Abstract
The rumen is known to harbor dense populations of bacteriophages (phages) predicted to be capable of infecting a diverse range of rumen bacteria. While bacterial genome sequencing projects are revealing the presence of phages which can integrate their DNA into the genome of their host to form stable, lysogenic associations, little is known of the genetics of phages which utilize lytic replication. These phages infect and replicate within the host, culminating in host lysis, and the release of progeny phage particles. While lytic phages for rumen bacteria have been previously isolated, their genomes have remained largely uncharacterized. Here we report the first complete genome sequences of lytic phage isolates specifically infecting three genera of rumen bacteria: Bacteroides, Ruminococcus, and Streptococcus. All phages were classified within the viral order Caudovirales and include two phage morphotypes, representative of the Siphoviridae and Podoviridae families. The phage genomes displayed modular organization and conserved viral genes were identified which enabled further classification and determination of closest phage relatives. Co-examination of bacterial host genomes led to the identification of several genes responsible for modulating phage:host interactions, including CRISPR/Cas elements and restriction-modification phage defense systems. These findings provide new genetic information and insights into how lytic phages may interact with bacteria of the rumen microbiome.
Keywords: bacteriophage, phage, rumen, Streptococcus, Bacteroides, Ruminococcus, Siphoviridae, Podoviridae
Introduction
The rumen contains dense and highly diverse populations of bacteriophages (phages). Rumen phage populations have been examined using culture-independent techniques such as morphological surveys (electron microscopy), genome length profiling (pulsed field gel electrophoresis) and more recently, viral metagenomics (sequencing of viral community DNA) (Paynter et al., 1969; Klieve and Bauchop, 1988; Klieve and Swain, 1993; Berg Miller et al., 2012; Ross et al., 2013). Culture-based techniques have also been employed for studying rumen phage populations, where successful phage infection and replication is indicated by the formation of clearing zones or plaques in otherwise confluent layers of bacterial growth (Klieve, 2005). This approach tends to detect phages able to undergo the lytic cycle of phage infection where phage particles attach to and replicate in susceptible bacteria without forming a stable genetic association with their host (lysogeny) (Ackermann and DuBow, 1987). Culture-based techniques cannot fully encompass the viral diversity of the rumen source material, instead they capture viable phages which can successfully infect and replicate in the bacterial strains employed in the study.
The first isolation of lytic phages from rumen fluid was reported in 1966 using rumen fluid from cattle as source material and rumen-derived isolates of Serratia and Streptococcus as bacterial hosts (Adams et al., 1966). Following this initial report, rumen fluid was used as a source of lytic phages able to infect a range of predominant rumen bacterial genera for example Streptococcus, Bacteroides, and Ruminococcus (Iverson and Millis, 1976a; Tarakanov, 1976; Klieve et al., 1991, 2004; Styriak et al., 1994). While many rumen phages were isolated and stored in culture collections, particularly in the 1970's and 1980's (Gilbert and Klieve, 2015), only those phages which could be potentially employed in applications such as genetic engineering and phage therapy were characterized beyond an initial assessment of morphology.
This study describes the genome sequences of five lytic phages capable of infecting rumen bacteria. These phages have all been previously described and classified as Caudovirales, having double-stranded DNA genomes and characteristic head-tail morphology. Three of the phages, ϕBrb01 and ϕBrb02 infecting strains of Bacteroides sp. (Klieve et al., 1991) and ϕSb01 (Klieve and Bauchop, 1991) infecting strains of Streptococcus classified within the Streptococcus bovis/Streptococcus equinus complex (SBSEC) (Jans et al., 2015), form particles with long, flexible tails characteristic of the family Siphoviridae. The further two phages, ϕRa02 and ϕRa04 infecting Ruminococcus albus AR67 (Klieve et al., 2004) form smaller particles with short tails and have been classified within the Podoviridae family.
The phages ϕBrb01 and ϕBrb02 have been the most genetically characterized, with restriction mapping experiments showing both genomes to be circularly permuted and terminally redundant (Klieve et al., 1991) resulting in the formation of phage particles containing variable lengths of genomic DNA. Rumen phage:host interactions have also been reported with investigations into the host range of the Bacteroides phages ϕBrb01 and ϕBrb02 and the Streptococcus phage ϕSb01, indicating that each of these Siphoviridae could utilize multiple strains within a single bacterial species as hosts (Klieve et al., 1991, 1999). Physical mechanisms for the development of phage resistance have also been observed, with the formation of a thick, polysaccharide capsule conferring Bacteroides resistance to infection by ϕBrb01 (Klieve et al., 1991).
This study addresses the paucity of information available for rumen phages in current sequence databases and represents the first report of complete genome sequences for lytic phages infecting rumen bacteria. It also provides the first genome sequences of lytic phages infecting the genus Ruminococcus. The genome sequences of the host bacteria were also analyzed in order to determine how these phages interact with their host and provide new insights into the defense mechanisms rumen bacteria may employ to counteract phage infection.
Materials and methods
Phage cultivation and phage DNA extraction
Phages and respective host bacteria are detailed in Table 1. All cultivation of bacteria and phage isolates was undertaken in a Physical Containment Level 2, Australian Government Office of the Gene Technology Regulator (OTGR) certified laboratory facility using biosecurity and institutional safety procedures required for these organisms and for products derived from these organisms, including DNA (Biohazard Risk Group 1 and 2—Animal origin). Although three of the phages have been previously shown to infect multiple host strains, S. equinus 2B (also classified as S. bovis 2B) was used as the host strain for cultivation of ϕSb01 and Bacteroides sp. AR20 was used as the host strain for ϕBrb01 and ϕBrb02 and R. albus AR67 was used as the host strain for ϕRa02 and ϕRa04. Rumen bacterial host strains were cultivated using anaerobic techniques (Hungate et al., 1964) and a rumen fluid-based growth medium (Klieve et al., 1989). Phages were propagated according to previously described methods (Klieve, 2005) with phage infection times adjusted to account for differential growth rates (2 h for S. equinus 2B and 4–6 h for R. albus AR67 and Bacteroides sp. AR20). Phage particles were harvested 24 h post-infection by differential centrifugation, filtration through 0.45 and 0.22 μm low protein binding filter units (Millipore) (Klieve, 2005) and precipitated with 20% (w/v) PEG6000 and 25 M NaCl at 4°C for a minimum of 1 h. Phage particles were pelleted by ultracentrifugation at 52,350 × g for 2 h at 4°C (50.2Ti rotor, OptimaTM L-100 XP Ultracentrifuge, Beckmann Coulter) and phage DNA extracted (Klieve and Gilbert, 2005).
Table 1.
Summary of original phage descriptions (classification according to viral particle morphology, genome lengths calculated by restriction enzyme digestion of phage genomic DNA and particle dimensions) and description of bacterial host strains.
| Phage | Classification | Genome length (kb) | Phage particle dimensions | Phage isolation source (Reference) | Bacterial host/s | Bacterial host/s isolation source (Reference) |
|---|---|---|---|---|---|---|
| ϕBrb01 | Siphoviridae | 33.91 ± 2.41 | Head 55–60 nm, tail 105 × 9 nm | Municipal sewage (Klieve et al., 1991) | Bacteroides sp. AR20***, Bacteroides (Prevotella) ruminicola ss brevis AR22, AR23 | Ovine rumen fluid (Klieve et al., 1989) |
| ϕBrb02 | Siphoviridae | 33.02 ± 1.16 | Head 55–60 nm, tail 105 × 9 nm | Municipal sewage (Klieve et al., 1991) | Bacteroides sp. AR20***, Bacteroides (Prevotella) ruminicola ss brevis AR7, AR22, AR23 | Ovine rumen fluid (Klieve et al., 1989) |
| ϕRa02 | Podoviridae | 12.8 | Head 25 nm, tail 18 nm* | Bovine fecal waste material (Klieve et al., 2004) | Ruminococcus albus AR67 | Ovine rumen fluid (Klieve et al., 1989) |
| ϕRa04 | Podoviridae | 14.0 | Head 25 nm, tail 18 nm* | Bovine faecal waste material (Klieve et al., 2004) | Ruminococcus albus AR67 | Ovine rumen fluid (Klieve et al., 1989) |
| ϕSb01 | Siphoviridae | 30.9 kb ± 4.4 | Head 60 nm, tail 110 × 7 nm | Bovine rumen fluid (Klieve and Bauchop, 1991) | Streptococcus bovis/equinus 2B, S. equinus Sb04, Sb17 | NS** (Iverson and Millis, 1976b), ovine rumen fluid; bovine rumen fluid (Klieve et al., 1999) |
Tail dimension estimated from TEM image presented in the Corrigendum to original phage isolation publication (Klieve et al., 2006);
Not stated (NS) in original isolation reference (Iverson and Millis, Personal Communication);
Bacteroides sp. strain AR20 was previously classified as Bacteroides ruminicola ss brevis, Prevotella ruminicola ss brevis in the Cytophaga-Flavobacterium-Bacteroides (CFB) group bacteria and Bacteroides thetaiotaomicro.
Sequencing and assembly
Genome sequences were generated at the U.S. Department of Energy, Joint Genome Institute (JGI). An Illumina standard shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform (2 × 151 bp read lengths with at least 50,000 reads generated per phage). All raw Illumina sequence data was quality filtered with DUK, a sequence filtering program developed at JGI, which removes Illumina sequencing and library preparation artifacts (Mingkun, L., Copeland, A. and Han, J., unpublished). Following the JGI standard protocol, genome assembly involved the following steps (1) quality filtered reads were assembled using Velvet (version 1.2.07) (Zerbino and Birney, 2008); (2) 1–3 kb simulated paired end reads were created from Velvet contigs using wgsim (https://github.com/lh3/wgsim); (3) reads were assembled with simulated read pairs using Allpaths–LG (version r46652) (Gnerre et al., 2011). For all phages sequenced, a minimum assembly input read coverage of 380 x was obtained (based on the number of quality-filtered reads) and each respective phage assembly resulted in one contig in one scaffold.
Annotation
Open reading frames were determined using Glimmer version 3 (Delcher et al., 2007) within GAMOLA2 (Altermann et al., 2017) and Prodigal version 2.6.2, (Hyatt et al., 2010) within Prokka version 1.1 (Seemann, 2014). Sequence homologies were identified by GAMOLA2 using the protein model databases Pfam-A (release 30.0), TIGRfam 15.0, and COG2014 (Haft et al., 2013; Galperin et al., 2015; Finn et al., 2016), and Prokka version 1.1 using the protein model databases Pfam, COG clusters, HAMAP, Resfams, TIGRfam 15.0, and dbCAN v4 (Yin et al., 2012; Haft et al., 2013; Galperin et al., 2015; Gibson et al., 2015; Pedruzzi et al., 2015; Finn et al., 2016). The putative function of predicted ORF's were also assigned based on homologies to proteins identified in BLASTP and BLASTX searches (BLAST+ version 2.2.31, Camacho et al., 2009) using the National Center for Bioinformatic Information (NCBI) virus reference sequence database (update 11th January, 2017) containing 8321 virus genome sequences and the NCBI-nr database (January 2017 update), with an e-value threshold of 10−3. Annotations generated by Prokka and GAMOLA2 and additional BLAST searches were merged using Geneious R9 (Kearse et al., 2012). Transmembrane proteins, signal peptides and tRNA elements were identified by GAMOLA2 and Prokka. Predicted transmembrane proteins were verified with the Transmembrane Helices Hidden Markov Model (TMHMM) 2.0 server (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001). In addition, signal peptides were verified using the SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011). Predicted tRNA regions were verified with the tRNAscan-SE server (http://lowelab.ucsc.edu/tRNAscan-SE/) (Lowe and Eddy, 1997). Inverted repeat sequences were determined using einverted (http://emboss.bioinformatics.nl/cgi-bin/emboss/einverted) (Rice et al., 2011).
Determination of nearest relatives
The best hit result obtained following BLASTP with an e-value threshold of 10−3 (BLAST+ version 2.2.31 (Camacho et al., 2009) matching of phage proteins against the NCBI virus reference sequence database (11 January 2017 update) was used to determine closest viral relatives. Using a classification system similar to that adopted by PHAST (Zhou et al., 2011), the most common phage name (name of the phage with the highest number of proteins most similar to those in the subject phage sequence) and most common phage number (the number of phages with the highest number of proteins most similar to those in the subject phage sequence) were determined for each phage. The bacterial hosts and environmental sources of the most closely related phages were determined using the Virus-host database (Mihara et al., 2016) and manual checks of original publications.
Phylogenetic analysis and genomic alignment
Phylogenetic analysis was conducted on the basis of the amino acid sequence of phage terminase large subunit genes (TerL) of the phages classified within the family Siphoviridae (ϕBrb01, ϕBrb02, and ϕSb01) and head-tail connector proteins for phages classified within the family Podoviridae (ϕRa01 and ϕRa02). Phylogenetic trees for respective phage genes were generated according to host genera (Bacteroides, Streptococcus, and Ruminococcus) using MEGA7 (Kumar et al., 2016) with ClustalW (Thompson et al., 1994) used for the initial amino acid sequence alignment. The model employed for each phylogenetic analysis was chosen following a model test incorporating a minimum of 56 amino acid substitution models, with the model with the lowest bayesian information criterion (BIC) score used for the final maximum likelihood phylogenetic analysis. The genetic distance presented on each phylogenetic tree was determined on the basis of 1,000 replicate trees (bootstraps). Phylogenetic analysis of the ϕBrb01 and ϕBrb02 TerL genes also included eight additional TerL genes of integrated prophage elements identified within the genomes of seven herbivore-gut associated Bacteroides isolates (Hungate 1000 project, JGI proposal ID: 612). Phylogenetic analysis of the ϕRa02 and ϕRa04 head-tail connector genes included 33 head-tail connector protein sequences obtained from the NCBI reference sequence protein database using the search terms head-tail connector and head-tail joining and an additional seven representative head-tail connector protein sequences from Podoviridae phages infecting hosts classified within the Firmicute genera Staphylococcus, Streptococcus, and Clostridium. Genomic plots were generated using Geneious R9 (Kearse et al., 2012) and alignments to identify homologous protein regions undertaken in Geneious R9 using Mauve (Darling et al., 2010) and LASTZ (Harris, 2007).
Analysis of bacterial host genomes: CRISPR analysis, identification of restriction: modification systems, and prophage detection
Host genomes (described in Table 1) included Bacteroides sp. AR20 (IMG ID 2596583541, 61 scaffolds), R. albus AR67 (IMG ID 2593339152, 103 scaffolds), and S. equinus strains 2B (IMG ID 2561511223, 9 scaffolds) Sb04 (IMG IDs 2651870306, 21 scaffolds) and Sb17 (IMG ID 2654588136, 13 scaffolds). The genome annotations from IMG (Markowitz et al., 2012) were used for the detection of genes associated with phage defense systems.
Host bacteria genomes were screened for integrated prophage elements using PHASTer (Arndt et al., 2016) and manual checking of host genome annotations from IMG. Predicted prophage elements were annotated with Prokka (Seemann, 2014) and BLASTP using the virus reference sequence database (January 2017 update). Closest phage relatives were determined using methods described above (section Determination of Nearest Relatives).
CRISPR associated Cas proteins were detected from the IMG genome annotation. CRISPR elements including direct repeats and spacer regions were identified using Prokka and CRISPRFinder (Grissa et al., 2007). Identified direct repeats and spacers were assessed for homology to known direct repeat regions using BLAST+ (version 2.2.31) BLASTN search against the NCBI nr database (January 2017 update) with 10−3 e-value threshold. Genome annotations from IMG were used for the detection of genes associated with restriction modification systems using custom word searches and on the basis of homology to restriction-modification system-associated proteins listed in the REBASE proteins database (28th October 2016 download, Roberts et al., 2015), using BLASTP (BLAST+ version 2.2.31), 10−3 e-value threshold.
Genome accession numbers
Whole genome sequence data for phages ϕBrb01, ϕBrb02, ϕRa02, ϕRa04, and ϕSb01 have the JGI genome portal project ID numbers 1035879, 1035881, 1035884, 1035887, and 1035872 respectively. Prophage elements identified during host genome analysis and designated ϕSb2Bpro1 and ϕBrbAR29pro1, were derived from the genome assemblies IMG ID 2561511223 (T517DRAFT_scaffold00001.1, nt 367,615–409,981) and IMG ID 2593339260 (IE59DRAFT_scaffold00020.20, nt 1 to 35,574) respectively.
Results and discussion
Each phage had a double-stranded DNA genome and their general features are presented in Table 2. Sequencing of phages was completed with a minimum of 380 x sequence coverage and sequence assembly resulted in all phage genomes being represented by a single contig and scaffold. All of the phage sequences were of a similar length to those predicted in the original isolation studies, where genome lengths were estimated on the basis of restriction enzyme digestion patterns (Table 1). The G+C content (%) varied, with the two phages infecting Bacteroides sp. AR20, ϕBrb01, and ϕBrb02, having higher GC content than the other phages. All phages had non-coding sequences associated with the complete genomes, and encoded for overlapping reading frames (Table 3). The majority of predicted open reading frames started with ATG, with exception being a single ORF of ϕBrb02 beginning with GTG and two ORFs of ϕSb01 beginning with GTG and a further two ORFs beginning with TTG. No complete tRNA elements were found in any of the lytic phage genomes examined and signal peptides were only identified in the genomes of ϕBrb01 and ϕBrb02.
Table 2.
Description of phage genomes and nearest relatives including the most common phage name.
| Phage | Assembled genome length (kb) | G + C content (%) | ORFs (no.) | Most common phage name (number of best hit homologous proteins)* | Additional common phage names (number of best hit homologous proteins >1)** |
|---|---|---|---|---|---|
| ϕBrb01 | 33.602 | 45.5 | 46 | Croceibacter phage P2559Y NC_023614.1 (4) | Bacteroides phage B40-8 NC_011222.1 (2) |
| ϕBrb02 | 34.687 | 45.7 | 48 | Croceibacter phage P2559Y NC_023614.1 (4) | Bacteroides phage B40-8 NC_011222.1 (1) |
| Bacteroides phage B124-14 NC_016770.1 (1) | |||||
| ϕRa02 | 12.985 | 35.6 | 16 | n.d. | Bacillus phage MG-B1 NC_021336.1 (1) |
| Clostridium phage phiCPV4 NC_018083.1 (1) | |||||
| Bacillus virus B103 NC_004165.1 (1) | |||||
| ϕRa04 | 12.977 | 35.3 | 15 | n.d. | Bacillus phage MG-B1 NC_021336.1 (1) |
| Clostridium phage phiCPV4 NC_018083.1 (1) | |||||
| Bacillus virus B103 NC_004165.1 (1) | |||||
| ϕSb01 | 33.595 | 37.2 | 46 | Streptococcus phage 315.5 NC_004588 (10) | Streptococcus phage 20617 NC_023503 (3) |
| Streptococcus phage Str-PAP-1 NC_028666.1 (2) | |||||
| Streptococcus phage TP-778L NC_022776.1 (2) | |||||
| Streptococcus phage T12 NC_028700.1 (2) |
Phage names and NCBI virus reference sequence IDs are listed except where the most common phage name was not determined (n.d.) as the number of best hit homologous proteins did not exceed more than one hit for any one phage.
For some phages the number of best hit homologous proteins did not exceed more than one hit for any one phage. The phage names for all ORF best hits are included for ϕRa02 and ϕRa04 and phage names for hits from the same host genus (Bacteroides) are reported for ϕBrb02.
Table 3.
Number of tRNA, signal peptides, and transmembrane (TM) proteins identified in phage genomes.
| Phage | Overlapping reading frames | tRNA | Signal peptides | Proteins with TM helices | Proteins with >2 TM helices |
|---|---|---|---|---|---|
| ϕBrb01 | 18 | neg | 2 | 6 | Holin protein |
| ϕBrb02 | 18 | neg | 1 | 5 | Holin protein |
| ϕRa02 | 7 | neg | 0 | 1 | – |
| ϕRa04 | 7 | neg | 0 | 1 | – |
| ϕSb01 | 21 | neg | 0 | 2 | Tail tape measure protein |
Genome organization and phylogeny of Bacteroides phages ϕBrb01 and ϕBrb02
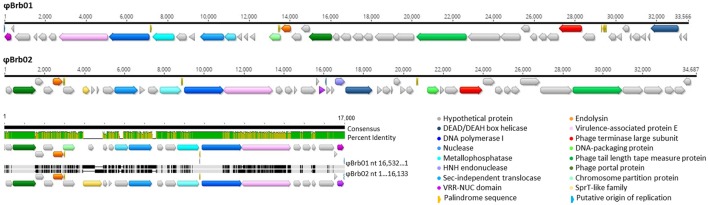
The two phages infecting the Bacteroides sp. strain AR20, ϕBrb01, and ϕBrb02 were previously shown to be of similar genome length and morphology (Table 1), and were isolated from the same source material (Klieve et al., 1991). The genome of ϕBrb02 encoded 48 ORF and was slightly longer in nt sequence length than ϕBrb01 which encoded only 46 ORF (Table 2). Alignment of the phage nt sequences indicated a 40.6% pairwise identity (14,301 identical sites, 1,146 nt gaps). While it had been reported that both of these phage genomes were circularly permuted and that terminal redundancy of the genome occurred during phage particle packaging (Klieve et al., 1991), only the ϕBrb01 genome sequence appeared to have a short (15 nt) overlapping region of terminal redundancy, adjacent to a 24 nt polyA region. This overlapping region was designated as a putative origin of replication, with an identical 15 nt sequence also identified within the ϕBrb02 genome, toward the middle of the genome sequence (Figure 1). Signal peptides and proteins with predicted transmembrane (TM) helices were identified in the genomes of ϕBrb01 and ϕBrb02, indicating that several of the proteins produced by these phages may undergo cleavage events and interact with the host bacterial cell membrane.
Figure 1.
Linear genome arrangement of the Bacteroides phages ϕBrb01 and ϕBrb02 showing predicted ORFs and alignment of the most highly homologous region. Predicted ORFs are depicted with arrows colored according to putative gene product names. The alignment was constructed using Mauve (Darling et al., 2010) and includes a consensus sequence (14,790 identical sites) and corresponding percentage identity graph ( 100%, >30%, <30%) with differences in the linear alignment of nucleotide sequences indicated with vertical shading.
Both of the Bacteroides phage genomes showed little similarity in overall genome sequence to the previously sequenced human gut-specific lytic phages of B. fragilis, B40-8 (Puig and Gironés, 1999) and B124-14 (Ogilvie et al., 2012) and showed greater genetic homology to the Croceibacter phage P2559Y (Kang et al., 2016) (Table 2). Only two genes found in the lytic B. fragilis phages, encoding for the phage tail length tape measure protein and a hypothetical protein preceding the endolysin protein, were related to the genes present in the ϕBrb01 and ϕBrb02 genomes.
This relative lack of genetic homology to any known viruses meant that for many of the open reading frames identified with the ϕBrb01 and ϕBrb02 genome sequences, gene function could not be predicted with confidence, particularly for expected phage structural proteins (phage tail and capsid proteins). Both ϕBrb01 and ϕBrb02 were isolated on the basis of their lytic ability, as evidenced by the formation of viral plaques within confluent lawns of the AR20 host strain, and no genes for phage integration and maintenance of lysogeny were identified within these phage genomes.
The ϕBrb01 and ϕBrb02 genomes showed some sequence homology to prophage-related gene sequences found within other Bacteroides strains (Table 4), including a prophage element found within a bacterial genome assembly of Bacteroides sp. AR29. This strain was isolated from ovine rumen fluid in the same laboratory as Bacteroides sp. AR20 and had previously been shown to harbor a prophage element which could be chemically induced to form intact phage particles (Klieve et al., 1989). The complete genome for this inducible prophage has been reported (Seet, 2005) although the corresponding prophage sequence obtained from the AR29 genome assembly (GenBank: FOBY01000020.1; IE59DRAFT_scaffold00020.20) was used in the current study (Figure 7). Alignment of this scaffold to the lytic Bacteroides phage genomes showed extensive homology to 13 genes including those involved in DNA replication and phage particle packaging (for example, the phage terminase large subunit, portal and tail length tape measure proteins). There were also phage proteins in the ϕBrb01 and ϕBrb02 genomes related to prophage elements within other species within the genus Bacteroides (Table 4) as well as Firmicutes such as Bacillus (B. licheniformis; B. subtilis) and Clostridium (C. botulinum).
Table 4.
Bacteria with prophage-associated genes homologous to the five lytic phages (ϕBrb01, ϕBrb02, ϕRa02, ϕRa04, and ϕSb01).
| Phage | Bacteria with homologous proteins (number of homologous phage proteins*) |
|---|---|
| ϕBrb01 | Bacteroides sp. HMSC067B03 (14); Bacteroides sp. AR29 (13); B. faecichinchillae (10); B. fragilis (8); uncultured Clostridium sp. (8) |
| ϕBrb02 | Bacteroides sp. AR29 (13); Bacteroides sp. HMSC067B03 (13); B. faecichinchillae (10); B. fragilis (8); uncultured Clostridium sp. (8) |
| ϕRa02 | Flavobacterium sp. Fl (6)**; Terriglobus sp. TAA 43 (3)**; Candidatus Moranbacteria GW2011_GWF2_35_39 (2); Eggerthella sp. CAG:1427 (2); Lucilia cuprina (2) |
| ϕRa04 | Flavobacterium sp. Fl (7)**; Terriglobus sp. TAA 43 (3)**; Candidatus Moranbacteria GW2011_GWF2_35_39 (2); Eggerthella sp. CAG:1427 (2); Lucilia cuprina (2) |
| ϕSb01 | Streptococcus parauberis (28); S. agalactiae (26); S. pyogenes (26); S. dysgalactiae (24); S. iniae SF1 (21) |
The bacterial names are listed for the top five most-related bacteria only.
Number of respective lytic phage proteins with homology to proteins of listed bacteria [number based on results of BLASTp analysis with the NCBI nr protein database (January 2017 update) with e-value threshold of 10−3].
Possible false positive result.
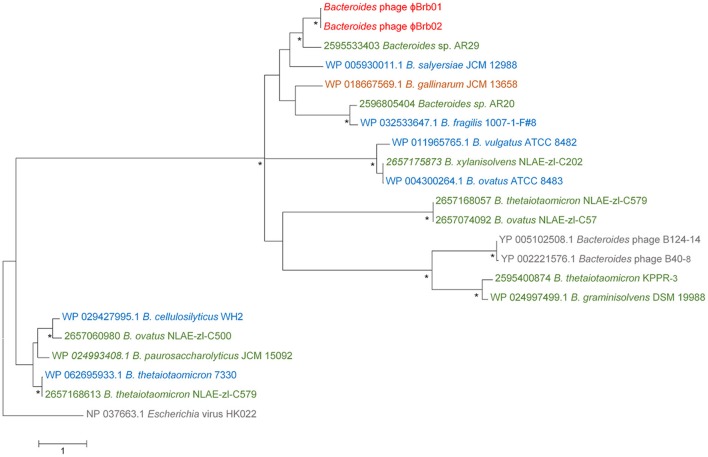
Phylogeny was also determined on the basis of the Siphoviridae terminase large subunit gene TerL, a hallmark gene for phages the order Caudovirales (Iranzo et al., 2016). This analysis incorporated TerL genes identified within the bacterial genome assemblies of Bacteroides strains isolated from ruminant gut and waste material and phage reference sequences. There was no clear distinction between either the isolation source or Bacteroides host-specific clustering of the TerL genes, however the three phages derived from ovine rumen isolates did show the greatest TerL protein sequence homology (Figure 2).
Figure 2.
Maximum likelihood phylogeny of the phage terminase large subunit (TerL) proteins of the Bacteroides phages ϕBrb01 and ϕBrb02, the virus reference phages B124-14 and B40-8 and TerL protein sequences obtained from the bacterial genome sequences of 17 Bacteroides isolates, including 10 sourced from ruminants. The tree with the highest log likelihood (−18,869.80) based on the Le Gascuel model (Le and Gascuel, 2008) and a gamma distribution with five categories is shown. This tree used a total of 1,094 amino acid positions and is drawn to scale, with branch lengths measured in the number of substitutions per site and is rooted with the TerL protein sequence of the Escherichia virus HK022. Bootstrap supports greater than 85%, based on 1,000 replicates, are indicated (*). The TerL proteins of the Bacteroides phages ϕBrb01 and ϕBrb02 are colored red (). For all other TerL protein sequences, the phage or bacterial genome names and accession numbers are detailed and colored according to the environmental isolation source, including ruminant origin (rumen fluid, feces and bovine waste); sewage (waste primarily of human origin); human origin (gut and patient samples, excluding sewage); and other gut material (chicken gut).
Genome organization and phylogeny of Ruminococcus phages ϕRa02 and ϕRa04
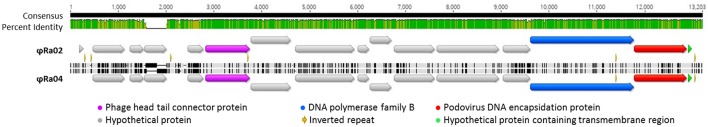
The two phages infecting R. albus strain AR67 were previously classified within the family Podoviridae (Klieve et al., 2004). The genomes of these phages were found to be genetically very similar (95.68% nt sequence homology) with identical genomic arrangement (Figure 3). The main difference between the two phages occurred at the start of the sequence, with ϕRa02 having an additional short predicted ORF (35 amino acid sequences in length) which was not found in the ϕRa04 sequence. The ϕRa02 sequence also encoded an additional five amino acids in the 4th predicted ORF with this protein being predicted to confer some outside transmembrane potential as determined by TMHMM analysis, thus potentially being able to interact with the bacterial cell membrane. The ϕRa02 sequence also included three inverted repeats, whereas the ϕRa04 sequence had only one, which exactly matched one of the inverted repeat sequences detected in ϕRa02.
Figure 3.
Genomes and alignment of the Ruminococcus phages ϕRa02 and ϕRa04. Predicted ORFs are depicted with arrows and putative gene products colored accordingly. The alignment was constructed using Mauve (Darling et al., 2010) and includes a consensus sequence and corresponding percentage identity graph ( 100%, >30%, <30%) with differences in the linear alignment of nucleotide sequences indicated with vertical shading.
The lack of any highly-related phage genomes limited the extent to which these genomes could be annotated and the most common phage name could not be determined (Table 2). The few phage genes for which identity could be predicted were all related to genes previously shown to be highly conserved in Podovirus genomes (Iranzo et al., 2016), including the head-tail connector protein, DNA polymerase B and Podovirus encapsidation proteins.
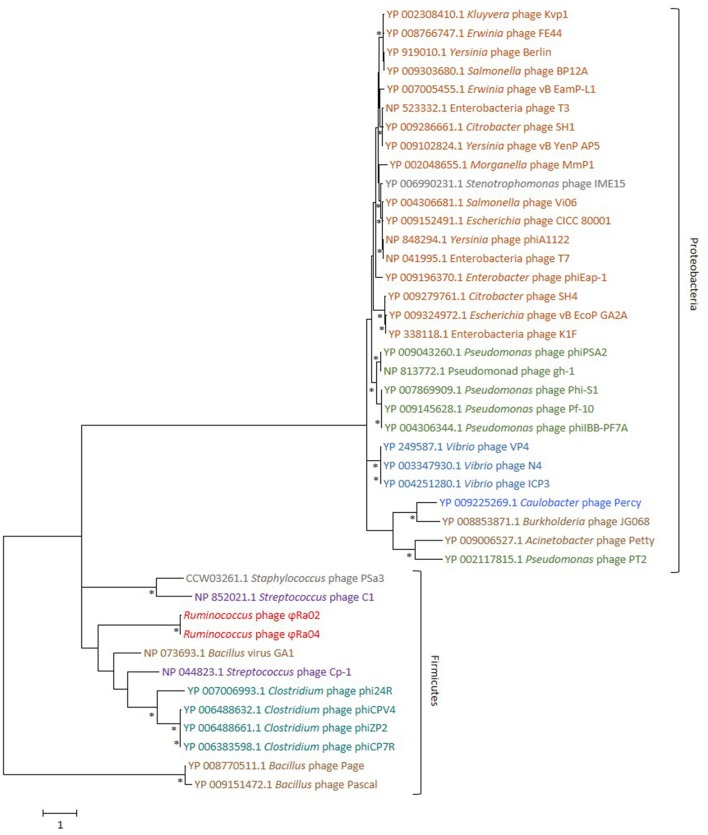
Phylogenetic analysis on the basis of the signature Podovirus gene, the head-tail connector protein (Iranzo et al., 2016), showed that phages infecting hosts of the same or highly related genera for example, phages infecting the Gram negative family Enterobacteriaceae, usually clustered together (Figure 4). In accordance with this, as the phages ϕRa02 and ϕRa04 infect a Ruminococcus host classified within the Gram positive bacterial phylum Firmicutes, the head-tail connector proteins of these phages clustered more closely with phages infecting the Firmicutes genera Bacillus, Clostridium, and Streptococcus (Figure 4). There was however a greater extent of genetic diversity in the head-tail connector genes of the phages infecting bacteria classified within the Gram positive phylum Firmicutes (for example Bacillus), compared to the Podoviruses infecting bacterial genera classified within the Gram negative phylum Proteobacteria (for example Yersinia, Pseudomonas, and Vibrio). The head-tail connector genes of phages ϕRa02 and ϕRa04 were distinctly different to those found in most other previously sequenced Podoviruses, being most like the head-tail connector protein of the phages such as GA1, infecting B. subtilis and classified with the Podovirus subfamily, Picovirinae, and Phi29 genus (Gascon et al., 2000).
Figure 4.
Maximum Likelihood phylogeny of the Podovirus head-tail connector protein sequences of the Ruminococcus phages ϕRa02 and ϕRa04 and 40 Podoviruses. The tree with the highest log likelihood (−19562.46) using the Le Gascuel model (Le and Gascuel, 2008) and a gamma distribution with five categories is shown. The rate variation model allowed for some sites to be evolutionarily invariable (0.02% sites). This tree used a total of 707 amino acid positions and is drawn to scale, with branch lengths measured in the number of substitutions per site. The tree is rooted with head-tail connector protein sequences of two Bacillus megaterium phages and bootstrap supports greater than 85%, based on 1,000 replicates, are indicated (*). Major phyla are bracketed and phage names and accession numbers are colored according to the phage-host family (Enterobacteriaceae ), genera (Pseudomonas , Vibrio and Ruminococcus , Clostridium ) or environmental source (soil-associated Bacillus and Acinetobacter , human respiratory tract associated Streptococcus and water-associated Caulobacter ). Names of phages with hosts classified outside of these taxons or of unknown isolation source (Stenotrophomonas and Staphylococcus) are colored gray .
An additional gene identified within the ϕRa02 and ϕRa04 phage sequences, the Podovirus encapsidation protein or DNA packaging ATPase (Pfam 05894.6) is also characteristic of the Phi29-like viruses (Garvey et al., 1985; Volozhantsev et al., 2012). This gene was flanked in the ϕRa02 and ϕRa04 genomes with a preceding DNA replication protein (DNA polymerase family B) and a terminal protein with predicted N-terminal signal sequence and transmembrane helix, indicating the presence of an N-terminally located transmembrane-spanning domain of 20 amino acids. This genetic arrangement is characteristic of Phi29-like viruses which have been shown to undergo membrane-associated DNA replication (Meijer et al., 2001b). In addition, the ϕRa02 and ϕRa04 genomes are of similar length, <20 kb and encoded approximately 16 ORF, similar to other Phi29-like phages (Meijer et al., 2001a; Redondo et al., 2013).
The ϕRa02 and ϕRa04 genomes were also unlike most prophage sequences associated with bacterial genomes (Table 4). The ϕRa02 and ϕRa04 genomes were however highly homologous (100% nt homology) to genomic scaffolds associated with the genome sequences of two bacteria isolated from aquatic sediment and soil (Eichorst et al., 2007; McTaggart et al., 2015), specifically two short contigs (6,985 and 5,502 nt) of the Flavobacterium sp. F1 assembly (NCBI Reference sequence scaffolds NZ_JQJY01000012.1 and NZ_JQJY01000014.1) and a single contig (12,818 nt) in the genome assembly of Terriglobus sp. TAA 43 (NCBI Reference sequence NZ_JUGR01000007.1). The lack of any genes relating to phage integration within these scaffolds and the very high nucleotide sequence homology observed, suggests that these findings be treated with caution. The differences in G+C content of these phage-related scaffolds compared to the remainder of the bacterial genome assembly also suggests possible sequence contamination. For example, the Terriglobus genome has seven scaffolds, six of which have G+C of 54–58% whereas the phage-related scaffold has G+C of 35%.
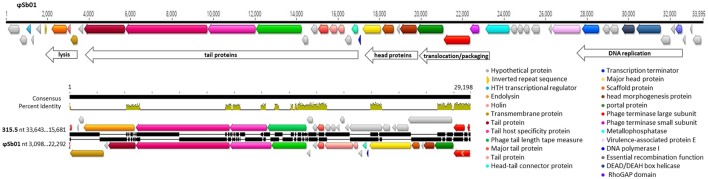
Genome organization and phylogeny of the Streptococcus phage ϕsb01
The Streptococcus phage ϕSb01 was the rumen phage most highly related to previously reported phages, with 34 of the 35 ORFs for which gene function could be assigned, having protein homology to previously characterized Streptococcus phage and prophage-associated genes (Tables 2, 4). The only exception was one ORF (essential recombination function protein YP_009216915.1) which was homologous to a protein from the Clostridium phage phiCDHM19 (Hargreaves and Clokie, 2015). The ϕSb01 genome was found to be organized into modular groups relating to gene function (Figure 5). For example, genes relating to phage particle structure including tail proteins (major and minor structural tail proteins, tail host-specificity protein, tail length tape measure protein, and head-tail connector proteins) were clustered together. These genes were also adjacent to a transcription regulator, indicating that phage genome replication was regulated and controlled through sequential transcription of phage genes (late and early gene replication).
Figure 5.
Genome arrangement of the Streptococcus phage ϕSb01 showing predicted ORFs and alignment with a homologous region of the Streptococcus phage 315.5 (NCBI virus reference sequence NC_004588). Modular groupings of phage genes with similar functional roles are described and predicted ORFs are depicted with arrows colored according to putative gene products. The alignment was constructed using Mauve (Darling et al., 2010) and includes a consensus sequence and corresponding percentage identity graph ( 100%, >30%, <30%) with differences in the linear alignment of nucleotide sequences indicated with vertical shading.
The nearest phage relative to ϕSb01 on the basis of protein homology was the S. pyogenes phage 315.5 (Beres et al., 2002) with a conserved module of 10 genes encoding proteins responsible for phage particle structure and morphogenesis, including tail proteins, a phage:host specificity minor tail fiber protein and phage head and head morphogenesis proteins (Figure 5). The major phage packaging genes (phage portal and terminase genes) were most like the Streptococcus phage T12 which is able to infect and form a lysogenic association with group A Streptococcus (GAS) strains (Zabriskie, 1964; McShan et al., 1997). When originally isolated from bovine rumen fluid, ϕSb01 particles were shown to replicate and produce clear plaques on lawns of bacterial host (Klieve and Bauchop, 1991) and pure preparations of this phage were subsequently shown to rapidly clear liquid cultures, indicating phage lytic replication. While the genome of this phage did contain several genes involved in gene regulation, no integrase genes were identified, further indicating that this phage was unable to integrate and form a stable, lysogenic associations with its bacterial host.
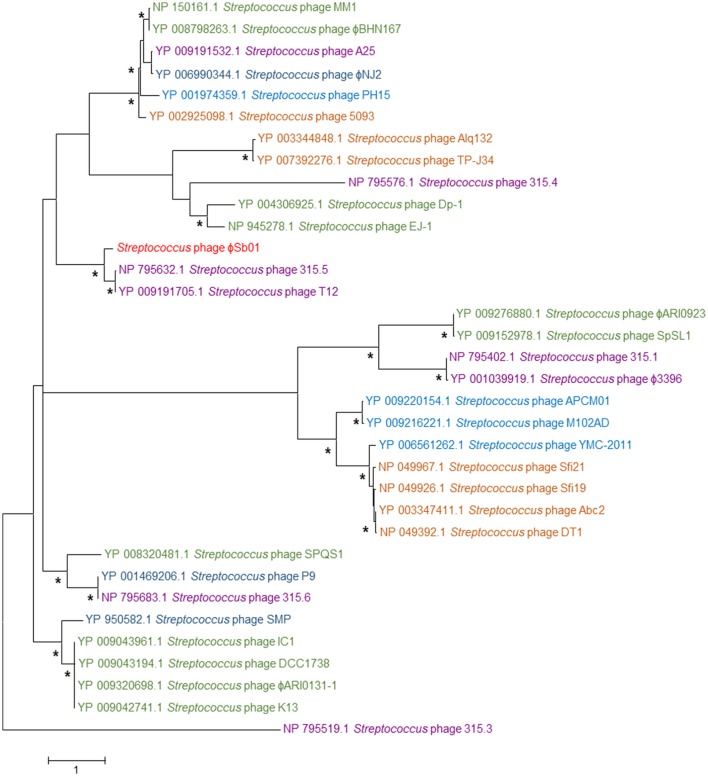
Phylogeny on the basis of the terminase large subunit gene of phages known to infect human, animal and environmental isolates of the genus Streptococcus (Figure 6), showed that the TerL protein of this phage was most similar to the TerL genes of phages infecting S. pyogenes of human respiratory tract origin.
Figure 6.
Maximum likelihood phylogeny of the phage terminase large subunit (TerL) proteins of the Streptococcus phage ϕSb01 and representative reference phages infecting the genus Streptococcus. The tree with the highest log likelihood (−21,218.68) based on the Whelan and Goldman model (Whelan and Goldman, 2001) with frequencies and a gamma distribution with five categories is shown. This tree used a total of 667 amino acid positions and is drawn to scale, with branch lengths measured in the number of substitutions per site and is rooted with the TerL protein sequence of the Streptococcus phage 315.3. Bootstrap supports greater than 85%, based on 1,000 replicates, are indicated (*). The TerL proteins are colored according to phage host species and environmental isolation source S. bovis/equinus 2B (rumen, gut-associated); S. pneumoniae (human respiratory tract); S. pyogenes, GAS strains and S. dysgalactiae subsp. equisimilis (human, skin flora, respiratory tract); S. suis and S. equi (non-human, mammal respiratory tract); S. mutans, S. salivarius, and S. gordonii (human dental and oral); S. thermophilus (dairy fermentations, whey).
Phage:host resistance mechanisms
Five whole genome sequences representing at least one bacterial host strain for the five lytic phages, were examined. Host strains included (1) Bacteroides sp. AR20, the Gram negative, anaerobic bacterial host for ϕBrb01 and ϕBrb02 and classified within the Fibrobacteres-Chlorobi-Bacteroidetes superphylum (FCB group), Bacteroidetes phylum; (2) R. albus AR67, the Gram positive, anaerobic bacterial host of ϕRa02 and ϕRa04 and classified within the Phylum Firmicutes, order Clostridiales; and (3) three known bacterial hosts of ϕSb01, S. equinus strains 2B, Sb04 and Sb17 which are all Gram positive, facultative anaerobes, classified within the Phylum Firmicutes, order Lactobacillales. All bacterial sequences were interrogated to identify genetic factors which may contribute to phage resistance including prophage elements, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and restriction modification (RM) systems. The five lytic phages reported in this study however, must be unaffected by, or be able to counteract, these resistance mechanisms as they can all successfully infect and replicate within these bacteria.
Phage-related genes were found to be present in all the bacterial host genomes examined (Table 5) and were often present as modules of genes resembling integrated prophage-like elements. The prophage-like elements identified in Bacteroides sp. AR20, the S. equinus strains Sb04 and Sb17 and R. albus AR67 incorporated phage integrase and DNA replication genes and were of variable lengths (predicted sequence length ranging between 4.7 and 32 kb). Genes for major structural proteins or phage particle morphogenesis were usually not associated with these prophage regions and given the relatively low number of ORFs identified, these regions most likely represent incomplete remnants of phage or horizontally-transferred genetic elements. Moreover, sequence alignment of these regions with each of the respective lytic phage genomes indicated that the majority of genes found in these incomplete phage-like regions, were not homologous to the genes found in the lytic phages.
Table 5.
Description of predicted prophage regions identified in the bacterial host strain genomes Bacteroides sp. AR20, R. albus AR67, and S. equinus strains 2B, Sb04, and Sb17.
| Prophage region | Region length (bp) | G + C content (%) | ORFs (no.) | Most common phage name (number of best hit homologous proteins)* | Additional common phage names (number of best hit homologous proteins)** |
|---|---|---|---|---|---|
| ϕBrbAR20pro1 | 27,691 | 38.6 | 32 | n.d. | Acanthocystis turfacea Chlorella virus 1 (1); Cafeteria roenbergensis virus BV-PW1 (1); Bathycoccus sp. RCC1105 virus BpV1 (1); Mycobacterium phage Kratio (1); Streptococcus virus 9872 (1); Staphylococcus phage phiN315 (1); Bacillus phage SP-15 (1); Only Syngen Nebraska virus 5 (1); Staphylococcus phage phiN315 (1); Cedratvirus A11 (1); Stx2-converting phage 1717 (1); Acanthamoeba polyphaga moumouvirus (1); Enterococcus phage EF62phi (1) |
| ϕBrbAR20pro2 | 29,595 | 46.7 | 28 | n.d. | Natrialba phage PhiCh1 (1); Gordonia phage Nyceirae (1); Bacillus phage BM5 (1); Bacillus phage vB_BanS-Tsamsa (1); Clostridium phage c-st (1); Arthrobacter phage vB_ArS-ArV2 (1); Pseudomonas phage YuA (1); Ostreococcus mediterraneus virus 1 (1) |
| ϕBrbAR20pro3 | 4,783 | 40.7 | 5 | Riemerella phage RAP44 NC_019490.1 (2) | Clostridium phage phiCT9441A (1) |
| ϕRaAR67pro1 | 5,340 | 41.5 | 9 | n.d. | Clostridium phage phiCT19406C (1); Geobacillus phage GBSV1 (1); Paenibacillus phage Vegas (1) |
| ϕRaAR67pro2 | 17,507 | 50 | 22 | n.d. | Clostridium phage phiSM101 (1); Brevibacillus phage Jenst (1); Deep-sea thermophilic phage D6E (1); Paramecium bursaria Chlorella virus FR483(1) |
| ϕRaAR67pro3 | 8,051 | 37.8 | 8 | n.d. | Bacillus phage phIS3501 (1); Thermus phage P2345 (1); Brevibacillus phage Jenst (1); Enterobacteria phage fiAA91-ss (1); Clostridium phage CDMH1 (1); Aureococcus anophagefferens virus (1) |
| ϕSb2Bpro1 | 42,367 | 38.5 | 52 | Streptococcus phage P9 NC_009819 (6) | Streptococcus phage 315.6 (6); Bacillus phage BCJA1c (4); Enterococcus phage EFC-1 (2); Staphylococcus phage X2 (2); Streptococcus phage SM1 (2); Streptococcus phage phiARI0468-2 (2) |
| ϕSb2Bpro2 | 10,673 | 38.5 | 15 | n.d. | Clostridium phage phi3626 (1); Clostridium phage phiMMP02 (1); Vibrio phage douglas 12A4 (1); Mycobacterium phage Squirty (1); Acidianus two-tailed virus (1); Geobacillus phage GBSV1 (1); Acinetobacter phage Acj9 (1) |
| ϕSeSb04pro1 | 16,394 | 37.1 | 15 | Bacillus phage SPBc2 NC_001884.1 (3) | Streptococcus phage Dp-1 (2); Bacillus virus G (1); Acanthocystis turfacea Chlorella virus 1 (1); Aureococcus anophagefferens virus (1); Cellulophaga phage phiSM (1); Bacillus phage BCD7 (1); Lactococcus phage bIL311 (1); Synechococcus phage S-SSM7(1) |
| ϕSeSb04pro2 | 6,961 | 37.9 | 9 | n.d. | Streptococcus phage 315.3 (1); Aeropyrum pernix spindle-shaped virus 1 (1); Escherichia phage phAPEC8 (1); Sphingomonas phage PAU (1); Enterobacteria phage phi92 (1); Streptococcus phage phiARI0460-1 (1); Acinetobacter phage ZZ1(1) |
| ϕSeSb04pro3 | 8,194 | 35.4 | 9 | Bacillus virus G NC_023719.1 (2) | Acanthamoeba polyphaga mimivirus (1); Planktothrix phage PaV-LD (1); Lactobacillus phage LfeInf (1); Staphylococcus phage phiIPLA-C1C(1) |
| ϕSeSb17pro1 | 12,602 | 33.1 | 21 | Streptococcus phage EJ-1 NC_005294.1 (3) | Lactococcus phage bIL310 (2); Streptococcus phage phi3396 (2); Lactococcus phage bIL311 (2); Streptococcus phage 315.3 (2) |
Most common phage names and NCBI Virus reference sequence IDs are listed except where the most common phage name was not determined (n.d.) as the number of best hit homologous proteins did not exceed more than one hit for any one phage.
Additional common names listed include those where the number of best hit homologous proteins >1. Where the number of best hit homologous proteins never exceeded more than one phage name, all phage names corresponding to each ORF best hit homologous protein match are listed.
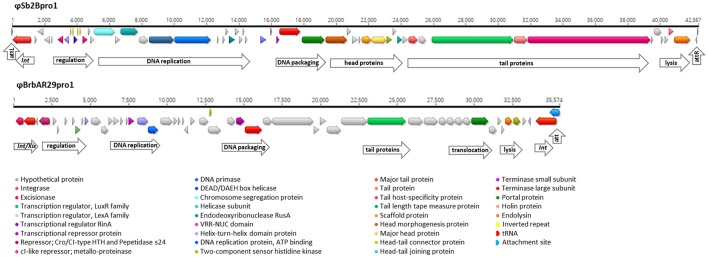
The exception to this was the TerL gene of ϕSb01 which was similar (69% pairwise nt identity) to the TerL gene present within the relatively long (42.3 kb) prophage element identified within S. equinus 2B. This prophage element, designated ϕSb2Bpro1 was the only host-related element identified in the study which contained a potentially complete complement of phage genes. Prophage ϕSb2Bpro1 encoded 52 ORFs and included genes with homology to both structural and non-structural phage proteins arranged in a modular sequence, interspaced by regulatory control genes (Figure 7). In contrast to the lytic phage ϕSb01, the genome of ϕSb2Bpro1 incorporated the key genes required for phage integration and maintaining lysogeny through regulation of gene expression.
Figure 7.
Genome arrangement of prophage regions identified in the bacterial genome sequences of Streptococcus equinus strain 2B (ϕSb2Bpro1) and Bacteroides sp. AR29 (ϕBrbAR29pro1). Modular groupings of phage genes with similar functional roles are described and predicted ORFs are depicted with arrows colored according to putative gene products. Predicted phage attachment sites (att), inverted repeat sequences and a tRNA element are also depicted.
CRISPR associated proteins (Cas proteins) were found to be present in all the bacterial strains examined (Table 6), with the exception of R. albus AR67, which had several relatively short CRISPR direct repeat (DR) regions present in multiple scaffolds of the AR67 genome assembly. This may be attributed to the relatively incomplete nature of this genome assembly (103 scaffolds), which may have hampered the detection of CRISPR-Cas regions with a full complement of genes. Interestingly, of all the CRISPR regions identified, none showed any spacer sequence homology to the lytic phage genome sequences reported in this study.
Table 6.
CRISPR/Cas elements identified in bacterial host genome sequences.
| Bacterial host strain | Cas proteins* | DR (no. repeats; repeat length), nt co-ordinates** | DR homology (10−3 e-value threshold)*** | Spacers (no.) | Spacer sequence homology (10−3 e-value threshold)**** |
|---|---|---|---|---|---|
| Bacteriodes sp. AR20 | None | DR1 (4, 23 nt) contig 52.52 nt 609–778 | No homology | 3 | R. albus plasmid pAR67 (D88665.1) |
| R. albus AR67 | None | DR1 (4; 24 nt) contig1.1 nt 261160–261418 | No homology | 3 | No homology |
| DR2 (4, 24 nt) contig 1.1 nt 261550–261807 | No homology | 3 | No homology | ||
| DR3 (4, 24 nt) contig 14.14 nt 9638–9973 | No homology | 4 | No homology | ||
| DR4 (12, 23 nt) contig 31.31 nt 1118–1998 | No homology | 11 | No homology | ||
| DR5 (4, 24) contig 44.44 nt 14202–14460 | No homology | 3 | No homology | ||
| S. equinus 2B | Cas2, Cas1, Csn1 | None | |||
| S. equinus Sb04 | Csn1, Cas1, Cas2, Csn2, Cas Csm6 | DR1 (21; 36) contig 101.101 nt 253157–254514 | S. thermophilus strain ND07 (CP016394.1); S. thermophilus strain KLDS (CP016877.1); S. thermophilus strain KLDS SM (CP016026.1) | 20 | No homology |
| S. equinus Sb17 | Cas Csm6 | None |
CRISPR associated (Cas) proteins from IMG genome annotation;
DR with >3 repeats;
DR homology, top 3 best hits results, BLASTn against the NCBI nr database (January 2017 update), e-value threshold of 10−3.
Spacer sequence homology top 3 best hits results, BLASTn against the NCBI nr database (January 2017 update), e-value threshold of 10−3.
The two Streptococcus strains, 2B and Sb17 had genes for Cas proteins however no CRISPR direct repeats (DR) were either associated with these genes or detected elsewhere within the genome assemblies. The Bacteroides sp. AR20 genome contained a CRISPR incorporating three spacer regions however there were no Cas proteins identified within this genome. Two of the AR20 CRISPR spacer regions were homologous (100% homology over the complete 32 nt spacer region) to a plasmid replication protein previously identified in R. albus AR67 (Ohara et al., 1998). The most complete CRISPR element was identified in the genome of S. equinus Sb04, with the DR sequences being highly related to those found in other Streptococcus (Table 6). This CRISPR region had all the elements of a type II-A CRISPR-Cas system with a genomic arrangement similar to those previously observed in S. thermophilus and S. pyogenes (Barrangou, 2013; Shmakov et al., 2017) incorporating the consecutive proteins, Csn1, Cas1, Cas2, and Csn2 followed by the CRISPR repeat region. The Cas proteins and two of the DR in this CRISPR element also showed extensive sequence homology to a complete CRISPR-Cas system present in the Streptococcus infantarius subsp. infantarius strain CJ18 (Jans et al., 2013) with > 94% nt homology to Cas and Csn proteins and 100% nt homology to two of the DRs of 35 and 36 nt in length.
Restriction enzyme and DNA methylase sequences were identified in all of the bacterial host genomes (Table 7) however the fragmented nature of the AR67 genome assembly made it difficult to identify the modules of consecutive genes characteristic of some restriction-modification systems. The most complete Type I RM systems were identified in the genomes of S. equinus Sb04 and S. equinus Sb17, recognizable on the basis of sequential genes representing multi-subunit systems of Type I restriction enzyme specificity (S) subunits, Type 1 methyltransferase (M) and Type I restriction (R) enzymes. These enzymes can act in combination to recognize bi-partite motifs and cleave at large distances from their binding site (Blow et al., 2016).
Table 7.
Number of bacterial host genes homologous to genes associated with restriction-modification systems (10−3 threshold BLASTp search against the REBASE protein database).
| Restriction-modification system related genes (REBASE classifications*) | Bacteroides sp. AR20 | R. albus AR67 | S. equinus 2B | S. equinus Sb04 | S. equinus Sb17 |
|---|---|---|---|---|---|
| Putative control protein | 5 | 11 | 7 | 5 | 9 |
| Putative homing endonuclease | 8 | 6 | 8 | 8 | 8 |
| Homing endonuclease | 2 | ||||
| Putative orphan methyltransferase | 1 | 1 | 1 | 1 | 1 |
| Putative Type I restriction enzyme | 3 | 7 | 7 | 6 | 6 |
| Type I specificity subunit | 3 | 9 | |||
| Putative Type I specificity subunit | 15 | 13 | 9 | 9 | 11 |
| Type I methyltransferase | 8 | 7 | 9 | 8 | 8 |
| Putative Type I methyltransferase | 10 | 10 | 6 | 9 | 8 |
| Putative Type II specificity subunit | 1 | 1 | 1 | 1 | 1 |
| Type II methyltransferase | 1 | ||||
| Putative Type II methyltransferase | 23 | 44 | 22 | 20 | 19 |
| Putativetype II nicking endonuclease | 2 | 2 | 3 | 2 | 2 |
| Putative Type II restriction enzyme | 41 | 30 | 12 | 13 | 17 |
| putative Type II helicase domain protein | 1 | ||||
| Type IIG restriction enzyme/methyltransferase | 1 | 1 | |||
| Putative Type IIG restriction enzyme/methyltransferase | 43 | 49 | 12 | 12 | 11 |
| Putative Type III restriction enzyme | 2 | ||||
| Type III methyltransferase | 1 | ||||
| Putative Type III methyltransferase | 1 | ||||
| Putative Type IV methyl-directed restriction enzyme | 32 | 33 | 26 | 26 | 23 |
Restriction modification type classifications from the REBASE protein database, October 2016 (Roberts et al., 2015) including biochemically characterized proteins and predicted proteins (putative).
Intact Type I RM systems were not identified in the other rumen bacterial host genomes, with the Bacteroides sp. AR20 and R. albus AR67 genomes instead containing multiple copies of Type II RM systems, Type IIG restriction/methyltransferase genes and Type IV methyl-directed restriction enzymes. These RM systems are simpler in structure with the Type II RM systems comprising restriction endonuclease and DNA methyltransferase enzymes which are expected to show identical DNA-binding specificity (Pingoud et al., 2014). The Type IIG enzymes which contain both DNA restriction and methylation activities (Blow et al., 2016), were more abundant and diverse in the Bacteroides and Ruminococcus genomes than the three Streptococcus genomes. The Streptococcus genomes Sb04 and Sb17 both encoded multiple copies of the putative Type IIG restriction enzyme/methyltransferase MboSP38ORF5300 (from Mycobacterium bovis strain SP38, Genbank CP015773). S. equinus 2B also encoded a second putative Type IIG restriction enzyme/methyltransferase, Sin18ORF1583, from S. infantarius subsp. infantarius CJ18 (AEZ62887.1). In contrast R. albus AR67 encoded multiple copies of seven different putative Type IIG restriction enzyme/methyltransferases (Ral7ORF3617, MboSP38ORF5300, UbaMSORFI6, SstMg1ORFD; AviDJORF22890, HauORF2540, and Ral7ORF3270) and Bacteroides sp. AR20 encoded multiple copies of 11 different putative Type IIG restriction enzyme/methyltransferases (Mpu984ORF12500, OrhH06ORF7025, Bxy1bORF28720, Bxy1bORF28730, PkoX141TORF16370, DxiR13ORFI, MspMAB1ORF830, Htr1232ORFE, HmgGUTORFU2, AspNJ1ORF21265, MboSP38ORF5300). The Type IV RM systems, distinguished by their ability to cut modified DNA without relying on an associated methyltransferase component (Loenen and Raleigh, 2014), were also more predominant in the AR20 and AR67 genomes than the Streptococcus genomes.
Conclusion
This study reports the first full genome sequences obtained for lytic phages that infect bacteria isolated from the rumen. These phages were sourced directly from the rumen and ruminant-housing and waste-water environments and infect Firmicutes (Ruminococcus and Streptococcus) and Bacteroidetes (Bacteroides). The genome sequences were novel in their genetic composition and while some more conserved phage genes could be identified, for example, DNA polymerase, head-tail connector proteins, and phage DNA packaging genes (terminase large subunit genes), the majority of phage genes encoded by these lytic phages could not be assigned functional roles on the basis of sequence homology. This was most apparent for the phages ϕRa02 and ϕRa04 infecting R. albus AR67, where only three genes could be annotated with confidence. Drawing on these few genes for phylogenetic analysis and the prediction of proteins with transmembrane potential, enabled these tailed phages to be further classified within the Podovirus subfamily, Picovirinae and the Phi29 genus.
The phages infecting Bacteroides sp. AR20, ϕBrb01, and ϕBrb02 were also genetically novel, showing little similarity to the two previously sequenced lytic phages infecting the Bacteroides genera, B. fragilis, B40-8 (Puig and Gironés, 1999) and B124-14 (Ogilvie et al., 2012). Despite the dominance and importance of the Bacteroides taxa in the human gut (Ndeh et al., 2017) and a history of lytic phages infecting the genera Bacteroides being employed for the purposes of tracking fecal contamination of waterways (Puig et al., 1999), very few prophage elements or lytic phages infecting the genus Bacteroides have been genetically described. Within the ϕBrb01 and ϕBrb02 genomes, conserved phage genes including those involved in phage DNA replication (DNA polymerase I and helicases), particle packaging and translocation (terminase large subunit and portal proteins) and host cell lysis (phage endolysin) could be identified. Phylogenetic analysis on the basis of the terminase large subunit gene and comparative analysis on the basis of predicted protein sequences, indicated that the ϕBrb01 and ϕBrb02 genomes were most closely related to prophage elements associated with bacterial isolates of Bacteroides and an intact prophage present in the genome of an ovine rumen Bacteroides isolate, strain AR29. The ϕBrb01 and ϕBrb02 genomes however lacked genes for phage integration and replication control, indicating that they could not establish a stable, lysogenic association with their bacterial host. These genomes however were also quite unique in their genetic composition, containing many genes of unknown function. Anticipated structural phage genes such as those encoding for tail and capsid proteins, homologous to any previously characterized phage structural proteins, were not found and further protein expression studies would be required to fully elucidate the functional roles of many of the ϕBrb01 and ϕBrb02 genes.
Of the five phage genomes described in this study, the genome sequence of the lytic phage ϕSb01 was found to have the greatest homology to previously sequenced prophages and lytic phages infecting bacteria of the Streptococcus genus. This greatly facilitated annotation of the phage genome and the functional, modular organization of the genome could be determined. Genes for phage integration and maintenance of lysogeny were absent from the genome indicating that this phage relied on the lytic cycle of phage replication to persist in the rumen environment. This lytic phage could also infect and successfully replicate in multiple hosts, being originally isolated with the ovine rumen isolate S. equinus 2B as host strain and later shown to also infect two additional bovine rumen isolates of S. equinus (strains Sb04 and Sb17) (Klieve and Bauchop, 1991; Klieve et al., 1999). This indicates that this phage uses cell wall receptors that are relatively common in S. equinus for initial phage attachment and encodes for proteins which can function in closely-related bacterial strains, including those of both ovine and bovine origin. Phylogenetic analysis on the basis of the TerL gene, and comparative analysis of predicted phage proteins, indicated that this phage was highly related to phages and prophages known to infect other Streptococcus species, including those of S. pyogenes, S. parauberis, and S. agalactiae. Modules of genes contained in related phages were also present in the ϕSb01 genome, suggesting an evolutionary history of genetic exchange and conservation of phage gene modules, which may facilitate the ability to replicate in a wider spectrum of Streptococcus hosts and contribute to an increased phage host range.
The availability of bacterial genome assemblies for the phage host strains has contributed to our understanding of phage:host interactions, enabling an assessment of the phage defense systems employed by these bacteria. Previous studies have noted the formation of extracellular polysaccharide capsules and clumping of cells contributing to host resistance to the lytic phages ϕBrb01 and ϕSb01 (Klieve and Bauchop, 1991; Klieve et al., 1991). Interestingly, whatever physical and enzymatic mechanisms the respective bacterial hosts employ to prevent phage attachment and intracellular replication, the lytic phages must be either unaffected by or be able to avoid or circumvent these mechanisms, in order to successfully replicate.
While the phages ϕBrb01 and ϕBrb02 were previously shown to infect three and four rumen-derived Bacteroides isolates (Klieve et al., 1991) respectively, only one of these bacterial strains has been genome sequenced (Bacteroides sp. AR20). Genomes of the rumen Bacteroides and Ruminococcus host strains AR20 and AR67 did not contain intact prophage elements and although short CRISPR regions were detected, these strains appeared to lack Cas proteins. The genomes of these bacteria however, did contain an extensive diversity and numbers of genes relating to RM systems, suggesting that these bacteria rely more on these mechanisms for specific and non-specific methylation and/or cleavage of incoming, foreign DNA and consequently protect against phage infection. In contrast the three Streptococcus genomes (strains 2B, Sb04, and Sb17) contained fewer genes relating to Type II, Type IIG, and Type IV RM systems than the Bacteroides and Ruminococcus genomes. The complex Type I RM systems identified in the strains Sb04 and Sb17 were absent from the genome of strain 2B. In addition, the genome for strain 2B was the only bacterial host found to contain a possibly intact prophage element (designated ϕSb2Bpro1). This prophage element did not appear to convey super-infection immunity to the lytic phage ϕSb01 and the lack of CRISPR elements and relatively low numbers of genes relating to RM systems, may explain the previous observation that this Streptococcus strain may be more receptive to lytic and lysogenic phage infection than other rumen-derived Streptococcus isolates (Klieve et al., 1999). The variety and extent of the phage defense strategies utilized by the five different bacteria examined in this study are likely to reflect those employed in the wider rumen microbial community, indicating that some bacterial strains, even within the same genus, may be more receptive to phage infection and sustaining phage replication than others.
With improved methods for DNA sequencing and the advent of metagenomic studies to comprehensively sequence phage particle fractions obtained from rumen fluid, the full extent of viral diversity within the rumen is gradually being revealed. Exploring metagenomic datasets relies on sequence homology to classify the large volumes of sequence information generated and explorations of the rumen viral metagenome have indicated extensive sequence homology to Caudovirales-like lytic phages and prophage elements originating from rumen bacteria and bacteria from other environments (Berg Miller et al., 2012; Ross et al., 2013). The genome sequencing of phage isolates that infect common gastrointestinal bacteria will complement and greatly enhance the accuracy of phage classification and bioinformatics analysis of viral metagenomics datasets, providing reference sequences and novel viral genes to which relevant sequence homology can be conferred.
Author contributions
RG, SL, WK, EA, DO, and AK: Contributed to the conception and design of the work; RG, CM, and DO: Contributed to the preparation of DNA; WK and SL: Contributed to aquisition of data; RG, WK, and EA: Contributed to data analysis. All authors contributed to the interpretation of data, critical revision and final approval of the version to be published. All authors are in agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge and thank Mrs. Kerri Chandra for assistance with data analysis and Mr. David Diggles and Mr. Lindsay Brebber of the Queensland Department of Natural Resources and Mines for assistance with software installation and high performance computing advice.
Footnotes
Funding. The Hungate1000 project was funded by the New Zealand Government in support of the Livestock Research Group of the Global Research Alliance on Agricultural Greenhouse Gases (http://www.globalresearchalliance.org) and was a global effort by members of the Rumen Microbial Genomics Network (http://www.rmgnetwork.org). The genome sequencing and analysis component of the project was supported by the US Department of Energy Joint Genome Institute (JGI), a DOE Office of Science User Facility supported under Contract No. DE-AC02-05CH11231, through their Community Science Program (CSP 612), and used resources of the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the US Department of Energy.
References
- Ackermann H. W., DuBow M. S. (1987). General Properties of Bacteriophages. Boca Raton, FL: CRC Press. [Google Scholar]
- Adams J. C., Gazaway J. A., Brailsford M. D., Hartman P. A., Jacobson N. L. (1966). Isolation of bacteriophages from the bovine rumen. Experentia 22, 717–718. 10.1007/BF01901335 [DOI] [Google Scholar]
- Altermann E., Lu J., McCulloch A. (2017). GAMOLA2, a comprehensive software package for the annotation and curation of draft and complete microbial genomes. Front. Microbiol. 8:346. 10.3389/fmicb.2017.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D., Grant J. R., Marcu A., Sajed T., Pon A., Liang Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, 16–21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R. (2013). CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 4, 267–278. 10.1002/wrna.1159 [DOI] [PubMed] [Google Scholar]
- Beres S. B., Sylva G. L., Barbian K. D., Lei B., Hoff J. S., Mammarella N. D., et al. (2002). Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. U.S.A. 99, 10078–10083. 10.1073/pnas.152298499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg Miller M. E., Yeoman C. J., Chia N., Tringe S. G., Angly F. E., Edwards R. A., et al. (2012). Phage–bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ. Microbiol. 14, 207–227. 10.1111/j.1462-2920.2011.02593.x [DOI] [PubMed] [Google Scholar]
- Blow M. J., Clark T. A., Daum C. G., Deutschbauer A. M., Fomenkov A., Fries R., et al. (2016). The epigenomic landscape of prokaryotes. PLoS Genet. 12:e1005854. 10.1371/journal.pgen.1005854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLOS ONE 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679. 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichorst S. A., Breznak J. A., Schmidt T. M. (2007). Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the Phylum Acidobacteria. Appl. Environ. Microbiol. 73, 2708–2717. 10.1128/AEM.02140-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, 279–285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Makarova K. S., Wolf Y. I., Koonin E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. 10.1093/nar/gku1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey K. J., Saedi M. S., Ito J. (1985). The complete sequence of Bacillus phage phi 29 gene 16: a protein required for the genome encapsidation reaction. Gene 40, 311–316. 10.1016/0378-1119(85)90054-X [DOI] [PubMed] [Google Scholar]
- Gascón I., Lazaro J. M., Salas M. (2000). Differential functional behavior of viral phi29, Nf and GA-1 SSB proteins. Nucleic Acids Res. 28, 2034–2042. 10.1093/nar/28.10.2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson M. K., Forsberg K. J., Dantas G. (2015). Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 9, 207–216. 10.1038/ismej.2014.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert R. A., Klieve A. V. (2015). Ruminal viruses (Bacteriophages, Archaeaphages), in Rumen Microbiology: From Evolution to Revolution, eds Puniya A. K., Singh R., Kamra D. N. (New Delhi: Springer; ), 121–141. [Google Scholar]
- Gnerre S., MacCallum I., Przybylski D., Ribeiro F. J., Burton J. N., Walker B. J., et al. (2011). High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. U.S.A. 108, 1513–1518. 10.1073/pnas.1017351108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D. H., Selengut J. D., Richter R. A., Harkins D., Basu M. K., Beck E. (2013). TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 41, D387–D395. 10.1093/nar/gks1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves K. R., Clokie M. R. (2015). A Taxonomic review of Clostridium difficile phages and proposal of a novel genus, “Phimmp04likevirus.” Viruses 7, 2534–2541. 10.3390/v7052534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. S. (2007). Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, The Pennsylvania State University. [Google Scholar]
- Hungate R. E., Bryant M. P., Mah R. A. (1964). The rumen bacteria and protozoa. Annu. Rev. Microbiol. 18, 131–166. 10.1146/annurev.mi.18.100164.001023 [DOI] [PubMed] [Google Scholar]
- Hyatt D., Chen G. L., Locascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J., Krupovic M., Koonin E. V. (2016). The double-stranded DNA virosphere as a modular hierarchical network of gene sharing. mBio 7:e00978–16. 10.1128/mBio.00978-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson W. J., Millis N. F. (1976a). Characterisation of Streptococcus bovis bacteriophages. Can. J. Microbiol. 22, 847–852. 10.1139/m76-122 [DOI] [PubMed] [Google Scholar]
- Iverson W. J., Millis N. F. (1976b). Lysogeny in Streptococcus bovis. Can. J. Microbiol. 22, 853–857. 10.1139/m76-123 [DOI] [PubMed] [Google Scholar]
- Jans C., Follador R., Hochstrasser M., Lacroix C., Meile L., Stevens M. J. (2013). Comparative genome analysis of Streptococcus infantarius subsp. infantarius CJ18, an African fermented camel milk isolate with adaptations to dairy environment. BMC Genomics 14:200. 10.1186/1471-2164-14-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans C., Meile L., Lacroix C., Stevens M. J. (2015). Genomics, evolution, and molecular epidemiology of the Streptococcus bovis/Streptococcus equinus complex (SBSEC). Infect. Genet. Evol. 33, 419–436. 10.1016/j.meegid.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Kang I., Kang D., Cho J.-C. (2016). Complete genome sequence of bacteriophage P2559Y, a marine phage that infects Croceibacter atlanticus HTCC2559. Mar Genomics 29, 35–38. 10.1016/j.margen.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieve A. V. (2005). 2.2. Bacteriophages in Methods in Gut Microbial Ecology for Ruminants, eds Makkar H. P. S., McSweeney C. S. (Dordrecht: International Atomic Energy Agency; Springer Academic Press; ), 39–46. [Google Scholar]
- Klieve A. V., Bain P. A., Yokoyama M. T., Ouwerkerk D., Forster R. J., Turner A. F. (2004). Bacteriophages that infect the cellulolytic ruminal bacterium Ruminococcus albus AR67. Lett. Appl. Microbiol. 38, 333–338. 10.1111/j.1472-765X.2004.01493.x [DOI] [PubMed] [Google Scholar]
- Klieve A. V., Bain P. A., Yokoyama M. T., Ouwerkerk D., Forster R. J., Turner A. F. (2006). Corrigendum. Lett. Appl. Microbiol. 42, 654–654. 10.1111/j.1472-765X.2006.01944.x [DOI] [PubMed] [Google Scholar]
- Klieve A. V., Bauchop T. (1988). Morphological diversity of ruminal bacteriophages from sheep and cattle. Appl. Environ. Microbiol. 54, 1637–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieve A. V., Bauchop T. (1991). Phage resistance and altered growth habit in a strain of Streptococcus bovis. FEMS Microbiol. Lett. 80, 155–160. 10.1111/j.1574-6968.1991.tb04653.x [DOI] [PubMed] [Google Scholar]
- Klieve A. V., Gilbert R. A. (2005). 4.2 Bacteriophage populations, in Methods in Gut Microbial Ecology for Ruminants, eds Makkar H. P. S., McSweeney C. S. (Dordrecht: International Atomic Energy Agency; Springer Academic Press; ), 129–137. [Google Scholar]
- Klieve A. V., Gregg K., Bauchop T. (1991). Isolation and characteristics of lytic phages from Bacteroides ruminicola ss brevis. Curr. Microbiol. 23, 183–187. 10.1007/BF02092277 [DOI] [Google Scholar]
- Klieve A. V., Heck G. L., Prance M. A., Shu Q. (1999). Genetic homogeneity and phage susceptibility of ruminal strains of Streptococcus bovis isolated in Australia. Lett. Appl. Microbiol. 29, 108–112. 10.1046/j.1365-2672.1999.00596.x [DOI] [PubMed] [Google Scholar]
- Klieve A. V., Hudman J. F., Bauchop T. (1989). Inducible bacteriophages from ruminal bacteria. Appl. Environ. Microbiol. 55, 1630–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klieve A. V., Swain R. A. (1993). Estimating ruminal bacteriophage numbers using pulsed field gel electrophoresis and laser densitometry. Appl. Environ. Microbiol. 59, 2299–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Q., Gascuel O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Raleigh E. A. (2014). The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 42, 56–69. 10.1093/nar/gkt747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a proGram for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz V. M., Chen I. M., Palaniappan K., Chu K., Szeto E., Grechkin Y., et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122. 10.1093/nar/gkr1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan W. M., Tang Y. F., Ferretti J. J. (1997). Bacteriophage T12 of Streptococcus pyogenes integrates into the gene encoding a serine tRNA. Mol. Microbiol. 23, 719–728. 10.1046/j.1365-2958.1997.2591616.x [DOI] [PubMed] [Google Scholar]
- McTaggart T. L., Shapiro N., Woyke T., Chistoserdova L. (2015). Draft genomes of two strains of Flavobacterium isolated from lake washington sediment. Genome Announc. 3, e01597–e01514. 10.1128/genomeA.01597-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer W. J., Serna-Rico A., Salas M. (2001b). Characterization of the bacteriophage ϕ29-encoded protein p16.7: a membrane protein involved in phage DNA replication. Mol. Microbiol. 39, 731–746. 10.1046/j.1365-2958.2001.02260.x [DOI] [PubMed] [Google Scholar]
- Meijer W. J., Horcajadas J. A., Salas M. (2001a). Phi29 family of phages. Microbiol. Mol. Biol. Rev. 65, 261–287. 10.1128/mmbr.65.2.261-287.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara T., Nishimura Y., Shimizu Y., Nishiyama H., Yoshikawa G., Uehara H., et al. (2016). Linking virus genomes with host taxonomy. Viruses 8:66. 10.3390/v8030066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndeh D., Rogowski A., Cartmell A., Luis A. S., Baslé A., Gray J., et al. (2017). Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70. 10.1038/nature21725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogilvie L. A., Caplin J., Dedi C., Diston D., Cheek E., Bowler L., et al. (2012). Comparative (meta)genomic analysis and ecological profiling of human gut-specific bacteriophage phiB124-14. PLoS ONE 7:e35053. 10.1371/journal.pone.0035053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara H., Miyagi T., Kaneichi K., Karita S., Kobayashi Y., Kimura T., et al. (1998). Structural analysis of a new cryptic plasmid pAR67 isolated from Ruminococcus albus AR67. Plasmid 39, 84–88. 10.1006/plas.1997.1324 [DOI] [PubMed] [Google Scholar]
- Paynter M. J. B., Ewert D. L., Chalupa W. (1969). Some morphological types of bacteriophages in bovine rumen contents. Appl. Microbiol. 18, 942–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedruzzi I., Rivoire C., Auchincloss A. H., Coudert E., Keller G., de Castro E., et al. (2015). HAMAP in 2015: updates to the protein family classification and annotation system. Nucleic Acids Res. 43, D1064–D1070. 10.1093/nar/gku1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pingoud A., Wilson G. G., Wende W. (2014). Type II restriction endonucleases–a historical perspective and more. Nucleic Acids Res. 42, 7489–7527. 10.1093/nar/gku447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig A., Queralt N., Jofre J., Araujo R. (1999). Diversity of Bacteroides fragilis strains in their capacity to recover phages from human and animal wastes and from fecally polluted wastewater. Appl. Environ. Microbiol. 65, 1772–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M., Gironés R. (1999). Genomic structure of phage B40-8 of Bacteroides fragilis. Microbiology 145, 1661–1670. 10.1099/13500872-145-7-1661 [DOI] [PubMed] [Google Scholar]
- Redondo R. A., Kupczok A., Stift G., Bollback J. P. (2013). Complete genome sequence of the novel phage MG-B1 infecting Bacillus weihenstephanensis. Genome Announc. 1, e00216–e00213 10.1128/genomeA.00216-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P. M., Bleasby A. J., Ison J. C., Mullan L., Bottu G. (2011). EMBOSS User's Guide: Practical Bioinformatics. Cambridge: Cambridge University Press. [Google Scholar]
- Roberts R. J., Vincze T., Posfai J., Macelis D. (2015). REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299. 10.1093/nar/gku1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E. M., Petrovski S., Moate P. J., Hayes B. J. (2013). Metagenomics of rumen bacteriophage from thirteen lactating dairy cattle. BMC Microbiol. 13:242. 10.1186/1471-2180-13-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Seet S. G. M. (2005). Genome Sequence of Bacetriophage Phi-AR29: A Basis for Integrative Plasmid Vectors. Ph.D. Thesis, Murdoch University. [Google Scholar]
- Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., et al. (2017). Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169–182. 10.1038/nrmicro.2016.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styriak I., Spanova A., Montagova H., Kmet V. (1994). Isolation and characterisation of a new ruminal bacteriophage lytic to Streptococcus bovis. Curr. Microbiol. 28, 355–358. 10.1007/BF01570201 [DOI] [Google Scholar]
- Tarakanov B. V. (1976). Biological properties of Streptococcus bovis bacteriophages isolated from lysogenic cultures and sheep rumen. Microbiologiia 45, 695–700. [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volozhantsev N. V., Oakley B. B., Morales C. A., Verevkin V. V., Bannov V. A., Krasilnikova V. M., et al. (2012). Molecular characterization of podoviral bacteriophages virulent for Clostridium perfringens and their comparison with members of the Picovirinae. PLoS ONE 7:e38283. 10.1371/journal.pone.0038283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S., Goldman N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. 10.1093/oxfordjournals.molbev.a003851 [DOI] [PubMed] [Google Scholar]
- Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y. (2012). dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, W445–W451. 10.1093/nar/gks479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabriskie J. B. (1964). The role of temperate bacteriophage in the production of erythrogenic toxin by group A Streptococci. J. Exp. Med. 119, 761–780. 10.1084/jem.119.5.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J., Wishart D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39, W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]