2920.12736 [DOI] [PubMed] [Google Scholar]
- Beck D. A., McTaggart T. L., Setboonsarng U., Vorobev A., Kalyuzhnaya M. G., Ivanova N., et al. (2014). The expanded diversity of Methylophilaceae from Lake Washington through cultivation and genomic sequencing of novel ecotypes. PLOS ONE 9:e102458. 10.1371/journal.pone.0102458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. (2011). Modularity of methylotrophy, revisited. Environ Microbiol 13 2603–2622. 10.1111/j.1462-2920.2011.02464.x [DOI] [PubMed] [Google Scholar]
- Chistoserdova L. (2016). Lanthanides: new life metals? World J. Microbiol. Biotechnol. 32:138. 10.1007/s11274-016-2088-2 [DOI] [PubMed] [Google Scholar]
- Chistoserdova L., Lidstrom M. E. (2013). “Aerobic methylotrophic prokaryotes,” in The Prokaryotes, 4th Edn, eds Rosenberg E., DeLong E. F., Thompson F., Lory S., Stackebrandt E. (Berlin: Springer; ), 267–285. [Google Scholar]
- Chu F., Beck D. A., Lidstrom M. E. (2016). MxaY regulates the lanthanide-mediated methanol dehydrogenase switch in Methylomicrobium buryatense. PeerJ 4:e2435. 10.7717/peerj.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F., Lidstrom M. E. (2016). XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J. Bacteriol. 198 1317–1325. 10.1128/JB.00959-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli F. R., Monlezun L., Coulthurst S. J. (2016). Aim, load, fire: the Type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24 51–62. 10.1016/j.tim.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Crevecoeur S., Vincent W. F., Comte J., Lovejoy C. (2015). Bacterial community structure across environmental gradients in permafrost thaw ponds: methanotroph-rich ecosystems. Front. Microbiol. 6:192. 10.3389/fmicb.2015.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedysh S. N., Dunfield P. F. (2014). “Cultivation of methanotrophs,” in Hydrocarbon and Lipid Microbiology Protocols, Springer Protocols Handbooks, eds McGenity T., Timmis K., Nogales B. (Berlin: Springer-Ferlag; ). [Google Scholar]
- Edgar R., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27 k2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate ITU sequences from microbial amplicon reads. Nat. Methods 10 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Fitriyanto N. A., Fushimi M., Matsunaga M., Pertiwiningrum A., Iwama T., Kawai K. (2011). Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J. Biosci. Bioeng. 111 613–617. 10.1016/j.jbiosc.2011.01.015 [DOI] [PubMed] [Google Scholar]
- Gu W., Farhan Ul Haque M., DiSpirito A. A., Semrau J. D. (2016). Uptake and effect of rare Earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol. Lett. 363:fnw129. 10.1093/femsle/fnw129 [DOI] [PubMed] [Google Scholar]
- Gu W., Semrau J. D. (2017). Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl. Microbiol. Biotechnol. 101 8499–8516. 10.1007/s00253-017-8572-2 [DOI] [PubMed] [Google Scholar]
- Hernandez M. E., Beck D. A., Lidstrom M. E., Chistoserdova L. (2015). Oxygen availability is a major factor in determining the composition of microbial communities involved in methane oxidation. PeerJ 3:e801. 10.7717/peerj.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A., Angel R., Veraart A. J., Daebeler A., Jia Z., Kim S. Y., et al. (2016). Biotic interactions in microbial communities as modulators of biogeochemical processes: methanotrophy as a model system. Front. Microbiol. 7:1285. 10.3389/fmicb.2016.01285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. E., Shrestha P. M., Walker D. J. F., Dang Y., Nevin K. P., Woodard T. L., et al. (2017). Metatranscriptomic evidence for direct interspecies electron transfer between Geobacter and Methanothrix species in methanogenic rice paddy soils. Appl. Environ. Microbiol. 83:e00223–17. 10.1128/AEM.00223-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi H., Yurimoto H., Sakai Y. (2011). Stimulation of methanotrophic growth in cocultures by cobalamin excreted by rhizobia. Appl. Environ. Microbiol. 77 8509–8515. 10.1128/AEM.05834-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G., Lamb A. E., McTaggart T. L., Oshkin I. Y., Shapiro N., Woyke T., et al. (2015). Draft genomes of gammaproteobacterial methanotrophs isolated from Lake Washington sediment. Genome Announc. 3:e00103–15. 10.1128/genomeA.00103-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G., Lapidus A., Ivanova N., Copeland A. C., McHardy A. C., Szeto E., et al. (2008). High-resolution metagenomics targets specific functional types in complex microbial communities. Nat. Biotechnol. 26 1029–1034. 10.1038/nbt.1488 [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G., Yang S., Rozova O. N., Smalley N. E., Clubb J., Lamb A., et al. (2013). Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 4:2785. 10.1038/ncomms3785 [DOI] [PubMed] [Google Scholar]
- Karwautz C., Kus G., Stöckl M., Neu T. R., Lueders T. (2017). Microbial megacities fueled by methane oxidation in a mineral spring cave. ISME J. 10.1038/ismej.2017.146 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaserer H. (1906). Uber die oxydation des wasserstoffes und des methans durch mikroorganismen. Zentr. Bakt. Parasitenk 15 573–576. [Google Scholar]
- Keltjens J. T., Pol A., Reimann J., Op den Camp H. J. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 98 6163–6183. 10.1007/s00253-014-5766-8 [DOI] [PubMed] [Google Scholar]
- Krause S. M. B., Johnson T., Samadhi Karunaratne Y., Fu Y., Beck D. A. C., Chistoserdova L., et al. (2017). Lanthanide-dependent cross-feeding of methane derived carbon is linked by microbial community interactions. Proc. Natl. Acad. Sci. U.S.A. 114 358–363. 10.1073/pnas.1619871114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus A., Clum A., LaButti K., Kalyuzhnaya M. G., Lim S., Beck D. A., et al. (2011). Genomes of three methylotrophs from a single niche reveal the genetic and metabolic divergence of the Methylophilaceae. J. Bacteriol. 163 3757–3764. 10.1128/JB.00404-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. D., Walworth N. G., McParland E. L., Fu F. X., Mincer T. J., Levine N. M., et al. (2017). The Trichodesmium consortium: conserved heterotrophic co-occurrence and genomic signatures of potential interactions. ISME J. 11 1813–1824. 10.1038/ismej.2017.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R. (2017). Happy together: microbial communities that hook up to swap electrons. ISME J. 11 327–336. 10.1038/ismej.2016.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyan S. K., Bhatia A., Kumar A., Gupta D. K., Singh R., Kumar S. S., et al. (2016). Methane production, oxidation and mitigation: a mechanistic understanding and comprehensive evaluation of influencing factors. Sci. Total Environ. 572 874–896. 10.1016/j.scitotenv.2016.07.182 [DOI] [PubMed] [Google Scholar]
- Martineau C., Whyte L. G., Greer C. W. (2010). Stable isotope probing analysis of the diversity and activity of methanotrophic bacteria in soils from the Canadian high Arctic. Appl. Environ. Microbiol. 76 5773–5784. 10.1128/AEM.03094-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gomez N. C., Vu H. N., Skovran E. (2016). Lanthanide chemistry: from coordination in chemical complexes shaping our technology to coordination in enzymes shaping bacterial metabolism. Inorg. Chem. 55 10083–10089. 10.1021/acs.inorgchem.6b00919 [DOI] [PubMed] [Google Scholar]
- McTaggart T. L., Beck D. A., Setboonsarng U., Shapiro N., Woyke T., Lidstrom M. E., et al. (2015a). Genomics of methylotrophy in Gram-positive methylamine-utilizing species. Microorganisms 3 94–112. 10.3390/microorganisms3010094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart T. L., Benuska G., Shapiro N., Woyke T., Chistoserdova L. (2015b). Draft genomes of five new strains of Methylophilaceae isolated from Lake Washington sediment. Gen. Announc. 3:e01511–14. 10.1128/genomeA.01511-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart T. L., Shapiro N., Woyke T., Chistoserdova L. (2015c). Draft genomes of two strains of Flavobacterium isolated from Lake Washington sediment. Gen. Announc. 3:e01597–14. 10.1128/genomeA.01597-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart T. L., Shapiro N., Woyke T., Chistoserdova L. (2015d). Draft genome of Janthinobacterium sp. RA13 isolated from Lake Washington sediment. Gen. Announc. 3:e01588–14. 10.1128/genomeA.01588-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart T. L., Shapiro N., Woyke T., Chistoserdova L. (2015e). Draft genome of Pseudomonas sp. 11/12A isolated from Lake Washington sediment. Gen. Announc. 3:e01587–14. 10.1128/genomeA.01587-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modin O. (2017). A mathematical model of aerobic methane oxidation coupled to denitrification. Environ. Technol. 10.1080/09593330.2017.1323961 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Modin O., Fukushi K., Yamamoto K. (2007). Denitrification with methane as external carbon source. Water Res. 41 2726–2738. 10.1016/j.watres.2007.02.053 [DOI] [PubMed] [Google Scholar]
- Morris J., Lenski R. E., Zinser E. R. (2012). The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. Mol. Biol. 3:e00036–12. 10.1128/mBio.00036-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Mitsui R., Tani A., Sasa K., Tashiro S., Iwama T., et al. (2012). A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLOS ONE 7:e50480. 10.1371/journal.pone.0050480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshkin I. Y., Beck D. A., Lamb A. E., Tchesnokova V., Benuska G., McTaggart T. L., et al. (2015). Methane fed microcosms show differential community dynamics and pinpoint specific taxa involved in communal response. ISME J. 9 1119–1129. 10.1038/ismej.2014.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada A. E., Needham D. M., Fuhrman J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18 1403–1414. 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- Pol A., Barends T. R., Dietl A., Khadem A. F., Eygensteyn J., Jetten M. S., et al. (2014). Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 16 255–264. 10.1111/1462-2920.12249 [DOI] [PubMed] [Google Scholar]
- Schuur E. A., McGuire A. D., Schädel C., Grosse G., Harden J. W., Hayes D. J., et al. (2015). Climate change and the permafrost carbon feedback. Nature 520 171–179. 10.1038/nature14338 [DOI] [PubMed] [Google Scholar]
- Singer E., Bushnell B., Coleman-Derr D., Bowman B., Bowers R. M., Levy A., et al. (2016). High-resolution phylogenetic microbial community profiling. ISME J. 10 2020–2032. 10.1038/ismej.2015.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B. K., Bardgett R. D., Smith P., Reay D. S. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 8 779–790. 10.1038/nrmicro2439 [DOI] [PubMed] [Google Scholar]
- Smalley N. E., Taipale S., De Marco P., Doronina N. V., Kyrpides N., Shapiro N., et al. (2015). Functional and genomic diversity of methylotrophic Rhodocyclaceae: description of Methyloversatilis discipulorum sp. nov. Int. J. Syst. Evol. Microbiol. 65 2227–2233. 10.1099/ijs.0.000190 [DOI] [PubMed] [Google Scholar]
- Söhngen N. L. (1906). Uber bakterien, welche methan als kohlenstoffnahrung energiequelle gebrauchen. Zentr. Bakt. Parasitenk. 15 513–517. [Google Scholar]
- Trotsenko Y. A., Murrell J. C. (2008). Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63 183–229. 10.1016/S0065-2164(07)00005-6 [DOI] [PubMed] [Google Scholar]
- Vu H. N., Subuyuj G. A., Vijayakumar S., Good N. M., Martinez-Gomez N. C., Skovran E. (2016). Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J. Bacteriol. 98 1250–1259. 10.1128/JB.00937-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P., Coates L., Mohammed F., Gill R., Erskine P., Bourgeois D., et al. (2006). The 1.6A X-ray structure of the unusual c-type cytochrome, cytochrome cL, from the methylotrophic bacterium Methylobacterium extorquens. J. Mol. Biol. 357 151–162. 10.1016/j.jmb.2005.12.055 [DOI] [PubMed] [Google Scholar]
- Williams P. A., Coates L., Mohammed F., Gill R., Erskine P. T., Coker A., et al. (2005). The atomic resolution structure of methanol dehydrogenase from Methylobacterium extorquens. Acta Crystallogr. D Biol. Crystallogr. 61 75–79. 10.1107/S0907444904026964 [DOI] [PubMed] [Google Scholar]
- Yu Z., Chistoserdova L. (2017). Communal metabolism of methane and the rare Earth element switch. J. Bacteriol. 10.1128/JB.00328-17 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Krause S. M., Beck D. A., Chistoserdova L. (2016). A synthetic ecology perspective: how well does behavior of model organisms in the laboratory predict microbial activities in natural habitats? Front. Microbiol. 7:946. 10.3389/fmicb.2016.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample map.
Raw read counts.