Abstract
Channelrhodopsins are light-gated ion channels that, via regulation of flagellar function, enable single-celled motile algae to seek ambient light conditions suitable for photosynthesis and survival. These plant behavioral responses were initially investigated more than 150 years ago. Recently, major principles of function for light-gated ion channels have been elucidated by creating channelrhodopsins with kinetics that are accelerated or slowed over orders of magnitude, by discovering and designing channelrhodopsins with altered spectral properties, by solving the high-resolution channelrhodopsin crystal structure, and by structural model–guided redesign of channelrhodopsins for altered ion selectivity. Each of these discoveries not only revealed basic principles governing the operation of light-gated ion channels, but also enabled the creation of new proteins for illuminating, via optogenetics, the fundamentals of brain function.
A light-gated ion pore
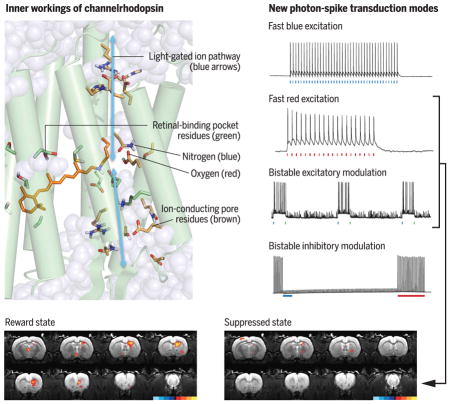
(Top) Left: Inner workings of channelrhodopsin. Right: New photon-spike transduction modes arising from structure-guided redesign. (Bottom) Discovering the causal underpinnings of depression-related symptomatology. Brain region–specific activity dynamics of the mammalian dopamine neuron–driven reward state (left) are suppressed by the prefrontal cortex (right) as shown, using the second and third photon-spike transduction modes.
The study of neural circuitry underlying adaptive and maladaptive animal behavior has become intertwined with the investigation of algae-derived light-gated ion channels (1, 2). Each of these two fields has unexpectedly, profoundly advanced the other, culminating in mutually illuminating discoveries (3). Curiosity regarding the nature and operation of these unique channels, combined with a long-standing impetus to bring precise cellular-resolution causality to basic and clinical neuroscience, together have resulted in the elucidation both of diverse fundamental processes in neuroscience [reviewed in (3)] and of underlying structural and functional mechanisms within the algae proteins themselves (explored here).
The natural role of channelrhodopsins (ChRs) in motile algae involves coupling irradiance information from ambient light to flagellar motion, allowing the organism to seek light optimal for photosynthesis and survival. ChR signaling must be fast enough to implement meaningful coupling to the flagella during helical swimming of the alga with its typical 2-Hz rotation, which continuously alters eye position relative to direction of incident light (4). Here, we explore how studies over the past 10 years have elucidated the atomic-scale protein structural underpinnings of this single polypeptide–mediated fast transduction of visible-wavelength photons into selective transmembrane ion flow. We describe how, within these compact proteins, even single amino acids play multiple roles in light-gated ion channel function, including key processes such as activation/deactivation gating, light adaptation, color specificity, and ion selectivity (Fig. 1). In doing so, we bring together structural and functional insights, consider general principles of selective ion channel design and evolution (subject to biophysical constraints that the light-activated ion-conducting pore satisfies), and highlight how the basic discovery process of designing and creating new classes of light-gated ion channels not only provided fundamental insight into channelrhodopsin pore function, but also enabled the discovery of principles underlying nervous system function and neuropsychiatric disease.
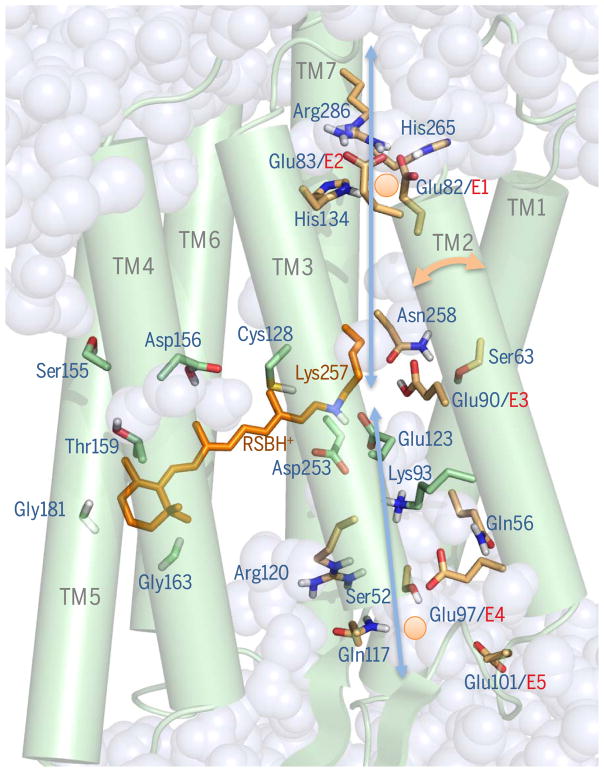
Fig. 1. The light-gated pore.
The crystal structure of C1C2 is shown as a 3UG913-derived pore snapshot calculated via molecular dynamics (MD). Protonated retinal Schiff base (RSBH+, orange) and key pore (brown) or RBP (green) residues showing protonation states of polar residues (81) are shown in stick form (red, oxygen; blue, nitrogen; white, polar protons). ChR2 residue numbering is used. Displayed waters invaded from both membranes during calculations (81). Two Na+ ions (orange circles) are shown at positions of preferred occupation within the access channel and near inner gate calculated by MD (81). Blue arrows denote the presumptive permeation pathway. The two-headed orange arrow shows the predicted (43) tilting dynamic of helix 2. Conserved glutamates in helix 2 are designated E1 to E5.
Deep roots in botany
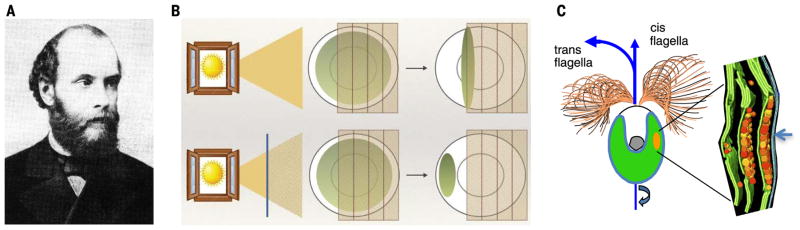
The channelrhodopsin story begins at the Neva River in Russia nearly 150 years ago, where the famed 19th-century botanist Andrei Sergeyevich Faminzin studied motile microalgae (Fig. 2, A and B). Faminzin provided the first comprehensive descriptions of single-celled motile algae moving toward or away from light (5). Despite intensive study over the next century of behavioral ecology among algal species, and of subcellular light detection structures that algae use to modulate flagellar beating (Fig. 2C, Chlamydomonas reinhardtii), the photoreceptor and molecular phototransduction mechanisms remained mysterious. Not until the late 20th century were light-evoked rhodopsin-type currents in Chlamydomonas-type chlorophyceae (6, 7), and the broader family of microbial rhodopsin-type ion conductance regulators, detected (1, 8, 9).
Fig. 2. Deep roots in botany.
(A) Faminzin, pioneer of algal behavior (4). (B) Faminzin’s landmark observation (5): At low light intensities (bottom), Chlamydomonas exhibits phototaxis toward light, whereas at higher light intensities (top) the algal cells accumulate at optimal irradiance. (C) Adapted cartoons showing subcellular Chlamydomonas structure: flagellar beating pattern (20) and eye structure reconstructed from high-resolution tomography (17). Eyespot overlaying part of plasma membrane (arrow) corresponds to ChR location. Layers of carotenoid-containing vesicles (colored spheres) held by chloroplast membranes (green) serve as an optical device (interference reflector) (4, 19).
In 1971, evidence for microbial retinal-binding membrane proteins was obtained, initially the proton pumping–type bacteriorhodopsin within its native archaeal system (Halobacterium salinarum) (8). Purified bacteriorhodopsin-mediated light-activated transmembrane pump currents were later demonstrated in artificial black lipid membrane preparations (9). More than 40 years of study revealed that rhodopsins produced by microbial organisms include several subclasses of single gene–single protein ion transporters. All are seven-transmembrane (7TM) retinal-binding proteins encoded by single opsin genes (1, 2). Unlike rhodopsins of the vertebrate retina, these microbial proteins bind all-trans retinal (ATR, rather than cis isomers) in the dark/inactive state (Fig. 3). Rather than triggering an effector cascade to elicit ion flux (like vertebrate rhodopsins), light sensation and ion flow regulation are implemented by a single polypeptide chain (1, 2).
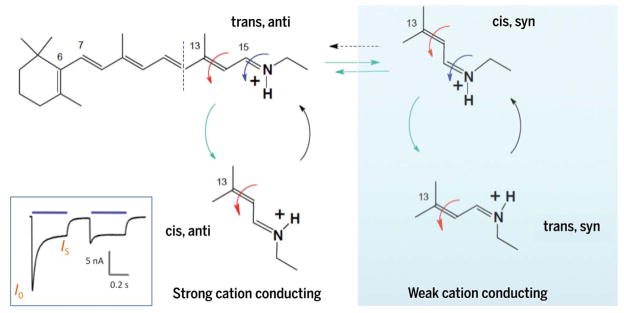
Fig. 3. Chromophore states and channel currents.
Chromophore configurations in RSB: all-trans, 15-anti (trans, anti) of dark-adapted state; 13-cis, 15-syn (cis, syn) of second dark state (increasingly occupied after blue illumination during photocurrent inactivation from initial I0 to stationary IS; inset) (1, 37). In corresponding open states occupied after 13C=14C photoisomerization, RSB is in the 13-cis, 15-anti (cis, syn) configuration in the open conducting state O1 and 13-trans, 15-syn (trans, syn) in O2 (1, 37). Cis-trans isomerization is indicated by red arrows, anti-syn isomerization by blue arrows, photochemical conversions by green arrows, and thermal conversions by black arrows.
The photocurrents of motile Chlorophyceae are mediated by the ChR subtype of this family; the prototype ChRs are ChR1 and ChR2 of Chlamydomonas. These algal rhodopsins were discovered via action spectroscopy of phototaxis/photoelectric responses and via restoration of photobehavior in blind mutant algae by supplementation with retinoids (7, 10). Characterization of algal photocurrents led to the suggestion that light-sensing and ion-conducting units derived from a single protein (11) conducting 10 to 100 charges per photocycle [initial 300-fS estimation assuming 10,000 channels in the algal eyespot (7) was quite consistent with 100-fS unitary conductances inferred by noise analysis years later for heterologously expressed ChR2]. After photoreceptor-like sequences were discovered in the Kazusa cDNA database, heterologous expression demonstrated passive transmembrane currents down electrochemical gradients (revealing channel-like behavior) (12, 13).
ChRs are larger than any previously identified rhodopsin, but only the N-terminal 40% [including all transmembrane domains with both light sensor and ion channel functionality (12)] is needed for optogenetics (14). As confirmed by crystal structure determination (15), the ATR chromophore appears to have inherited positioning within the retinal binding pocket (RBP) and covalent binding to the protein backbone (via lysine; Fig. 1) of its presumed rhodopsin-pump evolutionary forebears (1, 2).
Studies of motile photosynthetic microbes have continued along several directions. Not all such organisms use ChRs; light-activated nucleotide cyclases with flavin-based light sensors have been found in phototactic Euglenophyceae (16). In Chlamydomonas, structural and behavioral work is ongoing; ChRs have been localized to the eyespot (photosensitive organelle) overlaying part of the plasma membrane (17–19) (Fig. 2C), and light modulation of flagellar beating has recently been recorded with unprecedented precision (20) as it occurs via flagellar action potentials (upon large and abrupt changes in light intensity) or by gradual modulation of membrane voltage. As detailed next, studies of these behaving plants have also profoundly advanced the understanding of behaving animals (3, 14).
Impetus from neuroscience and basic principles of optogenetics
Francis Crick was the first to suggest that light might be a useful cell type–targeted neural control modality for the investigation of nervous system function, but did not see how to achieve this, terming the possibility “far-fetched” (21). Over the decade beginning in 2005, light-activated proton or Cl− pumps (bacteriorhodopsins or halorhodopsins) and light-activated cation or Cl− channels (ChRs) were all discovered to serve as high-speed optically activated regulators of action potential firing when genetically targeted to neurons of metazoa (14). Together with several additional (and necessary) technological innovations, this enabled precisely what Crick had sought (3, 14).
Targeted optical control of single neurons had been achieved earlier with spatially guided lasers but not broadly applied (22, 23). Early genetically guided efforts were elegant but limited as well, in part because their multicomponent design required multiple genes, or both exogenous chemicals and effector genes, to couple the chromophore to neuronal activity (24–26). Reports of the feasibility of a single-component microbial opsin approach to optical control of spiking were initially published only in transparent or spatially superficial systems (e.g., cultured neurons, isolated retina preparations, nervous system slices, and small invertebrates) (27–31). Justified skepticism remained for many years regarding the potential for general utility across intact neural systems.
These microbial opsin genes constitute only one feature of what became the optogenetic approach. Two other key developments—the fiberoptic neural interface and versatile targeting methodology (32–37)—would ultimately enable generalizable millisecond-precision genetically targeted neuronal control with light throughout the brain during behavior (3, 14). By 2007, optogenetic control of defined spiking patterns in specified neurons deep in the hypothalamus of freely moving adult mice had been achieved, along with resulting behavioral state transitions (33). Optogenetics had attained an early form of the fundamental methodology that neuroscientists use today: expression of microbial opsins with cell type (and even projection type) specificity and behavioral control potency by means of high-titer viral gene-targeting vectors and light targeting through implanted fiber optics (34). By 2009, generalizable gene-targeting strategies had been developed and shown to be suitable for mammalian behavioral control (35). From across the global scientific community, many thousands of discoveries have resulted, revealing the otherwise inaccessible cell-specific neural activity building blocks of behavior (3, 14).
Structural models and pore-gating kinetics
The light-activated membrane pore of ChRs is unique. Within the ChR family, however, many variants exist. ChR is a highly flexible protein heavily modified in action spectrum, photocycle kinetics, and ion selectivity by scientists over the past dozen years, but much more so by nature on the billion-year time scale.
There was initial confusion regarding the structure of the light-activated pore. Early characterization of ChRs had elucidated properties such as nonspecific cation selectivity (permeability to H+ ≫ Na+ > K+ ≫ Ca2+ with inward rectification) (12, 13). But it was unclear where the pore would be localized, except for perhaps a contribution from helix 2, with its prominent intramembranous glutamate residues. In the quest to find the light-gated pore, comparisons with microbial light-driven pump structures were of little help—for example, helices 1 and 2 show very low homology with presumptive pump counterparts— or even misleading [low-resolution cryo–electron microscopy (cryo-EM) had led to predictions that the pore would reside at the interface of two ChR monomers (36)—which turned out to not be the case (15)]. This level of uncertainty stalled pore engineering for design and creation of novel ion-selective ChRs. Many other functions, including the key kinetic properties of activation and inactivation (Fig. 3), nonetheless yielded to construction of structural models and detailed molecular design before the three-dimensional structure was solved.
Gradual inactivation of ChR photocurrents in strong light (Fig. 3) appears useful for algae in coping with high light intensities but poses challenges for neuroscience. This inactivation manifests as a probabilistic alternative photoisomerization around the ATR C15=NH bond (37), corresponding to transition from a syn-cycle to an anti-cycle (Fig. 3) as in animal rhodopsins during transitions from Meta-I to Meta-III (38) or in bacteriorhodopsin during dark adaptation (39). The syn-cycle open state is less permeable to protons and cations (versus the anti-cycle), contributing to photocurrent decline under prolonged illumination (Fig. 3) (40). Under dark adaptation, the ATR exists as all-trans/15-anti, but shortly after illumination a mixture of two similar but distinguishable states appears (all-trans/15-anti and 13-cis/15-syn), leading to conducting states in 13-cis/15-anti (O1) and 13-trans/15-syn (O2) configurations, respectively (37, 41). The O1/O2 ratio also depends on membrane voltage and pH gradient, and differs substantially among ChR variants, contributing to diversity of photocurrent properties (1, 2, 42).
The fast time constant for actuation of pump rhodopsins known from earlier work was concordant with later-observed ChR activation in oocyte membrane patches (τon < 250 μs) (13), and elucidation of molecular principles governing pore kinetics became a major goal, not only to obtain insight into the mechanics of pore operation but also to achieve optogenetic control over both short and long time scales of behavior and physiology. Retinal isomerization appears to cause rapid rearrangement of central gate residues and gate opening on the nanosecond time scale; helix hydration via water influx proceeds within 10 to 100 μs, which (in conjunction with helix 2 movement) opens an inner gate, allowing formation of the cation-conducting pore (43, 44) (Fig. 1). Under typical conditions, the submillisecond opening time constant (τon) of the ChR pore is fast enough not to influence or limit neuroscience applications. In contrast, photocurrent decay after light-off (τoff) is of great functional consequence in neuroscience and is broadly tunable. Pumps and channels will cease to enter the photocycle upon cessation of light, whereas observed photocurrents can outlast light because already activated proteins are completing their photocycle. The τoff for wildtype ChR2 (~10 ms) is impressively fast for a plant, but is slow for fast-spiking mammalian neurons and contributes to impaired fidelity in optogenetics (45). This parameter [which in many ChRs further slows upon membrane depolarization (42, 46)] elicits prolonged post-spike depolarization lasting tens of milliseconds, counteracting typical millisecond-scale repolarizations that terminate natural spikes; artifactual doublets rather than precise single spikes can result (42, 45), and spiking can even fail within higher-frequency spike trains (because repolarization de-inactivates native voltage-dependent channels required for subsequent spiking) (42, 45). These fidelity issues derived from long ChR τoff values (45) thus were addressable with redesigned shorter τoff once key molecular principles had been determined.
Mutations at Glu123 of ChR2, one of the negatively charged “counterion” residues stabilizing the obligate intramembranous positive charge of the protonated retinal Schiff base (RSBH+; Fig. 1 and Fig. 4, A and B), reduce τoff to 4 ms at −100 mV and also reduce voltage-dependent slowing, thus accelerating pore closure during spiking (45). These “ChETA” variants (45) exhibited the drawback of moderately reduced light sensitivity due to reduced photoisomerization efficiency and charge transfer per photon (45). However, accelerated deactivation addressed the key parameter limiting neuroscience application to fast-spiking cells and enabled firing at 200 Hz or more, along with reduced numbers of extra or missed spikes (42, 45) (Fig. 4B). These high-speed variants are often used when precise control and temporal stationarity is desired, particularly in fast-spiking cells [involved in sleep, fear, and feeding (46–49)]. Further engineered or naturally occurring ChRs were later identified that also exhibited fast deactivation and/or improved temporal stationarity [e.g., ChIEF, Chronos, and other variants; reviewed here (14, 41, 42, 50, 51)], and the fast-ChR toolkit grew further as ChETA and ChIEF modifications proved portable to other ChRs, including designer variants with larger photocurrents (51) and redshifted spectra (discussed below).
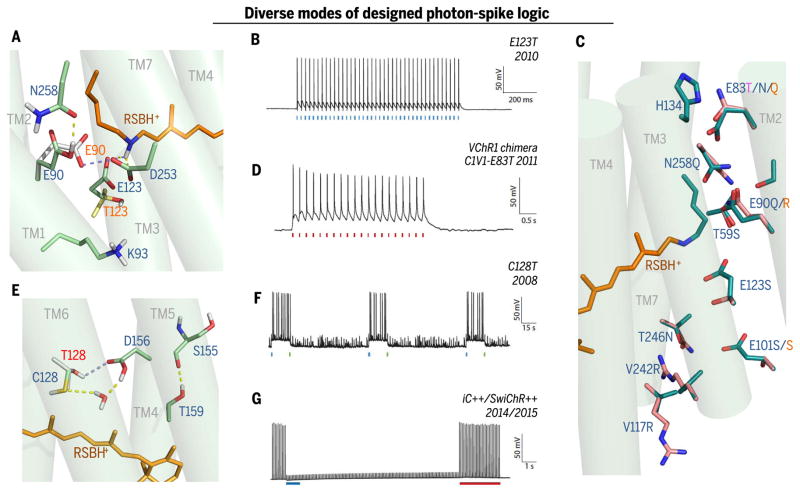
Fig. 4. Diverse modes of photon-spike transduction logic with underlying structural design.
(A) Snapshot of ChR active site (MD calculation from C1C2 structure 3UG9, with predicted repositioning of side chains resulting from Glu123 → Thr (E123T) mutation in red lettering; E123Tcauses inward flipping of Glu90 to compensate for the lost Glu123 counterion (43) of the RSBH+, additionally preventing inactivation. (B) Photon-spike transduction mode arising from ChETA mutation [ChR2-E123Tvariant (45)]: single blue flash–single spike coupling with high speed and high fidelity. Pore redesign implements faster closure after light-off, permitting rapid firing [e.g., 200-Hz trains in interneurons (45)]. (C) Pore residues (from C1C2 structure 3UG9) altered in spectral and selectivity variants. Modification of inner gate in red-activated C1V1-E83Tvariant (magenta letter–designated mutation; ChR2 numbering) (54). Selectivity variants are shown as original cation-conducting C1C2 pore residues and modifications to create the Cl−-selective iC++ [new pink side chains overlaid on original C1C2 green side chain positioning; blue letters denote iC++ mutations (62, 64)] or iChloC [orange letter–denoted mutations (93, 94)]. (D) Photon-spike transduction mode arising in Volvox-derived C1V1-E83T: single red flash–single spike coupling (54) with moderately high speed/fidelity; later Volvox derivative bReaChES exhibits faster responses with ChETA modification for accelerated channel closure (77) (not shown). (E) Snapshot of most likely structure of the DC-pair region in C1C2 (blue lettering) and the C128T (Cys128 → Thr) variant (red lettering) based on MD calculation (100) of restructured hydrogen-bonding network [yellow → blue dashed-line transition represents this SFO-mutation (52, 54) transition] and modified TM3-TM4 interaction (100), resulting in extension of open-state lifetime (52–54) and many-orders-of-magnitude-increased light sensitivity of expressing cells (42, 52, 54). (F) Photon-spike transduction mode arising from C128T (SFO) mutation is bistable, ultra–light-sensitive, two-color switchable, and excitatory [note blue light actuation and green light termination (52, 54)]. (G) Photon-spike transduction mode arising from adding the Cys128 SFO mutation (E) to Cl−-selective iC++ mutations (C) to create (64) SwiChR++ provides ultra–light sensitivity and is bistable, two-color switchable, and inhibitory under typical conditions (62, 64).
Bistable modes of pore operation
What about extending rather than shortening photocycles? In a surprising discovery, slowing τoff was even more useful than accelerating τoff in achieving kinetic pore bistability (52). These modified ChRs did not require continuous light for continuous function, but instead were bistable as a result of precisely reversible photocycle arrest (Fig. 4F) (52). Similarly arresting pump photocycles would mean termination of photocurrent, but for channels, precision kinetic locking into an open-pore state enables stable current for tens of minutes after light-off (52). Modification of helix 3–helix 4 interaction at the DC pair (Cys128-Asp156 in ChR2; Fig. 4, E and F) extends open-state lifetimes by up to 6 log units and reduces inactivation (52–54). Interestingly, redshifted light excitation of the resulting stable conducting state reconverts ATR back to the dark-state conformation, causing channel closure (52). Thus, high temporal precision for both onset and offset is preserved, with intervening step-like performance stable enough to make continuous light unnecessary (Fig. 4F); these variants were thus termed step-function opsins (SFOs) (52).
Several advantages accrue from application of SFOs to optogenetics: (i) Light need not be provided continuously. The many-orders-of-magnitude reduction (42, 52) in energy delivered can eliminate any intense-light toxicity from long-term experiments. (ii) If complex behaviors (e.g., large group social interactions) or experimental settings [e.g., naturalistic environments or magnetic resonance imaging (MRI) scanners] are incompatible with certain devices, cells can be stably modulated with a light pulse, followed by characterization of physiology or behavior over prolonged periods without light delivery hardware (54). (iii) Continuous mild depolarization can simply favor excitability of targeted cells [rather than provide precise user-defined spikes (52, 54)], an approach often leveraged to maintain or enhance native rhythm/timing relationships (54–61). (iv) Cells expressing these bistable ChRs exhibit vastly greater light sensitivity (42, 52–54) due to photon integration; physiologically relevant excitatory photocurrents of >100 pA can be elicited with wild-type ChRs at safe expression levels using ~1 mW/mm2 irradiance at the expressing neurons, but cells expressing the SFO ChR2-C128A/S variants (τoff up to three orders of magnitude longer) are 300 times as light-sensitive (52) for stationary photocurrents. Because bistable channels are slow to close, photocurrent accumulates even with orders-of-magnitude weaker light. Even further reduced energy delivery (over time and instantaneously), as well as greater tissue volumes recruited at a given light intensity in large-brained subjects, can result from the use of DC double mutations [the most stable SSFO (stabilized SFO) variant (54) with τoff ≥ 30 min; Fig. 5A]. This phenomenon has been leveraged to minimize heating (and heating artifacts) during optogenetic functional MRI (fMRI) (Fig. 5B) (56).
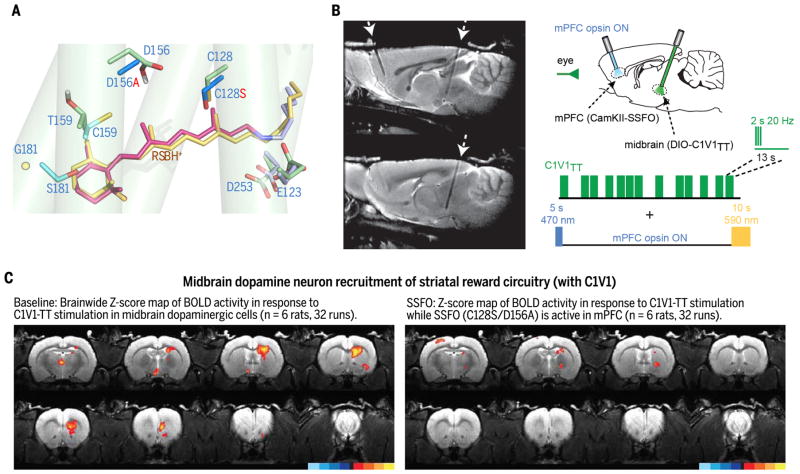
Fig. 5. Causal underpinnings of depression-related symptomatology identified via algal channel structure discovery and redesign.
(A) Design of major opsin classes used together to identify brainwide dynamics of anhedonia (C1V1TT/SSFO) (56): Model of red-shifted C1V1 RBP (red RSBH+; C1V1-specific side chains in cyan, based on 3UG9 structure) overlying modeled blue-responsive RBP (yellow RSBH+; green side chains, energy-optimized/calculated structure). Nearby DC-pair (SSFO) double mutant (D156A/C128S; new side chains in dark blue) shown (to illustrate relative positioning) that in the blue-responsive RBP confers stable activity shifts and blood oxygen level– dependent (BOLD) signal acquisition in MRI without heating or other artifacts. Expected further rearrangements and water influx are not shown. (B) Left: Tracks in rat brain for opsin injection and fiber-optic light access to mPFC (left arrow) and midbrain dopamine neurons (right arrow). Right: Testing causal influence of elevated mPFC activity (with blue-on/yellow-off SSFO) over communication within reward circuitry (from midbrain dopamine neurons controlled with C1V1). (C) Probing second-order brainwide dynamics. Natural prominent BOLD signal in dorsal and ventral striatum (left) recruited by dopamine neurons is potently suppressed (right) by mPFC excitability shift (56).
Numerous applications leverage these SFO capabilities (54–61). Indeed, SFO mutations, like the ChETA/CHIEF mutations described above, are portable to certain other ChR backbones for new functionality. For example, combined with His134 → Arg (13), a mutation that increases photocurrents [as do Thr159 → Cys (51), Thr59 → Ser (62), and Thr246 → Asn (62)], the SFO strategy creates two-photon illumination–recruited bistable ChRs (2PSFOs) (63). Furthermore, when combined with Cl−-conducting ChRs as discussed below, bistable inhibition results (Fig. 4G) (62, 64), with versatile utility [e.g., in studying pain circuitry (60)].
Spectral properties
Fast, efficient structural changes require tight protein-chromophore contact and thus RBP structural rigidity; such fast changes are important not only for kinetics, but also for spectral properties of ChR pore gating. Absorption peaks for known ChRs span 440 to 590 nm (1, 41, 50, 65), and 630- to 644-nm peaks are seen in certain other retinal-binding proteins (66, 67). Absorption spectra are determined by factors including RBP polarity, ATR planarity [in particular, coplanarity of the β-ionone ring C6=C7 bond with the polyene chain in the 6-s conformation (68)], and connection of negatively charged RSBH counterions with long-range hydrogen-bonding networks (68–70) (Figs. 1 and 4).
ChR spectra continue to drive basic and applied investigation. Certain bands of lower-energy (red) light can penetrate somewhat more deeply and safely into biological tissue than blue light. Combinatorial control is also enabled using two-color strategies (54); moreover, all-optical activity-guided/closed-loop experiments become more straightforward when redshifted control tools can be integrated with robust blue light–activated optical readouts (3, 50, 71). But the extent to which RBP-level understanding from other redshifted rhodopsins (69) could drive insights into ChR itself, or into development of color-shifted optogenetic tools, was initially unclear. Despite the recent emergence of additional RBP structural information from a color-shifted ChR (68), attempts over years in many laboratories to build and leverage structural models to engineer or evolve usefully redshifted (by ~100 nm) ChRs were unsuccessful, perhaps because of the predominance of multifactorial and long-range contributions to spectral properties evolved over hundreds of millions of years.
The first discovery (70) of a substantially redshifted ChR, VChR1, finally came in the alga Volvox carteri (via searching for opsins in U.S. Joint Genome Institute databases). Initial characterization in culture (70) revealed that a previously inaccessible window was now available, >560 nm, where Chlamydomonas ChRs (peaking at ~470 to 490 nm) exhibit almost zero photocurrent. Universally, ChRs (including VChR1 and all other ChRs identified or engineered since) retain robust responsivity in a short-wavelength/blue shoulder of the action spectrum (50) (~470 nm). Persistent blue actuation appears to be an intrinsic property of the retinal polyene system [relating to activation of higher electronic state transitions beyond the lowest-energy transition available (71)]. Although VChR1 photocurrents were small (<100 pA) (70), several modifications in combination (Fig. 1)—including provision of membrane-trafficking/endoplasmic reticulum export motifs identified earlier for enabling halorhodopsin optogenetics (72, 73), chimerization with ChR1 elements (74), and in some cases mutations reducing the blue shoulder and/or ChETA mutation (45)—resulted in diverse members of the new C1V1 ChR family (54). In 2011, C1V1 enabled the first red light–driven spiking (Fig. 4D) (54) as well as in vivo combinatorial optogenetics [two populations separably controlled with red and blue light (54)], which allowed the long-sought demonstration in mouse experiments a causal role for excitation-inhibition balance in governing gamma oscillations and social behavior (54). Another application emerged with (i) the discovery of high responsivity of red light–driven opsins to two-photon illumination, enabling single-cell resolution optogenetics in brain tissue (63), and (ii) integration of red light–excited control with blue light–excited readout (via genetically encoded activity sensors such as GCaMP Ca2+ reporters). Enabling these in vivo all-optical play-in/read-out experiments (50, 75–78) has opened the door to tuning optogenetic control in order to match timing and amplitude of naturally occurring activity in the same circuit elements (3, 76), and more broadly to keeping stimulation attuned to native dynamics and events in real time through closed-loop and activity-guided strategies (50, 74–78). Many opsins are now available for redshifted excitation, including not just VChR1 and C1V1 but also the VChR1-based ReaChR (79) as well as MChR1 (80), Chrimson (65), and bReaCHES (77) (with ChETA modifications for speed). As with earlier insights into kinetics, the initial VChR backbones and C1V1 modifications provided not only insight into ChR structure-function relationships (Fig. 4), but also new neuroscience functionality [for example, enabling discovery of the causal role of the medial prefrontal cortex (mPFC) in regulating midbrain–nucleus accumbens interactions and reward-related behavior (56) (Fig. 5)].
High-resolution crystal structure
Although the above innovations were enabled by ChR modeling without complete structural knowledge, major domains of discovery and design remained difficult to address without high-resolution information. After many years and the creation of numerous constructs for expression and crystallization, in 2012 the 2.3 Å crystal structure was obtained for a truncated functional chimera between Chlamydomonas ChR1 and ChR2 (C1C2) (15). This structure revealed many of the deepest mysteries of the channel, including its long-sought internal light-activated pore (Fig. 1); as anticipated, questions were also raised and new ideas emerged.
Although the structure revealed a covalently linked dimer of 7TM proteins (associated by N termini and helices 3 and 4) (15), no pore was present at the interface (15), contrary to prior prediction (36). Rather, monomers displayed internal pores formed from four of the seven transmembrane helices (TMs 1, 2, 3, and 7; Fig. 1), with two tilted to create space for a partially water-filled polar channel with two closed-state pore gates requiring repositioning for creation of ion-conducting states. Close to the intracellular membrane, the inner gate is formed by two highly conserved glutamates (E1 and E2; Fig. 1), which interact with His134 and His265, closing the channel to both water and ions in the dark. Interestingly, this site is also a key determinant of cation/proton selectivity (severely impaired by E1/E2 or His134 mutation). The central gate [structurally separated from the inner by a water-deprived vestibule, as also supported by QM/MM (quantum mechanics/molecular mechanics) calculations (81) (Fig. 1)] consists of Ser63, Asn258, and E3 of Fig. 1, locking the protein from the extracellular bulk phase with respect to water/proton/cation flux and serving as a second selectivity filter even though separated from the surface (15) by 8 Å.
Other key insights included revelation of the RBP and exact side chain positioning defining the chromophore electrostatic environment (15). RBP structure has numerous implications for understanding the channel, including gating regulation between conducting and nonconducting states, ion selectivity, and spectral properties. ChR photocurrents exhibit a transient peak (Ip) decaying to stationary (Is) under continuous illumination (Fig. 3); for conducting states (O1, O2), most channels reside in O1 during Ip, but under prolonged stimulation, low-conduction O2 accumulates and photocurrent decreases. These transitions likely involve protonations and partial charge-localization shifts among chromophore-proximal residues, overlapping with potential targets for shifting spectral properties.
In natural and artificial ChRs with bathochromic (red-shifted) absorption, the RBP is made more polar near the RSBH β-ionone ring primarily by Gly181 → Ser replacement (as in VChR1, ReaChR, and Chrimson), and/or Thr159 → Cys (or Met), Leu162 → Cys, and Phe178 → Tyr replacements (ChR2 numbering, Fig. 1), and/or by polarity reduction near the RSBH as in C1V1TT (Fig. 1 and Fig. 4, A and B). In the C1C2 crystal structure “frozen” snapshot, counterions Glu123 and Asp253 are directly connected to the RSBH+ proton without a water bridge, but molecular dynamics (MD) calculations reveal an active site with highly dynamic sampling of hydrogen-bonding patterns, with and without water bridges, separated only by tiny energetic differences (81). Red (lower-energy) photons suffice to induce deprotonation/isomerization in RBPs with high polarity near the β-ionone and low polarity at the RSBH [as positive charge shifts toward the ring (69, 70); Figs. 4 and 5]. Although this principle has now been demonstrated, color tuning is still challenging, because high polarity near the β-ionone may reduce retinal-binding affinity, whereas low polarity near the RSBH lowers Schiff-base pKa (favoring deprotonation), both of which could impair function. The ChR pore complex engages in long-range coupling interactions from the RBP to distant residues (Fig. 5); among other examples, substantial pH-dependent bathochromic color shifts were found to be associated with the presence of Glu48 (using ChR2 numbering) at the N-terminus of TM1 in redshifted ChR1, PsChR1, VChR1, DChR1, and relatives (Fig. 1). Precise mechanisms, although not known, could involve long-range coupling or tilting of the flexible TM1; causality is supported by observations that Glu48 → Gln inhibited pH dependence of red shift, whereas Glu48 → Ala (corresponding to blue-responsive ChR2; Fig. 1) instead stabilized the blue-absorbing form (82, 83). Future advances in long-range molecular modeling methodology will build on these observations alongside current and emerging high-resolution structural and MD work.
The precise dynamical sequence of pore side chain repositioning will continue to serve as a subject of intense investigation and modeling. RBP amino acid replacements are expected to change energy profiles of both the electronic ground and excited states, with consequences for dynamics (influencing, besides spectrum and photocycle kinetics, the efficiency of retinal isomerization as with Glu123 and Cys128). Several new structures, including additional red-shifted ChRs, are likely to emerge that will add finer-grained details to this intriguing landscape.
Selectivity variants
ChR ion selectivity is important in understanding the behavioral ecology of motile algae (10) and in achieving bidirectional impact on neuronal activity. Nonselective cation flux elicits excitation in neural systems via membrane depolarization, whereas anion or K+ selectivity typically provides inhibition by clamping membrane potentials below the spike-firing threshold and reducing membrane resistance. Chlamydomonas ChRs naturally conduct cations, with modest inward rectification and cation species preference: Li+>Na+> K+ > Cs+ ≫ Ca2+>Mg2+ (13, 40, 83). Most ChRs are highly selective for protons, with PH+/PNa+~ 2 × 106 to 6 × 106 for ChR2 (13, 51), comparable to that of highly selective mammalian proton channels (e.g., H+Hv1; PH+/PNa+ ≥ 106) (84). For proton selectivity in algal ChRs, this level of preference suffices (85) because Na+ becomes essentially irrelevant for freshwater algae in low-Na+ environments. However, in mammalian brains with ~130mMNa+ at pH7.5, Na+ becomes a dominant component, modulated by voltage-dependent competition among protons/Na+/K+ and by weak Mg2+ block of proton permeation (40).
The C1C2 structure (15) alone was necessary but not sufficient for achieving insight into selective permeation, which required not only a crystal structure but also subsequent detailed structure-guided mechanistic studies. The magnitude of the mystery appeared even greater upon structure determination because few water molecules and no internally bound ions or high-affinity ion-binding sites were seen (15); moreover, the pore was large and disordered (relative to highly structured pores of Shaker and other K+ channels) in the crystal (despite high-resolution refinement of the rigid RBP) (15), raising questions about the mechanism of cation conductance/selectivity. Because proton conductances can involve proton transfer reactions along residues forming a biological “proton wire,” candidate protonatable pore-flanking residues were tested by replacement, with attention to five glutamates of helix 2 (E1 to E5, Fig. 1). Replacement of E3 [hydrogen-bonded to Ser63 and Asn258 forming the central gate (86, 87); ChR2 residue numbering] had the largest impact. Replacement of inner-gate His134/E2 or access-channel E4/E5 also reduced H conductance (62, 64, 88, 89). Further support for this proton-wire hypothesis is derived from cryptophyte anion-conducting ChRs (90), which show reduced H+ conductance with fewer helix-2 glutamates (90); interestingly, these glutamates have little influence on kinetics, and only mutation of E1 decelerates closing (88, 91).
ChR Ca2+ flux is presumed to be important for flagellar regulation, whereas Na+/K+ permeance could be incidental to more functionally relevant Ca2+/H+ flux in cation-conducting ChRs. Indeed, some ChRs (e.g., Dunaliella salina) are virtually H+-selective (1). Incidental Na+ flux might bring little energetic cost in freshwater algae (for pumping to restore small Na+ gradients) and may arise as channels evolve within nonchannel gene families such as the pump-type microbial rhodopsins (perhaps after gene transfer in niches wherein archaea and algae are intimately linked, as with Halobacteria salinarum and Dunaliella salina in high-salt environments). The versatile ChR pore without high-affinity ion-binding sites (15) contrasts with naturally occurring K+- or Ca2+-channel selectivity mechanisms, which use tetrameric and symmetric structures to achieve coordinated ion-binding sites that mimic hydration shells or direct symmetric chelation of ions, respectively.
How, then, do ChRs achieve cation selectivity? The C1C2 crystal structure (15) revealed a preponderance of intracellular vestibule basic residues (64, 92), but attempts to modify conduction pathway access here and elsewhere (although successful in quantitatively shifting ion permeation ratios) did not alter cation selectivity (13). However, the predicted electronegative environment of the entire conduction pathway in cation selective C1C2 (64, 92) suggested a pore-lining surface electrostatic model and new possibilities for testing mechanisms of selective permeation. In 2014, a direct test (remodeling the C1C2 pore for surface electropositivity) confirmed this new model (62), resulting in a Cl−-selective ChR (iC1C2) that was inhibitory in neural systems (62). At the same time, different mutations (including negative-to-positive replacement of the central gate Glu90 with Lys+ or Arg+) were used to create a Cl−-selective ChR2 (93), with the unifying theme of similar-direction effects on predicted internal pore electrostatics (93). Further mutagenesis of both variants [guided by the new model, in which complete elimination of proton conductance also required mutation of one or two putative proton wire pore glutamates (64, 94)] resulted in highly Cl−-selective ChRs [iChloC and iC++; Fig. 4G (64, 94)] now widely used for studies of animal behavior (conferring optogenetic inhibition in typical situations wherein internal Cl− concentration is low) (64, 95, 96).
Naturally occurring Cl−-selective ChRs (GtACR1 and GtACR2) were described from cryptophyte algae the year after the first designed Cl−-selective ChR was reported; these were termed anion channelrhodopsins (ACRs) (90). Even though ACRs are evolutionarily remote from cation-conducting ChRs with more amino acid replacements than the designed Cl−-conducting ChRs, these new ChRs independently fit the earlier pore model as well, with the unifying principle across all Cl−-selective ChRs corresponding to a net effect on pore-lining electrostatics (92). The universality of the selectivity mechanism is underscored by the fact that no single charge seems crucial for Cl− selectivity, as long as net pore electrostatics are preserved (92). ChR Glu90 is invariant in all naturally occurring ChRs (Glu64 in GtACR2 numbering), but this negative charge is compensated in anion-conducting GtACR2 via numerous charge-altering replacements of other pore-facing glutamate residues found in cation-conducting ChRs (changed in GtACR2 to non-charged residues: Ser57, Thr67, Ala71, Asn75, and Ser93) (92). Moreover, Glu90 is not essential for Cl− flux; both families of engineered Cl−-selective ChRs (iC1C2, ChloC) lack glutamate at this position (62, 93). Finally, even altering this glutamate in naturally occurring GtACR1 (Glu68 → Gln) has no detectable effect on photocurrent magnitude or kinetics under physiological conditions (91). Although the ACR primary sequence suggests rearrangement at both gates, an E3 at the central gate is retained (Fig. 1). The comparative value of natural variants is substantial, but structural and spectroscopic data remain to be acquired.
The intense interest that led to resolving this fundamental question of selectivity had the additional benefit of driving the creation of new optogenetic tools, enabling the first round of studies using Cl−-selective ChRs for inhibitory optogenetics in animal behavior (64, 95, 96). Moreover, integration of diverse structural insights has begun; the SFO kinetic principle was discovered to be portable to inhibitory ChRs, creating bistable and extremely light-sensitive inhibitory ChRs (SwiChR and SwiChR++) (62, 64) that have found utility in studies of mouse behavior (60).
Outlook
The ChR light-gated pore is unique among biological structures. The investigation, modeling, and redesign of atomic-scale structure-function relationships governing this light-to-current converter led not only to advances in neuroscience but also to sophisticated understanding of the underlying chemical processes. These light-gated channels will continue to fascinate (and occupy a privileged position within ion-channel research) because only light-gated systems allow structure-function analyses over time scales of femtoseconds (10−15 s) to seconds or more (97).
The role of ChR in neuroscience may continue to be special as well, building on precision redesign for new roles (Fig. 4). Diverse modes of photon-spike logic are now accessible (four examples in Fig. 4, each alongside key pore-modeling and pore-redesign features required to create the new logic). Even psychiatry has yielded some of its deepest mysteries to ChR pore structural insights (Fig. 5), as exemplified by studies of circuit dynamics underlying the core depression symptom of anhedonia (59) in which natural reward responses central to the behavior of all animals are lost. The presence of anhedonia allows a diagnosis of major depressive disorder even without depressed mood, but causal circuit dynamics–level understanding had remained elusive.
The experiments summarized in Fig. 5 used a combination of designed opsins arising from structure-function ChR discoveries (56). The blue light–activated SSFO (DC pair–modified for bistable behavior on a 30-min time scale) allowed causal analysis of brainwide dynamics in awake rats during fMRI scanning (56). SSFO modulation in mPFC was combined with modulation of neural elements across the brain using C1V1TT [containing no ChR2 sequence but with chimeric elements of VChR1/ChR1, RBP point mutations for redshifted compatibility (54) with SSFO, and the ChETA (45) mutation] (56). Figure 5A shows elements of the pore redesign that enabled this approach; Fig. 5B shows the strategy for brain-spanning interrogation of reward circuitry. Elevated mPFC activity (to mimic a pattern seen in depression with anhedonia) altered the ability of midbrain dopamine neurons to recruit striatal reward circuitry (Fig. 5C)—a higher-order brainwide form of modulation by which the evolutionarily advanced frontal neocortex is capable of modifying interactions between two distant subcortical structures—thereby controlling reward-mediating physiology and behavior (56).
In addition to these and other insights into adaptive and maladaptive behavior, basic insights into ion channel biophysics and evolution have emerged. Since the resolution of the crystal structure of ChR and the demonstration of the pore surface electrostatic mechanism for Cl− selectivity, two recent cryo-EM structures subsequently emerged for unrelated Cl− channels that (like ChR) have arisen within larger nonchannel protein families (98, 99). The cystic fibrosis transmembrane conductance regulator (CFTR) is a Cl− channel within the nonchannel ABC transporter family. Cryo-EM (98) revealed a putative Cl− conduction pathway with striking parallels to the unrelated 7TM Cl−-conducting ChRs with its wide pore, lack of high-affinity identified Cl− binding sites (as detectable in lower-resolution cryo-EM), and largely continuous positive surface electrostatics along the conduction pathway. Similarly, CLC-K is a Cl− channel within a broad family of 12TM CLC proteins that includes nonchannel Cl−/H+ exchangers. CLC-K, like CFTR, is distinct from ChRs in topology (12-TM protein dimer). Cryo-EM (99) for CLC-K revealed a pattern of positively charged arginines in the conduction pathway (with no high-affinity binding sites detectable in cryo-EM). Together with the earlier ChR discoveries, a model emerges by which Cl−-conducting channels can evolve using low-affinity surface electrostatic pathways, within nonchannel families that may not otherwise be well suited (by symmetry or other properties) to give rise to highly selective channels.
Much more remains to be uncovered regarding the operation of these unparalleled biological machines, including mechanisms of long-range molecular tuning of color, kinetics, and selectivity. Understanding will continue to be refined with new structural information and ongoing advances in modeling of molecular dynamics, which will also likely continue to drive neuroscience. For light-activated ChR pores, studying the interplay of form and function has illuminated not only the governing principles of the channel, but also the basic behavioral ecology of algae, the evolution of ion channels, and the fundamental circuit underpinnings of animal behavior in health and disease—remarkable achievements for a light-activated molecule from a single-celled plant.
Acknowledgments
We thank our co-workers for contributions to the analysis of ChR structure and function over the years, and for the interdisciplinary exchange of knowledge among the communities of algal biology, membrane biophysics, structural biology, neuroscience, and medicine. We also thank E. A. Lysenko (Institute of Plant Physiology, RAS Moscow) for the photograph of Faminzin; F. Beyle for the ChR2 structure homology file; A. Berndt, E. Ferenczi, H. Kato, Y. Kim, F. Schneider, J. Wietek, and O. Yizhar for comments; and S. Kelterborn for Fig. 2B. Supported by the Deutsche Forschungsgemeinschaft, European Research Council, and Hertie Foundation (P.H.) and by NIH, NSF, DARPA, the NOMIS Foundation, and the Else Kroner Fresenius Foundation (K.D.).
REFERENCES AND NOTES
- 1.Zhang F, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ernst OP, et al. Microbial and animal rhodopsins: Structures, functions, and molecular mechanisms. Chem Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim CK, Adhikari A, Deisseroth K. Integration of optogenetics with complementary methodologies in systems neuroscience. Nat Rev Neurosci. 2017;18:222–235. doi: 10.1038/nrn.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster KW, Smyth RD. Light antennas in phototactic algae. Microbiol Rev. 1980;44:572–630. doi: 10.1128/mr.44.4.572-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faminzin A. Die Wirkung des Lichtes auf die Bewegung der Chlamidomonas pulvisculus Ehr., Euglena viridis Ehr. und Oscillatoria insignis Tw. Melanges Biologiques tires du Bulletin de l’Ácademie Imperial des Sciences De St-Petersbourg. 1866:73–93. [Google Scholar]
- 6.Litvin FF, Sineshchekov OA, Sineshchekov VA. Photoreceptor electric potential in the phototaxis of the alga Haematococcus pluvialis. Nature. 1978;271:476–478. doi: 10.1038/271476a0. [DOI] [PubMed] [Google Scholar]
- 7.Harz H, Hegemann P. Rhodopsin-regulated calcium currents in Chlamydomonas. Nature. 1991;351:489–491. doi: 10.1038/351489a0. [DOI] [Google Scholar]
- 8.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 9.Herrmann TR, Rayfield GW. A measurement of the proton pump current generated by bacteriorhodopsin in black lipid membranes. Biochim Biophys Acta. 1976;443:623–628. doi: 10.1016/0005-2736(76)90482-X. [DOI] [PubMed] [Google Scholar]
- 10.Foster KW, et al. A rhodopsin is the functional photoreceptor for phototaxis in the unicellular eukaryote Chlamydomonas. Nature. 1984;311:756–759. doi: 10.1038/311756a0. [DOI] [PubMed] [Google Scholar]
- 11.Braun FJ, Hegemann P. Direct measurement of cytosolic calcium and pH in living Chlamydomonas reinhardtii cells. Eur J Cell Biol. 1999;78:199–208. doi: 10.1016/S0171-9335(99)80099-5. [DOI] [PubMed] [Google Scholar]
- 12.Nagel G, et al. Channelrhodopsin-1: A light-gated proton channel in green algae. Science. 2002;296:2395–2398. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 13.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–1225. doi: 10.1038/nn.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato HE, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iseki M, et al. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 17.Engel BD, et al. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife. 2015;4:e04889. doi: 10.7554/eLife.04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berthold P, et al. Channelrhodopsin-1 initiates phototaxis and photophobic responses in Chlamydomonas by immediate light-induced depolarization. Plant Cell. 2008;20:1665–1677. doi: 10.1105/tpc.108.057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mittelmeier TM, Boyd JS, Lamb MR, Dieckmann CL. Asymmetric properties of the Chlamydomonas reinhardtii cytoskeleton direct rhodopsin photoreceptor localization. J Cell Biol. 2011;193:741–753. doi: 10.1083/jcb.201009131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan KY, Leptos KC, Goldstein RE. Lag, lock, sync, slip: The many ‘phases’ of coupled flagella. J R Soc Interface. 2014;11:20131160. doi: 10.1098/rsif.2013.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crick F. The impact of molecular biology on neuroscience. Philos Trans R Soc B. 1999;354:2021–2025. doi: 10.1098/rstb.1999.0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farber IC, Grinvald A. Identification of presynaptic neurons by laser photostimulation. Science. 1983;222:1025–1027. doi: 10.1126/science.6648515. [DOI] [PubMed] [Google Scholar]
- 23.Fork RL. Laser stimulation of nerve cells in Aplysia. Science. 1971;171:907–908. doi: 10.1126/science.171.3974.907. [DOI] [PubMed] [Google Scholar]
- 24.Zemelman BV, Lee GA, Ng M, Miesenböck G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/S0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 25.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lima SQ, Miesenböck G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 28.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 31.Bi A, et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aravanis AM, et al. An optical neural interface: In vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–S156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 33.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller M, Bamann C, Bamberg E, Kühlbrandt W. Projection structure of channelrhodopsin-2 at 6 Å resolution by electron crystallography. J Mol Biol. 2011;414:86–95. doi: 10.1016/j.jmb.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 37.Bruun S, et al. Light-dark adaptation of channelrhodopsin involves photoconversion between the all-trans and 13-cis retinal isomers. Biochemistry. 2015;54:5389–5400. doi: 10.1021/acs.biochem.5b00597. [DOI] [PubMed] [Google Scholar]
- 38.Vogel R, et al. Deactivation of rhodopsin in the transition from the signaling state meta II to meta III involves a thermal isomerization of the retinal chromophore C=D. Biochemistry. 2003;42:9863–9874. doi: 10.1021/bi034684+. [DOI] [PubMed] [Google Scholar]
- 39.Harbison GS, et al. Dark-adapted bacteriorhodopsin contains 13-cis, 15-syn and all-trans, 15-anti retinal Schiff bases. Proc Natl Acad Sci USA. 1984;81:1706–1709. doi: 10.1073/pnas.81.6.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider F, Gradmann D, Hegemann P. Ion selectivity and competition in channelrhodopsins. Biophys J. 2013;105:91–100. doi: 10.1016/j.bpj.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider F, Grimm C, Hegemann P. Biophysics of channelrhodopsin. Annu Rev Biophys. 2015;44:167–186. doi: 10.1146/annurev-biophys-060414-034014. [DOI] [PubMed] [Google Scholar]
- 42.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2011;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuhne J, et al. Early formation of the ion-conducting pore in channelrhodopsin-2. Angew Chem Int Ed. 2015;54:4953–4957. doi: 10.1002/anie.201410180. [DOI] [PubMed] [Google Scholar]
- 44.Lórenz-Fonfría VA, et al. Temporal evolution of helix hydration in a light-gated ion channel correlates with ion conductance. Proc Natl Acad Sci USA. 2015;112:E5796–E5804. doi: 10.1073/pnas.1511462112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gunaydin LA, et al. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 46.Assaf F, Schiller Y. The antiepileptic and ictogenic effects of optogenetic neurostimulation of PV-expressing interneurons. J Neurophysiol. 2016;116:1694–1704. doi: 10.1152/jn.00744.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jego S, et al. Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat Neurosci. 2013;16:1637–1643. doi: 10.1038/nn.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klavir O, Prigge M, Sarel A, Paz R, Yizhar O. Manipulating fear associations via optogenetic modulation of amygdala inputs to prefrontal cortex. Nat Neurosci. 2017;20:836–844. doi: 10.1038/nn.4523. [DOI] [PubMed] [Google Scholar]
- 49.Carus-Cadavieco M, et al. Gamma oscillations organize top-down signalling to hypothalamus and enable food seeking. Nature. 2017;542:232–236. doi: 10.1038/nature21066. [DOI] [PubMed] [Google Scholar]
- 50.Grosenick L, Marshel JH, Deisseroth K. Closed-loop and activity-guided optogenetic control. Neuron. 2015;86:106–139. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berndt A, et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci USA. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 53.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 54.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmitt LI, et al. Thalamic amplification of cortical connectivity sustains attentional control. Nature. 2017;545:219–223. doi: 10.1038/nature22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferenczi EA, et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351:aac9698. doi: 10.1126/science.aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan CL, et al. Warm-sensitive neurons that control body temperature. Cell. 2016;167:47–59.e15. doi: 10.1016/j.cell.2016.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorokin JM, et al. Bidirectional control of generalized epilepsy networks via rapid real-time switching of firing mode. Neuron. 2017;93:194–210. doi: 10.1016/j.neuron.2016.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makino H, Komiyama T. Learning enhances the relative impact of top-down processing in the visual cortex. Nat Neurosci. 2015;18:1116–1122. doi: 10.1038/nn.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iyer SM, et al. Optogenetic and chemogenetic strategies for sustained inhibition of pain. Sci Rep. 2016;6:30570. doi: 10.1038/srep30570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haikala V, Joesch M, Borst A, Mauss AS. Optogenetic control of fly optomotor responses. J Neurosci. 2013;33:13927–13934. doi: 10.1523/JNEUROSCI.0340-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344:420–424. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prakash R, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berndt A, et al. Structural foundations of optogenetics: Determinants of channelrhodopsin ion selectivity. Proc Natl Acad Sci USA. 2016;113:822–829. doi: 10.1073/pnas.1523341113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marshall J, Cronin TW, Kleinlogel S. Stomatopod eye structure and function: A review. Arthropod Struct Dev. 2007;36:420–448. doi: 10.1016/j.asd.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 67.Wang W, et al. Tuning the electronic absorption of protein-embedded all-trans-retinal. Science. 2012;338:1340–1343. doi: 10.1126/science.1226135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kato HE, et al. Atomistic design of microbial opsin-based blue-shifted optogenetics tools. Nat Commun. 2015;6:7177. doi: 10.1038/ncomms8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kloppmann E, Becker T, Ullmann GM. Electrostatic potential at the retinal of three archaeal rhodopsins: Implications for their different absorption spectra. Proteins. 2005;61:953–965. doi: 10.1002/prot.20744. [DOI] [PubMed] [Google Scholar]
- 70.Zhang F, et al. Red-shifted optogenetic excitation: A tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buss V. Inherent chirality of the retinal chromophore in rhodopsin—A nonempirical theoretical analysis of chiroptical data. Chirality. 2001;13:13–23. doi: 10.1002/1520-636X(2001)13:1<13::AID-CHIR4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 72.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, et al. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from chlamydomonas. J Biol Chem. 2009;284:5685–5696. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- 74.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rickgauer JP, Deisseroth K, Tank DW. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat Neurosci. 2014;17:1816–1824. doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim CK, et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat Methods. 2016;13:325–328. doi: 10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajasethupathy P, et al. Projections from neocortex mediate top-down control of memory retrieval. Nature. 2015;526:653–659. doi: 10.1038/nature15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carrillo-Reid L, Yang W, Bando Y, Peterka DS, Yuste R. Imprinting and recalling cortical ensembles. Science. 2016;353:691–694. doi: 10.1126/science.aaf7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: A red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Govorunova EG, Spudich EN, Lane CE, Sineshchekov OA, Spudich JL. New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. MBio. 2011;2:e00115–e11. doi: 10.1128/mBio.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo Y, et al. Active site structure and absorption spectrum of channelrhodopsin-2 wild-type and C128T mutant. Chem Sci. 2016;7:3879–3891. doi: 10.1039/C6SC00468G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsunoda SP, Hegemann P. Glu 87 of channelrhodopsin-1 causes pH-dependent color tuning and fast photocurrent inactivation. Photochem Photobiol. 2009;85:564–569. doi: 10.1111/j.1751-1097.2008.00519.x. [DOI] [PubMed] [Google Scholar]
- 83.Govorunova EG, Sineshchekov OA, Li H, Janz R, Spudich JL. Characterization of a highly efficient blue-shifted channelrhodopsin from the marine alga Platymonas subcordiformis. J Biol Chem. 2013;288:29911–29922. doi: 10.1074/jbc.M113.505495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Demaurex N, et al. Proton currents in human granulocytes: Regulation by membrane potential and intracellular pH. J Physiol. 1993;466:329–344. [PMC free article] [PubMed] [Google Scholar]
- 85.Ehlenbeck S, Gradmann D, Braun F-J, Hegemann P. Evidence for a light-induced H+ conductance in the eye of the green alga Chlamydomonas reinhardtii. Biophys J. 2002;82:740–751. doi: 10.1016/S0006-3495(02)75436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eisenhauer K, et al. In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J Biol Chem. 2012;287:6904–6911. doi: 10.1074/jbc.M111.327700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruffert K, et al. Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem Biophys Res Commun. 2011;410:737–743. doi: 10.1016/j.bbrc.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 88.Plazzo AP, et al. Bioinformatic and mutational analysis of channelrhodopsin-2 protein cation-conducting pathway. J Biol Chem. 2012;287:4818–4825. doi: 10.1074/jbc.M111.326207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wietek J, Broser M, Krause BS, Hegemann P. Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. J Biol Chem. 2016;291:4121–4127. doi: 10.1074/jbc.M115.699637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Govorunova EG, Cunha SR, Sineshchekov OA, Spudich JL. Anion channelrhodopsins for inhibitory cardiac optogenetics. Sci Rep. 2016;6:33530. doi: 10.1038/srep33530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berndt A, Deisseroth K. Expanding the optogenetics toolkit. Science. 2015;349:590–591. doi: 10.1126/science.aac7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wietek J, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 94.Wietek J, et al. An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo. Sci Rep. 2015;5:14807. doi: 10.1038/srep14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim H, Ährlund-Richter S, Wang X, Deisseroth K, Carlén M. Prefrontal parvalbumin neurons in control of attention. Cell. 2016;164:208–218. doi: 10.1016/j.cell.2015.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Takahashi N, Oertner TG, Hegemann P, Larkum ME. Active cortical dendrites modulate perception. Science. 2016;354:1587–1590. doi: 10.1126/science.aah6066. [DOI] [PubMed] [Google Scholar]
- 97.Nango E, et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science. 2016;354:1552–1557. doi: 10.1126/science.aah3497. [DOI] [PubMed] [Google Scholar]
- 98.Liu F, Zhang Z, Csanády L, Gadsby DC, Chen J. Molecular structure of the human CFTR ion channel. Cell. 2017;169:85–95.e8. doi: 10.1016/j.cell.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 99.Park E, Campbell EB, MacKinnon R. Structure of a CLC chloride ion channel by cryo-electron microscopy. Nature. 2017;541:500–505. doi: 10.1038/nature20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Watanabe HC, Welke K, Sindhikara DJ, Hegemann P, Elstner M. Towards an understanding of channelrhodopsin function: Simulations lead to novel insights of the channel mechanism. J Mol Biol. 2013;425:1795–1814. doi: 10.1016/j.jmb.2013.01.033. [DOI] [PubMed] [Google Scholar]