Abstract
Actinomycetes, a large group of filamentous bacteria account for 70–80% of secondary metabolites available commercially. The present investigation was undertaken with an aim to identify and characterize pigment from actinomycetes. Actinomycetes were isolated from rhizosphere soil samples collected from different regions of Madhya Pradesh state. Out of 85 actinomycetes, only 5 actinomycetes showed pigment production and based on diffusible pigment production ability one actinomycete ARITM02 was selected. The extraction of pigment was done by solvent extraction method using methanol and purified by TLC and column chromatography. The pigment was characterized by UV–Vis spectroscopy which showed the lamda maximum of 277.44. FTIR spectroscopy suggested various functional groups like amino group, amide group, hydroxide, benzene and lactone group. The Mass spectroscopy and NMR spectroscopy showed that the molecular mass of pigment is 621.7 and molecular formula is C34H43N3O8. The pigment was also tested for Antimicrobial activity against broad spectrum human pathogens, antioxidant test and toxicity test for safe use as a natural colorant in cosmetic, food, pharmaceutical and textile industries. The conclusion of study suggested that this novel pigment could be a versatile natural, safe and multipurpose.
Keywords: FTIR, Uv–vis spectrophotometer, NMR, Pigment, Antioxidant
Highlights
-
•
This study clearly describes about the characterization of pigment extracted from actinomycete ARITM02 which was isolated from rhizosphere soil.
-
•
The selected isolate showed antimicrobial activity against bacteria, fungi and molds but no activity against dermatophytes.
-
•
The study confirmed the basic biochemical nature of pigment which is similar to previous reported pigments.
-
•
The pigment was characterized by UV–Vis spectroscopy, FTIR spectroscopy, Mass spectroscopy and NMR spectroscopy.
-
•
The natural pigment is tested for Antimicrobial, antioxidant, anticancer and Allergy tests.
1. Introduction
Colours have been widely used in many industries such as textiles, food, painting, cosmetic and pharmaceuticals and they play an important role in providing an attractive look to the product. As a food colorant, they used as an additive in food industries and play a significant role due to safety and serious environmental problems caused by artificial and synthetic pigments research has been focused on research of new natural pigments to use in food and pharma industries [4].
Before the modern era, the natural organic pigments were an important and historical part particularly for ornamentation, cosmetic and textile dying. Natural pigments are generally extracted from different parts of plants like seeds, fruits, vegetables and roots. Sometimes they are also called Biocolours because they were originated from biological things. [7]. Natural pigments have many other functions other than imparted beauty. They are useful in photosynthesis. Hemoglobin is the key transporter of oxygen and carbon dioxide in animals and human beings [2], [5].
One of the most important quality of food is color and most often the buyer did not care about the kind of pigments used (natural or synthetic). But recently with reference to food colorant there is an aversion towards synthetic pigments that they are engaged with several harmful elements while natural pigments are safe from pharmacological view. However, there are very limited natural pigments which are permitted for human foods [10]. Recently there have been changes in the legislation also causing a significant reduction in number of synthetic colours used in foods [1].
A major number of Streptomycetes are neutrophilic and isolated at neutral pH. In case of acidophilic strains, the pH of medium can be adjusted accordingly and alkalophilic strain than pH could be changed [9]. 16S rRNA sequencing is used as a most powerful technique to sequence and the phylogenetic studies of actinomycetes ([12]) by using 16s rRNA technique [11]. Phylogenetic analysis of actinomycetes is done by studying the sequence for molecular identification using computer software. The main benefit of 16S rDNA generally allows the identification of actinomycetes up to genus level.
Actinomycetes are largest group of antibiotic producers in the pharmaceutical industries. Despite the production of antibiotics is of great benefit in medicine, some actinomycetes are pathogens and some cause allergic reactions. Actinomycetes sometimes cause biodeterioration of materials and are often responsible for spoilage of hay, straw, cereal grains, seeds, wood, paper, wool, hydrocarbons. Biologically actinomycetes can play an important role in waste management and its recycling of materials in nature [3], [8].
2. Materials and methods
2.1. Isolation and characterization of actinomycetes
The isolation of actinmycete ARITM02 was done on the basis of morphological, biochemical and molecular characterization [6].
The optimization of various parameters for pigment production by actinomycete isolate was carried out. Parameters namely temperature, pH, carbon sources, nitrogen sources and incubation time were carried out using the protocol of with minor modifications.
2.2. Extraction of pigment
The starch casein broth was prepared for extraction of pigment. Actinomycete isolate was inoculated into medium and incubated under standard optimized conditions.
Parameters namely temperature, pH, carbon sources, nitrogen sources and incubation time were carried out using the protocol of with minor modifications.
2.3. Extraction of pigment
The starch casein broth was prepared for extraction of pigment. Actinomycete isolate was inoculated into medium and incubated under standard optimized conditions. As maximum production was observed on the 4th day of incubation, fermentation was terminated after 96 h and stored in BOD at 2–3 °C temperature. The broth was centrifuged and separated by solvent extraction method using different solvents from culture supernatant. The crude pigment was separated, collected and dried in vacuum oven at 40 °C overnight.
2.4. Purification of crude pigment
The crude pigment was screened for number of fragments by Thin-Layer Chromatography (TLC) plates using Methanol: acetone: water: (4:4:2), Chloroform: methanol (9:1), Chloroform: methanol (6:4), Ethanol: water: chloroform (4:4:2) and Ethanol: water: chloroform (4:2:4) solvent system. The chromatographic chamber with solvent was kept for 15 mints for equilibration. The sample was spotted on readymade silica gel sheet (Merk) with the help of capillary tube and air dried. A control sheet without spot was also used as a blank. The TLC sheets now dipped in solvent system and allowed to run. TLC sheet was carefully removed and air dried. The TLC plates were exposed to iodine vapors, sprayed with vanillin and ninhydrin separately. Retention factor (Rf) value was calculated according to the following equation from the chromatogram.
Purification of the pigment was carried out by column chromatography using silica gel (60–120 mesh). Fractions were collected at 20 min interval. TLC of each fraction was performed. The fractions having same Rf value were mixed together and the solvent was evaporated at 40 °C in a vacuum oven. These fractions were tested for their antimicrobial activity by using the well agar diffusion method. The pure compound obtained was stored in an ampoule at 4 °C.
2.5. Characterization of purified pigment
2.5.1. UV–Vis spectrophotometer analysis
The UV–Visible absorption spectra of the bioactive component in solvent extracts were determined with a UV–Vis spectrophotometer (PerkinElmer) at 200–600 nm to determine the Lambda maximum of the band.
2.6. Fourier transform infra red (FTIR) spectroscopy
Fourier transform infrared (FTIR) spectroscopy, analysis, is a technique that provides information about the chemical bonding or molecular structure of materials, whether organic or inorganic. The technique works on the fact that bonds and groups of bonds vibrate at characteristic frequencies. A molecule that is exposed to infrared rays absorbs infrared energy at frequencies, which are characteristic to that molecule. The purified pigment sample was subjected to FT –IR spectroscopic analysis (Perkin Elmer Lambda), equipped with KBr beam splitter with DTGS (Deuterated triglycine sulfate) detector.
2.7. Mass spectroscopy analysis
Mass spectrometry (MS) is an analytical technique which identifies compounds based on the atomic sample composition of the molecules and their charge state. Therefore, “blind” analysis of unknown samples is possible since MS does not require detailed prior knowledge of the sample composition. Ideally, the chemical identification by MS is not limited by analyte pre-selection as, for example, in analysis techniques based on fluorescent or radioactive labeling. This bears the advantage that the analysis technique itself does not make any functional changes to the molecules under investigation. The applications of mass spectrometry range from among others environmental analysis, isotope dating and tracking, trace gas analysis, proteomics, lipidomics, metabolomics to clinical applications.
2.8. Nuclear magnetic resonance (NMR) spectroscopy analysis
Nuclear magnetic resonance spectroscopy most commonly known as NMR spectroscopy is the technique which exploits the magnetic properties of certain nuclei. The pigment was analyzed by NMR spectrophotometer.
2.9. Antimicrobial activity
For evaluation of antimicrobial activity of pigment was done by well agar diffusion method, wells were drilled using a sterile cork borer in fresh test microbial lawn cultures on nutrient agar medium for bacteria. The pigment with dissolved in methanol was then administered to fullness in each well. The plates were incubated at 35 °C for 24–48 h. Bioactivity was determined by measuring the diameter of inhibitory zones (mm) of test microorganisms around the well after incubation. The antibiotic tetracycline was used as a control (7 µg/ml) for bacteria and nystatine (10 µg/ml) used for fungi.
2.10. Toxicity test of pigment
HDF (Human dermal fibroblast) and U-87 (Human Glial cell) cell lines were procured from National Centre for Cell Sciences (NCCS), Pune, India. HDF and U-87 stock cells were cultured in Ham's F12 medium supplemented with 10% inactivated Fetal Bovine Serum (FBS), penicillin (100 IU/ml), streptomycin (100 μg/ml) and amphotericin B (5 μg/ml) in an humidified atmosphere of 5% CO2 at 37 °C until confluent. The cells were dissociated with TPVG solution (0.2% trypsin, 0.02% EDTA, 0.05% glucose in PBS). The stock cultures were grown in 25 cm2 culture flasks and all experiments were carried out in 96 microtitre plates (Tarsons India Pvt. Ltd., Kolkata, India).
The absorbance was measured using a microplate reader at a wavelength of 540 nm. The percentage growth inhibition was calculated using the following formula and concentration of test drug needed to inhibit cell growth by 50% (CTC50) values is generated from the dose-response curves for each cell line.
2.11. Antioxidant test
The DPPH free radical is reduced to a corresponding hydrazine when it reacts with hydrogen donors. The DPPH radical is purple in color and upon reaction with hydrogen donor changes to yellow color. It is a discoloration assay, which is evaluated by the addition of the antioxidant to a DPPH solution in ethanol or methanol and the decrease in absorbance was measured at 490 nm.
2.12. Procedure
The assay was carried out in a 96 well microtitre plate. To 200 μl of DPPH solution, 10 μl of each of the test sample or the standard solution was added separately in wells of the microtitre plate. The final concentration of the test and standard solutions used were 1000, 500, 250, 125, 62.5, 31.25 and 15.625, 7.812 μg/ml. The plates were incubated at 37° C for 30 min and the absorbance of each solution was measured at 490 nm, using a microplate reader.
3. Results
3.1. Separation and purification of pigment
The pigment was extracted by using different solvents. The methanol was found suitable solvent for extraction of pigment. The crude pigment compound was screened for number of components by Thin-Layer Chromatography (TLC) plate. The Methanol:acetone:water: (4:4:2) was found best solvent system for TLC. A dark red single spot along with very light spot was found after spray of ninhydrin (Fig. 4). The Rf value was found 0.76 which indicates protein nature of pigment. The pigment obtained was stored in an ampoule at 4 °C.
Fig. 4.
NMR spectrum of pigment.
3.2. Characterization of pigment
3.2.1. UV–Vis spectroscopy
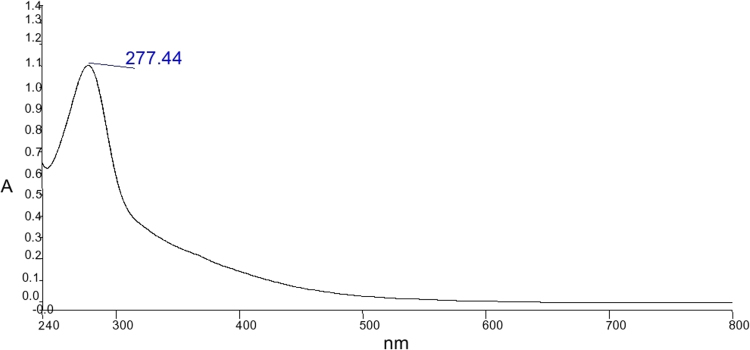
The UV–Vis spectrum was found by UV–Vis spectrophotometer (Modal: PerkinElmer-650). The spectra showed lambda max at 277.44 (Fig. 1).
Fig. 1.
UV-Vis spectrum of pigment, extracted from selected isolate ARITM02
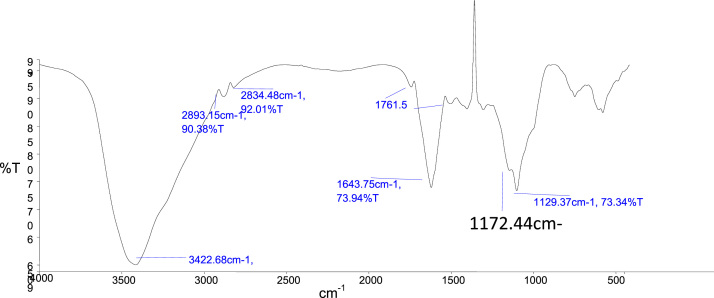
FTIR spectroscopy is important for the interpretation of the structure binding capacity, affinity and sites of metal ions in pigment. The spectrum obtained from Fourier-Transform Infrared Spectroscopy (FT-IR Spectroscopy, Modal no. L125000P) indicates strong bands at 3422 cm−1 (Hydroxide group or amin group), 2893 and 2834 cm−1 (Aromatic or benzene group), 1761 cm−1 (Lacton group) and 1643 cm−1 showed aliphatic group (Fig. 2).
Fig. 2.
The FTIR spectrum of pigment.
3.3. Mass spectroscopy
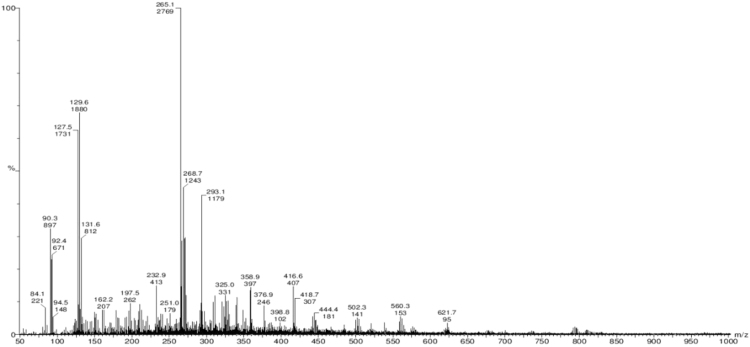
Mass spectrometry has the ability to provide highly accurate molecular mass measurements for compound. The mass spectrum was analyzed and the molecular mass of the pigment was found 621.7 Da (Fig. 3).
Fig. 3.
Mass spectrum of pigment.
3.4. Nuclear magnetic resonance spectroscopy
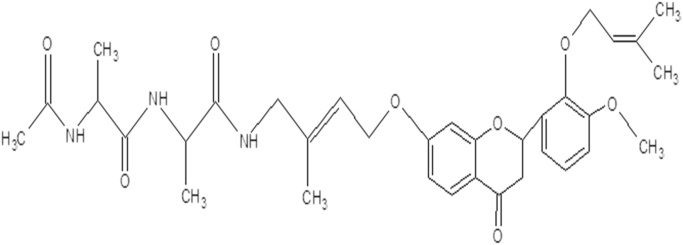
The 1HNMR spectrum is obtained by NMR spectroscopy. The interpretation of NMR spectrum concluded that the expected molecular formula of pigmented compound is C34H43N3O8. The suggested expected structure of pigment is shown in Fig. 4, Fig. 5.
Fig. 5.
The expected molecular structure of pigment.
3.5. Antioxidant assay
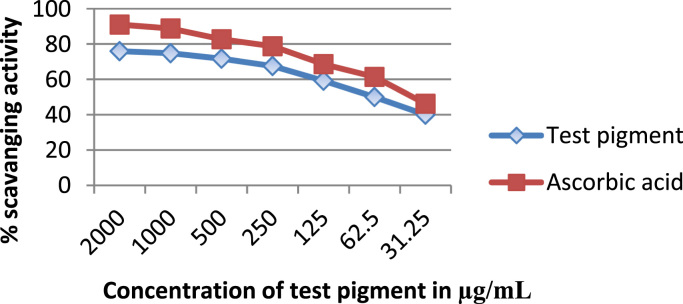
DPPH is reduction of purple colored DPPH in the presents of hydrogen donating antioxidant, by formation of yellow colored non radical form of DPPH. The scavenging activity was increased when increasing of concentration of pigment and IC50 value found 62.5 µg/ml. This result shows that pigment also have good antioxidant activity and could be used further as an antioxidant compound (Fig. 6).
Fig. 6.
Showing antioxidant activity of pigment by DPPH assay.
3.6. Toxicity test by MTT assay
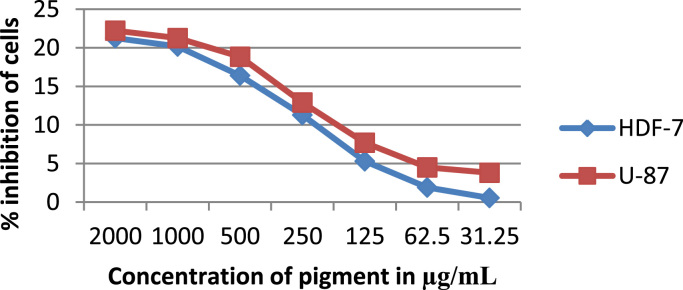
The toxicity effect was studied on different concentration was tested from 31.25, 62.50,150,250,500,1000 and 2000 µg/ml. It is found that cell inhibition is increasing as dose of pigmented compound increasing on both cell lines. The cytotoxic effect of pigmented compound showed CTC50 > 1000 μg/ml which indicates that pigmented compound has very low cytotoxicity or mildly toxic against both cell lines. The study confirmed that the natural pigment has very less cytotoxic effect and probably used in food and pharma industries as a natural colorant agent. Compound is more effective against cancer cell lines as compared to normal cell lines (Fig. 7).
Fig. 7.
Cytotoxic properties of test drugs against cell lines, HDF and U-87.
3.7. Antimicrobial activity of pigment
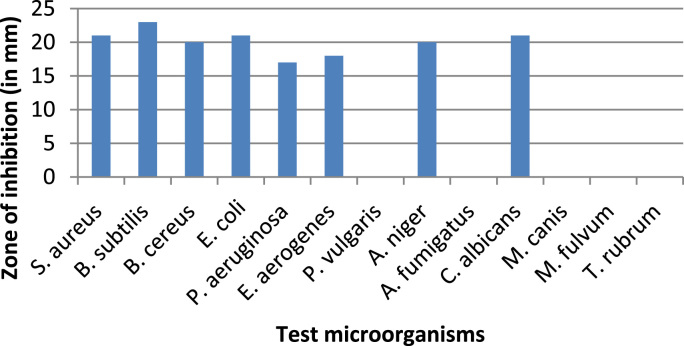
In well agar diffusion method, purified pigment was used and dissolved in methanol. The result indicates that the pigment have antagonistic activity against microorganisms including bacteria, yeast and molds but didn't showed any activity against dermetophytes (Fig. 8).
Fig. 8.
Graph represents antibiogram of pigment against various microorganisms.
Acknowledgments
Authors are very thankful to Department of Life Sciences, ITM University, Gwalior, India for providing necessary facilities.
Acknowledgments
Ethical statement
This article does not contain any studies with human participants and/or animals performed any of the authors. Formal consent is not required in this study.
Conflict of interest
We authors declare that we have no conflict of interest.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.11.002.
Appendix A. Transparency document
Supplementary material
References
- 1.Downham A., Collins P. Coloring our foods in the last and next millennium. Int. J. Food Sci. Technol. 2000;35:5–22. [Google Scholar]
- 2.Hari R.K., Pate T.R., Martin A.M. An overview of pigment production in biological systems: functions, biosynthesis, and applications in food industry. Food Rev. Int. 1994;10:49–70. [Google Scholar]
- 3.Kim B., Sahin N., Tan G.Y.A., Zakrzewska-Czerwinska J., Kiruthika P., Nisshanthini S., Angayarkanni J. In vitro antimicrobial and antioxidant profile of Streptomyces Sp. isolated from Coromandel coast region, India. Int. J. Pharma BioSci. 2013;4(4):127–136. (B) [Google Scholar]
- 4.Lu Y., Wang L., Xue L., Zhang C., Xing X.H., Lou Z.Z., Li Y., Zhang G., Bi J., Su Z. Production of violet pigment by a newly isolated psychrotrophic bacterium from a glacier in Xinjiang, China. Biochem. Eng. J. 2009;43:135–141. [Google Scholar]
- 5.Mol I., Jenkins G., Schafer E., Weiss D. Signal perception, transduction and gene expression involved in anthocyanin biosynthesis. Crit. Rev. Plant Sci. 1996;155:525–557. [Google Scholar]
- 6.Parmar R.S., Singh C., Saini P., Kumar A. Isolation and screening of antimicrobial and extracellular pigment producing actinomycetes from chambal territory of Madhya pradesh region, India. Asian J. Pharm Clin. Res. 2016;9(1):157–160. (1) [Google Scholar]
- 7.Pattnaik P., Roy U., Jain P. Biocolours: new generation additives for food. Indian Food Ind. 1997;16(5):21–32. [Google Scholar]
- 8.Petinate S.D.G., Martins R.M., Coelho R.R.R., Meirelles M.N.L., Branquinha M.H., Vermelho A.B. Influence of growth medium in proteinase and pigment production by Streptomyces cyaneus. Mem. Inst. Oswaldo Cruz. 1999;94(2):173–177. doi: 10.1590/s0074-02761999000200008. [DOI] [PubMed] [Google Scholar]
- 9.Suutari M., Lignell U., Hirvonen M.R., Nevalainen A. Growth pH ranges of Streptomyces sp. ASM News. 2000;66(10):588. [Google Scholar]
- 10.Wissgot U., Bortlik K. Prospects for new food colorants. Trends Food Sci. Technol. 1996;7:298–302. [Google Scholar]
- 11.Xu L.H., Jin X., Mao P.M., Lu Z.F., Cui X.L., Jiang C.L. Three new species of the genus Actinobispora of the family Pseudonocardiaceae, Actinobispora alaniniphila sp. nov., Actinobispora aurantiaca sp. nov. and Actinobispora xinjiangensis sp. nov. Int. J. Syst. Bacteriol. 1999;49:881–886. doi: 10.1099/00207713-49-2-881. [DOI] [PubMed] [Google Scholar]
- 12.Yokota A. Phylogenetic relationship of actinomycetes. Atlas of actinomycetes. Soc. Actinomycetes Jpn. 1997;194 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material