Abstract
Among the intestinal pathogenic Escherichia coli, enteroinvasive E. coli (EIEC) are a group of intracellular pathogens able to enter epithelial cells of colon, multiplicate within them, and move between adjacent cells with a mechanism similar to Shigella, the ethiological agent of bacillary dysentery. Despite EIEC belong to the same pathotype of Shigella, they neither have the full set of traits that define Shigella nor have undergone the extensive gene decay observed in Shigella. Molecular analysis confirms that EIEC are widely distributed among E. coli phylogenetic groups and correspond to bioserotypes found in many E. coli serogroups. Like Shigella, also in EIEC the critical event toward a pathogenic life-style consisted in the acquisition by horizontal gene transfer of a large F-type plasmid (pINV) containing the genes required for invasion, intracellular survival, and spreading through the intestinal mucosa. In Shigella, the ample gain in virulence determinants has been counteracted by a substantial loss of functions that, although important for the survival in the environment, are redundant or deleterious for the life inside the host. The pathoadaptation process that has led Shigella to modify its metabolic profile and increase its pathogenic potential is still in infancy in EIEC, although maintenance of some features typical of E. coli might favor their emerging relevance as intestinal pathogens worldwide, as documented by recent outbreaks in industrialized countries. In this review, we will discuss the evolution of EIEC toward Shigella-like invasive forms going through the epidemiology, including the emergence of new virulent strains, their genome organization, and the complex interactions they establish with the host.
Keywords: pathogenic E. coli, enteroinvasive E. coli (EIEC), Shigella, bacterial evolution, emerging EIEC
Introduction
Escherichia coli is not only a harmless commensal of the human and animal intestine but also a major cause of morbidity and mortality (Kaper et al., 2004; Wirth et al., 2006). Indeed, many pathogenic E. coli have been described as cause of diseases both in healthy and immunocompromised individuals. Based on the specific virulence factors and pathogenicity processes, pathogenic E. coli have been subdivided into different pathogroups, that can be broadly grouped as DEC (or intestinal) or extraintestinal E. coli (ExPEC) (Kaper et al., 2004; Croxen and Finlay, 2009; Gomes et al., 2016). DEC include at least six major pathotypes differing in virulence mechanisms, infectious processes, and damages provoked to the target cells: enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), diffusely adherent E. coli (DAEC), as well as adherent invasive E. coli (AIEC), a recently identified pathotype. As for ExPEC, the most common strains belong to two different pathotypes targeting different body compartments: uropathogenic E. coli (UTI) and neonatal meningitis E. coli (NMEC).
The presence of so many different pathotypes exemplifies the remarkable plasticity of E. coli genome, which is characterized by an extremely large pangenome of approximately 20,000 genes in contrast to a common core of about 1700 genes (Rasko et al., 2008; Touchon et al., 2009). Those that vary among different pathogenic E. coli strains have been acquired by intense HGT and are often conveyed by mobile genetic elements (Touchon et al., 2009; Dobrindt et al., 2010; van Elsas et al., 2011).
Among the DEC pathotypes, EIEC are etiological agents of bacillary dysentery in humans, particularly in low-income countries (Croxen et al., 2013; Gomes et al., 2016). The pathogenesis of EIEC infection is characterized by the ability of bacteria to invade the human colonic mucosa, conferred by the expression of chromosomal and plasmid-borne genes (Harris et al., 1982; Sansonetti et al., 1982; Hale et al., 1983; Kaper et al., 2004). Following penetration into colonic epithelial cells, EIEC replicate intracellularly and spread to adjacent cells causing the inflammatory destruction of the intestinal epithelial barrier. This provokes the characteristic dysentery syndrome, usually self-limiting, characterized by the presence of blood, mucus, and leukocytes in stools (DuPont et al., 1971; O’Brien et al., 1979; Taylor et al., 1988). The clinical illness caused by EIEC is similar to that induced by Shigella spp. (Formal and Hornick, 1978; Small and Falkow, 1988), with whom they are closely related in their virulence and other phenotypic properties (Kopecko et al., 1985; Lan et al., 2004). Notwithstanding the similarities in the invasion mechanisms, the infectious dose of EIEC has been observed to be much higher than that of Shigella and the diseases caused by EIEC appear in some cases to be milder (DuPont et al., 1971).
Despite several studies, whether EIEC are precursors of the “full blown” pathogen Shigella, or not is still under debate. In this review, we will attempt at tracing the evolutionary pathway of EIEC considering their epidemiology, the complex mechanisms of their interaction with host cells, the key steps that could have characterized their evolution from a commensal life style toward pathogenicity, and the organization of their genome, including the description of the major traits of emerging EIEC clones.
Epidemiology of Enteroinvasive E. coli (EIEC)
The first report of an EIEC strain dates back at 1947 (Ewing and Gravatti, 1947). At that time, it was defined as “paracolon bacillus” but the strain was later identified as an O124 E. coli. In the 1950s and 1960s, other E. coli strains, isolated from dysentery and initially classified as Shigella manolovi, S. sofia, Shigella strain 13, and S. metadysenteriae, due to their ability to cause experimental keratoconjunctivitis in guinea pigs, were later renamed as EIEC (Manolov, 1959; Rowe et al., 1977; Edwards and Ewing, 1986). Their biochemical characters were first described in 1967 (Sakazaki et al., 1967; Trabulsi et al., 1967).
Enteroinvasive E. coli and Shigella spp. share several phenotypic and genotypic characteristics, often making the discrimination between the two genera challenging (Silva et al., 1980; Toledo and Trabulsi, 1983; Bando et al., 1998; Lan and Reeves, 2002; Pavlovic et al., 2011; van den Beld and Reubsaet, 2012), especially in case of shared serogroups. This difficulty biases the interpretation of the epidemiological information available, hindering the evaluation of the real burden of EIEC infections. As a matter of fact, both EIEC and Shigella spend much of their life cycle within the eukaryotic cells, possessing the ability to use nutrients coming from the host environment. Similarly to Shigella, most EIEC strains are unable to decarboxylate lysine, lack the ability to ferment lactose, and are generally non-motile, with the exception of strains belonging to a few serogroups (Silva et al., 1980; Farmer et al., 1985; Bando et al., 1998; Casalino et al., 2003; Tozzoli and Scheutz, 2014).
A limited set of serotypes have been assigned to EIEC, namely O28ac:H-, O29:H-, O112ac:H-, O115:H-, O121:H-, O124:H-, O124:H7, O124:H30, O124:H32, O135:H-, O136:H-, O143:H-, O144:H-, O144:H25,O152:H-, O159:H-, O159:H2, O164:H-, O167:H-, O167:H4, O167:H5, O173:H-, and recently O96:H19 (Voeroes et al., 1964; Silva et al., 1980; Gomes et al., 1987, 2016; Orskov et al., 1991; Matsushita et al., 1993; Escher et al., 2014; Tozzoli and Scheutz, 2014; Michelacci et al., 2016; Newitt et al., 2016). Some of these EIEC-associated O antigens, such as O28, O112ac, O121, O124, O143, O144, O152, and O167, are identical to O antigens present in Shigella spp. (Cheasty and Rowe, 1983; Tozzoli and Scheutz, 2014).
Enteroinvasive E. coli-infected humans seem to be the major source of infection, as no animal reservoirs have been identified, and transmission uses mainly the oral–fecal route. Although EIEC infections occur worldwide, these are particularly common in low-income countries where poor general hygiene favors their spreading (Chatterjee and Sanyal, 1984; Beutin et al., 1997; Kaper et al., 2004; Vieira et al., 2007).
Enteroinvasive E. coli incidence has been estimated in several countries, and it differs depending on the region (Gomes et al., 2016). Discrepancies among some of the reports can be observed, probably due to the difficulty in discriminating between Shigella and EIEC. In certain countries of Latin America and Asia, namely Chile, Thailand, India, and Brazil, EIEC were found to be common diarrheagenic pathogens (Chatterjee and Sanyal, 1984; Faundez et al., 1988; Echeverria et al., 1992; Blake et al., 1993; Levine et al., 1993), with frequent reports of asymptomatic infected subjects excreting the pathogen (Beutin et al., 1997). In industrialized countries, EIEC infections have been mainly described as travel-related, being reported in returning travelers from high-incidence countries (Wanger et al., 1988; Beutin et al., 1997; Svenungsson et al., 2000). Occasionally, food and water sources have been identified as vehicles of infection, but usually as a secondary contamination by a human source (Tozzoli and Scheutz, 2014).
Enteroinvasive E. coli cause sporadic cases of infection but have been implicated in outbreaks as well, sometimes involving large numbers of cases. In the 1970s a huge outbreak, affecting 387 patients and linked to cheese contaminated with an O124 E. coli strain, occurred in United States (Marier et al., 1973). Recently, an increase of cases of infections linked to an emerging EIEC clone has been observed in Europe, where in 2012 a large and severe outbreak of bloody diarrhea in Italy involving more than 100 individuals was reported (Escher et al., 2014; Pettengill et al., 2015). An EIEC O96:H19 strain, a serotype never described before for EIEC, was isolated and the suspected source of infection was traced to cooked vegetables (Escher et al., 2014). During the outbreak investigation an EIEC O96:H19 strain was also isolated from two asymptomatic food handlers working in the canteen linked with the outbreak, supporting the hypothesis of a secondary contamination of the vegetables during post-cooking handling procedures (Escher et al., 2014). In 2014, two linked outbreaks of gastrointestinal disease occurred in the United Kingdom, involving more than 100 cases of infection. One of the episodes was associated to the consumption of contaminated salad vegetables and, again, an O96:H19 EIEC was isolated from some of the patients and from vegetable samples (Newitt et al., 2016). Finally, an EIEC belonging to the same serotype was isolated in a case of traveler’s diarrhea in Spain in 2013 (Michelacci et al., 2016). Pheno-genotypic characterization of the strains involved in the three episodes suggests that the EIEC O96:H19 could be emerged as a result of the recent acquisition of the invasion plasmid by an E. coli strain (Michelacci et al., 2016).
The Invasive Process
Similarly to Shigella, EIEC are responsible of bacillary dysentery (Taylor et al., 1988). However, the disease caused by EIEC is usually less severe than that induced by Shigella (DuPont et al., 1971). Following the discovery that EIEC strains carry a pINV plasmid identical to that of Shigella (Harris et al., 1982; Sansonetti et al., 1982; Hale et al., 1983) and that they can display a Shigella-like invasive behavior (Hale et al., 1985; Small and Falkow, 1988; Taylor et al., 1988), in vitro and in vivo studies have been extensively focused on Shigella, providing in-depth knowledge about its pathogenicity/virulence mechanisms. In recent years, the pathogenicity of EIEC has gained new interest and comparative analyses between EIEC and Shigella have been performed, aimed at understanding the different clinical outcome severity of the two infections (Moreno et al., 2009; Bando et al., 2010; Sanchez-Villamil et al., 2016). Here we first present the invasive process as it has been inferred from studies on S. flexneri. Then, we address what it is known about the difference between these two enteroinvasive bacteria.
In order to gain access to intestinal epithelia, bacteria first transit from the lumen to the submucosa by preferentially entering M cells in Peyer’s patches. After endocytosis by M cells bacteria are transcytosed toward the M cell pocket, where they meet, and are phagocytosed by resident macrophages. Shigella infection of macrophages is accompanied by the release of T3SS effectors and components that are recognized as PAMPs by NLRs, ultimately leading to pyroptosis with the release of proinflammatory cytokines, IL-1β and IL-18 (Ashida et al., 2011). The induction of macrophage cell death is pivotal for bacteria to invade enterocytes, though pyroptosis is a form of cell death that induces a massive inflammatory response. Once released from dying macrophages, invasive bacteria infect the neighboring enterocytes by entering through the basolateral surface. Here they are enclosed into a vacuole that is rapidly disrupted freeing them into the cytosol. Subsequently, the bacteria multiply and, using actin-based motility, spread to adjacent cells (Schroeder and Hilbi, 2008).
Inside epithelial cells, bacterial PAMPs and DAMPs are detected by various PRRs, including TLRs and NLRs, which stimulate host defense signal pathways such as those involving MAPKs and NF-κB leading to the secretion of proinflammatory cytokines (e.g., IL-8 and TNF-α) (Takeuchi and Akira, 2010). These molecules induce the recruitment of phagocytic cells to the infection site, initially facilitating the invasion process and eventually clearing the bacterial pathogens. In order to maximize invasion and permanence and save the replicative niche in epithelial cells, invading Shigella modulate host cell responses throughout the infection process by secreted effectors (Killackey et al., 2016). Induction of a very early inflammatory response upon invasion of epithelial cells is functional to bacterial spreading as it results in recruitment of polymorphonuclear leucocytes (PMNL), which migrate across the epithelium destabilizing the intercellular junctions and increasing the surface available for bacterial entry into target cells (Ashida et al., 2011). Several T3SS effectors, such as OspB, OspC1, and OspZ (Zurawski et al., 2009; Ambrosi et al., 2015; Mattock and Blocker, 2017), contribute to promote inflammation at early stages of the infection process. They mainly act by enhancing activation of MAPK and NF-κB pathways, which are involved in the control of the production of PMNL chemoattractants, including IL-8, whose secretion triggers PMNL migration in a basolateral to apical direction causing epithelial barrier disruption. However, though this early inflammatory response is essential to initiate infection, it would also contribute toward rapidly clearing the infecting agents. Thus, to establish infection, at later stages Shigella must overcome the host innate response. This is achieved by delivering T3SS effectors, whose function is aimed mainly at inhibiting MAPK and NF-κB signaling pathways with the consequent decrease of inflammatory chemokine and cytokine production (Killackey et al., 2016; Mattock and Blocker, 2017).
An important obstacle Shigella must tackle during the invasion of the epithelial tissue is host cell targeting and degradation by autophagy. Several studies have demonstrated that Shigella are particularly exposed to autophagy targeting only when they are associated to cell membranes. Two bacterial factors, IcsB and VirA, have been implicated in bacterial evasion of autophagy targeting by interfering with LC3 recruitment and by allowing bacteria to escape from LC3-positive vacuoles (Ogawa et al., 2005; Baxt and Goldberg, 2014; Campbell-Valois et al., 2015).
Typically, intracellular pathogens need to save their host to establish a successful infection. As part of their pathogenic mechanism Shigella employ several countermeasures to avoid premature cell death to maintain their epithelial replicative niche. The early stage of infection is characterized by induction of DNA damage and genotoxic stress, which lead to activation of p53 and stimulation of apoptosis. Apoptotic cell death is prevented by the activity of the T3SS effectors VirA and IpgD, which promote p53 degradation and activate the PI3K/Akt pro-survival pathway, respectively, and by the pilus component protein FimA, which inhibits cytochrome c release by mitochondria (Mattock and Blocker, 2017).
As discussed above, EIEC share many aspects of the Shigella infection process that involves crossing of intestinal epithelial barrier, killing of resident macrophage cells, invasion of enterocytes, intra-cellular replication, and dissemination from cell to cell without extracellular steps (Croxen and Finlay, 2009). Moreover, EIEC express the same virulence factors found in Shigella (Parsot, 2005). However, the infectious dose required for EIEC to cause disease is higher than that of Shigella and the disease caused by EIEC appears to be milder (DuPont et al., 1971), suggesting differences between EIEC and Shigella in sensing and shaping the host environment, which, in turn, would influence the pathways toward virulence. To date only few studies have investigated the differences in the infectiveness between EIEC and Shigella. Moreno et al. (2009) detailed for the first time the relationship between the expression of some genes crucial for the infection process and the reduced ability of EIEC to cause disease. This is well supported by their Serény tests in guinea pigs, showing how the signs of keratoconjunctivitis induced by Shigella appear earlier and are more severe as compared to those caused by EIEC. Using an epithelial cell model, the authors also demonstrate that, although Shigella and EIEC display similar invasion ability, EIEC disseminates less efficiently, producing smaller plaques in plaque assays. As compared to Shigella the overall behavior of EIEC apparently reflects a reduced expression of key virulence genes, during both invasion and cell-to-cell spreading, except for virF that is expressed at higher levels by intracellular EIEC than Shigella during the dissemination step. This apparent discrepancy may be explained in the light of recent results showing that Shigella virF is transcribed into two mRNAs, with the shortest one encoding a smaller protein that negatively regulates transcription of full-length mRNA and, consequently, the expression of the VirF regulator (Di Martino et al., 2016b). Since in the real-time PCR experiments carried out by Moreno et al. (2009) virF expression was assayed by using primers that did not discriminate between the two mRNAs, comparative virF expression studies between Shigella and EIEC deserve further investigations to deeper analyze potential differences.
A more recent work has compared the host cell response to infection by different E. coli pathotypes, including EIEC, and by Shigella. The kinetic of NF-κB and ERK1/2 activation in HT-29 epithelial cells shows only a slightly higher p65 phosphorylation after 4 h of infection with Shigella as compared with EIEC. Conversely, although following a similar kinetics, the accumulation of phosphorylated ERK1/2 is much higher in cells infected with EIEC at 4 h post-infection. Despite these differences, HT-29 cells infected with EIEC or Shigella release comparable amounts of cytokines, as IL-8 and TNF-α with similar kinetics (Sanchez-Villamil et al., 2016). The phosphorylation of ERK1/2 and p38 is controlled by the phosphothreonine lyase activity of OspF, to which both anti-inflammatory (Arbibe et al., 2007) and pro-inflammatory roles (Reiterer et al., 2011) have been attributed. Since both Shigella and EIEC express OspF, it is reasonable that additional factors are involved in determining the different ERK1/2 phosphorylation profile and the outcome of MAPK activation.
The key step in invasion of the epithelial cells resides in the ability of EIEC and Shigella to escape from macrophages after phagocytosis by induction of caspase 1-dependent cell death. It has been reported that, as compared to Shigella, EIEC have a decreased capacity to escape from murine J774 macrophages and are less efficient in cell killing during the first 4 h of infection (Bando et al., 2010). This likely depends on differences in the expression of some virulence genes. In particular, as compared to Shigella the expression of the ipaC gene is reduced in intracellular EIEC at all the time points after infection. As for the release of pro- and anti-inflammatory cytokines (as TNF-α, IL-1, and IL-10) by infected cells, contrasting results exist. While no significant differences between EIEC and Shigella-infected J774 cells (Bando et al., 2010) have been reported, other studies carried out using human THP-1 cells differentiated into macrophages (Sanchez-Villamil et al., 2016) have shown that Shigella infection results in higher secretion of both pro-inflammatory and anti-inflammatory cytokines.
To date, banking on the modest amount of data available from in vitro infection of macrophage-like cells and epithelial cells, the milder disease caused by EIEC appears to be mainly associated to a lower expression of key virulence genes involved in phagosomal escape inside host cells and in dissemination among epithelial cells (Moreno et al., 2009; Bando et al., 2010). There are no obvious differences in the inflammatory response by epithelial cells, at least as far as the secretion of IL-8 and TNF-α is concerned, neither at early nor at late times of infection. Despite this cytokine profile, the activation state of ERK1/2 MAPK seems to be more elevated in epithelial cells infected with EIEC than in those infected with Shigella (Sanchez-Villamil et al., 2016). Deeper investigations will clarify to what extent this may depend on differences in manipulating certain cell signaling pathways and on differences in the activity of bacterial factors involved therein.
The Major Virulence Trait of EIEC: The Large Virulence Plasmid pINV
The evolution of E. coli toward pathogenic phenotypes has been determined, as in many other bacterial pathogens, mainly by two mechanisms: the acquisition of virulence genes by HGT as parts of plasmids, phages, transposons, or PAI and the loss or modification of genes of the core genome. While the first mechanism plays a crucial role in the colonization of a new host environment, the latter, known as pathoadaptation, strongly contributes to drive the evolution of bacteria toward a more pathogenic phenotype (Kaper et al., 2004; Dobrindt et al., 2010).
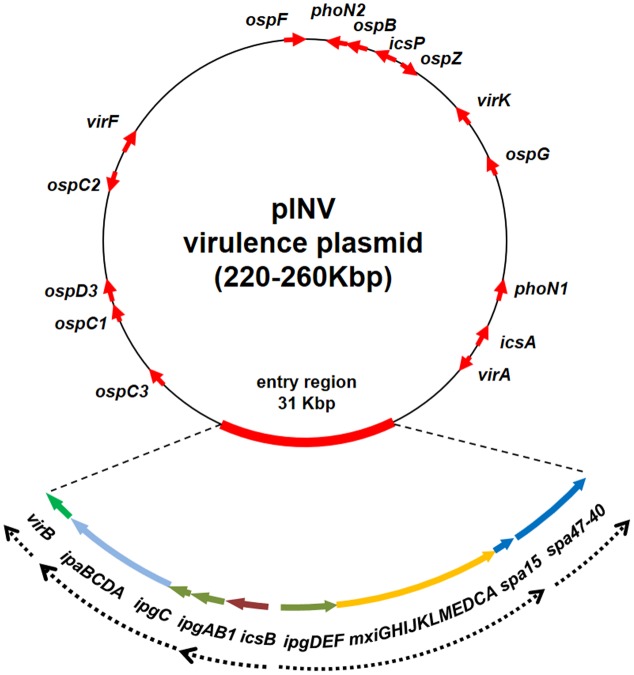
It is widely acknowledged that, as in Shigella, in EIEC the critical event in the transition toward a pathogenic lifestyle has been the acquisition of a large F-type plasmid (pINV) which encodes the molecular machinery required for invasion, survival, and diffusion of the bacterium within the host (Harris et al., 1982; Sansonetti et al., 1982; Hale et al., 1983; Small and Falkow, 1988; Figure 1). The pINV plasmid has been found only in the Shigella/EIEC pathotype and its loss is a very rare event, which determines an avirulent phenotype.
FIGURE 1.
Genetic map of the pINV of Shigella and EIEC strains. The red arrows indicate major virulence determinants. Due to the variability in position and number, the ipaH genes are not shown. The genetic organization of the entry region is shown in detail, with dashed arrow lines indicating known transcriptional units. The entry region organization is based on the sequence of plasmid pWR100 (Venkatesan et al., 2001) while the entire plasmid is freely drawn to provide the layout of a typical pINV plasmid (the figure is not to scale).
The genetic organization of the pINV is very complex (Johnson and Nolan, 2009). As a matter of fact these plasmids are made up of a mosaic of genes of various origins and harbor traces of four different plasmids (Buchrieser et al., 2000; Venkatesan et al., 2001; Escobar-Páramo et al., 2003). pINV isolated from EIEC share wide regions of high structural and functional homology and are interchangeable with those isolated from Shigella strains (Hale et al., 1983; Lan et al., 2001; Johnson and Nolan, 2009). pINV share with IncFIIA plasmids high homology in the regions involved in replication (rep) and conjugation (tra) (Makino et al., 1988) and stable inheritance of pINV is ensured by the presence of several plasmid segregation and maintenance systems (Lan et al., 2001). Due to large deletions in the tra region, pINV are not capable of self-transfer by conjugation, but they can be mobilized by other conjugative plasmids. All over the plasmid genome, an astonishing number of ISs is present as a mixture of complete and incomplete IS elements repeated several times, confirming the relevant role played by ISs in pINV assembly and evolution (Buchrieser et al., 2000; Venkatesan et al., 2001). Most ISs are related to known elements while others represent novel ISs. Among the latter, ISEc11, an IS belonging to the IS1111 family, is widespread and functional in pINV from EIEC while only defective copies are present in the Shigella pINV plasmids (Prosseda et al., 2006).
In the pINV there is only one large (31 kb) region, which does not host any IS elements. This is the so-called entry region, which displays a PAI-like structure (Buchrieser et al., 2000; Venkatesan et al., 2001). It is composed by two large, divergently transcribed gene clusters coding for a T3SS apparatus (Mxi and Spa), for most of its effectors (IpaB, IpaC, and IpaD) with their chaperons (IpgA, IpgC, IpgE, and Spa15), and for two transcriptional regulators (VirB and MxiE), both required for the activation of most virulence genes (Schroeder and Hilbi, 2008; Figure 1). The entry region is extremely conserved among Shigella and EIEC pINV plasmids (Lan et al., 2001). Albeit it had been initially proposed as a PAI, likely acquired in a single recombination event, it lacks the presence of flanking tRNA sequences and at least remnants of a recombinase-encoding gene. It is therefore unclear if the acquisition of the entry region has occurred independently from its insertion into tRNA sequences or if the absence of tRNA genes may have resulted from rearrangement following gene transfer. The latter hypothesis is supported by the fact that the entry region is flanked by truncated IS elements, suggesting that rearrangements may have occurred after its acquisition en bloc by the plasmid (Buchrieser et al., 2000). The T3SS encoded by the entry region plays a critical role in the bacterial invasive process, since it delivers a large number of effectors involved in the reorganization of the host cell actin cytoskeleton and in the modulation of cell signaling pathways to evade the host immune response (Mattock and Blocker, 2017). With the exception of few proteins of the IpaH family, which are chromosomally encoded, all T3SS effectors are encoded by pINV genes located within or outside the entry region. Since the entry region is highly conserved, the phylogenetic analysis of three of its genes (ipgD, mxiC, and mxiA) has allowed differentiating pINV from Shigella spp. and EIEC into two forms, A and B, with the first one predominantly associated with EIEC strains (Lan et al., 2001, 2004).
Besides the large PAI-like region, a small islet carries the genes coding for IcsA (a protein responsible for the bacterial motility inside the cytoplasm), VirA (a GTPase-activating protein), and RnaG (a regulatory sRNA negatively controlling icsA expression) (Giangrossi et al., 2010; Tran et al., 2011; Dong et al., 2012). Other genes encoding proteins crucial for the invasive process cover the pINV plasmid including the OspG and OspF proteins which interfere with the host innate immune response (Kim et al., 2005; Arbibe et al., 2007), the PhoN2 protein required for IcsA localization (Scribano et al., 2014), and the IpaH proteins which interfere with the host protein degradation (Ashida and Sasakawa, 2015; Figure 1). Moreover, in contrast to the other two virulence regulatory genes (virB and mxiE), the virF gene, coding for the primary virulence regulator, is located on a “desert island” surrounded by several IS sequences and far away from all other virulence genes, including those under its direct control, virB and icsA (Di Martino et al., 2016a). While the CG content of virF is only slightly lower as compared to that of the entry region (Buchrieser et al., 2000), its position suggests that it has been acquired independently to promote the expression of the virulence genes. VirF is also involved in the activation of some chromosomal genes, indicating that it acts as global regulator and that its acquisition by HGT has contributed to a reshaping of the core genome, easing the adaptation of bacteria to the host environment (Barbagallo et al., 2011; Leuzzi et al., 2015).
The mechanisms involved in the activation of the pINV virulence genes have been extensively studied both in EIEC and in Shigella (Dagberg and Uhlin, 1992; Prosseda et al., 2002). They rely on a sophisticated regulatory cascade involving global and specific regulators, encoded by both, pINV and the chromosome. Outside the human host, the nucleoid-associated protein H-NS represses each of the key promoters of the pINV virulence genes (Dorman, 2004). In response to environmental conditions found in the human intestine, the transcriptional activation of the invasive operons is triggered by an increased level of VirF counteracting H-NS repression at the icsA and virB promoters (Prosseda et al., 2004). Then VirB activates most operons within the entry region, including the gene for the last regulator (mxiE), as well as all other virulence genes scattered along the pINV genome, except icsA. Finally, MxiE, assisted by IpgC, activates the transcription of genes encoding the late effectors (Schroeder and Hilbi, 2008).
As in other pathogenic E. coli, also in EIEC the virulence genes are stably maintained on an extrachromosomal element (Johnson and Nolan, 2009). Nevertheless, it has been reported that the pINV of EIEC strain HN280 is able to integrate into the host chromosome and that integration results in silencing of all pINV-encoded virulence genes also under host temperature conditions (Zagaglia et al., 1991). Silencing was shown to depend on a severe reduction of virB transcription, likely dependent on the inability of VirF to counteract the negative control of H-NS at the virB promoter when it is chromosomally located (Colonna et al., 1995). This has led to the hypothesis that the presence of virulence genes on the pINV is the result of an evolutionary pathway toward the optimization of gene expression.
Evolution of EIEC
The studies on the evolutionary origin of the Shigella/EIEC pathovar have led to two major hypotheses. The pINV could have been acquired only once by an ancestral E. coli that subsequently gave rise to the different Shigella/EIEC lineages (Escobar-Páramo et al., 2003; Zuo et al., 2013), as suggested by the inability of the plasmid to autonomously undergo horizontal transmission. Alternatively, the different Shigella/EIEC strains could have arisen from different E. coli that had acquired the pINV independently, e.g., from an unknown donor or from other Shigella/EIEC that already harbored it. This view is supported by the diversity of the genotypes within the Shigella/EIEC pathovar, revealed by phylogenetic analyses of chromosomal genes and by genome comparison (Pupo et al., 2000; Hazen et al., 2016; Pettengill et al., 2016). Besides the large pINV, several virulence genes have been acquired on the chromosome of Shigella and EIEC as part of PAIs (Figure 2). The PAIs described so far for Shigella (SHI islands) carry genes encoding different traits, including an enterotoxin and a cytotoxic protease (SHI-1) and systems involved in iron uptake and evasion of immune response (SHI-2 and SHI-3 in S. boydii), in the modification of O antigens (SHI-O) or in multi-drug resistance (SRL) (Schroeder and Hilbi, 2008). Recently, 20 genomes from EIEC belonging to different serotypes have been compared with those of reference strains belonging to diverse E. coli pathovars and Shigella species. This comparison highlights the existence of at least three distinct lineages containing only EIEC strains and suggests a convergent evolution of non-pathogenic E. coli toward invasive phenotype (Hazen et al., 2016). An in silico search for protein-encoding genes of SHI-1, SHI-2, SHI-3, SHI-O, and SRL indicates that, with the exception of SHI-O, portions of the other PAIs are present in EIEC genomes in a lineage-specific manner (Hazen et al., 2016). Interestingly, while a whole SHI-1 Island has never been detected in EIEC, SHI-1 fragments of different length have been found in all EIEC genomes. However, the ShET1 toxin genes, typically harbored by SHI-1 in S. flexneri genomes, were found only in EIEC strains of lineage 2. In the case of virulence genes associated with SHI-2, the shiA gene, involved in the reduction of the host inflammatory response, is absent in all EIEC lineages, while shiD, which provides immunity to colicins, is present in all EIEC of lineages 1 and 2. An entire SHI-3 PAI, typically associated with S. boydii strains, has been detected only in few EIEC strains of lineages 1 and 2, while portions of it, including the genes encoding for aerobactin-mediated iron uptake, are found in all three lineages. As for the large SRL PAI, widely distributed among Shigella spp. and containing a cluster of multiple antibiotic resistance determinants (Turner et al., 2003), only a few of its genes are present in EIEC genomes.
FIGURE 2.
Genetic events contributing to the evolution of EIEC from ancestral commensal E. coli. The acquisition of the pINV by HGT is a major evolutionary event toward pathogenicity. This can be accompanied by the sporadic acquisition of entire or incomplete SHI-1 PAI and incomplete SHI-2 and SHI-3 PAIs. Rarely, also incomplete SRL PAI are acquired by EIEC genomes. The absence of ompT and the loss of cadaverine synthesis (usually resulting from cadC silencing) counterbalance the gain of virulence-associated determinants. The inactivation of speG (involved in spermidine acetylation) and nad (involved in NAD biosynthesis) is regarded as emergent pathoadaptive mutations in EIEC.
The variable presence of the PAIs in EIEC confirms the phylogenetic diversity among EIEC and Shigella and further supports the hypothesis that the EIEC pathovar has not a single origin but rather stems from multiple lineages (Hazen et al., 2016; Michelacci et al., 2016; Pettengill et al., 2016).
A significant complementary step toward the pathogenic lifestyle has been pathoadaptation, the inactivation, or loss of several chromosomal genes, which negatively interfere with the expression of virulence factors required for survival within the host. The antivirulence loci identified encode a broad spectrum of functions, confirming that adaptation to the new host environments is the result of long and ordered process targeting core genome determinants (Casalino et al., 2003; Di Martino et al., 2013b; Campilongo et al., 2014).
Despite the close similarity of the Shigella and EIEC pathogenicity process, it is well known that EIEC have a metabolic activity more similar to E. coli and have not undergone the intense gene decay observed in Shigella (Silva et al., 1980; Pettengill et al., 2016). It is therefore not surprising that the pathoadaptation in EIEC has not reached a level comparable to Shigella (Prosseda et al., 2012) and that most of the antivirulence loci characterized in Shigella are still encoding functional products in EIEC. One of the pathoadaptive mutations conserved both in EIEC and in Shigella is the deletion of the ompT gene, located within the defective lambdoid prophage DLP12 (Nakata et al., 1993; Figure 2). The OmpT protease triggers the degradation of IcsA protein and therefore negatively interferes with host cell invasion by drastically reducing the ability of Shigella to spread into adjacent epithelial cells. Considering that the loss of OmpT is widespread, it is as yet unclear if E. coli lineages that gave rise to the Shigella/EIEC pathovar have not hosted DLP12 ab initio or if the prophage has been excised during the pathoadaption process (Bliven and Maurelli, 2012; Leuzzi et al., 2017).
Another typical pathoadaptive mutation of Shigella spp. is the inability to catabolise lysine, due to the silencing of lysine decarboxylase (LDC) activity (Prosseda et al., 2007). The LDC- phenotype, which is found also in most EIEC, is determined by mutations in the cad locus, which hamper the synthesis of cadaverine. Cadaverine is a polyamine that interferes with pathogenicity by blocking the release of Shigella into the cytoplasm of the infected cells and inhibiting the migration of PMNL across the intestinal epithelium (Bliven and Maurelli, 2012). A detailed analysis of the molecular rearrangements occurred in the cad operon of several EIEC strains belonging to different serotypes (Casalino et al., 2003) has shown that, similarly to Shigella, the silencing of the cad locus has been accomplished through convergent evolution. In contrast to Shigella, in EIEC the cad region is colinear with the E. coli K12 chromosome and the lack of cadaverine synthesis is mainly due to the inactivation of the gene encoding the CadC transcriptional regulator (Casalino et al., 2010). By comparing the cad loci of EIEC and Shigella, it appears that the rearrangements occurred in EIEC are less severe compared to the complete erosion of the locus observed in several Shigella strains (Casalino et al., 2005; Prosseda et al., 2007; Figure 2). Indeed, despite the antivirulence role played by cadaverine (Fernandez et al., 2001), emerging O96:H19 EIEC strains still maintains an integer cad operon and exhibits a LDC+ phenotype (Michelacci et al., 2016).
As compared to the commensal E. coli the polyamine profile of Shigella is affected not only by the lack of cadaverine but also by the marked accumulation of spermidine and by the loss of N-acetyl spermidine, the inert form of spermidine (Di Martino et al., 2013a). The increased spermidine content depends on the loss of the spermidine acetyltransferase (SAT), the enzyme encoded by the speG gene and responsible for the conversion of spermidine into N-acetylspermidine. In Shigella it has been demonstrated that a higher level of spermidine increases survival within macrophages and confers bacteria a higher resistance to oxidative stress (Barbagallo et al., 2011). Similarly to how observed for the cad locus, also speG silencing is the result of convergent evolution. A comparison of the polyamine profiles of several EIEC strains with those of Shigella and E. coli K12 has revealed that in EIEC major polyamines attain levels in-between those observed in E. coli and Shigella. Indeed, as compared to commensal E. coli, in EIEC intracellular putrescine is significantly increased and spermidine tends to be higher. Nevertheless, in contrast to Shigella, N-acetylspermidine is still present in most EIEC strains (Campilongo et al., 2014), indicating that the loss of speG is an emerging trait. However, when spermidine accumulation is induced in EIEC through deletion of the speG gene, survival within macrophages, as well as resistance to oxidative stress are increased (Campilongo et al., 2014). This confirms that the absence of SAT activity confers to intracellular bacteria like EIEC and Shigella an increased capability to defy antagonistic host environment. Moreover, the analysis of the polyamine profiles has revealed that the higher level of putrescine in EIEC is determined by increased transcription of speC, promoted by the lack of cadaverine. The speC gene encodes the enzyme converting L-ornithine into putrescine. On the basis of these observations it has been suggested (Campilongo et al., 2014) that during the transition toward the pathogenic phenotype, the modification of the polyamine profile might have been triggered by the loss of cadaverine, with the double effect of favoring the invasive process and increasing the putrescine level. Since putrescine is an important intermediate in the synthesis of spermidine and, consequently, of N-acetylspermidine, its increase may in turn have caused higher levels of both polyamines. In this scenario the silencing of speG, which appears completed in Shigella but can be regarded as an ongoing process in EIEC, would represent the last step favoring further accumulation of spermidine and the disappearance of N-acetylspermidine.
Another noteworthy pathoadaptive mutation in Shigella is the requirement for exogenous nicotinic acid (NAD) due to inactivation of the nad genes (Prunier et al., 2007), required for de novo synthesis of NAD. Also in this case the inability to synthesize NAD is not a generalized feature among EIEC strains (Di Martino et al., 2013b). In those EIEC strains requiring NAD it has been shown that the preferential target in the pathoadaptation process is the nadB gene, inactivated through diverse strategies, involving point mutations or IS insertions.
Altogether, the picture emerging from the observations on pathoadaptive mutations suggests that EIEC might represent intermediates in the evolution toward a full-blown phenotype, with some mutational events still confined to Shigella (Figure 2). However, a recent whole-genome comparative analysis (Pettengill et al., 2016), performed on a large number of Shigella and EIEC genomes, indicates that Shigella and EIEC evolved independently. Nevertheless, the same authors proposed that, while EIEC as a group cannot be considered the ancestor to Shigella, some EIEC lineages might have been the Shigella ancestor.
Emerging Enteroinvasive Escherichia coli
The recent outbreaks occurred in Europe caused by the EIEC O96:H19 led the scientific community to reconsider the role of EIEC infection in industrialized countries (Escher et al., 2014; Michelacci et al., 2016; Newitt et al., 2016). Such EIEC serotype had never been reported before 2012 and represents a new virulent emergent clone. The EIEC O96:H19 isolated from two outbreaks occurred in Italy and United Kingdom and from a sporadic case of disease reported in Spain were studied by whole genome sequencing (Pettengill et al., 2015; Michelacci et al., 2016). The genomic analysis confirmed that all the isolates belonged not only to the same unprecedented EIEC serotype, but also to the same sequence type (ST-99), never observed before in EIEC strains (Michelacci et al., 2016). The analysis of the distribution of virulence genes typical of EIEC and Shigella highlighted the presence in the three strains of the plasmid genes encoding the T3SS system and its effectors, as well as the master transcriptional regulators genes virF and virB. As for the chromosomally located virulence genes, the three isolates showed the presence of the genetic determinants of a T2SS and were all negative for those encoding the aerobactin system involved in iron uptake. Interestingly, none of the O96:H19 isolates was found to have undergone the process of pathoadaptation through accumulation of the mutations described in the literature for EIEC and Shigella (Bliven and Maurelli, 2012; Prosseda et al., 2012). Nevertheless, the three isolates were shown to display minor differences. The plasmid profiles obtained through the genomic analysis highlighted the presence of five plasmids in the strains isolated in Spain and United Kingdom and three plasmids in that responsible of the Italian outbreak, with three plasmids in common in the three strains. Altogether, these observations strengthen the hypothesis of the emergence of a new virulent EIEC clone circulating in Europe.
Phenotypic analysis also highlighted peculiar properties of this EIEC clone, when compared to reference EIEC and Shigella strains. Biochemical characterization showed that the isolates displayed the LDC activity, confirming the lack of the related pathoadaptive mutations observed through genome analysis, and interestingly showed that the isolates retained the ability to ferment lactose (Michelacci et al., 2016), usually lacking in Shigella and in the majority of EIEC strains (Tozzoli and Scheutz, 2014). Generally, a better fitness was observed for the O96:H19 strains when comparing the growth curves with those of Shigella and reference EIEC strains (Michelacci et al., 2016). Moreover, swimming motility was observed for the strains from Italian and Spanish cases, which was instead completely absent in the strain from United Kingdom and in all the other EIEC and Shigella strains tested. Such phenotypic traits are not typical of intracellular pathogens such as EIEC and Shigella, while they are more common in E. coli strains, contributing to their great ability in surviving and adapting in different ecological niches.
These findings support the hypothesis of the evolution of EIEC and Shigella after the acquisition of the pINV by multiple lineages of commensal E. coli, followed by a multi-step adaptation process. Such an evolutionary pathway could be exemplified by EIEC ST-280 Clonal Complex, which could have been generated with the acquisition of the pINV plasmid by a commensal E. coli eventually evolving toward Shigella belonging to related clonal complexes (ST-149, 152, 243, 245, 250) (Wirth et al., 2006; Michelacci et al., 2016). The mechanism could have involved multiple events of pathoadaptive mutations, giving origin to the existing Shigella clones, specialized for intracellular survival with detriment of the ability to persist outside the host. A similar paradigm could also explain the emergence of other EIEC clones following the acquisition of pINV by other commensal E. coli. This event in some cases could be followed by the accumulation of pathoadaptive mutation, as it is the case of the EIEC strains belonging to ST-6 clonal complex, while some other clones could have maintained all the functions granting an efficient extracellular persistence, such as the EIEC O96:H19 belonging to ST-99 (Michelacci et al., 2016). The observed better fitness of EIEC O96:H19 in comparison with that of the other reference EIEC and Shigella strains could have favored its survival in the extracellular environment and allowed its overgrowth in the food vehicles, granting it a high potential as a foodborne pathogen, as demonstrated in the two large episodes occurred in Italy and United Kingdom (Michelacci et al., 2016; Newitt et al., 2016).
Conclusion and Perspectives
Genomics approaches in combination with phenotypic analyses have a strong potential toward the formulation of new intriguing hypotheses on the ongoing evolution of EIEC. Currently available comparisons between EIEC and Shigella genomes support the need for a taxonomical revision moving the Shigella genus back within the E. coli species (Michelacci et al., 2016; Pettengill et al., 2016). As a matter of fact, Shigella clades are interspersed in clusters of E. coli genomes regardless of the bioinformatics approach used for the phylogenetic analysis (Sahl et al., 2015; Pettengill et al., 2016). In the light of recent studies, the organization of the EIEC genome appears to have been originated from multiple independent events (Hazen et al., 2016; Pettengill et al., 2016). This hypothesis finds even stronger evidence in the emergence of a novel EIEC clone belonging to O96:H19 serotype, which exhibits phenotypic traits more typical of E. coli than of reference EIEC or Shigella (Michelacci et al., 2016).
The acquisition of the plasmid may represent the first step in the emergence of new EIEC clones, but it is well known to be not sufficient for establishing the full pathogenicity (Sansonetti et al., 1983). In this context, it is of great interest to deeper investigate on the role and relevance of functions that Shigella has lost in its route toward an intracellular life-style but that are still retained by most EIEC strains.
Author Contributions
BC, VM, SM, and GP proposed the idea of the review; MP, VM, MG, and RT wrote the review draft; MP, MM, and GP design the figures; BC, MM, GP, MG, and SM wrote the final version of the review. The final text has been read and approved by all the authors of the review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- DAMP
damage-associated molecular pattern
- DEC
diarrheagenic E. coli
- HGT
horizontal gene transfer
- H-NS
heat-stable nucleoid-structuring protein
- IL
interleukin
- IS
insertion sequence
- MAPK
mitogen-activated protein kinase
- NLR
Nod-like receptor
- PAI
pathogenicity island
- PAMP
pathogen-associated molecular pattern
- pINV
virulence plasmid
- PMNL
polymorphonuclear leukocytes
- PRR
pattern recognition receptor
- SHI
Shigella pathogenicity island
- SRL
Shigella resistance locus
- sRNA
small RNA
- T2SS
type II secretion system
- T3SS
type III secretion system
- TLR
Toll-like receptor
- TNF
tumor necrosis factor.
Footnotes
Funding. This research was supported by grants from Sapienza Università di Roma and from Institut Pasteur (PTR-24-16).
References
- Ambrosi C., Pompili M., Scribano D., Limongi D., Petrucca A., Cannavacciuolo S., et al. (2015). The Shigella flexneri OspB effector: an early immunomodulator. Int. J. Med. Microbiol. 305 75–84. 10.1016/j.ijmm.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Arbibe L., Kim D. W., Batsche E., Pedron T., Mateescu B., Muchardt C., et al. (2007). An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8 47–56. 10.1038/ni1423 [DOI] [PubMed] [Google Scholar]
- Ashida H., Ogawa M., Kim M., Suzuki S., Sanada T., Punginelli C., et al. (2011). Shigella deploy multiple countermeasures against host innate immune responses. Curr. Opin. Microbiol. 14 16–23. 10.1016/j.mib.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Ashida H., Sasakawa C. (2015). Shigella IpaH family effectors as a versatile model for studying pathogenic bacteria. Front. Cell. Infect. Microbiol. 5:100 10.3389/fcimb.2015.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando S. Y., do Valle G. R., Martinez M. B., Trabulsi L. R., Moreira-Filho C. A. (1998). Characterization of enteroinvasive Escherichia coli and Shigella strains by RAPD analysis. FEMS Microbiol. Lett. 165 159–165. 10.1111/j.1574-6968.1998.tb13141.x [DOI] [PubMed] [Google Scholar]
- Bando S. Y., Moreno A. C. R., Albuquerque J. A. T., Amhaz J. M. K., Moreira-Filho C. A., Martinez M. B. (2010). Expression of bacterial virulence factors and cytokines during in vitro macrophage infection by enteroinvasive Escherichia coli and Shigella flexneri: a comparative study. Mem. Inst. Oswaldo Cruz 105 786–791. 10.1590/S0074-02762010000600009 [DOI] [PubMed] [Google Scholar]
- Barbagallo M., Di Martino M. L., Marcocci L., Pietrangeli P., De Carolis E., Casalino M., et al. (2011). A new piece of the Shigella pathogenicity puzzle: spermidine accumulation by silencing of the speG gene [corrected]. PLOS ONE 6:e27226. 10.1371/journal.pone.0027226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxt L. A., Goldberg M. B. (2014). Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLOS ONE 9:e94653. 10.1371/journal.pone.0094653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Gleier K., Kontny I., Echeverria P., Scheutz F. (1997). Origin and characteristics of enteroinvasive strains of Escherichia coli (EIEC) isolated in Germany. Epidemiol. Infect. 118 199–205. 10.1017/S0950268897007413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake P. A., Ramos S., MacDonald K. L., Rassi V., Gomes T. A., Ivey C., et al. (1993). Pathogen-specific risk factors and protective factors for acute diarrheal disease in urban Brazilian infants. J. Infect. Dis. 167 627–632. 10.1093/infdis/167.3.627 [DOI] [PubMed] [Google Scholar]
- Bliven K. A., Maurelli A. T. (2012). Antivirulence genes: insights into pathogen evolution through gene loss. Infect. Immun. 80 4061–4070. 10.1128/IAI.00740-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C., Glaser P., Rusniok C., Nedjari H., D’Hauteville H., Kunst F., et al. (2000). The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38 760–771. 10.1046/j.1365-2958.2000.02179.x [DOI] [PubMed] [Google Scholar]
- Campbell-Valois F.-X., Sachse M., Sansonetti P. J., Parsot C. (2015). Escape of actively secreting Shigella flexneri from ATG8/LC3-positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. mBio 6:e02567-14. 10.1128/mBio.02567-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campilongo R., Di Martino M. L., Marcocci L., Pietrangeli P., Leuzzi A., Grossi M., et al. (2014). Molecular and functional profiling of the polyamine content in enteroinvasive E. coli: looking into the gap between commensal E. coli and harmful Shigella. PLOS ONE 9:e106589. 10.1371/journal.pone.0106589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino M., Latella M. C., Prosseda G., Ceccarini P., Grimont F., Colonna B. (2005). Molecular evolution of the lysine decarboxylase-defective phenotype in Shigella sonnei. Int. J. Med. Microbiol. 294 503–512. 10.1016/j.ijmm.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Casalino M., Latella M. C., Prosseda G., Colonna B. (2003). CadC is the preferential target of a convergent evolution driving enteroinvasive Escherichia coli toward a lysine decarboxylase-defective phenotype. Infect. Immun. 71 5472–5479. 10.1128/IAI.71.10.5472-5479.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino M., Prosseda G., Barbagallo M., Iacobino A., Ceccarini P., Carmela Latella M., et al. (2010). Interference of the CadC regulator in the arginine-dependent acid resistance system of Shigella and enteroinvasive E. coli. Int. J. Med. Microbiol. 300 289–295. 10.1016/j.ijmm.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Chatterjee B. D., Sanyal S. N. (1984). Is it all shigellosis? Lancet 2 574 10.1016/S0140-6736(84)90783-9 [DOI] [PubMed] [Google Scholar]
- Cheasty T., Rowe B. (1983). Antigenic relationships between the enteroinvasive Escherichia coli O antigens O28ac, O112ac, O124, O136, O143, O144, O152, and O164 and Shigella O antigens. J. Clin. Microbiol. 17 681–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna B., Casalino M., Fradiani P. A., Zagaglia C., Naitza S., Leoni L., et al. (1995). H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J. Bacteriol. 177 4703–4712. 10.1128/jb.177.16.4703-4712.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M. A., Finlay B. B. (2009). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8 26–38. 10.1038/nrmicro2265 [DOI] [PubMed] [Google Scholar]
- Croxen M. A., Law R. J., Scholz R., Keeney K. M., Wlodarska M., Finlay B. B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26 822–880. 10.1128/CMR.00022-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagberg B., Uhlin B. E. (1992). Regulation of virulence-associated plasmid genes in enteroinvasive Escherichia coli. J. Bacteriol. 174 7606–7612. 10.1128/jb.174.23.7606-7612.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino M. L., Campilongo R., Casalino M., Micheli G., Colonna B., Prosseda G. (2013a). Polyamines: emerging players in bacteria-host interactions. Int. J. Med. Microbiol. 303 484–491. 10.1016/j.ijmm.2013.06.008 [DOI] [PubMed] [Google Scholar]
- Di Martino M. L., Falconi M., Micheli G., Colonna B., Prosseda G. (2016a). The multifaceted activity of the VirF regulatory protein in the Shigella lifestyle. Front. Mol. Biosci. 3:61. 10.3389/fmolb.2016.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino M. L., Fioravanti R., Barbabella G., Prosseda G., Colonna B., Casalino M. (2013b). Molecular evolution of the nicotinic acid requirement within the Shigella/EIEC pathotype. Int. J. Med. Microbiol. 303 651–661. 10.1016/j.ijmm.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Di Martino M. L., Romilly C., Wagner E. G. H., Colonna B., Prosseda G. (2016b). One gene and two proteins: a leaderless mRNA supports the translation of a shorter form of the Shigella VirF regulator. mBio 7:e01860-16. 10.1128/mBio.01860-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrindt U., Chowdary M. G., Krumbholz G., Hacker J. (2010). Genome dynamics and its impact on evolution of Escherichia coli. Med. Microbiol. Immunol. 199 145–154. 10.1007/s00430-010-0161-2 [DOI] [PubMed] [Google Scholar]
- Dong N., Zhu Y., Lu Q., Hu L., Zheng Y., Shao F. (2012). Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150 1029–1041. 10.1016/j.cell.2012.06.050 [DOI] [PubMed] [Google Scholar]
- Dorman C. J. (2004). H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2 391–400. 10.1038/nrmicro883 [DOI] [PubMed] [Google Scholar]
- DuPont H. L., Formal S. B., Hornick R. B., Snyder M. J., Libonati J. P., Sheahan D. G., et al. (1971). Pathogenesis of Escherichia coli diarrhea. N. Engl. J. Med. 285 1–9. 10.1056/NEJM197107012850101 [DOI] [PubMed] [Google Scholar]
- Echeverria P., Sethabutr O., Serichantalergs O., Lexomboon U., Tamura K. (1992). Shigella and enteroinvasive Escherichia coli infections in households of children with dysentery in Bangkok. J. Infect. Dis. 165 144–147. 10.1093/infdis/165.1.144 [DOI] [PubMed] [Google Scholar]
- Edwards P. R., Ewing W. H. (1986). Edwards and Ewing’s Identification of Enterobacteriaceae, 4th Edn. New York, NY: Elsvier Science. [Google Scholar]
- Escher M., Scavia G., Morabito S., Tozzoli R., Maugliani A., Cantoni S., et al. (2014). A severe foodborne outbreak of diarrhoea linked to a canteen in Italy caused by enteroinvasive Escherichia coli, an uncommon agent. Epidemiol. Infect. 142 2559–2566. 10.1017/S0950268814000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Páramo P., Giudicelli C., Parsot C., Denamur E. (2003). The evolutionary history of Shigella and enteroinvasive Escherichia coli revised. J. Mol. Evol. 57 140–148. 10.1007/s00239-003-2460-3 [DOI] [PubMed] [Google Scholar]
- Ewing W. H., Gravatti J. L. (1947). Shigella types encountered in the mediterranean area. J. Bacteriol. 53 191–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer J. J., Davis B. R., Hickman-Brenner F. W., McWhorter A., Huntley-Carter G. P., Asbury M. A., et al. (1985). Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 21 46–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faundez G., Figueroa G., Troncoso M., Cabello F. C. (1988). Characterization of enteroinvasive Escherichia coli strains isolated from children with diarrhea in Chile. J. Clin. Microbiol. 26 928–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I. M., Silva M., Schuch R., Walker W. A., Siber A. M., Maurelli A. T., et al. (2001). Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: a connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 184 743–753. 10.1086/323035 [DOI] [PubMed] [Google Scholar]
- Formal S. B., Hornick R. B. (1978). Invasive Escherichia coli. J. Infect. Dis. 137 641–644. 10.1093/infdis/137.5.641 [DOI] [PubMed] [Google Scholar]
- Giangrossi M., Prosseda G., Tran C. N., Brandi A., Colonna B., Falconi M. (2010). A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 38 3362–3375. 10.1093/nar/gkq025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes T. A., Toledo M. R., Trabulsi L. R., Wood P. K., Morris J. G. (1987). DNA probes for identification of enteroinvasive Escherichia coli. J. Clin. Microbiol. 25 2025–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes T. A. T., Elias W. P., Scaletsky I. C. A., Guth B. E. C., Rodrigues J. F., Piazza R. M. F., et al. (2016). Diarrheagenic Escherichia coli. Braz. J. Microbiol. 47 3–30. 10.1016/j.bjm.2016.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale T. L., Oaks E. V., Formal S. B. (1985). Identification and antigenic characterization of virulence-associated, plasmid-coded proteins of Shigella spp. and enteroinvasive Escherichia coli. Infect. Immun. 50 620–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale T. L., Sansonetti P. J., Schad P. A., Austin S., Formal S. B. (1983). Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect. Immun. 40 340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. R., Wachsmuth I. K., Davis B. R., Cohen M. L. (1982). High-molecular-weight plasmid correlates with Escherichia coli enteroinvasiveness. Infect. Immun. 37 1295–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen T. H., Leonard S. R., Lampel K. A., Lacher D. W., Maurelli A. T., Rasko D. A. (2016). Investigating the relatedness of enteroinvasive Escherichia coli to other E. coli and Shigella isolates by using comparative genomics. Infect. Immun. 84 2362–2371. 10.1128/IAI.00350-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Nolan L. K. (2009). Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73 750–774. 10.1128/MMBR.00015-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J. B., Nataro J. P., Mobley H. L. T. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2 123–140. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- Killackey S. A., Sorbara M. T., Girardin S. E. (2016). Cellular aspects of Shigella pathogenesis: focus on the manipulation of host cell processes. Front. Cell. Infect. Microbiol. 6:38 10.3389/fcimb.2016.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. W., Lenzen G., Page A.-L., Legrain P., Sansonetti P. J., Parsot C. (2005). The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U.S.A. 102 14046–14051. 10.1073/pnas.0504466102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecko D. J., Baron L. S., Buysse J. (1985). Genetic determinants of virulence in Shigella and dysenteric strains of Escherichia coli: their involvement in the pathogenesis of dysentery. Curr. Top. Microbiol. Immunol. 118 71–95. 10.1007/978-3-642-70586-1_5 [DOI] [PubMed] [Google Scholar]
- Lan R., Alles M. C., Donohoe K., Martinez M. B., Reeves P. R. (2004). Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 72 5080–5088. 10.1128/IAI.72.9.5080-5088.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R., Lumb B., Ryan D., Reeves P. R. (2001). Molecular evolution of large virulence plasmid in Shigella clones and enteroinvasive Escherichia coli. Infect. Immun. 69 6303–6309. 10.1128/IAI.69.10.6303-6309.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R., Reeves P. R. (2002). Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect. 4 1125–1132. 10.1016/S1286-4579(02)01637-4 [DOI] [PubMed] [Google Scholar]
- Leuzzi A., Di Martino M. L., Campilongo R., Falconi M., Barbagallo M., Marcocci L., et al. (2015). Multifactor regulation of the MdtJI polyamine transporter in Shigella. PLOS ONE 10:e0136744. 10.1371/journal.pone.0136744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzzi A., Grossi M., Di Martino M. L., Pasqua M., Micheli G., Colonna B., et al. (2017). Role of the SRRz/Rz1 lambdoid lysis cassette in the pathoadaptive evolution of Shigella. Int. J. Med. Microbiol. 307 268–275. 10.1016/j.ijmm.2017.03.002 [DOI] [PubMed] [Google Scholar]
- Levine M. M., Ferreccio C., Prado V., Cayazzo M., Abrego P., Martinez J., et al. (1993). Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago. Chile. Am. J. Epidemiol. 138 849–869. 10.1093/oxfordjournals.aje.a116788 [DOI] [PubMed] [Google Scholar]
- Makino S., Sasakawa C., Yoshikawa M. (1988). Genetic relatedness of the basic replicon of the virulence plasmid in shigellae and enteroinvasive Escherichia coli. Microb. Pathog. 5 267–274. 10.1016/0882-4010(88)90099-X [DOI] [PubMed] [Google Scholar]
- Manolov D. G. (1959). A new type of the genus Shigella –“Shigella 13”. J. Hyg. Epidemiol. Microbiol. Immunol. 3 184–190. [PubMed] [Google Scholar]
- Marier R., Wells J., Swanson R., Callahan W., Mehlman I. (1973). An outbreak of enteropathogenic Escherichia coli foodborne disease traced to imported French cheese. Lancet 302 1376–1378. 10.1016/S0140-6736(73)93335-7 [DOI] [Google Scholar]
- Matsushita S., Yamada S., Kai A., Kudoh Y. (1993). Invasive strains of Escherichia coli belonging to serotype O121:NM. J. Clin. Microbiol. 31 3034–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattock E., Blocker A. J. (2017). How do the virulence factors of Shigella work together to cause disease? Front. Cell. Infect. Microbiol. 7:64 10.3389/fcimb.2017.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelacci V., Prosseda G., Maugliani A., Tozzoli R., Sanchez S., Herrera-León S., et al. (2016). Characterization of an emergent clone of enteroinvasive Escherichia coli circulating in Europe. Clin. Microbiol. Infect. 22 287.e11–287.e19. 10.1016/j.cmi.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Moreno A. C. R., Ferreira L. G., Martinez M. B. (2009). Enteroinvasive Escherichia coli vs. Shigella flexneri: how different patterns of gene expression affect virulence. FEMS Microbiol. Lett. 301 156–163. 10.1111/j.1574-6968.2009.01815.x [DOI] [PubMed] [Google Scholar]
- Nakata N., Tobe T., Fukuda I., Suzuki T., Komatsu K., Yoshikawa M., et al. (1993). The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol. Microbiol. 9 459–468. 10.1111/j.1365-2958.1993.tb01707.x [DOI] [PubMed] [Google Scholar]
- Newitt S., MacGregor V., Robbins V., Bayliss L., Chattaway M. A., Dallman T., et al. (2016). Two linked enteroinvasive Escherichia coli outbreaks, Nottingham, UK, June 2014. Emerg. Infect. Dis. 22 1178–1184. 10.3201/eid2207.152080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien A. D., Gentry M. K., Thompson M. R., Doctor B. P., Gemski P., Formal S. B. (1979). Shigellosis and Escherichia coli diarrhea: relative importance of invasive and toxigenic mechanisms. Am. J. Clin. Nutr. 32 229–233. [DOI] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. (2005). Escape of intracellular Shigella from autophagy. Science 307 727–731. 10.1126/science.1106036 [DOI] [PubMed] [Google Scholar]
- Orskov I., Wachsmuth I. K., Taylor D. N., Echeverria P., Rowe B., Sakazaki R., et al. (1991). Two new Escherichia coli O groups: O172 from “Shiga-like” toxin II-producing strains (EHEC) and O173 from enteroinvasive E. coli (EIEC). APMIS 99 30–32. 10.1111/j.1699-0463.1991.tb05114.x [DOI] [PubMed] [Google Scholar]
- Parsot C. (2005). Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol. Lett. 252 11–18. 10.1016/j.femsle.2005.08.046 [DOI] [PubMed] [Google Scholar]
- Pavlovic M., Luze A., Konrad R., Berger A., Sing A., Busch U., et al. (2011). Development of a duplex real-time PCR for differentiation between E. coli and Shigella spp. J. Appl. Microbiol. 110 1245–1251. 10.1111/j.1365-2672.2011.04973.x [DOI] [PubMed] [Google Scholar]
- Pettengill E. A., Pettengill J. B., Binet R. (2015). Phylogenetic analyses of Shigella and enteroinvasive Escherichia coli for the identification of molecular epidemiological markers: whole-genome comparative analysis does not support distinct genera designation. Front. Microbiol. 6:1573. 10.3389/fmicb.2015.01573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettengill E. A., Pettengill J. B., Binet R. (2016). Phylogenetic analyses of Shigella and enteroinvasive Escherichia coli for the identification of molecular epidemiological markers: whole-genome comparative analysis does not support distinct genera designation. Front. Microbiol. 6:1573. 10.3389/fmicb.2015.01573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosseda G., Carmela Latella M., Barbagallo M., Nicoletti M., Al Kassas R., Casalino M., et al. (2007). The two-faced role of cad genes in the virulence of pathogenic Escherichia coli. Res. Microbiol. 158 487–493. 10.1016/j.resmic.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Prosseda G., Di Martino M. L., Campilongo R., Fioravanti R., Micheli G., Casalino M., et al. (2012). Shedding of genes that interfere with the pathogenic lifestyle: the Shigella model. Res. Microbiol. 163 399–406. 10.1016/j.resmic.2012.07.004 [DOI] [PubMed] [Google Scholar]
- Prosseda G., Falconi M., Giangrossi M., Gualerzi C. O., Micheli G., Colonna B. (2004). The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 51 523–537. 10.1046/j.1365-2958.2003.03848.x [DOI] [PubMed] [Google Scholar]
- Prosseda G., Falconi M., Nicoletti M., Casalino M., Micheli G., Colonna B. (2002). Histone-like proteins and the Shigella invasivity regulon. Res. Microbiol. 153 461–468. 10.1016/S0923-2508(02)01346-3 [DOI] [PubMed] [Google Scholar]
- Prosseda G., Latella M. C., Casalino M., Nicoletti M., Michienzi S., Colonna B. (2006). Plasticity of the P junc promoter of ISEc11, a new insertion sequence of the IS1111 family. J. Bacteriol. 188 4681–4689. 10.1128/JB.00332-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier A.-L., Schuch R., Fernandez R. E., Mumy K. L., Kohler H., McCormick B. A., et al. (2007). nadA and nadB of Shigella flexneri 5a are antivirulence loci responsible for the synthesis of quinolinate, a small molecule inhibitor of Shigella pathogenicity. Microbiology 153 2363–2372. 10.1099/mic.0.2007/006916-0 [DOI] [PubMed] [Google Scholar]
- Pupo G. M., Lan R., Reeves P. R. (2000). Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U.S.A. 97 10567–10572. 10.1073/pnas.180094797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D. A., Rosovitz M. J., Myers G. S. A., Mongodin E. F., Fricke W. F., Gajer P., et al. (2008). The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190 6881–6893. 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterer V., Grossniklaus L., Tschon T., Kasper C. A., Sorg I., Arrieumerlou C. (2011). Shigella flexneri type III secreted effector OspF reveals new crosstalks of proinflammatory signaling pathways during bacterial infection. Cell. Signal. 23 1188–1196. 10.1016/j.cellsig.2011.03.006 [DOI] [PubMed] [Google Scholar]
- Rowe B., Gross R. J., Woodroof D. P. (1977). Proposal to recognise seroovar 145/46 (synonyms: 147, Shigella 13, Shigella sofia, and Shigella manolovii) as a New Escherichia coli O group, O164. Int. J. Syst. Bacteriol. 27 15–18. 10.1099/00207713-27-1-15 [DOI] [Google Scholar]
- Sahl J. W., Morris C. R., Emberger J., Fraser C. M., Ochieng J. B., Juma J., et al. (2015). Defining the phylogenomics of Shigella species: a pathway to diagnostics. J. Clin. Microbiol. 53 951–960. 10.1128/JCM.03527-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakazaki R., Tamura K., Saito M. (1967). Enteropathogenic Escherichia coli associated with diarrhea in children and adults. Jpn. J. Med. Sci. Biol. 20 387–399. 10.7883/yoken1952.20.387 [DOI] [PubMed] [Google Scholar]
- Sanchez-Villamil J., Tapia-Pastrana G., Navarro-Garcia F. (2016). Pathogenic lifestyles of E. coli pathotypes in a standardized epithelial cell model influence inflammatory signaling pathways and cytokines secretion. Front. Cell. Infect. Microbiol. 6:120. 10.3389/fcimb.2016.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P. J., Hale T. L., Dammin G. J., Kapfer C., Collins H. H., Formal S. B. (1983). Alterations in the pathogenicity of Escherichia coli K-12 after transfer of plasmid and chromosomal genes from Shigella flexneri. Infect. Immun. 39 1392–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansonetti P. J., Kopecko D. J., Formal S. B. (1982). Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder G. N., Hilbi H. (2008). Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21 134–156. 10.1128/CMR.00032-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scribano D., Petrucca A., Pompili M., Ambrosi C., Bruni E., Zagaglia C., et al. (2014). Polar localization of PhoN2, a periplasmic virulence-associated factor of Shigella flexneri, is required for proper IcsA exposition at the old bacterial pole. PLOS ONE 9:e90230. 10.1371/journal.pone.0090230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva R. M., Toledo M. R., Trabulsi L. R. (1980). Biochemical and cultural characteristics of invasive Escherichia coli. J. Clin. Microbiol. 11 441–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small P. L., Falkow S. (1988). Identification of regions on a 230-kilobase plasmid from enteroinvasive Escherichia coli that are required for entry into HEp-2 cells. Infect. Immun. 56 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenungsson B., Lagergren A., Ekwall E., Evengård B., Hedlund K. O., Kärnell A., et al. (2000). Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clin. Infect. Dis. 30 770–778. 10.1086/313770 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2010). Pattern recognition receptors and inflammation. Cell 140 805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Taylor D. N., Echeverria P., Sethabutr O., Pitarangsi C., Leksomboon U., Blacklow N. R., et al. (1988). Clinical and microbiologic features of Shigella and enteroinvasive Escherichia coli infections detected by DNA hybridization. J. Clin. Microbiol. 26 1362–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo M. R., Trabulsi L. R. (1983). Correlation between biochemical and serological characteristics of Escherichia coli and results of the Serény test. J. Clin. Microbiol. 17 419–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M., Hoede C., Tenaillon O., Barbe V., Baeriswyl S., Bidet P., et al. (2009). Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLOS Genet. 5:e1000344. 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozzoli R., Scheutz F. (2014). “Diarrheagenic Escherichia coli infections in humans,” in Pathogenic Escherichia coli: Molecular and Cellular Microbiology, ed. Morabito S. (Norfolk: Caister Academic Press; ). [Google Scholar]
- Trabulsi L. R., Fernandes M. R., Zuliani M. E. (1967). New intestinal bacteria pathogenic to man. Rev. Inst. Med. Trop. Sao Paulo 9 31–39. [PubMed] [Google Scholar]
- Tran C. N., Giangrossi M., Prosseda G., Brandi A., Di Martino M. L., Colonna B., et al. (2011). A multifactor regulatory circuit involving H-NS, VirF and an antisense RNA modulates transcription of the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 39 8122–8134. 10.1093/nar/gkr521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. A., Luck S. N., Sakellaris H., Rajakumar K., Adler B. (2003). Molecular epidemiology of the SRL pathogenicity island. Antimicrob. Agents Chemother. 47 727–734. 10.1128/AAC.47.2.727-734.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Beld M. J. C., Reubsaet F. A. G. (2012). Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. Eur. J. Clin. Microbiol. Infect. Dis. 31 899–904. 10.1007/s10096-011-1395-7 [DOI] [PubMed] [Google Scholar]
- van Elsas J. D., Semenov A. V., Costa R., Trevors J. T. (2011). Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. 5 173–183. 10.1038/ismej.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M. M., Goldberg M. B., Rose D. J., Grotbeck E. J., Burland V., Blattner F. R. (2001). Complete DNA sequence and analysis of the large virulence plasmid of Shigella flexneri. Infect. Immun. 69 3271–3285. 10.1128/IAI.69.5.3271-3285.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira N., Bates S. J., Solberg O. D., Ponce K., Howsmon R., Cevallos W., et al. (2007). High prevalence of enteroinvasive Escherichia coli isolated in a remote region of northern coastal Ecuador. Am. J. Trop. Med. Hyg. 76 528–533. [PMC free article] [PubMed] [Google Scholar]
- Voeroes S., Redey B., Csizmazia F. (1964). Antigenic structure of a new enteropathogenic E. coli strain. Acta Microbiol. Acad. Sci. Hung. 11 125–129. [PubMed] [Google Scholar]
- Wanger A. R., Murray B. E., Echeverria P., Mathewson J. J., DuPont H. L. (1988). Enteroinvasive Escherichia coli in travelers with diarrhea. J. Infect. Dis. 158 640–642. 10.1093/infdis/158.3.640 [DOI] [PubMed] [Google Scholar]
- Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L. H., et al. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60 1136–1151. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagaglia C., Casalino M., Colonna B., Conti C., Calconi A., Nicoletti M. (1991). Virulence plasmids of enteroinvasive Escherichia coli and Shigella flexneri integrate into a specific site on the host chromosome: integration greatly reduces expression of plasmid-carried virulence genes. Infect. Immun. 59 792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo G., Xu Z., Hao B. (2013). Shigella strains are not clones of Escherichia coli but sister species in the genus Escherichia. Genomics Proteomics Bioinformatics 11 61–65. 10.1016/j.gpb.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski D. V., Mumy K. L., Faherty C. S., McCormick B. A., Maurelli A. T. (2009). Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol. Microbiol. 71 350–368. 10.1111/j.1365-2958.2008.06524.x [DOI] [PMC free article] [PubMed] [Google Scholar]