Abstract
We present a 3-D mapping in WI38 human diploid fibroblast cells of chromosome territories (CT) 13,14,15,21, and 22, which contain the nucleolar organizing regions (NOR) and participate in the formation of nucleoli. The nuclear radial positioning of NOR-CT correlated with the size of chromosomes with smaller CT more interior. A high frequency of pairwise associations between NOR-CT ranging from 52% (CT13-21) to 82% (CT15-21) was detected as well as a triplet arrangement of CT15-21-22 (72%). The associations of homologous CT were significantly lower (24–36%). The arrangements of each pairwise CT varied from CT13-14 and CT13-22, which had a majority of cells with single associations, to CT13-15 and CT13-21 where a majority of cells had multiple interactions. In cells with multiple nucleoli, one of the nucleoli (termed “dominant”) always associated with a higher number of CT. Moreover, certain CT pairs more frequently contributed to the same nucleolus than to others. This nonrandom pattern suggests that a large number of the NOR-chromsomes are poised in close proximity during the postmitotic nucleolar recovery and through their NORs may contribute to the formation of the same nucleolus. A global data mining program termed the chromatic median determined the most probable interchromosomal arrangement of the entire NOR-CT population. This interactive network model was significantly above randomized simulation and was composed of 13 connections among the NOR-CT. We conclude that the NOR-CT form a global interactive network in the cell nucleus that may be a fundamental feature for the regulation of nucleolar and other genomic functions.
Keywords: NOR, Nucleolar Organizing Region, Chromosome Territories, Interactive network, Nucleolus
Introduction
The human genome is composed of 23 pairs of chromosomes. During interphase, chromosomes are organized in the nucleus as discrete chromosome territories (CT) with well distinguishable boundaries (Bickmore, 2013; Cremer and Cremer, 2010; Cremer et al., 2006; Cremer et al., 2000; Meaburn and Misteli, 2007b; Visser and Aten, 1999). The three dimensional position of chromosomes in the cell nucleus is considered to be important for determining the overall nuclear architecture of eukaryotes, the maintenance of genome stability and the epigenetic regulation of gene expression (Berezney, 2002; Berezney et al., 2005; Bickmore, 2013; Cremer and Cremer, 2010; Kumaran et al., 2008; Malyavantham et al., 2010; Meaburn and Misteli, 2007a; Misteli, 2007; Stein et al., 2003; Stein G, 2008; T, 2005; Zaidi et al., 2007). A generally accepted notion postulates that the three dimensional positions of chromosomes in the cell nucleus are probabilistically non-random. For instance, non-randomness in CT positioning is correlated with gene density, where gene-rich chromosomes are found more interior than gene-poor chromosomes within the nucleus (Boyle et al., 2001; Heride et al., 2010; Kreth et al., 2004). The radial positions of chromosomes also correlate with their sequence length, where longer chromosomes tend to associate with the nuclear periphery, while shorter chromosomes are usually localized more internally relative to the cell nucleus (Bolzer et al., 2005; Marella et al., 2009a; Sun et al., 2000; Zeitz et al., 2009).
In addition to nonrandom radial positioning of CT within the nucleus, specific chromosome neighborhood arrangements have been determined (Bolzer et al., 2005; Nagele et al., 1999) which are different between cell and tissue types (Marella et al., 2009a; Mayer et al., 2005; Neusser et al., 2007; Parada et al., 2004a; Zeitz et al., 2009), and are altered during cell differentiation, development (Kuroda et al., 2004; Marella et al., 2009b), and in cancer cells (Brianna Caddle et al., 2007; Fritz et al., 2014; Parada et al., 2002). Interestingly, increased translocation frequencies have been determined between specific CT which are in closer proximity (Brianna Caddle et al., 2007; Folle, 2008; Parada et al., 2002; Roix et al., 2003; Soutoglou et al., 2007) and those that are actually overlapping or intermingling (Branco and Pombo, 2006). At the same time, the intra-nuclear locations of chromosomes are probabilistic rather than absolute, and, therefore, an understanding of CT distribution patterns is difficult to attain.
In this investigation we determined the interchromosomal and nucleolar associations among the acrocentric CT 13, 14, 15, 21 and 22. During nucleolar biogenesis, these chromosomes congregate in late telophase/early G1 to form large sub-nuclear compartments (Prieto and McStay, 2005; Raska et al., 2006). This clustering of acrocentric chromosomes is thought to occur because they bear the nucleolar organizing regions (NOR), which contain tandem arrays of ribosomal RNA (rRNA) genes. The NOR-chromosomes together harbor approximately 400 ribosomal genes per human diploid genome (Zentner et al., 2011). Synthesis and processing of ribosomal RNA and formation of ribosomes is an exceptionally intense biosynthetic activity which is morphologically expressed in the formation of the nucleolus, the largest structure-function compartment of the cell nucleus (Busch and Smetana, 1970; Koberna et al., 2002; Raska et al., 2006).
Not all the NOR-CT are active at any given time and they can be grouped into transcriptional competent and non-competent NOR (Grob et al., 2014; Kalmarova et al., 2008a; Kalmarova et al., 2007; Smirnov et al., 2006). Moreover, particular NOR-CT have greater numbers of transcriptionally competent NOR and concomitantly a greater number of those CT are adjacent to nucleoli (Kalmarova et al., 2007). Kamarova et. al. (2007, 2008) further found nonrandom patterns of NOR-CT associated with nucleoli in HeLa cells and in daughter cells following mitosis (Kalmarova et al., 2008b; Kalmarova et al., 2007).
Despite this progress, there is still only limited information on how the diverse pairs of NOR-CT cooperate in the formation of nucleoli, as well as the relationships between the distribution of NOR-CT and the number, size and intranuclear positions of the nucleolar domains. Moreover, it is unclear as to whether there are specific positional patterns of arrangement at both the pairwise level of interactions and globally among the entire set of NOR-CT. To a large extent, comprehensive studies on the intranuclear distribution patterns of the NOR-CT and nucleolar associations are scarce because it is technically challenging to perform simultaneous localization of all five pairs of these CT.
In this investigation of the NOR-CT, we took a different approach. The spatial organization and associations of chromosomes 13, 14, 15, 21 and 22 were performed together in the same cell nuclei using repetitive cycles of fluorescence in situ hybridization (re-FISH). We report highly non-random pair-wise association patterns of the NOR-CT and their proximity to nucleoli. Application of a novel computational geometric and data mining approach enables us to demonstrate for the first time a highly preferred, albeit probabilistic, interconnected network of the NOR-CT in the cell nucleus.
Materials and Methods
Cell Culture
Labeling of chromosomes was performed in the WI38 diploid human lung fibroblast cell line (ATTC, Manassas, VA, USA). Cells were grown in Advanced DMEM (Life Technologies, Carlsbad, CA, USA) supplemented with 2.5% fetal bovine serum (FBS) and 1% antibiotic-antimycotic solution (Sigma-Aldrich, St. Louis, MO, USA) in 5% CO2 at 37°C. Prior to chromosome labeling, cells were placed on gridded CeLLocate coverslips (Eppendorf, Hamburg, Germany) and synchronized in G0 by serum starvation for 48 h in medium containing 0.1% FBS. This protocol yielded a nearly 100% synchronization in G0 as judged by the absence of Ki67 and BrdU staining or mitotic cells in the treated culture. The viability of treated cells was monitored by a live-dead cytotoxicity assay based on combined staining with calcein AM and ethidium homodimer (LifeTechnologies, Carlsbad, CA, USA). In this test, the cells with low activity of intracellular esterases and/or compromised integrity of membranes are recognized by a negative calcein AM signal (in the green spectral channel) and a positive ethidium staining (in the red spectral channel). Such cells were negligibly rare in the synchronized cell cultures.
Antibodies and FISH probes
Immunolabeling was performed with mouse anti- digoxigenin, anti-BrdU, anti Ki67 and rabbit anti-nucleolin antibodies (Abcam, Cambridge, England). Chromosome staining was performed by chromosome-specific aquarius paint probes (Cytocell, Windsor, CT), The hybridized segment of rDNA was prepared from a pA plasmid construct (Erickson et al., 1981) which codes for downstream 200 nucleotides of 18 S rRNA, internal transcribed spacer 1, sequence for 5.8 S rRNA, internal transcribed spacer 2 and upstream ~4,500 nucleotides of 28 S rRNA. The probe was labeled with digoxygenin-11-dUTP by nick translation.
Three Dimensional FISH
An approach for 3-D mapping of CT in the cell nucleus termed “re-FISH” (Walter et al., 2006) was previously adapted by our group (Marella et al., 2009a; Zeitz et al., 2009). This method involves a sequential series of FISH labeling and imaging as follows: (i) two selected pairs of CT are labelled by commercial fluorescence probes, and mapped in a group of cells by high resolution 3D imaging. (ii) The fluorescence probes are striped and two new probes recognizing another two pairs of CT are applied. (iii) Cells studied in previous imaging session are identified and the positions of the new CT are mapped in these cells. Steps (ii) and (iii) can be repeated to map locations of additional CT in the cell nucleus. (iv) Stacks of optical sequences acquired in the same cells at subsequent imaging sessions are aligned in x-, y- and z- planes, CT are segmented and their mutual positions are analyzed. We have validated this technique for confidently mapping up to eight or nine pairs of CT in the cell nucleus (Fritz et al., 2014; Marella et al., 2009a; Zeitz et al., 2009). In order to combine FISH and immunofluorescence detection, we introduced minor alterations to the above approach. The WI38 cells synchronized in G0 were first fixed with 4% paraformaldehyde in PBS for 12 min and permeabilized in 0.5 % Triton in PBS for 5 min. The nucleoli were labeled with anti-nucleolin rabbit antibody (Abcam, Cambridge, UK) followed by Cy-5 anti-rabbit conjugate (Jackson Immuno Research, West Grove, PA, USA). After immunolabeling, the cells were post-fixed overnight with methanol:acetic acid (3:1) at −20°C to crosslink antibodies to the cellular structure. In experiments involving rDNA hybridization, cells were then incubated with 100 μg/ml RNAse A (Roche, Basel, Switzerland) for 2 h at 37°C, to eliminate rRNA and avoid cross hybridization with the rDNA probe. Then cells were incubated in 20% glycerol/PBS (30min), freeze-thawed in liquid nitrogen three times, treated with 0.1N HCl for 5 min and subjected to DNA denaturation (70% deionized formamide/2X SSC, pH 7.0) at 75°C for 3 min. Aquarius paint probes for two chromosome pairs (Cytocell, Windsor, CT) or rDNA probes were then denatured for 8 min at 75°C. Labeling of rDNA was usually performed in the same series of experiments as immunolabeling of nucleolin. Probe solution was applied to coverslips and sealed with rubber cement. Hybridization was carried out at 37°C for 48h. Three consecutive post-hybridization washes each for 30 min at 37°C were: 50% formamide/2X SSC/0.05% Tween-20; 2X SSC/0.05% Tween-20 and 1X SSC. Coverslips were then mounted on slides in Vectashield. Following image collection, chromosome paints were stripped by immersion of the coverslips in 50% formamide/2X SSC for 1 min at 65°C. Another pair of denatured chromosome paint probes were then immediately added to cells and hybridized at 37°C for 48h. This process was repeated one more time to obtain images of all five NOR chromosome pairs. Before mounting of the coverslips on microscopy slides, the genomic DNA was counterstained with 10 μM DAPI (Sigma-Aldrich, St. Louis, MO, USA).
Microscopy and Image analysis
The images were collected on an Olympus BX51 fluorescence microscope equipped with a Sensicam QE (Cooke Corporation, Romulus, MI) digital CCD camera, a motorized z-axis controller (Prior, Rockland, MA) and Slidebook 4.0 software (Intelligent Imaging Innovations, Denver, CO). Stacks of optical sections at 0.5 μm intervals were collected to map nucleoli, CT and rDNA signals. 3-D volume rendering of the collected optical sections for visualization was performed using Slidebook 4.0. The exact position of each cell in coordinates of the CeLLocate grid was documented by 3D phase contrast imaging. These phase-contrast images were used to localize the same cells in subsequent imaging cycles. Finally, z-stacks of the optical sections were aligned in 3D using registration software developed in our laboratory by selecting reference points from corresponding optical sections of phase contrast images as previously described (Zeitz et al., 2009). Following the alignment, optical stacks were merged and subjected to segmentation and quantitative analysis by an in-house developed computer program running on Matlab termed eFISHent (Fritz et al., 2014). This program determines a wide range of spatial distances in 3-D between the segmented objects (Fritz et al., 2014; Zeitz et al., 2009). One feature of this suite of 3-D distance measurements enabled determination of the nearest neighbor 3-D distances (edge to edge distances) among the labeled CT in each image set as well as among the nucleoli and CT. A positive pair-wise association between 2 CT was then scored if the nearest distance was ≤ 8 pixels which corresponds to ≤ 0.56 μm. Similarly, a positive association between a CT and a nucleolus was scored when the nearest edge to edge 3-D distance was ≤ 1.0 μm. For CT-nucleolar interactions, the CT were assigned an association with the nearest nucleolus. In the few rare cases where an NOR-CT was spaced an equal distance between two nucleoli, the neighborhood association was assigned to the nucleolus having the largest surface contact with that particular CT. Thirty nine image sets which contained all 5 pairs of NOR-bearing chromosome territories were analyzed in this study.
Computer simulations
Computer simulations were run to determine to what extend the NOR-CT pair-wise interchromosomal associations and associations with the same nucleoli can be accounted for by random interactions of the NOR-CT. A constrained random simulation was performed as previous described for CT that are positioned in close association with the nuclear periphery (Zeitz et al, 2009). In this case, however, the NOR-CT are constrained to be placed at a distance of ≤ 1.0 μm from one of the nucleoli. To best mimic the experimental conditions, each image set was individually simulated. In the first step, the NOR-CT are removed from the each image set while the nuclear boundary and nucleoli remain fixed in position. Then the 5 pairs of NOR-CT are randomly placed back into the nucleus with the constraint that each NOR-CT is placed in close association with one or more of the nucleoli (see Fig 5). The pairwise association patterns and their pairwise associations with individual nucleoli are then calculated from the population of individual simulations. All computer simulations were run using Matlab.
Figure 5. Constrained random simulations of NOR-CT and nucleoli.
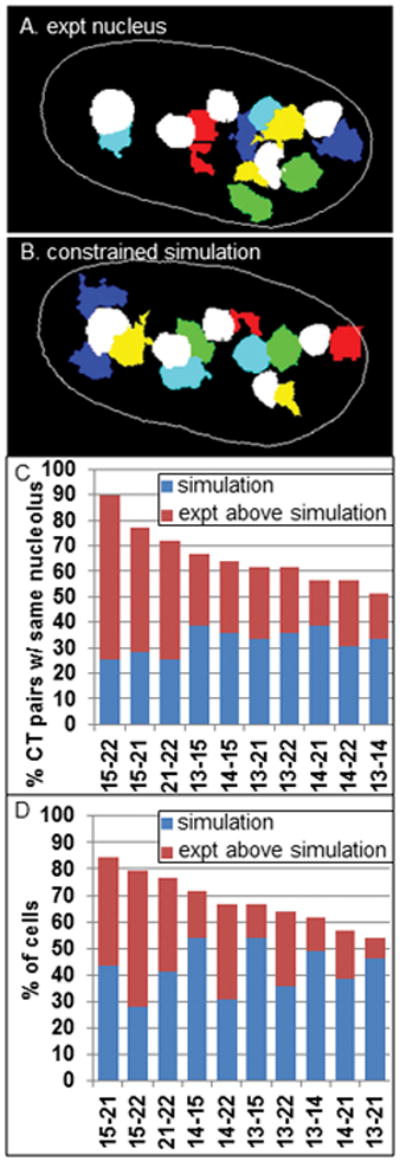
Each individual experimental nucleus was simulated by randomly placing the CT within a constrained ring of 1 micron surrounding each nucleolus while keeping the nucleoli fixed in position. This process is demonstrated in A-B for an experimental data set that contained 5 discrete nucleoli (white labeling) and the 5 pairs of NOR-CT in dark blue, light blue, red, green and yellow; (C) the percent of pairs of NOR-CT that are interacting with the same nucleolus in these constrained simulations are shown in blue bars, while the red bars demonstrate how much greater experimental values are than the simulations; (E) the percent of cells with CT-CT interactions are represented in the same manner (blue bars- simulations, red bars- experimental above simulations).
Chromatic Median Analysis
Previously our group developed a computational geometric approach termed the Generalized Median Graph (GMG) to determine the best fit probabilistic model of chromosome territory interactions in G0 WI38 fibroblasts (Mukherjee et al., 2009; Zeitz et al., 2009). The GMG considered all CT associations (i.e., all permutations of the association graphs) and simultaneously identifies the association patterns of all CT. Recently we reported an improved data mining pattern recognition algorithm termed the chromatic median (CM) which uses combinatorial optimization to infer the common chromosome interaction pattern for the cell population (Ding et al., 2013). The CM is capable of determining the corresponding homolog of the same chromosome across all cells based upon the interactions that homolog has with other CT. In the mathematical formulation of the problem, we use a cell representation which is suitable for describing interactions of CT. The interactions within each nucleus are represented as a matrix where a zero indicates no interaction and a one indicates an interaction (Fig 1). The objective is to find the best permutation (relabeling from a to b and vice versa for all chromosome pairs within each cell) which aligns the association matrix of each input cell with that of the common pattern.
Figure 1. Diagram illustrating the chromatic median analysis.
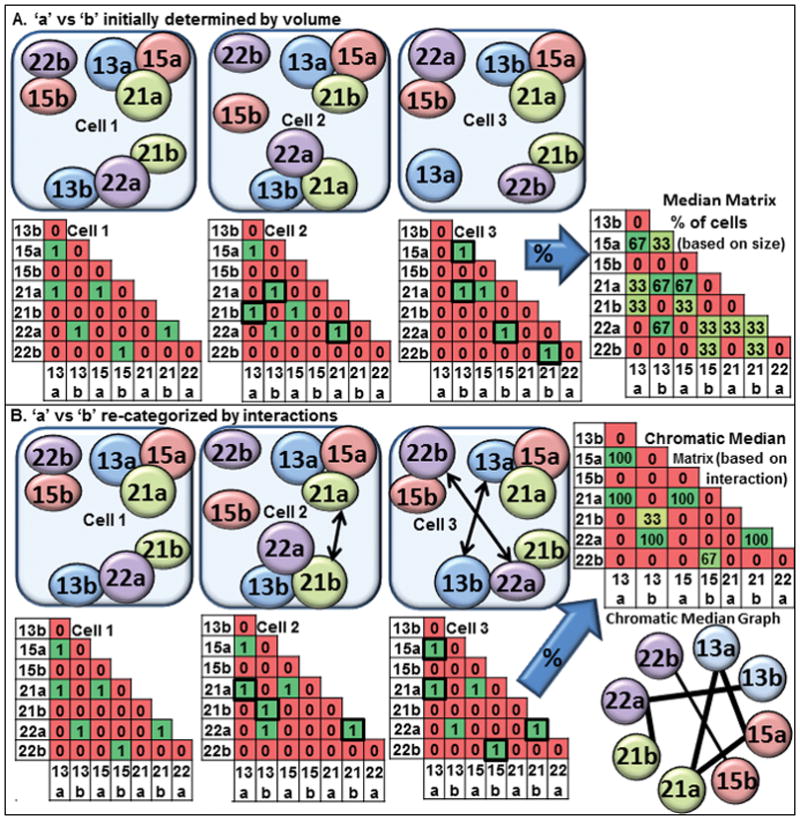
(A) Three schematic drawings of CT associations in nuclei are shown with the larger homolog defined as copy ‘a’ and the smaller one as copy ‘b’. The associations are represented in binary matrices wherein a 1 indicates an interaction and a 0 the absence of an interaction; (B) The chromatic median program determines which homolog is “copy a” versus “copy b” based upon which other CT are associated and switches “a” for “b” to match the best fit model for the population. The percent of cells with an interaction at any given position within the matrix is calculated. Using a threshold, a chromatic median graph enriches for those connections which are greater than randomizations of the input matrices.
This new CM algorithm considers all possible permutations of the interactions and simultaneously optimizes the interactions of all pairs of heterologs. For example, if there is a high frequency of nuclei wherein a homolog of CT13 associates with CT15 and 21 while the other homolog of CT13 associates with CT22, it will classify the first as “CT13a” and the second as “CT13b” across all cells (Fig 1). This process is performed simultaneously for all CT studied to identify similarity across the population. Once this process is complete, we then determine how many input nuclei have an interaction for each of the possible pairwise combinations. After permutation analysis, the CM generates an output matrix in which each excel cell contains the percent of nuclei that have that given interaction (Fig 1b). Using Excel’s conditional formatting, each interaction is filled in with a color ranging from green (high) to red (low). Yellow indicates moderate values. By taking a threshold that is greater than randomizations of the input matrices, we can generate the most probable graph of interactions above random interactions which is the chromatin median graph (Fig 1b). The CM represents the state-of-the-art for determining the most common pattern among a population of cells and yields almost optimal solutions. It has been validated extensively by empirical comparison analysis on both random and real datasets with various data sizes (Ding et al., 2013).
Results
Imaging NOR-CT and nucleoli: volume and radial position analysis
In this communication we characterize the interchromosomal interactions of the NOR-CT and their associations with nucleolar domains in WI38 normal human diploid lung fibroblast cells. To reduce experimental variations, all studies were performed in G0 synchronized cells. Nucleoli were visualized by immunofluorescence labeling of the abundant nucleolar protein nucleolin and the boundaries of the cell nucleus were identified using DAPI staining of genomic DNA. The CT were labeled in a sequential series of re-FISH procedures. In some experiments, in addition to CT, the rDNA genes were labeled, as described in Materials and Methods.
Representative 3-D volume rendering images following this sequence of immunolabeling and re-FISH are shown in Figure 2. Accumulative mapping of fluorescence signals are illustrated where the nucleolus, the five pairs of NOR-CT (# 13, 14, 15, 21 and 22), rDNA and genomic DNA were stained in several steps in the same cell (Fig 2A). Figure 2B displays images of cells with different numbers of nucleoli and all 5 NOR-CT. These multiplex sets of imaging data obtained in 39 cells were then used for computerized analysis of the NOR-CT and nucleoli.
Figure 2. 3-D visualization of NOR-CT, rDNA genes and nucleoli in WI38 fibroblasts following re-FISH.

(A) A panel of 3-D reconstructed images illustrates the progressive labeling from top left to bottom right of nucleoli (nucleolin labeling, top left image), 3 cycles of re-FISH to label all five NOR-CT and rDNA (bottom right image). The nucleoli are shown in white/gray and DAPI staining of genomic DNA in semi-transparent cyan. Note that the size of individual nucleoli may not correlate with the number of associated CT. In the displayed images, the two smaller nucleoli contain most of the rDNA signal and are associated with most of the NOR-CT; (B) 3-D reconstructed images of NOR-CT in nuclei displaying different numbers of nucleoli ranging from 1N to 6N. Note that some of the NOR-CT are in positions distal to nucleoli (white arrows). Orange arrows mark the “dominant nucleolus”. Color codes for the signals are listed in the upper right panel; scale bar = 4 microns.
The number of nucleoli in WI38 cells varied from one to six with an average of ~3 nucleoli per cell. As expected of diploid genomes, chromosome painting indicated the presence of only two copies of each labeled NOR-CT in almost all studied cells (> 99%). Consistent with the localization of ribosomal genes within nucleoli (Fig 2A), the majority of the NOR-CT were in close apposition to the nucleoli. A small portion of the NOR-CT, however, were separated from the nucleoli by variable distances that extended up to 4 μm (Fig. 2C and E). Since the rDNA gene clusters on the p-arms of the NOR-CT are completely associated with the nucleolar interior with no detectable extranucleolar signal (Fig 2A, bottom right image), the NOR-CT that are more distal from the nearest nucleoli must also have their rDNA clusters associated with the interior of nucleoli. To allow specificity of chromosome labeling, highly repetitive sequences are removed from the chromosome specific DNA libraries during chromosome paint preparation. Large portions of the NOR-CT p-arms (rDNA clusters and other repetitive sequences) are removed from the paints. These paints therefore predominantly label the q-arms (76–86% of the NOR-CT; see http://genome.ucsc.edu/). It is likely that those NOR-CT more distal to nucleoli are actually in close apposition via connections to the p-arm regions of the CT that are not well visualized with this approach as reported previously (Kalmarova et al., 2007).
For quantitative analysis, the acquired images were segmented based on the signal threshold intensity. The 3D coordinates of the segmented regions and a large number of distance measurements were then determined with our suite of computer programs (see Material and Methods and ((Zeitz et al., 2009). By analyzing the 3D segments, we measured the average volume of the WI38 cell nucleus as ~1300 μm3. Within the nucleus, the average sizes of labeled NOR-CT ranged from ~14 μm3 for the smallest CT 21, to ~25 μm3 for the larger CT 13, 14, and 15 (Fig 3A). Altogether, the NOR-CT occupied about 16% of the overall nuclear volume and there was a strong linear correlation [r2 = 0.94], between mbp DNA lengths and their volumes (Fig 3B).
Figure 3. Volumes and radial positioning of NOR-CT.

(A) Absolute CT volumes are shown; (B) the sequence lengths of the NOR-CT show a strong linear correlation with their CT volumes; (C) total nuclear and nucleolar volumes decrease in coordinate fashion with increases in the number of nucleoli per nucleus; (D) total nucleolar volume (% of total nuclear volume) is plotted against the number of nucleoli per nucleus; (E) the total surface area (μm2) is plotted against the number of nucleoli per nucleus; (F) a linear relationship was determined between sequence length and the minimal peripheral distance (NOR-CT border to nuclear border distance) with larger NOR-CT being closer to the nuclear periphery; (G) peripheral radial positioning of NOR-CT was not related to gene density. Error bars = SEM.
We documented a direct correlation between the volumes of the nucleoli and the nuclei in WI38 cells. While the total volume of the nucleoli decreased strikingly as the number of nucleoli in each cell increased (~2.4 fold between 1 to 5 nucleoli; Fig 3C), there was a corresponding decrease in total nuclear volume so that the overall nucleolar volume decreased only slightly when expressed as the % of total nuclear volume (8% to 6%, Fig 3C and D). Moreover, the total nucleolar surface area increased only slightly as the number of nucleoli increased (Fig 3E). Thus regardless of the number of nucleoli in different cells, the NOR-CT are associated with approximately the same nucleolar surface area for potential interactions. In addition, the size of the nucleoli did not correlate with the number of associated NOR-CT or with the intensity of the rDNA signal inside the nucleolar domains (Fig 2).
A number of previous studies have shown a linear correlation between chromosome size and their radial position where the smallest chromosomes occupy the nuclear interior and the largest chromosomes are closer to the nuclear periphery (Fritz et al., 2014; Marella et al., 2009a; Sun et al., 2000; Zeitz et al., 2009). It is remarkable to note that although the positions of all NOR-CT are influenced by their associations with nucleoli, a similar linear correlation was preserved for the radial positioning of these CT (Fig 3F). Previous studies of non-NOR-CT demonstrated that gene dense CT are more interior than gene poor CT (Boyle et al., 2001). This was dependent on the shape of nuclei (Cremer et al., 2001). However, we did not detect a correlation between the radial position of CT and gene density for NOR-CT (Fig 3G). For example, despite being of similar mbp length and volume, CT22 is twice the gene density of CT21, yet both share similar radial positioning (Fig 3F–G).
Nucleolar associations of NOR-CT
The inherent proximity of NOR-CT to nucleoli provides a convenient experimental model to study chromosomal neighborhood associations using re-FISH in combination with our computerized image analysis and computational geometric approaches. In one approach, the levels of interchromosomal associations as defined by a certain nearest distance threshold are determined for each CT pair. In a second approach the association of the NOR-CT with the nucleoli are measured using the same threshold method.
To determine NOR-CT-nucleolar associations, the nucleoli and NOR-CT were segmented and the nearest edge-to-edge 3-D distances were determined between individual CT and nucleoli. Nearest distances of ≤ 1.0 μm (14 pixels) were counted as positive associations. In cases where a CT was in association with more than one nucleolus, the association was assigned to the nucleolus with the largest contact area. Based on these criteria, we screened the frequency of pair-wise associations of specific NOR-CT with the same nucleolus. A large range of associations was measured for the heterologous CT pair from 51% of cells for the CT13-14 pair to 90% of cells for the CT15-22 pair (Fig 4A). Associations of homologous CT with the same nucleolus also showed a high frequency and wide range from 48% for the CT14 pair to 72 % for the CT21 pair (Fig 4B). In contrast, simulations where the NOR-CT are placed back into the nucleus randomly constrained (see Materials and Methods) to associate with the nucleoli (Fig 5A and B), revealed much lower levels of association and a much narrower range of values (26–38%, Fig 5C). All the individual experimental values were much higher (1.5–3.5 fold) than the values following simulation.
Figure 4. Association of NOR-CT with nucleoli.

(A) The percent of pairs of heterologous NOR-CT that are interacting with the same nucleolus (nearest 3-D border distances ≤ 1.0 μm); (B) the percent of pairs of homologous NOR-CT that are interacting with the same nucleolus; (C) cells with different number of nucleoli were assigned to three measurement groups: those with 1, 2, and ≥ 3 nucleoli. The numbers of NOR-CT associated with each nucleolar domain were sorted into descendent order to the nucleoli with the most CT (dominant), intermediate levels of CT (intermediate) and the least number of CT (minor) for each cell and then averaged for the entire measurement group. In cells with ≥ 3 nucleoli, the nucleoli with the three highest numbers of associated NOR-CT are shown. All values are statistically significant (p< 0.05) with the exception of comparisons of the intermediate to minor nucleoli in cells containing ≥ 3 nucleoli. The percent of total interaction of each specific NOR-CT with nucleoli are shown for cells with one nucleoli (D), two nucleoli (E), and three or more nucleoli (F). Error bars = SEM
Next, we determined the number of CT associated with each nucleolar domain. Using the same criteria as above, we determined associations between individual NOR-CT and discrete nucleoli. We observed that NOR-CT do not associate with different nucleoli in equal or near-equal amounts as would be implied by a random pattern of associated CT. Instead we report a highly nonrandom phenomenon in the chromosome distribution pattern, where one of the nucleoli in each cell is always association with the greatest number of NOR-CT (Fig 2 and 4C). Nucleoli associated with the greatest number of NOR-CT in each cell were termed “dominant” nucleoli. Those with the lowest and intermediate numbers of CT were termed “minor” and “intermediate” nucleoli, respectively. While the “dominant” nucleolus (indicated by orange arrows in Fig 2) is often the largest nucleolus in the cell (Fig 2D and E), this is not always the case (Figure 2A and F). Figure 2G shows a cell with only one nucleolus and all of the NOR-CT in close proximity.
Depending on the number of nucleoli, a wide range of nucleolar associations were found for individual NOR-CT (Fig 4D–E). In cells containing a single nucleolus, the association frequency ranged from 60% for CT14 to 82% for CT21 (Fig 4D). In cells with two nucleoli, CT13, CT14 and CT22 were predominantly associated with the dominant nucleolus while CT21 was predominantly associated with the intermediate nucleolus. With three or more nucleoli only CT13, CT14 and CT15 were preferential in their association with the dominant nucleolus. Thus while one nucleolus in each cell preferentially contains the greatest number of associated NOR-CT, the distribution of individual NOR-CT among the nucleolar population is more complex.
Interchromosomal associations of NOR-CT
NOR-CT are defined as “neighbors” and assigned with a positive pair-wise association, when the nearest edge-to-edge 3-D distance between the segmented CT is ≤0.56 μm (8 pixels). This distance is large enough to distinguish boundaries between two homologous CT stained in the same fluorescence channel. A wide range of pairwise associations (Fig 6A,B) were determined for both heterologous (52–82%) and homologous (24–36%) CT pairs. Increasing the threshold distance between 8 to 20 pixels only slightly altered the levels of association (Fig S1). Importantly, the levels of pairwise associations at zero pixel distance (38–72%) were still very high (Fig S1). Moreover, the triplet cluster CT15-21-22 was observed in 72 % of the cells indicating a highly non-random association pattern.
Figure 6. Pair-wise association profiles of NOR- CT.
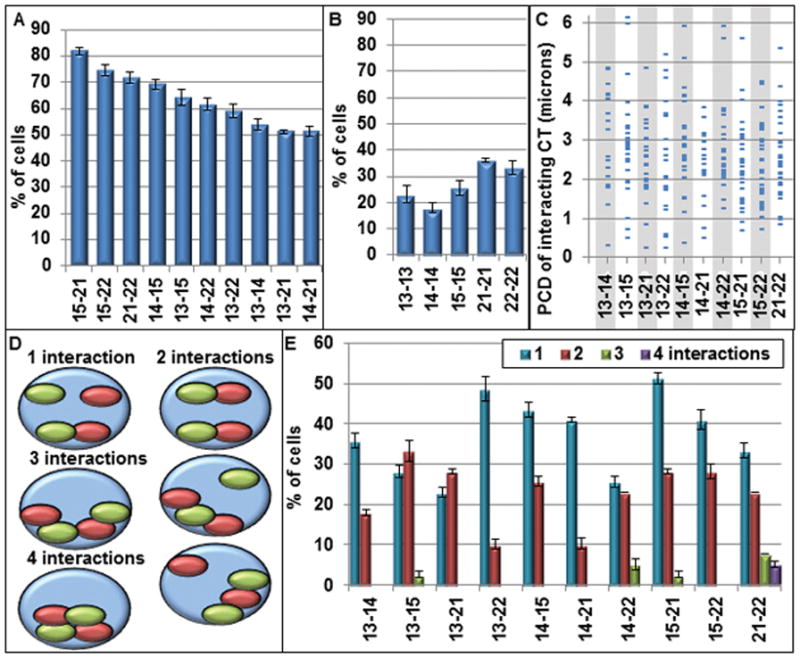
The percent of cells with pairwise interactions between CT (nearest 3-D border distances ≤ 0.56 μm) are shown for heterologous (A) and homologous (B) CT pairs; (C) the corresponding pairwise center to center distances for the pairwise associations displayed in (A) range from 0.2 to >6 μm; (D) Since each CT has two homologs there are four possible interactions in each nucleus. The percent of cells that have 1, 2, 3 and 4 interactions are shown including 3 types with 2 interactions; (E) the percent of cells with single and multiple interactions are displayed for each heterologous pair of NOR-CT. Error bars = SEM
Since the NOR-CT are predominantly confined to the crowded volume of the perinucleolar space, we applied our random constrained simulation program to determine to what extent these high associations values can be explained by a random association of the NOR-CT within this confined space. In this simulation (see Materials and Methods) the 5 pairs of NOR-CT are randomly placed into the nucleus with the constraint that each NOR-CT is positioned ≤ 1.0 μm of one or more of the nucleoli (Fig 5A–B). All of the association frequencies following this simulation were lower than the corresponding experimental results (Fig 5D). 6 of the 10 associations were 1.5–2.8 fold higher than the random simulation values while the remaining 4 associations displayed lower levels of differences (1.2 –1.3 fold).
In most previous studies, distances between centers of gravity have been used to study the organization of CT in the cell nucleus which may not be reflective of the interactions between CT (Fritz et al., 2014). Our analysis demonstrates that NOR-CT which are in close association as measured by the nearest edge-to-edge approach, have widely varying center-to-center distances (Fig 6C). Thus center-to-center distances, while providing important positional information, cannot accurately identify many of the close associations among the CT population in the nucleus.
As illustrated in Figure 6D, a CT pair may produce up to four pairwise interactions with another CT pair. We, therefore, determined the distribution for each NOR-CT pair of single and multiple pairwise interchromosomal associations (Fig 6E). While some CT pairwise associations consisted largely of singular interactions (CT13-14, CT13-22, CT14-15, CT14-21, CT15-21), others had approximately equal amounts of singular and multiple interactions (CT13-15, CT13-21, CT14-22, CT21-22). The great majority of the multiple interactions involved two interactions, while three and four interactions were infrequent and present mainly with the smaller CT21 and CT22 (Fig 6E).
A probabilistic model of NOR-CT interactions
The differences in the frequencies of pairwise associations (Fig 6A and B), the variations in distributions of singular and multiple interactions (Fig 6E), and the association of particular NOR-CT with nucleoli (Fig 4), suggest a complex and nonrandom pattern to the overall organization of NOR-CT in the cell nucleus. To investigate this further we used a novel computational data mining and pattern recognition program termed the chromatic median (CM). The CM is designed to determine the most probable overall pattern of CT associations across the population of nuclei (Ding et al. 2013). A simple illustration of this approach is presented in Figure 1 and is detailed in Materials and Methods. In brief, this program determines the corresponding homologs across all nuclei based upon that homolog’s interactions. It then represents each nucleus as a 10x10 (5 CT, 2 homologs per CT) binary matrix wherein a value of 1 indicates an interaction and a value of 0 indicates the absence of an interaction. Subsequently we determine the percent of nuclei that contain an interaction for each of the 45 positions in the matrices. Within these resulting median matrices, hot and cold-spots were found that ranged from 6 to 47% of input nuclei (Fig 7A) compared to 6–23% following randomizations of the input matrices (Fig 7B). This demonstrates that the process of determining corresponding homologs in a population of nuclei with a high degree of interactions does not artificially create a pattern of those interactions. Chi test analysis determined that median matrices from experimental and randomized input are significantly different from each other (p<0.001).
Figure 7. Overall association map of the 5 pairs of NOR- CT.

(A) The chromatic median algorithm determines correspondence between homologs across nuclei based upon which other CT it closely associates. This algorithm determined a median matrix for NOR CT- interactions in WI38 cells in which each position in the matrix represents the percent of input nuclei that have an interaction between those NOR-CT. The percent of chromosomal associations is represented in a color-coded spectrum from red (minimal) to green (maximal); (B) the median matrix following randomization of the input matrices; (C) thresholding above a level of 24% interactions (where there are no connections in randomization of input matrices) revealed 13 connections among the 5 pairs of NOR-CT. Thick lines represent connections that are within the top third of percent pairwise associations within the model, intermediate thickness are connections in the middle third of pairwise associations while thin dashed lines are within the bottom third. Any given nucleus contains on average 32.2% of the connections within these models (Sorenson, 1948).
Using the CM algorithm we were able to determine corresponding homologs based on which other CT each homolog is interacting. Our results demonstrate that homolog a does interact with certain other CT with higher frequency than homolog b. These differences between homologs in experimental nuclei were greater than randomizations. For example, CT15a interacts with CT21a in 47% of nuclei, but CT15b and 21b interact in only 11%. This observation suggests that functional neighborhoods of interaction extend to the homolog level. This could be a result of certain NOR-CT interacting with particular nucleoli.
To determine a preferred probabilistic model of NOR-CT interactions, thresholding was performed on the matrices at 24% association. This enriched for the CT interactions found at the higher levels among the total population. Moreover at 24%, there are no connections in randomizations of input matrices or random simulations. The CM program revealed a global interactive network composed of 13 connections between the 5 pairs of NOR-CT (Fig 7C). Interestingly, CT21 had the largest number of connections (7) despite being the smallest human chromosome. CT14, CT15 and CT22 had 5 connections each, while CT13 had only 3. This lower level was likely due to the absence of any predicted connections at this probabilistic level for the CT13b homolog (Fig 7C). The connections with the highest % associations in the model are indicated as thick connecting lines as CT21-22 and CT15-21 (Fig 7C) Moreover, the two smallest chromosomes in the human genome CT21 and CT22 were the only NOR-CT displaying homologous connections. Sorenson’s analysis (Sorenson, 1948) revealed that each individual nucleus in the population analyzed contains on average 32% of the network connections displayed in this probabilistic model.
Discussion
The role of the nucleolus in cell function extends beyond its central position as a ribosome factory (Busch and Smetana, 1970; Koberna et al., 2002). A plethora of functionally related properties have been ascribed to this sub-nuclear compartment which have largely baffled our understanding (Boisvert et al., 2007; Martelli et al., 2001; Olson et al., 2000; Pederson, 1998; Raska et al., 2006; Rubbi and Milner, 2003). Proteomics has demonstrated many hundreds of nucleolar proteins the great majority of which are not known to be involved in ribosomal RNA transcription or metabolism (Andersen et al., 2005; Pederson and Tsai, 2009). Genomics has elucidated nucleolar associated sequences (NADs) that are not limited to the NOR-bearing chromosomes but distributed throughout the genome (Nemeth et al., 2010; van Koningsbruggen et al., 2010). It is this latter property that captured our attention and led us to analyze the interchromosomal and nucleolar associations of the NOR-CT.
NOR-CT Nucleolar Interactions
There is growing emphasis on deciphering the relationship of higher level genomic organization and function to the regulation of gene expression (Berezney, 2002; Berezney et al., 2005; Bickmore, 2013; Cremer and Cremer, 2010; Kumaran et al., 2008; Malyavantham et al., 2010; Meaburn and Misteli, 2007a; Misteli, 2007; Stein et al., 2003; Stein G, 2008; T, 2005; Zaidi et al., 2007). It is now well established that the mitotic chromosomes are arranged in the interphase cell nucleus as discrete 3-D structures termed chromosome territories (CT), (Bickmore, 2013; Cremer and Cremer, 2001; Cremer and Cremer, 2010; Cremer et al., 2006; Meaburn and Misteli, 2007b). Recent findings suggesting an involvement of intra- and inter-chromosomal interactions in gene expression and regulation (Berezney, 2002; Berezney et al., 2005; Bickmore, 2013; Clowney et al., 2012; Kumaran et al., 2008; Lanctot et al., 2007; Malyavantham et al., 2008; Malyavantham et al., 2010; Misteli, 2004; Misteli, 2007; Osborne et al., 2004; Osborne et al., 2007; Parada et al., 2004b; Spilianakis et al., 2005; Stein G, 2003; Stein G, 2008; T, 2005; Zaidi et al., 2007) has brought to the forefront the need for a better understanding of the 3-D organization of CT and their interactions in the cell nucleus. By combining the techniques of re-FISH with 3-D microscopy, computer image analysis and novel computational geometric data mining and pattern recognition algorithms, progress has been made in deciphering the 3-D arrangement of large subsets of human CT (Fritz et al., 2014; Marella et al., 2009a; Zeitz et al., 2009). In this study we use these tools to investigate the interchromosomal arrangements of the five pairs of nucleolar associated NOR-CT (CT 13,14,15, 21 and 22) in WI38 human diploid lung fibroblasts and the relationships of NOR-CT and nucleoli associations.
Analysis of NOR-CT associations with nucleoli in G0 synchronized WI38 fibroblasts demonstrated that one nucleolus was always associated with more NOR-CT than the other nucleoli (Fig 4). Smetena et. al. (1999, 2006) previously observed this in cells from human patients using the silver staining procedure for NORs and termed these the “dominant nucleoli” (Smetana et al., 2006a; Smetana et al., 1999; Smetana et al., 2006b). We further show that while many of the individual NOR-CT are predominantly associated with the dominant nucleolus, others are preferentially associated with nucleoli with intermediate levels of NOR-CT (Fig 4).
A wide range of high level associations (50–90%) were found for heterologous pairs of NOR-CT with the same nucleolus which exceeded those demonstrated by random simulations by 1.5–3.5 fold (Fig 4, 5). There was also an unexpectedly high level of homologous NOR-CT pairs associated with the same nucleolus (48–72%). Random simulations showed a size dependency with the larger NOR-CT (e.g. CT 13–15) at higher association levels than the smaller CT (e.g., CT 21–22, Fig 5C). In contrast, the experimental data had no clear size dependency, e.g., CT 21–22, CT 21–21 and CT 22–22 showing much higher levels of association than CT 14-14 and 13-14 (Fig 4A, B). Together these results indicate the presence of non-random association patterns of NOR-CT with individual nucleoli. Further supporting this conclusion, it was reported that a high percentage of the NOR-CT are associated with nucleoli and that at least a portion of NOR-CT associated with the same nucleolus were preserved in daughter cells (Cvackova et al., 2009; Kalmarova et al., 2008b; Kalmarova et al., 2007).
These findings also have implications for nucleolar biogenesis in the cell cycle. It is well established that the nucleoli are formed in late telophase/ early G1-phase, around the sites of ribosomal genes expression, engulfing or fusing the material of several adjacent NORs protruded from their CT (Prieto and McStay, 2005; Raska et al., 2006). The discovery of high levels of specific NOR-CT pairs associated with the same nucleoli suggests that ordered clusters of certain combinations of NOR-CT may form by the time of exit from mitosis and subsequently contribute their NORs to the formation of the same nucleolus. In some cells this clustering involves all NOR-bearing chromosomes which consequently leads to formation of a single nucleolus. In other cells there is one larger cluster of chromosomes that gives a rise to a “dominant nucleolus” and several smaller sub-clusters or separate chromosomes that produce their own nucleoli (Fig 4). It is important to note that the nucleolar patterns measured in these investigations were performed in cells synchronized in G0. Thus differences in the cell cycle and particularly the observed phenomenon of fusion of smaller nucleoli into single larger ones that occur during the S and G2 phase (Sister Paula and Nardone, 1968; Wachtler et al., 1984; Wachtler and Stahl, 1993), do not contribute to these patterns.
By linking specific NOR-CT with non-NOR CT, the post-mitotic locations of NOR-CT and their associations with different nucleoli could have a major impact on the biogenesis of the overall chromosomal landscape of the cell nucleus. In this regard high levels of interchromosomal associations were detected between NOR-CT 15 and 21 and several non-NOR CT (Fritz et al., 2014). This is consistent with recent genomic findings suggesting that many CT aside from the NOR-CT associate with the nucleolus (Nemeth et al., 2010; van Koningsbruggen et al., 2010) where they presumably contribute to the perinucleolar heterochromatin region (Cvackova et al., 2009).
Interchromosomal Interactions and a global interactive network of NOR-CT
A high level and wide range (52–82%) were measured for heterologous interchromosomal pairwise associations (Fig 6) while homologous pairwise associations were much lower (24–36%). 6 of the 10 heterologous pairwise combinations were significantly above random simulations when the NOR-CT were rigorously constrained to positions surrounding the nucleoli (Fig 5). There were also major differences in the levels of multiple interactions among the CT pairs. For homologous associations, the smaller CT 21 and 22 pairs had significantly higher levels of association (Fig 4).
The above findings support a high degree of non-randomness to the arrangement of the NOR-CT. We, therefore, applied a novel pattern recognition algorithm termed the chromatic median (Ding et al., 2013) to determine if there was a preferred arrangement of interchromosomal associations for the entire population of NOR-CT. The analysis revealed an interconnected network of associations that encompassed all the individual NOR-CT except one copy of CT13 (Fig 7C). Interestingly, the highest levels of associations within this global interactive network involved the two smallest chromosomes CT 21 and 22. Global interactive networks of CT were previous reported for other CT subsets which showed cell type and tissue specificity (Marella et al., 2009a; Zeitz et al., 2009). Moreover, a recent study revealed major alterations in the global interactive network in malignant breast cancer cells (Fritz et al., 2014).
The analysis of NOR-CT positional associations not only provides valuable information about this subset of CT, but also represents an important approach for investigating the NOR-CT in relation to the rest of the genome. The NOR regions of these CT are anchored inside the nucleoli but the bulk volumes of these CT are located outside the nucleoli and can thus create pair-wise associations with different nucleolar and non-nucleolar CT. In this regard, a study of CT associations among a subset of 9 CT pairs including 2 NOR-CT, showed significant levels of association among the NOR and non-NOR CT which were also present in their global interactive networks (Fritz et al., 2014).
Recent genomic studies demonstrate that many if not all human chromosomes are associated with nucleoli via nucleolar associating domains (NADs) including a host of gene sequences unrelated to ribosomal RNA metabolism and ribosome biogenesis (Nemeth et al., 2010; van Koningsbruggen et al., 2010). This raises the possibility that the nucleolus may be directly involved in the regulation of a variety of genes other than rDNA (Nemeth et al., 2010; van Koningsbruggen et al., 2010). By sequestering inactive genes in the perinucleolar heterochromatin and protrusion of active genes into the extranucleolar regions for expression, the nucleolus may play a fundamental role in the global organization and genomic function of CT (van Koningsbruggen et al., 2010). Interestingly, NADs share partial sequence similarity with lamina-associated domains (LADs) and as a result some of this inactive chromatin may be repositioned to the nuclear periphery (Guelen et al., 2008; van Koningsbruggen et al., 2010).
The nucleoli and nuclear envelope may also be part of an overall architectural system in the cell nucleus for CT positioning and function. Previous studies have demonstrated a possible role of nucleoli and the nuclear periphery in constraining chromatin positions (Chubb et al., 2002). In this regard, the nuclear matrix was originally defined as a structural framework in the cell nucleus composed of residual components of the nucleoli, the peripheral nuclear envelope and a fibrogranular internal matrix (Berezney and Coffey, 1974; Berezney and Coffey, 1977; Berezney et al., 1995). Moreover, appropriate salt extraction of whole cells results in a remarkable preservation of chromosome territory organization but only if nuclear matrix organization is maintained (Ma et al., 1999). Treatments that result in nuclear matrix extraction lead to a corresponding disruption of chromosome territories (Ma et al., 1999).
In summary, our study identifies non-random associations for the subset of NOR-CT and demonstrates their arrangement into a global interactive network. Similar networks in other subsets of CT suggest that the NOR-CT are part of an overall genome wide network extending throughout the cell nucleus. We previously proposed that this interactive CT network may function as a probabilistic chromosome code that mediates genomic function at the global level (Fritz et al., 2014; Marella et al., 2009a; Zeitz et al., 2009). Deciphering the interactions of the NOR-CT in this genome wide network may, therefore, significantly contribute to our understanding of the multi-functional nucleolus.
Supplementary Material
Acknowledgments
This research was supported by grants from the National Institutes of Health (GM-072131) to R.B, the National Science Foundation (IIS-0713489 and IIS-1115220) to J.X. and R.B. and the University at Buffalo Foundation (9351115726) to R.B.
Abbreviations
- rDNA
ribosomal DNA
- NOR
nucleolar organizer region
References
- Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433(7021):77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- Berezney R. Regulating the mammalian genome: the role of nuclear architecture. Advan Enzyme Regul. 2002;42(2002):39–52. doi: 10.1016/s0065-2571(01)00041-3. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochemical and biophysical research communications. 1974;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. The Journal of cell biology. 1977;73(3):616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Malyavantham KS, Pliss A, Bhattacharya S, Acharya R. Spatio-temporal dynamics of genomic organization and function in the mammalian cell nucleus. Advances in enzyme regulation. 2005;45:17–26. doi: 10.1016/j.advenzreg.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. International review of cytology. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Bickmore WA. The spatial organization of the human genome. Annual review of genomics and human genetics. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nature reviews Molecular cell biology. 2007;8(7):574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS biology. 2005;3(5):e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Human molecular genetics. 2001;10(3):211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS biology. 2006;4(5):e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brianna Caddle L, Grant JL, Szatkiewicz J, van Hase J, Shirley BJ, Bewersdorf J, Cremer C, Arneodo A, Khalil A, Mills KD. Chromosome neighborhood composition determines translocation outcomes after exposure to high-dose radiation in primary cells. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2007;15(8):1061–1073. doi: 10.1007/s10577-007-1181-7. [DOI] [PubMed] [Google Scholar]
- Busch H, Smetana K. The nucleolus. New York: Academic Press; 1970. [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Current biology : CB. 2002;12(6):439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151(4):724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2001;9(7):541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nature reviews Genetics. 2001;2(4):292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harbor perspectives in biology. 2010;2(3):1–22. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories--a functional nuclear landscape. Current opinion in cell biology. 2006;18(3):307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kreth G, Koester H, Fink RH, Heintzmann R, Cremer M, Solovei I, Zink D, Cremer C. Chromosome territories, interchromatin domain compartment, and nuclear matrix: an integrated view of the functional nuclear architecture. Critical reviews in eukaryotic gene expression. 2000;10(2):179–212. [PubMed] [Google Scholar]
- Cvackova Z, Masata M, Stanek D, Fidlerova H, Raska I. Chromatin position in human HepG2 cells: although being non-random, significantly changed in daughter cells. Journal of structural biology. 2009;165(2):107–117. doi: 10.1016/j.jsb.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Stojkovic B, Berezney R, Xu JH. Gauging Association Patterns of Chromosome Territories via Chromatic Median. Proc Cvpr Ieee. 2013:1296–1303. [Google Scholar]
- Erickson JM, Rushford CL, Dorney DJ, Wilson GN, Schmickel RD. Structure and variation of human ribosomal DNA: molecular analysis of cloned fragments. Gene. 1981;16(1–3):1–9. doi: 10.1016/0378-1119(81)90055-x. [DOI] [PubMed] [Google Scholar]
- Folle GA. Nuclear architecture, chromosome domains and genetic damage. Mutation research. 2008;658(3):172–183. doi: 10.1016/j.mrrev.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Fritz AJ, Stojkovic B, Ding H, Xu J, Bhattacharya S, Gaile D, Berezney R. Wide-scale alterations in interchromosomal organization in breast cancer cells: defining a network of interacting chromosomes. Human molecular genetics. 2014 doi: 10.1093/hmg/ddu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grob A, Colleran C, McStay B. Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes & development. 2014;28(3):220–230. doi: 10.1101/gad.234591.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453(7197):948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Heride C, Ricoul M, Kieu K, von Hase J, Guillemot V, Cremer C, Dubrana K, Sabatier L. Distance between homologous chromosomes results from chromosome positioning constraints. Journal of cell science. 2010;123(Pt 23):4063–4075. doi: 10.1242/jcs.066498. [DOI] [PubMed] [Google Scholar]
- Kalmarova M, Kovacik L, Popov A, Testillano SP, Smirnov E. Asymmetrical distribution of the transcriptionally competent NORs in mitosis. Journal of structural biology. 2008a;163(1):40–44. doi: 10.1016/j.jsb.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmarova M, Smirnov E, Kovacik L, Popov A, Raska I. Positioning of the NOR-bearing chromosomes in relation to nucleoli in daughter cells after mitosis. Physiological research / Academia Scientiarum Bohemoslovaca. 2008b;57(3):421–425. doi: 10.33549/physiolres.931430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmarova M, Smirnov E, Masata M, Koberna K, Ligasova A, Popov A, Raska I. Positioning of NORs and NOR-bearing chromosomes in relation to nucleoli. Journal of structural biology. 2007;160(1):49–56. doi: 10.1016/j.jsb.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K, Malinsky J, Pliss A, Masata M, Vecerova J, Fialova M, Bednar J, Raska I. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. The Journal of cell biology. 2002;157(5):743–748. doi: 10.1083/jcb.200202007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth G, Finsterle J, von Hase J, Cremer M, Cremer C. Radial arrangement of chromosome territories in human cell nuclei: a computer model approach based on gene density indicates a probabilistic global positioning code. Biophysical journal. 2004;86(5):2803–2812. doi: 10.1016/S0006-3495(04)74333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Thakar R, Spector DL. Chromatin dynamics and gene positioning. Cell. 2008;132(6):929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Tanabe H, Yoshida K, Oikawa K, Saito A, Kiyuna T, Mizusawa H, Mukai K. Alteration of chromosome positioning during adipocyte differentiation. Journal of cell science. 2004;117(Pt 24):5897–5903. doi: 10.1242/jcs.01508. [DOI] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nature reviews Genetics. 2007;8(2):104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. The Journal of cell biology. 1999;146(3):531–542. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Alonso WD, Acharya R, Berezney R. Spatio-temporal dynamics of replication and transcription sites in the mammalian cell nucleus. Chromosoma. 2008;117(6):553–567. doi: 10.1007/s00412-008-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Berezney R. The architecture of functional neighborhoods within the mammalian cell nucleus. Advances in enzyme regulation. 2010;50(1):126–134. doi: 10.1016/j.advenzreg.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella NV, Bhattacharya S, Mukherjee L, Xu J, Berezney R. Cell type specific chromosome territory organization in the interphase nucleus of normal and cancer cells. Journal of cellular physiology. 2009a;221(1):130–138. doi: 10.1002/jcp.21836. [DOI] [PubMed] [Google Scholar]
- Marella NV, Seifert B, Nagarajan P, Sinha S, Berezney R. Chromosomal rearrangements during human epidermal keratinocyte differentiation. Journal of cellular physiology. 2009b;221(1):139–146. doi: 10.1002/jcp.21855. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Zweyer M, Ochs RL, Tazzari PL, Tabellini G, Narducci P, Bortul R. Nuclear apoptotic changes: an overview. J Cell Biochem. 2001;82(4):634–646. doi: 10.1002/jcb.1186. [DOI] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC cell biology. 2005;6:44. doi: 10.1186/1471-2121-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Cell biology - Chromosome territories. Nature. 2007a;445(7126):379–381. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007b;445(7126):379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Misteli T. Spatial positioning; a new dimension in genome function. Cell. 2004;119(2):153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128(4):787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mukherjee L, Singh V, Peng JM, Xu JH, Zeitz MJ, Berezney R. Generalized median graphs and applications. J Comb Optim. 2009;17(1):21–44. [Google Scholar]
- Nagele RG, Freeman T, McMorrow L, Thomson Z, Kitson-Wind K, Lee H. Chromosomes exhibit preferential positioning in nuclei of quiescent human cells. Journal of cell science. 1999;112( Pt 4):525–535. doi: 10.1242/jcs.112.4.525. [DOI] [PubMed] [Google Scholar]
- Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G. Initial genomics of the human nucleolus. PLoS genetics. 2010;6(3):e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neusser M, Schubel V, Koch A, Cremer T, Muller S. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma. 2007;116(3):307–320. doi: 10.1007/s00412-007-0099-3. [DOI] [PubMed] [Google Scholar]
- Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10(5):189–196. doi: 10.1016/s0962-8924(00)01738-4. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nature genetics. 2004;36(10):1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS biology. 2007;5(8):e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome biology. 2004a;5(7):R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Munson PJ, Misteli T. Conservation of relative chromosome positioning in normal and cancer cells. Current biology : CB. 2002;12(19):1692–1697. doi: 10.1016/s0960-9822(02)01166-1. [DOI] [PubMed] [Google Scholar]
- Parada LA, Sotiriou S, Misteli T. Spatial genome organization. Experimental cell research. 2004b;296(1):64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic acids research. 1998;26(17):3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. The Journal of cell biology. 2009;184(6):771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. Nucleolar biogenesis: the first small steps. Biochemical Society transactions. 2005;33(Pt 6):1441–1443. doi: 10.1042/BST0331441. [DOI] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Current opinion in cell biology. 2006;18(3):325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nature genetics. 2003;34(3):287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. The EMBO journal. 2003;22(22):6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sister Paula G, Nardone RM. Cyclic nucleolar changes during the cell cycle. I. Variations in number, size, morphology and position. Experimental cell research. 1968;50(3):599–615. doi: 10.1016/0014-4827(68)90422-9. [DOI] [PubMed] [Google Scholar]
- Smetana K, Klamova H, Mikulenkova D, Pluskalova M, Hrkal Z. On the nucleolar size and density in human early granulocytic progenitors, myeloblasts. European journal of histochemistry : EJH. 2006a;50(2):119–124. [PubMed] [Google Scholar]
- Smetana K, Likovsky Z, Jiraskova I, Cermak J. The asymmetric distribution of interphasic silver-stained nucleolus organizer regions in human and rat proerythroblasts. Folia biologica. 1999;45(6):243–246. [PubMed] [Google Scholar]
- Smetana K, Mikulenkova D, Jiraskova I, Klamova H. A karyometric note on nucleoli in human early granulocytic precursors. Folia biologica. 2006b;52(1–2):16–20. [PubMed] [Google Scholar]
- Smirnov E, Kalmarova M, Koberna K, Zemanova Z, Malinsky J, Masata M, Cvackova Z, Michalova K, Raska I. NORs and their transcription competence during the cell cycle. Folia biologica. 2006;52(3):59–70. [PMC free article] [PubMed] [Google Scholar]
- Sorenson T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species and its application to analyses of the vegetation on Danish commons. Biol Skr Dan Vid Sel. 1948;5:1–34. [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nature cell biology. 2007;9(6):675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Stein G, Zaidi S, Braastad C, Montecino M, van Wijnen A, Choi J, Stein J, Lian J, Javed A. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. TRENDS in Cell Biology. 2003;13(11):584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Stein GZS, Braastad C, Montecino M, van Wijnen A, Choi J, Stein J, Lian J, Javed A. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. TRENDS in Cell Biology. 2003;13(11):584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Stein GZS, Stein J, Lian J, van Wijnen A, Montecino M, Young D, Javed A, Pratap J, Choi J, Ali S, Pande S, Hassan M. Genetic and Epigenetic Regulation in Nuclear Microenvironments for Biological Control in Cancer. Journal of Cellular Biochemistry. 2008;104:2016–2026. doi: 10.1002/jcb.21813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HB, Shen J, Yokota H. Size-dependent positioning of human chromosomes in interphase nuclei. Biophysical journal. 2000;79(1):184–190. doi: 10.1016/S0006-3495(00)76282-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TM Concepts in nuclear architecture. BioEssays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Molecular biology of the cell. 2010;21(21):3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser AE, Aten JA. Chromosomes as well as chromosomal subdomains constitute distinct units in interphase nuclei. Journal of cell science. 1999;112( Pt 19):3353–3360. doi: 10.1242/jcs.112.19.3353. [DOI] [PubMed] [Google Scholar]
- Wachtler F, Schwarzacher HG, Smetana K. On the fusion of nucleoli in interphase. European journal of cell biology. 1984;34(1):190–192. [PubMed] [Google Scholar]
- Wachtler F, Stahl A. The nucleolus: A structural and functional interpretation. Micron. 1993;24:473–505. [Google Scholar]
- Walter J, Joffe B, Bolzer A, Albiez H, Benedetti PA, Muller S, Speicher MR, Cremer T, Cremer M, Solovei I. Towards many colors in FISH on 3D-preserved interphase nuclei. Cytogenetic and genome research. 2006;114(3–4):367–378. doi: 10.1159/000094227. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nature reviews Cancer. 2007;7(6):454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zeitz MJ, Mukherjee L, Bhattacharya S, Xu J, Berezney R. A probabilistic model for the arrangement of a subset of human chromosome territories in WI38 human fibroblasts. Journal of cellular physiology. 2009;221(1):120–129. doi: 10.1002/jcp.21842. [DOI] [PubMed] [Google Scholar]
- Zentner GE, Saiakhova A, Manaenkov P, Adams MD, Scacheri PC. Integrative genomic analysis of human ribosomal DNA. Nucleic acids research. 2011;39(12):4949–4960. doi: 10.1093/nar/gkq1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


