Abstract
Adenosine-to-inosine (A-to-I) RNA editing is a conserved post-transcriptional mechanism mediated by ADAR enzymes that diversifies the transcriptome by altering selected nucleotides in RNA molecules1. Although many editing sites have recently been discovered2–7, the extent to which most sites are edited and how the editing is regulated in different biological contexts are not fully understood8–10. Here we report dynamic spatiotemporal patterns and new regulators of RNA editing, discovered through an extensive profiling of A-to-I RNA editing in 8,551 human samples (representing 53 body sites from 552 individuals) from the Genotype-Tissue Expression (GTEx) project and in hundreds of other primate and mouse samples. We show that editing levels in non-repetitive coding regions vary more between tissues than editing levels in repetitive regions. Globally, ADAR1 is the primary editor of repetitive sites and ADAR2 is the primary editor of non-repetitive coding sites, whereas the catalytically inactive ADAR3 predominantly acts as an inhibitor of editing. Cross-species analysis of RNA editing in several tissues revealed that species, rather than tissue type, is the primary determinant of editing levels, suggesting stronger cis-directed regulation of RNA editing for most sites, although the small set of conserved coding sites is under stronger trans-regulation. In addition, we curated an extensive set of ADAR1 and ADAR2 targets and showed that many editing sites display distinct tissue-specific regulation by the ADAR enzymes in vivo. Further analysis of the GTEx data revealed several potential regulators of editing, such as AIMP2, which reduces editing in muscles by enhancing the degradation of the ADAR proteins. Collectively, our work provides insights into the complex cis- and trans-regulation of A-to-I editing.
The prevalence and importance of A-to-I RNA editing have been illuminated in recent years largely owing to the rapid adoption of high-throughput sequencing technologies11,12. Separate laboratories have examined the RNA editome across many tissues or developmental stages in human and other mammals13–17. However, the published studies are limited in the number of samples and tissues examined and do not systematically compare the editing landscape across species or thoroughly dissect the regulation of editing. In this work, we performed multidimensional analyses of thousands of new and publicly available sequencing libraries to address major gaps in our fundamental knowledge of A-to-I editing.
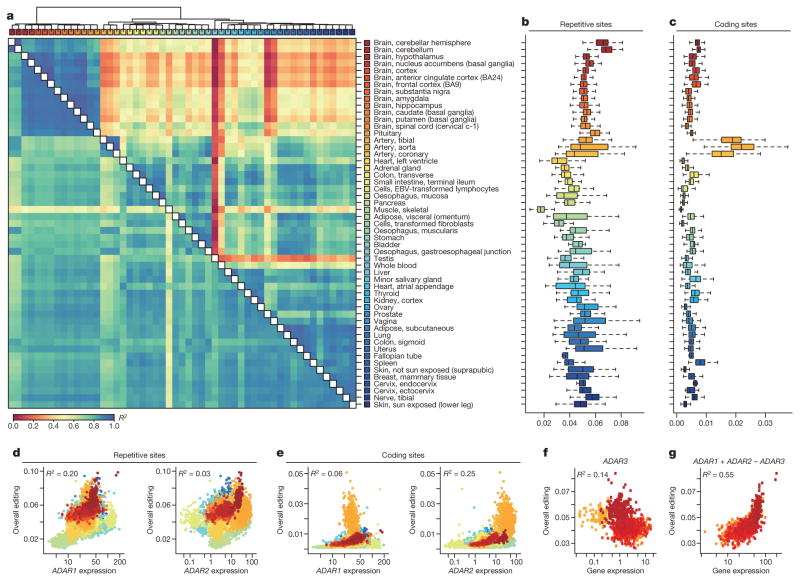
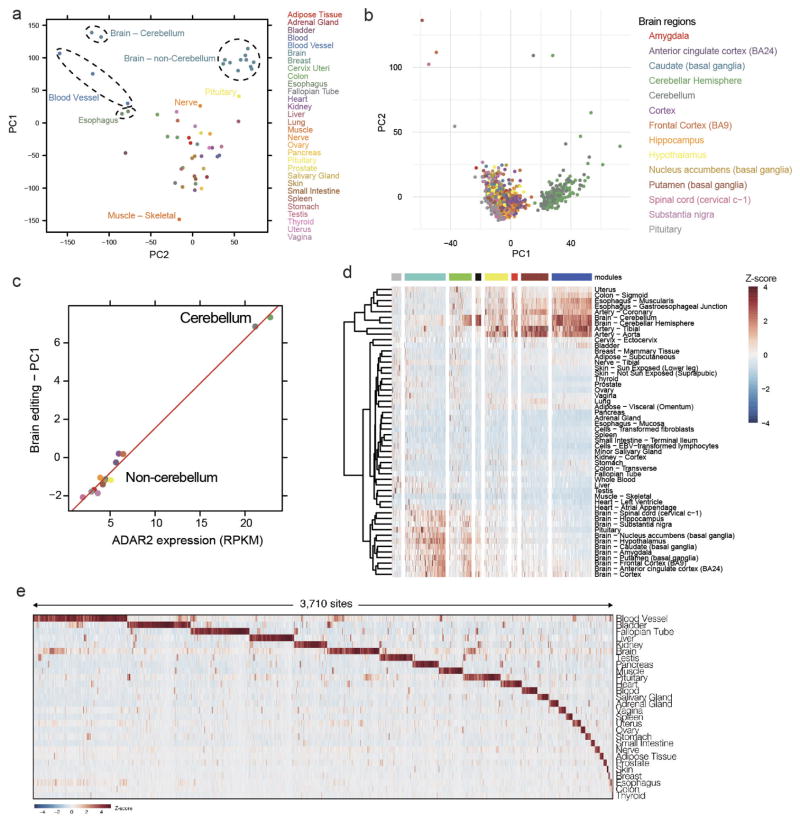
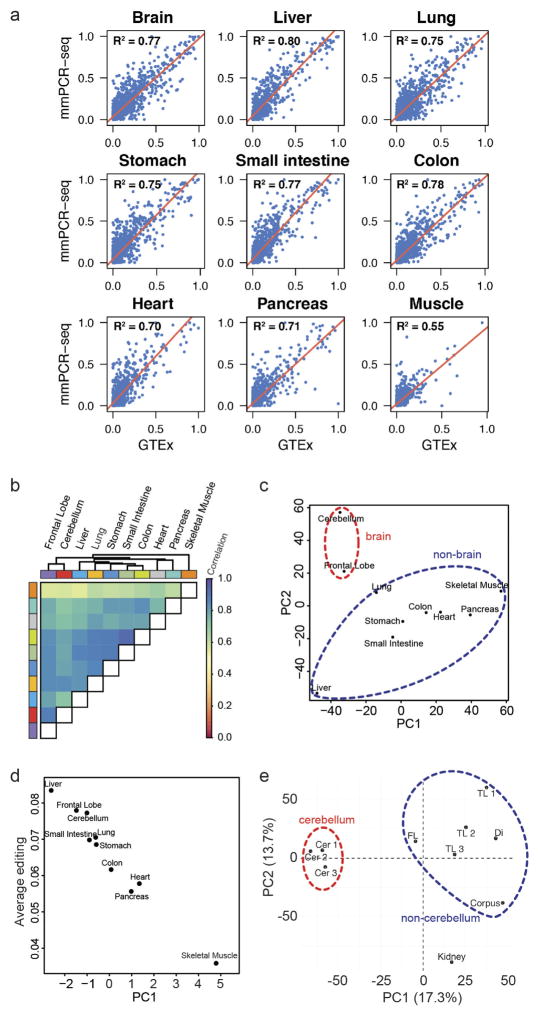
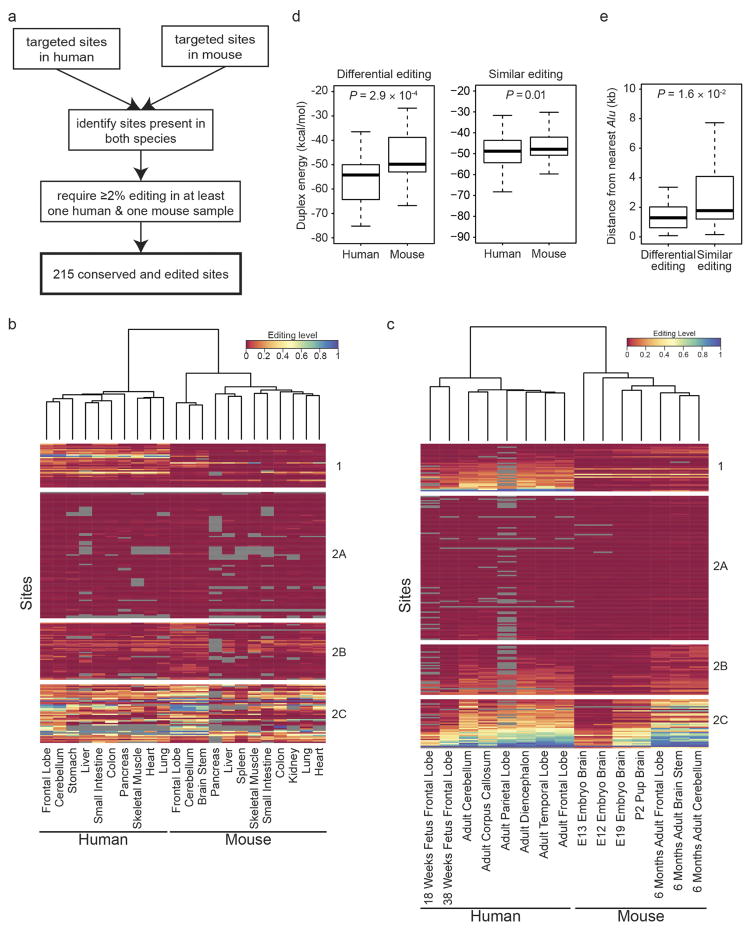
To construct a mammalian reference atlas of A-to-I editing, we first compiled a comprehensive list of editing sites in human and mouse (Supplementary Note 1) and then examined the RNA editome across tissues using 8,551 RNA-sequencing (RNA-seq) samples derived from 552 donors in the GTEx project (Supplementary Information 1). Notably, the editing profiles across different tissues were highly correlated (Fig. 1a) and the overall editing activities were also generally similar, except for skeletal muscle, in which editing was significantly lower than in other tissues (P <2.2 ×10−16, Wilcoxon rank sum test; Fig. 1b). Nevertheless, principal component analysis (PCA) showed that the brain regions could still be resolved from non-brain tissues (Extended Data Fig. 1a). Within the brain, the cerebellum was clearly segregated from other brain parts (Extended Data Fig. 1b), possibly owing to higher expression of ADAR2 (also known as ADARB1) (Extended Data Fig. 1c). When we examined non-repetitive sites in coding regions only, the editing levels became more distinct among the various tissues (Fig. 1a). The different brain regions clustered together, as did heart and skeletal muscle. Unexpectedly, the artery was the most highly edited tissue type (Fig. 1c). The importance of RNA editing in vascular disease was demonstrated in a recent study18. We further validated the results obtained from the GTEx data by applying a targeted sequencing approach (microfluidics-based multiplex PCR and deep sequencing; mmPCR–seq)19 (Supplementary Note 2) to examine 12,871 exonic sites in 672 loci (Supplementary File 2) on independent tissue samples from two individuals (Extended Data Fig. 2).
Figure 1. The GTEx multi-tissue RNA editome.
a, Heat map and dendrogram of Pearson correlations on the editing levels of 53 tissues calculated using all sites (below diagonal) or non-repetitive coding sites only (above diagonal). The dendrogram was drawn based on the distance metric computed by non-repetitive coding sites. The colour codes for GTEx tissues are the same as in a throughout, unless otherwise specified. b, c, Overall editing levels of repetitive (b) or non-repetitive (c) coding sites in various human tissues. Each box plot represents samples from one tissue type. Tissues are in the same order as in a (top to bottom). The overall editing level is defined as the percentage of edited nucleotides at all known editing sites. d, e, Correlations between expression levels of ADAR1 (quantified as the number of RNA-seq reads per kilobase of transcript per million mapped reads (RPKM)) and overall editing levels of either repetitive (d) or non-repetitive (e) coding sites in 8,551 GTEx samples. R2 values were calculated by robust linear regressions on overall editing levels and logarithmic transformed RPKM values. f, Correlation between the expression level of ADAR3 and overall editing level in various brain tissues. g, Correlation of ADAR1 and ADAR2 expression with overall editing of all sites in the brain tissues when the negative influence of ADAR3 was taken into account.
The extent to which variation in editing may be attributed to the expression of each ADAR enzyme is not well understood. From the GTEx data, we found that the expression of ADAR1 (also known as ADAR) accounted for approximately 20% of the variation in overall editing of repetitive sites (Fig. 1d), which represented 97.7% of all known editing sites. By contrast, ADAR2 expression explained 2.8% of the variation (Fig. 1d). However, when non-repetitive protein-coding sites were considered instead, ADAR1 expression accounted for only 6% of the variation, whereas ADAR2 expression accounted for 25% (Fig. 1e). The expression of ADAR3 (also known as ADARB2), which localizes exclusively to the brain and has no enzymatic activity, was negatively correlated with editing levels in brain (Fig. 1f). When the negative influence of ADAR3 was taken into account, ADAR1 and ADAR2 were able to explain better the variation in editing (Fig. 1g), supporting the hypothesis that ADAR3 served predominantly as an inhibitor of editing in the brain, possibly by competing for double-stranded RNA (dsRNA) substrates20.
We next sought to identify groups of individual editing sites that share similar patterns across different tissues. We performed a co-editing network analysis by focusing on 2,094 sites that exhibited higher variation of editing across tissues, and revealed 8 distinct clusters of sites (Supplementary Note 3 and Extended Data Fig. 1d). Additionally, we specifically searched for tissue-specific editing sites and identified 3,710 sites that were edited exclusively or preferentially in only one tissue type (Supplementary Note 3, Extended Data Fig. 1e and Supplementary File 3).
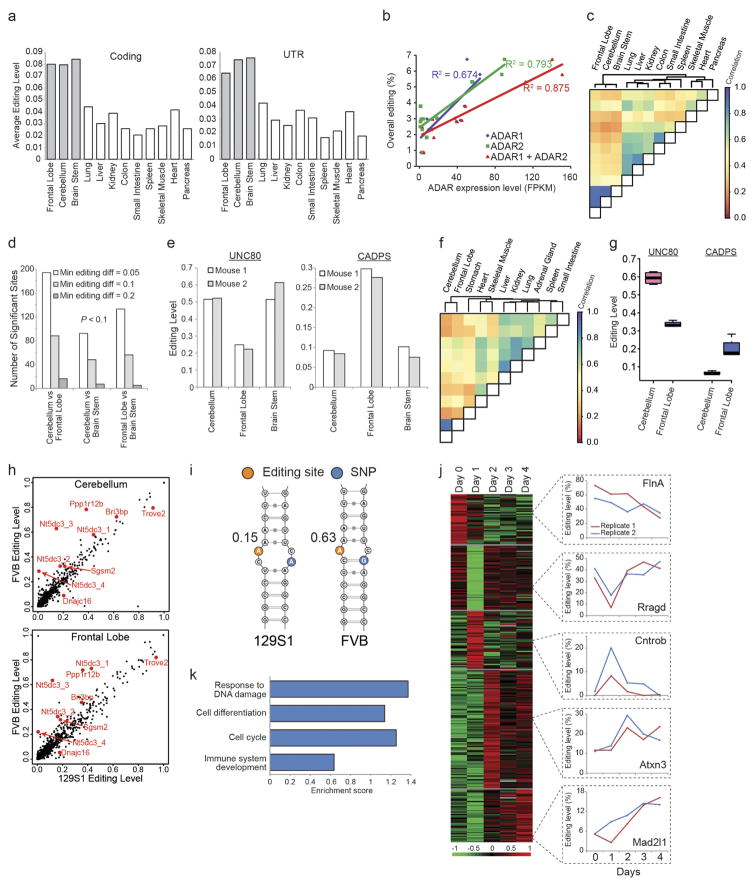
To obtain an expanded view of the A-to-I editing landscape in mammals, we applied mmPCR–seq by interrogating 11,103 exonic sites in 557 loci (Supplementary File 2) to 12 tissue types from several adult mice, and constructed a spatial map of editing in mouse that has both similar and distinct features to that in human (Extended Data Fig. 3a–i, Supplementary Note 4). Overall, we observed comparable spatial editing patterns between human and mouse, although there is less variation among human tissues mainly owing to the presence of Alu repeats. Furthermore, the editing landscape was plastic and responded to external stimuli, as demonstrated by a mouse liver injury model (Extended Data Fig. 3j, k).
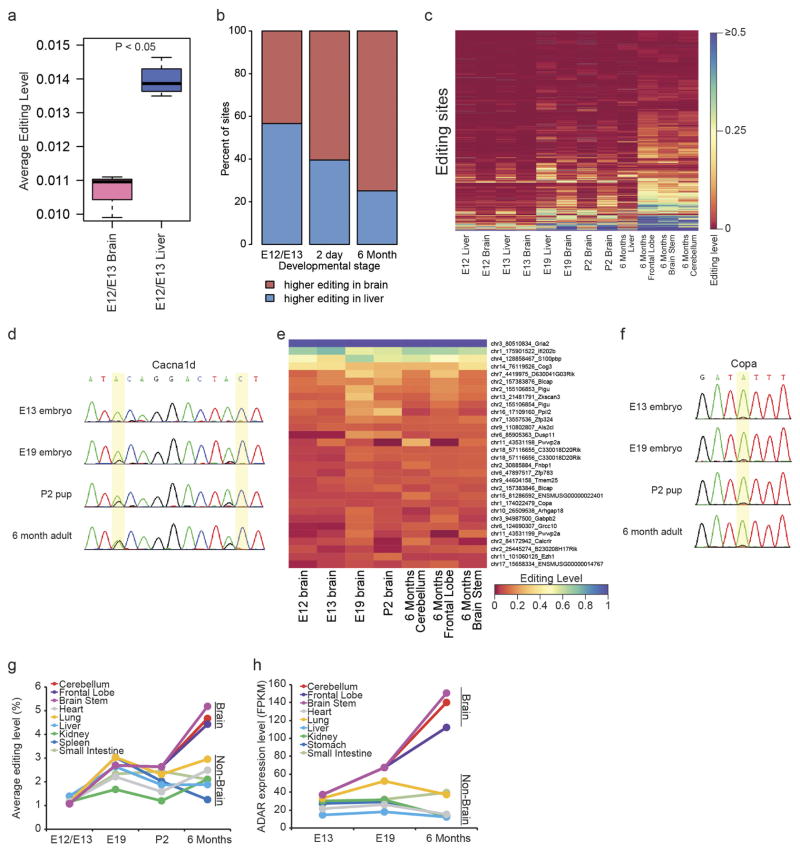
We next assessed the dynamics of RNA editing over mouse development of several tissues using mmPCR–seq. Although the brain was the most highly edited organ in the adult mouse, we found that the fetal liver was more highly edited than the brain during mid-embryogenesis (embryonic day (E) 12.0–E13.0) (Extended Data Fig. 4a). This is consistent with previous findings that an editing-deficient Adar1 mutant mouse dies at around E13.5 owing to failed haematopoiesis in the fetal liver21. Furthermore, we observed that the editing activity mostly increased over development in the brain but not in non-brain tissues (Extended Data Fig. 4b–f), which could be largely explained by expression levels of ADARs (Extended Data Fig. 4g, h). This is consistent with a recent study examining RNA editing in brain development17.
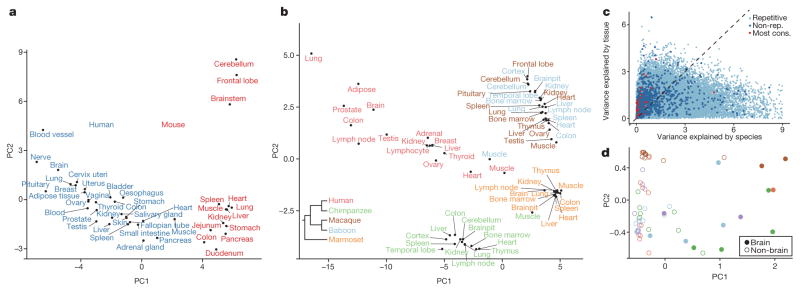
To compare RNA editing between human and mouse, we focused on sites that were conserved between both species. Notably, PCA revealed that the samples were grouped by species rather than by tissue type (Fig. 2a). A similar pattern was observed using mmPCR–seq data (Extended Data Fig. 5a–c). The differentially edited sites between the two species (often higher in human than in mouse) can be explained by the stability of the dsRNA structures (Extended Data Fig. 5d), possibly owing to the proximity of the human sites to Alu repeats (Extended Data Fig. 5e), which often form long double-stranded stem–loops22.
Figure 2. Comparison of A-to-I editing between different mammals.
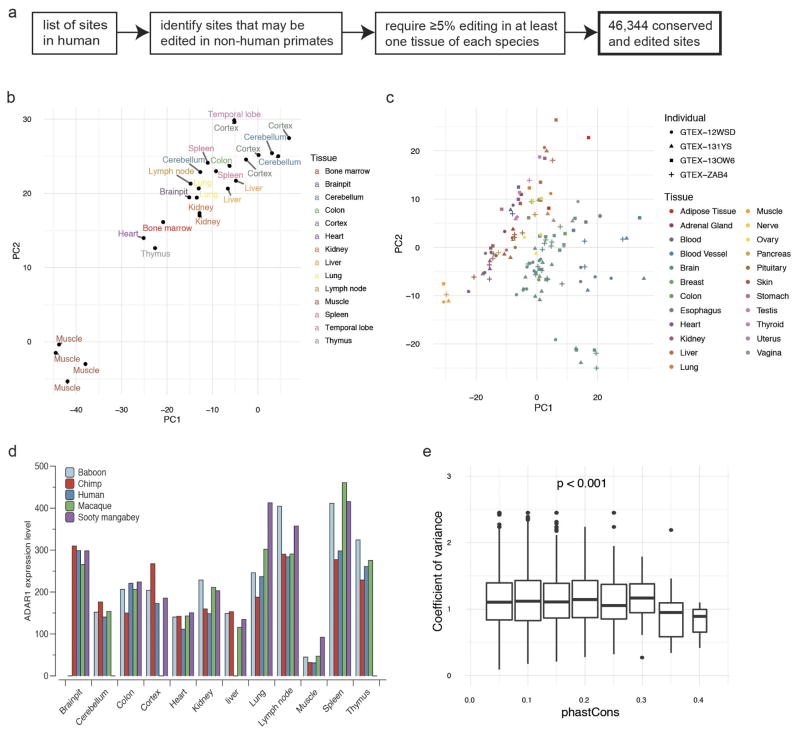
a, PCA on editing levels of various human (red) and mouse (blue) tissues. The editing levels for human were determined by averaging across GTEx samples of the same tissue type for each site. The editing levels for mouse were measured by RNA-seq data of various tissues (see Methods). b, PCA on editing levels of various human and non-human primate tissues. In total, editing levels measured from 68 samples spanning 26 tissue types were used for PCA. We averaged the editing levels for samples of the same tissue type from the same species. Inset, phylogenetic tree of the five primate species under consideration. The editing levels for human and non-human primates were all measured using the RNA-seq data from NHPRTR34 for consistency. c, Estimation of the extent to which variation in editing level of each site could be attributed to differences in tissues or species. The light and dark blue dots represent repetitive and non-repetitive sites, respectively. The red dots represent the 59 conserved sites identified in a recent study23. The variation in editing of these sites was mostly explained by tissue differences, with the exception of an intronic site in BLCAP, which is not edited in non-human primates. d, PCA on editing levels of various human and non-human primate tissues using only the 59 conserved sites. Colours around the circles denote species, using the same colour scheme as in b. The brain samples segregated from the non-brain samples.
Subsequently, we performed cross-species comparisons using datasets from the Non-human Primate Reference Transcriptome Resource (NHPRTR) (Extended Data Fig. 6a, Methods). Again, different NHPRTR samples were largely grouped by species and not by tissue types (Fig. 2b). However, we also found that the editing variance of non-repetitive sites, including most of the 59 highly conserved sites23, were mainly explained by tissue differences (Fig. 2c). When we performed PCA on the highly conserved sites only, we observed separation by tissue types (Fig. 2d). The overall grouping by species was not due to individual-to-individual variability or measurement limitations (Extended Data Fig. 6b, c). In addition, the expression of the ADAR enzymes was similar among species (Extended Data Fig. 6d), suggesting that the pattern was unlikely to be due to species-specific trans-acting factors. We also showed that sites edited similarly between species had more conserved flanking sequences than sites edited differentially (Extended Data Fig. 6e). Collectively, our data suggest that cis-acting elements exert a greater effect on RNA editing than trans-acting factors, consistent with our recent observations in Drosophila24,25, although non-repetitive sites are more directed by trans-acting factors. These results parallel recent findings that RNA splicing is primarily cis-directed26,27 and are in sharp contrast to gene expression programs, which exhibit tissue-specific signatures26,27.
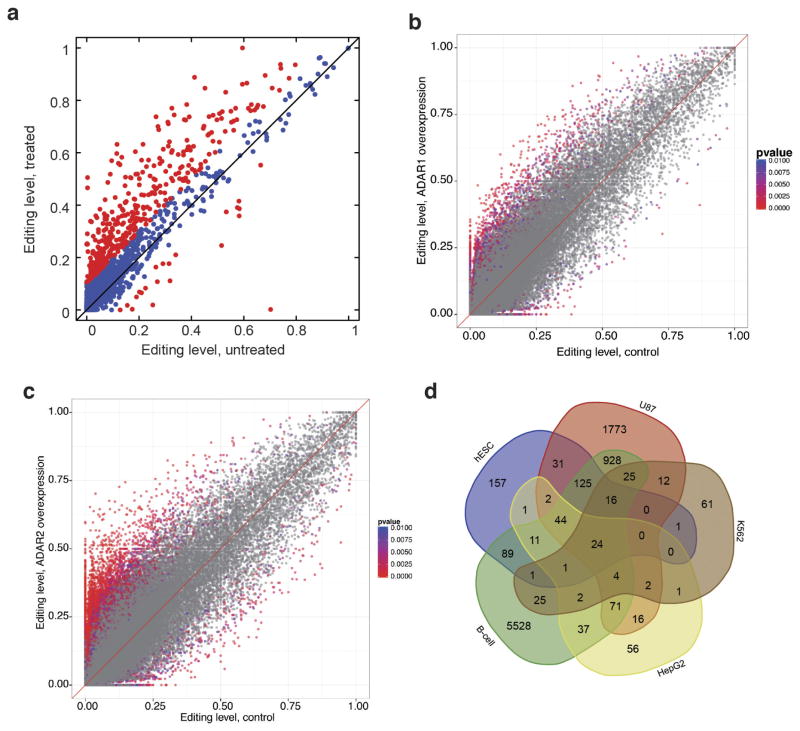
RNA editing in mammals is catalysed by ADAR1 and ADAR2, but their substrates are poorly defined. By perturbing ADAR enzymes in human cells, we curated 9,352 and 1,403 sites that are edited by ADAR1 and ADAR2, respectively, including 262 sites that are edited by both (Extended Data Fig. 7a–d, Supplementary File 4 and Supplementary Note 5). In addition, the editing levels of 73% of ADAR1 targets and 78% of ADAR2 targets are significantly correlated with ADAR1 and ADAR2 expression levels, respectively, in the GTEx data.
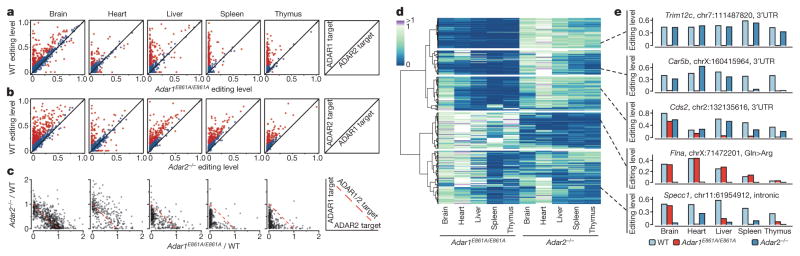
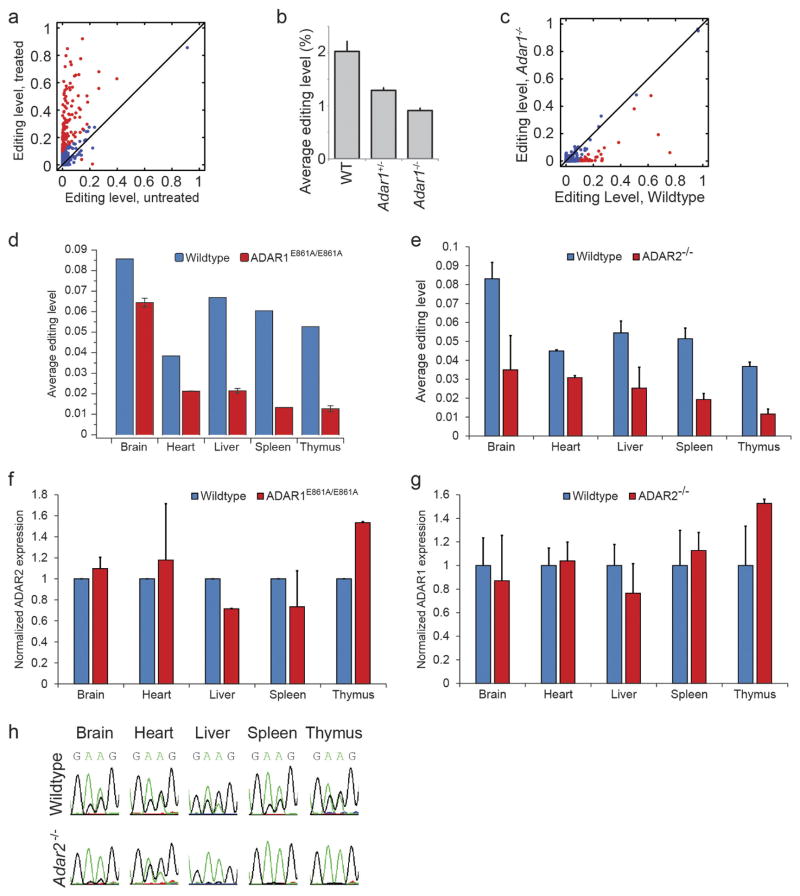
Next, we sought to identify the targets of each ADAR enzyme in mouse using mmPCR–seq not only in cells (Extended Data Fig. 8a) but also in vivo by using various mouse models in which ADAR1 or ADAR2 activity is depleted. To determine ADAR1 targets in vivo, we analysed the Adar1−/− mouse model at E12.0 (Extended Data Fig. 8b, c, Methods) and also several adult tissues from wild-type and Adar1E861A/E861A Ifih1−/− mice21 (Fig. 3a). To determine ADAR2 targets in vivo, we examined multiple adult tissues from wild-type and Adar2−/−Gria2R/R mice (Fig. 3b, Methods). In either ADAR1 or ADAR2 editing-deficient tissue, the average editing level was lower than in the wild-type tissues (P <0.05, Student’s t-test; Extended Data Fig. 8d, e), and expression of the other active ADAR enzyme remained largely unchanged (Extended Data Fig. 8f, g). In total, we curated 1,457 and 976 sites that are edited by ADAR1 and ADAR2, respectively, in mouse, including 698 sites that are edited by both (Supplementary File 5).
Figure 3. Dynamic regulation of RNA editing by ADAR1 and ADAR2.
a, b, Comparison of editing levels in five tissues between wild-type (WT) and Adar1E861A/E861AIfih1−/− adult mice (a), and between wild-type and Adar2−/−Gria2R/R adult mice (b). Sites that are differentially edited (≥10% editing level difference between wild-type and mutant mouse) are marked in red. The diagram on the right illustrates how ADAR1 and ADAR2 targets are determined by comparing their editing levels between wild-type and mutant mice. c, Comparison of editing ratios between Adar1E861A/E861AIfih1−/− and wild-type mice, and between Adar2−/−Gria2R/R and wild-type mice. The editing ratio is defined as the editing level in the ADAR editing-deficient mutant divided by the editing level in the wild-type mouse. The diagram on the right illustrates how ADAR1 and ADAR2 targets are determined by comparing their editing ratios. d, Heat map and dendrogram of hierarchical clustering of editing sites using editing ratios between either Adar1E861A/E861AIfih1−/− or Adar2−/−Gria2R/R and wild-type mice in five adult mouse tissues. The editing sites could be divided into five distinct clusters (1 to 5, top to bottom), three of which exhibit tissue-specific regulation by ADAR1 and ADAR2. Cluster 1, sites mostly affected by ADAR1 but unaffected by ADAR2 in all five tissues; cluster 2, sites mostly affected by ADAR1 in all five tissues while also being affected by ADAR2 in some tissues; cluster 3, sites mostly affected by both ADAR1 and ADAR2 in brain, while mostly by ADAR1 in the other four tissues; cluster 4, sites mostly affected by ADAR2 in all five tissues; cluster 5, sites affected by both ADAR1 and ADAR2 at different levels in five tissues. e, Exemplary sites for each cluster highlighting the complex regulation of A-to-I editing by the ADAR enzymes.
To dissect the interaction in regulation between ADAR1 and ADAR2, we compared the editing ratio of Adar1E861A/E861AIfih1−/− to wild-type mice with the ratio of Adar2−/−Gria2R/R to wild-type mice for each site (Fig. 3c). Globally, we observed that the dependency on different ADAR enzymes varied from tissue to tissue. In the brain, ADAR1 and ADAR2 performed comparable roles, whereas in the liver, spleen and thymus, ADAR1 was the dominant editing enzyme, possibly owing to lower expression levels of ADAR2 in non-brain tissues. In the heart, although ADAR1 functioned as the key enzyme, ADAR2 could also repress the editing of 66 ADAR1 targets. Clustering analysis of the ratios further revealed that the editing sites could be separated into five main groups of regulation that differed in their tissue-specific dependencies on ADAR1 and ADAR2 (Fig. 3d), as illustrated by sites in the Trim12c, Car5b, Cds2, Flna and Specc1 genes (Fig. 3e and Extended Data Fig. 8h). Notably, the dependency of most (62%) of the sites on the editing enzymes varied from tissue to tissue. Collectively, our results revealed an unexpectedly dynamic tissue-specific control of A-to-I editing by ADAR1 and ADAR2, which was not appreciated in previous studies.
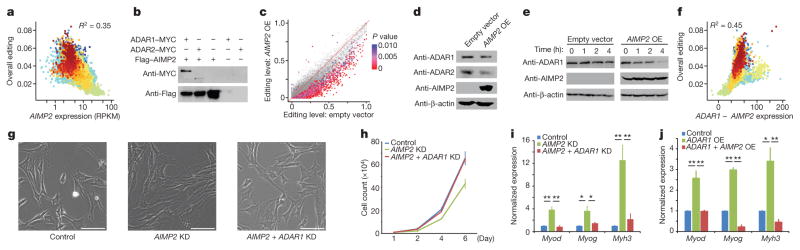
Our work uncovered many spatiotemporal patterns of editing that could not be fully explained by the ADAR enzymes, prompting us to identify factors that help to account for these diverse patterns. Although previous work reported editing effects of the fragile X mental retardation protein FMRP28 and the PIN1 isomerase29, we detected few notable editing changes using several tissues from the knockout mice (Extended Data Fig. 9a, b). Hence, to search for regulators of editing in human, we performed linear regression analysis on the GTEx datasets and identified 144 or 147 genes that had expression levels that were either positively or negatively correlated, respectively, with overall editing levels (Methods, Fig. 4a, Extended Data Fig. 9c and Supplementary File 6). Gene Ontology (GO) analysis revealed that they were significantly enriched for genes with functions in RNA metabolism (Extended Data Fig. 9d). From co-immunoprecipitation experiments in HEK293T cells, we showed that four out of the top six candidates interacted biochemically with either ADAR1 or ADAR2 (Fig. 4b and Extended Data Fig. 9e).
Figure 4. Identification of AIMP2 as a negative regulator of A-to-I editing.
a, Correlation of AIMP2 expression with overall editing of all sites in the GTEx samples. R2 values were calculated by robust linear regressions on overall editing levels and logarithmic transformed RPKM values. b, Co-immunoprecipitation experiment with either MYC-tagged ADAR1 or MYC-tagged ADAR2 and 3×Flag-tagged AIMP2 in HEK293T cells. Anti-Flag M2 beads were used to immunoprecipitate the regulator, and anti-MYC was then used to probe whether the relevant editing enzyme was pulled down together with AIMP2. c, Comparison of editing levels between control cells and cells with AIMP2 overexpressed (OE). The red-purple coloured dots indicate the differentially edited sites (P <0.01, Fisher’s exact test). d, Western blot analysis of ADAR1 and ADAR2 protein levels with or without overexpression of AIMP2 in HEK293T cells. Only the p110 isoform of ADAR1 was detected. e, Cycloheximide-chase analysis followed by western blotting was used to determine the rate at which the ADAR1 p110 protein was degraded with or without AIMP2 overexpression. f, Correlation of ADAR1 expression with overall editing of all sites in the GTEx samples when the negative influence of AIMP2 was taken into account. R2 values were calculated by robust linear regressions on overall editing levels and logarithmic transformed RPKM values. g–i, Effect of knocking down (KD) either AIMP2 alone or both AIMP2 and ADAR1 concurrently in undifferentiated C2C12 myoblasts. Morphology (g), proliferation rate (h) and expression (i) of muscle-specific markers. Myh3, myosin heavy chain; Myod, myogenic differentiation 1; Myog, myogenin. Scale bars, 100 μm. j, Effect of overexpressing ADAR1 alone or both ADAR1 and AIMP2 together in C2C12 myoblasts on the expression of muscle-specific markers. *P <0.05, **P <0.01, Student’s t-test. Error bars denote s.e.m. from three biological replicates.
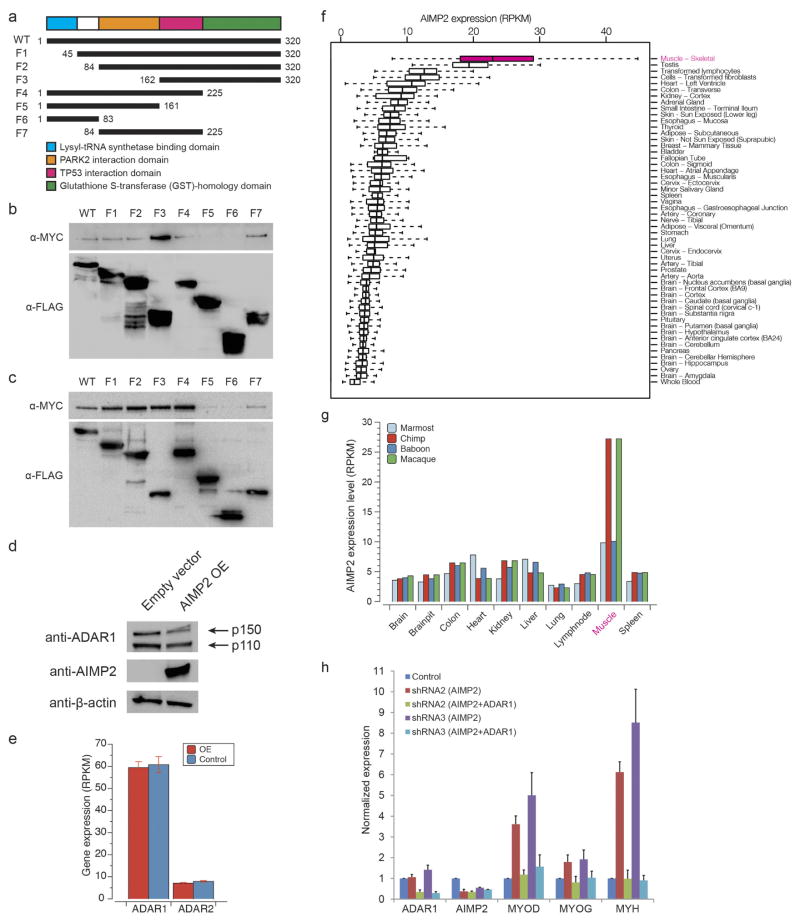
The top candidate negative regulator of editing was AIMP2 (Fig. 4a), which encodes a component of the aminoacyl-tRNA synthetase complex30. AIMP2 interacted with both ADAR1 and ADAR2 (Fig. 4b). Deletion mapping experiments revealed that residues 162–225 of AIMP2 were essential for the interaction (Extended Data Fig. 10a–c). Overexpression of AIMP2 led to a significant reduction in editing at 1,565 sites (P <0.01, Fisher’s exact test; Fig. 4c and Supplementary File 7), and a decrease in ADAR1 and ADAR2 protein levels (Fig. 4d and Extended Data Fig. 10d), although their transcript levels were unaffected (Extended Data Fig. 10e). In addition, when protein synthesis was inhibited by cycloheximide, levels of ADAR1 protein decreased more rapidly in AIMP2-overexpressed cells than in control cells (Fig. 4e). Hence, our results indicated that AIMP2 promotes the degradation of the editing enzymes, consistent with previous work that shows a non-canonical function of AIMP2 in regulating protein stability30.
Our survey of the editing landscape in mammals revealed unusually low editing in skeletal muscle. Intriguingly, of the tissues profiled, the expression level of AIMP2 was highest in skeletal muscle (Extended Data Fig. 10f, g). Furthermore, the expression of ADAR1 and AIMP2 together accounted for 45% of the overall editing differences (Fig. 4f), whereas ADAR1 alone accounted for 20% (Fig. 1d). To investigate the role of AIMP2–ADAR interactions in skeletal muscles, we performed gene perturbation experiments in the C2C12 mouse myoblast cell line. Knockdown of Aimp2 using short hairpin RNAs (shRNAs) altered the cell morphology from fusiform or star-shaped to a more elongated appearance (Fig. 4g), reduced the proliferation of C2C12 cells (Fig. 4h) and promoted the expression of markers normally associated with the transition from myoblasts to myotubes (Fig. 4i). Notably, these phenotypes could be rescued by the simultaneous knockdown of Adar1 with Aimp2 (Fig. 4g–i). Similar results were obtained using other independent shRNAs (Extended Data Fig. 10h). We further confirmed the results by overexpression of Adar1 with or without concomitant over-expression of Aimp2 (Fig. 4j). Hence, our analysis suggests that AIMP2 functions in myoblasts, at least in part, by blocking ADAR1-mediated RNA editing, which has recently been shown to be important for the myoblast-to-myotube transition31.
In summary, our work has afforded an unprecedented view of the dynamic landscape and regulation of RNA editing in mammals. We have demarcated major editing trends across tissues and over development and highlighted key differences in editing between human, non-human primates and mouse. We have identified a new regulator of editing, AIMP2, and determined its role in shaping the RNA editome in mammals. Future studies aimed at uncovering additional cis- and trans-regulators of A-to-I editing are necessary to determine how precise control of editing is achieved in a myriad of biological contexts10,22,32,33.
METHODS
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to allocation during experiments and outcome assessment.
Tissues
All human fetal and adult genomic DNA and RNA were purchased from BioChain Institute. The human tissues were collected post-mortem from individuals with no known medical history. For donor N37, we obtained RNAs for 10 somatic tissues (cerebellum, frontal lobe, heart, lung, liver, stomach, pancreas, colon, small intestine and skeletal muscle) and DNA for 2 somatic tissues (frontal lobe and small intestine). For donor N6, we obtained RNAs for 10 somatic tissues (cerebellum, corpus callosum, diencephalon, frontal lobe, parietal lobe, temporal lobe, kidney, adrenal gland, stomach and small intestine) and DNA for 3 tissues (cerebellum, frontal lobe and stomach). We also purchased additional RNAs and DNAs for adult lung, liver and small intestine, with each sample coming from a different individual. For fetuses F120 and F122, we obtained RNAs for five somatic tissues each (frontal lobe, lung, liver, small intestine and skeletal muscle) and DNA for two somatic tissues each (frontal lobe and small intestine). The RNA integrity numbers (RINs) of all human samples were at least 6.0.
Mouse samples were obtained as follows. Inbred FVB/N mice were purchased from Jackson Laboratory and maintained at Stanford University until they were 30 months old. One-month-old inbred C57BL/6J were purchased from Jackson Laboratory. Tissues from inbred 129S1/SvImJ 6 months old mice were provided by L. Attardi. Additional inbred 129S1/SvImJ male and female mice were purchased from Jackson Laboratory and crossed to obtain embryos and pups. Fmrp-null (also known as Fmr1-null) mice and the corresponding control wild-type mice were purchased from Jackson Laboratory. Pin1-null mice and the corresponding control wild-type mice were genotyped and provided by G. Del Sal and A. Rustighi. Adar1+/− male and female mice35 were crossed to obtain Adar1+/+, Adar1+/− and Adar1−/− embryos, which were genotyped36. Tissues were also obtained from previously published 6-week-old Adar1E861A/E861AIfih1−/− and Adar2−/−Gria2R/R mouse models21,37. To induce acute liver injury, 8-week-old male BALB/cJ mice were administered with a single dose CCl4 (0.4 mg g−1, Sigma) suspended in olive oil and liver biopsies were taken daily for four days. Tissues were flash frozen in liquid nitrogen or dry ice immediately after dissection. All care and procedures were in accordance with the Guide for the Care and Use of Laboratory Animals. All animal experiments were approved by Stanford’s Administrative Panel on Laboratory Animal Care (APLAC) or by the Animal Ethics Committee (AEC) of the athenaeum of the University of Trieste, the Institutional Animal Care and Use Committee of the Wistar Institute, and St Vincent’s Hospital (Melbourne) AEC.
High-quality RNA is important for constructing RNA-seq and mmPCR–seq libraries. Mouse tissues were kept frozen until they were immersed in Trizol or Qiazol and rapidly grounded using a handheld disposable pestle grinder system. After chloroform treatment, cold centrifugation, and retrieval of upper aqueous phase, each sample was purified through an RNeasy column (Qiagen). Concentrations were measured using Nanodrop (Thermo Scientific) and RNA qualities were checked using BioAnalyzer (Agilent). The RIN values of all mouse samples were at least 8.0.
Cell lines and transfection
Cell lines were obtained as follows. HEK293T cells were provided by H. H. Ng. C2C12 cells were provided by S.-C. Ng. 2fTGH cells were from G. Stark. The cell lines were routinely checked by PCR for myco-plasma contamination using the following primers: forward, 5′-GGGAGC AAACAGGATTAGATACCCT-3′; reverse, 5′-TGCACCATCTGTCAC TCTGTTAACCTC-3′.
HEK293T and C2C12 mouse myoblast cells were cultured in DMEM supplemented with 10% fetal bovine serum and penicillin/streptomycin (Life Technologies). Cells were incubated at 37 °C in a humidified 5% CO2 air incubator. For transfection of HEK293T cells, the cells were seeded at 50–60% confluency and next day 1 μg of AIMP2 (either full-length or its fragments) along with 1 μg of ADAR1 or ADAR2 were co-transfected using JETPRIME transfection reagent. The cells were collected 2 days after transfection for protein lysate or RNA preparation.
The samples for our interferon studies were prepared as follows. 2fTGH (wild-type) human cells were seeded at the rate of 5 ×105 cells per well in 6-well plates in DMEM with 10% fetal bovine serum. Interferon treatment (IFNα A/D) was carried out 24 h after seeding at a final concentration of 1,000 U ml−1. After incubation with interferon for 24 h, total RNA was isolated using Trizol (Ambion) following the manufacturer’s protocol. In brief, 1 ml Trizol was added to each well, mixed by pipetting, and collected into 15 ml polypropylene tubes. Chloroform (200 μl) was added, mixed vigorously for 1 min, and allowed to stand at room temperature for 5 min. Samples were centrifuged at 4,000 r.p.m. for 10 min at 4 °C and the aqueous phase was collected. Isopropanol (500 μl) was added to the aqueous fraction, mixed, and allowed to stand at room temperature for 10 min. The RNA precipitate was pelleted by centrifuging at 4,000 r.p.m. for 10 min at 4 °C. The pellet was rinsed with 1 ml 70% ethanol, air dried, and dissolved in RNase-free water. Total RNA was prepared from untreated and IFN-treated mouse embryonic fibroblasts in a similar way. Quantification was carried out using a nanodrop UV spectrophotometer.
Construction of RNA-seq and exome-seq libraries
The Illumina mRNA-seq library preparation workflow was followed with some modifications, as described previously38. The library amplification step was performed with SYBR Green I on a real-time PCR machine to prevent over-amplification. All libraries were quantified using the Qubit dsDNA High Sensitivity Assay Kit (Invitrogen) and sequenced on HiSeq 2000 (Illumina) to produce paired 100-bp reads. For the 1-month-old mouse samples, N6 human samples, as well as the non-N6 and non-N37 human samples, we used custom 3-bp barcodes that were inserted at the ligated end of the adapters. For all the other samples, we used the standard 6-bp Illumina barcodes that were added to each library in the final PCR step.
Genomic DNA from the frontal lobe and small intestine of N37, F120 and F122, as well as N6 cerebellum, N6 frontal lobe, N6 stomach, and all the non-N6 and non-N37 adult human tissues were prepared for exome sequencing. The enrichment of targeted regions was performed using the Agilent SureSelect Human All Exon 50Mb Kit (Agilent Technologies) following manufacturer’s instructions. We also prepared an additional library from the non-N6, non-N37 lung using Nextera DNA sample preparation kit (Epicentre). As with the RNA-seq libraries, the final PCR step was performed on the real-time thermocycler with SYBR Green I in the reaction mix to prevent over-amplification of libraries. Details of all the samples are provided in Supplementary File 8.
Construction of mmPCR–seq libraries
We have previously described our mmPCR–seq method in detail19. In brief, RNAs were reverse transcribed using either SuperScript III (Invitrogen) or iScript advanced reverse transcriptase (Bio-Rad). The cDNAs were purified using the MinElute PCR Purification Kit (Qiagen), with an elution volume of 15 μl or less. For brain samples, at least 200 ng cDNA was loaded into each well of an Access Array microfluidic chip (Fluidigm). For non-brain samples, at least 400 ng cDNA was loaded. The PCR reactions were performed on the Access Array System (Fluidigm) using 5× KAPA2G Multiplex PCR Mix (Kapa Biosystems). The primer sequences for both human and mouse are provided in Supplementary File 2. Barcodes were added in a second round of PCR using Phusion DNA polymerase (Finnzymes). Samples were sequenced on HiSeq 2000 (Illumina) to produce paired 101-bp reads. Details of all the samples are provided in Supplementary File 8.
Pre-amplification of low quantity samples
In some biological models in which material is limited, such as RNA from specific cell types or diseased samples, the samples have to be pre-amplified before loading into the Fluidigm chip. We tested different complexities of pre-amplification (number of pooled primers), different PCR protocols, different amounts of templates used for pre-amplification, different clean-up procedures, and different quantities of cDNA loaded into the Fluidigm chip. We found that the following protocol produces the least amount of undesired PCR products (based on gel electrophoresis) and the highest mapping rates. The low quantity RNAs were reverse transcribed using iScript advanced kit (Biorad) according to the manufacturer’s instructions. Next, the multiplex PCR primers were divided equally into three pools, so that there were approximately 200 primer pairs per pool. Hence, for each sample, three separate pre-amplification reactions have to be carried out. Each pre-amplification reaction consisted of 6 μl 5× KAPA2G Multiplex PCR Mix (Kapa Biosystems), 3 μl cDNA (typically 50–200 ng), and 21 μl pooled primers. The PCR program used was: 95 °C for 10 min, followed by 10–12 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 1 min 30 s, and lastly followed by 72 °C for 2 min. We used the MinElute PCR Purification Kit (Qiagen) to clean up the pre-amplification reactions with a slight modification: after DNA binding using buffer PB from the kit, the columns were washed once with 35% guanidine hydrochloride before the wash with buffer PE. Alternatively, AMPure XP beads can also be used to remove the smaller undesirable by-products. The concentrations of the pre-amplified cDNA were subsequently measured by Nanodrop. For loading into the Fluidigm chip, in contrast to unamplified cDNA where we used 200–2,000 ng per sample, here we loaded only 20–30 ng for each pre-amplified cDNA. After mmPCR–seq, we found that for neuronal samples (such as brain tissues or differentiated neurons), there were minimal undesired amplicons and we could simply use the Qiagen PCR Purification Kit for clean-up. For non-neuronal samples (such as lung tissues), there were still some additional undesired PCR products, which we had to remove by gel extraction. The editing level measurements from pre-amplified samples were highly reproducible and also highly correlated with results obtained from the same samples without pre-amplification (data not shown).
Validations by Sanger sequencing
To validate whether the newly identified editing sites are bona fide and to confirm the editing levels measured by mmPCR–seq, we performed regular PCR to amplify a selection of sites. We used either iQ SYBR Green Supermix (Bio-Rad) or KAPA SYBR FAST Master Mix Universal (Kapa Biosystems) for the PCR reactions. To ensure that even low abundant transcripts can be amplified and sequenced, a touch down PCR program was employed: 95 °C for 3 min, followed by 24 cycles of 95 °C for 15 s, 72 °C to 60 °C (decrement of 0.5 °C every cycle) for 30 s, and 72 °C for 45 s, then followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 45 s, and lastly followed by 72 °C for 2 min. For a handful of sites with low editing levels, the PCR product was inserted into a vector using the TOPO TA Cloning Kit (Invitrogen) and then transformed into Top10 Escherichia coli cells (Invitrogen). At least 30 colonies were picked for each site. All Sanger sequencing was carried out by Sequetech, Eurofins MWG Operon, AITbiotech, or Axil Scientific. Validations are available at http://lilab.stanford.edu/atlas.
Mapping of RNA-seq and mmPCR–seq reads
We adopted our previously published pipeline to accurately map RNA-seq reads onto the genome6,7. In brief, we used BWA39 to align RNA-seq reads to a combination of the reference genome and exonic sequences surrounding known splicing junctions from available gene models. We mapped each of the paired-end reads separately using the commands ‘bwa aln fastqfile’ and ‘bwa samse -n4’. We chose the length of the splicing junction regions to be slightly shorter than the RNA-seq reads to prevent redundant hits (that is, 95 bp for reads of 100 bp length). The reference genomes used were: human (hg19) and mouse (mm9). Gene models were obtained through the UCSC Genome Browser for Gencode, RefSeq, Ensembl, and UCSC Genes. We only considered uniquely mapped reads with mapping quality q >10 and used samtools rmdup40 to remove identical reads (PCR duplicates) that mapped to the same location. Of these identical reads, only the read with the highest mapping quality was retained for further analysis. Unique reads were subjected to local realignment and base score recalibration using the IndelRealigner and Table Recalibration tools from the Genome Analysis Toolkit (GATK)41. The above steps were applied separately to each of our RNA-seq samples for mouse and human. In addition, we downloaded and mapped publicly available RNA-seq data for brain tissues of 10 mouse strains (each with two biological replicates) (ENA: ERP000614) and six tissues for one mouse strain (each with six biological replicates) (ENA: ERP000591). Subsequently, the mapped reads from all samples were combined into one human and one mouse dataset (pooled) for variant calling.
Reads that were produced using the mmPCR–seq protocol were mapped to the genome and splicing junctions in the same way as RNA-seq reads. However, mmPCR–seq samples were not subjected to duplicate removal before local realignment and samples were treated separately (rather than pooled) for the subsequent steps.
GTEx data processing
The GTEx expression data used in this study was obtained from dbGap release of provisional analysis data (12 January 2015 version), which contained 8,555 postmortem samples. The editing level was called on the GTEx v6p release (study accession phs000424.v6.p1; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000424.v6.p1), which contained a total of 9,547 postmortem samples. For all the analyses, we used 8,551 samples that had both expression and editing data available. To call editing level of each site from GTEx RNA-seq, we computed the ratio of G reads divided by the sum of A and G reads for each of the know editing site and only kept the sites of which the sum of A and G reads was higher than 20 for downstream analysis. For each tissue type, we calculated the mean editing level of each site from different individuals and required the mean value should be computed from at least 10 samples. After applying above filters, in total there were 408,580 sites used for downstream analysis of the GTEx data. In addition, we analysed a representative subset of GTEx samples to identify new editing sites. For each tissue type, we chose six samples, with three datasets from males and three datasets from females, and for each gender, one from a young donor (20–35 years old), one from a middle-aged donor (35–55 years old), and one from an old donor (55–70 years old). We chose the RNA-seq datasets that had the deepest sequencing coverage. The whole-genome sequencing (WGS) data for each donor was also obtained from GTEx v6p release to minimize false discoveries. Details of chosen samples are presented in Supplementary File 9.
Identification of editing sites from RNA-seq data
We used the UnifiedGenotyper tool from the Genome Analysis Toolkit (GATK)41 to call variants from the mapped RNA-seq reads. In contrast to the usual practice of variant calling, we identified variants with very loose criteria by using the UnifiedGenotyper tool with options stand_call_conf 0, stand_emit_conf 0, and output mode EMIT_VARIANTS_ ONLY. Human variants at non-repetitive and repetitive non-Alu sites were required to be supported by at least three mismatch reads. A support of one mismatch read was required for variants in human Alu regions. Mouse variants at non- repetitive and repetitive non-Alu sites had to be supported by at least two and three mismatched reads, respectively. This set of variant candidates was subject to several filtering steps that increased the accuracy of editing site discovery. For humans, we removed all known single nucleotide polymorphisms (SNPs) present in the SNP database (dbSNP; except SNPs of molecular type ‘cDNA’; database version 135; http://www.ncbi.nlm.nih.gov/SNP/), the 1000 Genomes Project, and the University of Washington Exome Sequencing Project (http://evs.gs.washington.edu/EVS/). For GTEx samples, we also removed SNPs from the GTEx v6p release WGS data. For mouse, we removed all known SNPs based on annotations from dbSNP, the Sanger Institute42,43, and a recent in-house sequencing study of 10 mouse genomes (M.T.O. et al., unpublished observations). In addition, we removed all mouse RNA-seq variants if they showed any evidence for the same type of variation in the genome of any of the 11 inbred strains sequenced by the Sanger institute42,43. To remove false positive RNA-seq variant calls due to technical artefacts in both human and mouse, further variant filters were applied as previously described6,7. In brief, we (1) required a variant call quality q >20; (2) discarded variants if they occurred in the first six bases of a read; (3) removed variants in repetitive regions; (4) removed intronic variants if they were within 4 bp of splice junctions; and (5) discarded variants in homopolymer runs. Moreover, we removed sites in regions highly similar to other parts of the genome by BLAT44. Finally, variants were annotated using ANNOVAR45 based on gene models for Gencode, RefSeq, Ensembl, and UCSC Genes. The resulting sets of sites identified from our RNA-seq data were combined with all sites available in the RADAR database11 and were subsequently referred to as ‘known’ sites for further analysis by mmPCR-seq.
Identification of editing sites from mmPCR–seq
To identify novel editing sites from our mmPCR–seq samples, we called variants using the GATK UnifiedGenotyper41 and applied the same filters to remove technical artefacts as for the discovery of editing sites from RNA-seq (see above). We applied this procedure to each mmPCR–seq sample individually. Subsequently, we selected variants from each sample by applying a variable minimum variant frequency threshold that resulted in an A-to-G mismatch fraction of >80% per sample. Assuming all non-A-to-G mismatches are false and the error rate for all 12 mismatch types is equal, this resulted in a false discovery rate of (20%/11)/80% =2.3%. Finally, all A-to-G variants found in each sample separately were pooled and reported as ‘novel’ sites.
Expression analysis
The expression of known genes (that is, expected fragments per kilobase of transcript per million fragments mapped (FPKM)) was quantified using Cufflinks246 (parameter -G) on the basis of Tophat247 mappings for all RNA-seq libraries. Gene models for human and mouse were obtained from Ensembl for human (release 72) and mouse (release 67). If a variant overlapped with several gene models, the average of the FPKM values for all overlapping genes was calculated.
Comparison of editing levels in human and mouse mmPCR–seq samples
To determine the overall similarities in editing between samples, we selected sites that were covered by at least 100 reads in mouse samples and at least 300 reads in human samples and that were edited to at least 1% in any of the mouse or human samples respectively. Similarities in editing levels between samples were quantified using a Pearson correlation and are reported as R2 values. Statistical analyses were performed using the R package.
Overall editing levels
To identify the relation between editing levels and expression of the editing enzymes, we determined the overall editing by using the RNA-seq data for each tissue sample in human and mouse. We determined the overall editing as the total number of reads with G at all known editing positions as compared to all reads covering the position (that is, containing A and G nucleotides at the editing position). We did not impose any sequencing coverage criteria, but instead took all sites into account that were used in this study (including sites from the RADAR database11, sites discovered by our own and GTEx RNA-seq, and sites found in the data generated by mmPCR–seq) to obtain the total amount of editing in each sample.
PCA analysis
To identify the major sources of variation in mouse and human tissue samples, we performed PCA. In addition to the criteria that were imposed for correlation analysis (see above), we removed all sites that were missing editing value measurements in more than one-third of the samples. The missing values of the remaining sites were imputed using missForest R package48 with default settings. Then the ‘prcomp’ function in R was used to determine the principal components on the complete dataset.
Co-editing network analysis
Editing levels of individual site were quantified as the number of G reads divided by the total number of A and G reads mapped to an editing site when the latter was higher than 20 reads in more than 10 samples. By applying this criterion, we had 408,580 editing sites quantifiable in the GTEx data, including 369,797 Alu sites, 13,612 repetitive non-Alu sites and 25,171 non-repetitive sites including 2,642 sites in coding regions. To identify sites that are co-edited in different groups of tissues, several criteria were applied for preprocessing of data. We (1) removed sites with too many missing values of samples (≥4 samples, 20,726 sites remained); (2) removed samples with too many missing values of sites (≥50%, cervix–endocervix removed); (3) constructed a sample tree by hierarchical clustering (method =‘average’) and cut the tree (cutHeight =16, minSize =10) to remove outlier(s) from the sample tree (muscle removed); and (4) removed sites with low variance (coefficient of variance <0.8, 2,094 sites remained). We used the WGCNA R package49 to estimate the best soft-thresholding power for the co-editing network construction. The minimum power tested that reached the R2 cut-off of 0.8 for topology model fit was determined as the optimal value. We then calculated the adjacencies with the optimal soft-thresholding power estimated above and transformed the adjacency into a topological overlap matrix to calculate the corresponding dissimilarity. Next, we were able to use hierarchical clustering on the dissimilarity to produce a dendrogram of sites and identified 6 co-editing modules with minModuleSize =30. A heat map was used to show how sites were clustered into the modules and their editing patterns in different tissues (Extended Data Fig. 1f).
Tissue-specific editing analysis
To identify editing sites that are specifically edited in only one tissue, we focused on sites in which the editing level can be detected in at least 10 major tissues with at least 20 sequencing reads in the GTEx data. We then applied the ROKU R package50 to rank the sites by their overall tissue specificity using Shannon entropy and detected tissues specific to each site, if any exists, using an outlier detection method. We required the editing level range (maximum editing level minus minimum editing level) to be higher than 10% and Shannon entropy lower than 0.4 to generate a list of tissue-specific editing sites.
Comparison of editing at conserved sites between human and mouse
To compare editing sites directly between human and mouse, we identified positions that are conserved between human and mouse. For that purpose, we converted the coordinates of all targeted human sites to positions on the mouse reference genome using the liftOver tool and the ‘hg19.ToMm9.over.chain’ file provided by the UCSC Genome Browser (http://genome.ucsc.edu). We repeated the same procedure by converting all mouse sites to positions on the human reference genome using the ‘mm9ToHg19.over.chain’ file. For positions that were successfully lifted over we determined the nucleotide in the query and target genomes using the pairwise alignments in axt format that are provided by the UCSC Genome Browser. We repeated the same procedure 100 times using randomly chosen ‘A’ positions from edited genes to obtain a control for the substitution rates between the two species.
To ensure that all selected positions were truly edited, we chose only sites that were edited in at least one human and one mouse sample by more than 2% for further analysis. The correlations between human and mouse tissues and developmental stages were quantified using Pearson’s R2 value. These correlations served as similarities between samples for a hierarchical clustering using Ward’s minimum variance method as metric. Duplex energies were obtained using the RNAduplex program provided in the Vienna RNA package51.
Comparison of editing at conserved sites between human and non-human primates
To identify positions that are both present and edited in the human and non-human primates genomes, we started with the list of human editing sites and performed a ‘liftover’ process following the order of species in the phylogenetic tree (human, chimpanzee, baboon, rhesus, marmoset), and then repeated in the reverse direction. To calculate the editing levels of the conserved sites, we used the RNA-seq data for human, chimpanzee, baboon, rhesus and marmoset from NHPRTR34,51 and computed the ratio of G reads divided by the sum of A and G reads for each site and kept only the sites with the sum of A and G reads higher than 20. We further required that each site should be edited ≥5% in at least one sample of each tissue. After applying the above filters, in total there were 46,344 conserved and edited sites used for downstream analysis (Extended Data Fig. 10a). For each editing site, to test whether its usage changes more strongly between tissues or between species, we quantified the explained variance by fitting an ANOVA model to each site with tissue and species as explanatory variables and used the sum of squares as the measure of variation52.
Identification of variability in editing between different mouse strains
To identify sites that are differentially edited between the 129S1/SvImJ and FVB/N mouse strains, we applied a strategy that relied on the discovery of editing sites that exhibit consistent differences between the two strains in more than one tissue. More precisely, we first identified editing sites that were reproducible in technical replicates of 129S1/SvImJ cerebellum, 129S1/SvImJ frontal lobe, FVB/N cerebellum, and FVB/N frontal lobe. Reproducibility was determined using Fisher’s exact test with P >0.05 when comparing the numbers of edited and unedited nucleotides between two replicates. Subsequently, we calculated the average editing level between technical replicates at reproducible sites and compared them between the two strains. This comparison was performed independently for both cerebellum and frontal lobe tissues. Sites were required to show a difference of >10% between strains in both tissues to be reported as candidates. To determine structural differences in RNA secondary structures that may be caused by variation between mouse strains and that may affect editing levels, we used the following procedure: First, the RNAduplex software was used to determine candidate editing complementary sequences in a window of ±5 kb of the candidate sites. Second, for regions determined by RNAduplex we created sequences that were specific for each mouse strain by replacing reference nucleotides with genomic variants annotated in each mouse strain. Third, we used IPknot53 to predict a more accurate secondary structure (including pseudoknots) for both sequences separately and investigated the differences in structure that were caused by genomic variants in the two strains.
Identification of ADAR1 and ADAR2 target sites
To discover editing sites that are targets of ADAR1 in mouse, we measured editing levels in wild-type (Adar1+/+) (2 replicates), heterozygous (Adar1+/−) (7 replicates) and null (Adar1−/−) (5 replicates) mouse embryos (E12.0). In addition, we measured editing levels in Adar1+/+Ifih1−/− (5 tissues) and Adar1E861A/E861AIfih1−/− (5 tissues, 2 replicates) adult mouse samples. We required a minimum coverage of 100 reads in each replicate and reported sites to be ADAR1 targets if the editing level measurements between the wild-type (Adar1+/+) replicates and the null (Adar1−/−) replicates were significantly different (P <0.1, Student’s t-test), and if the average editing levels between wild-type and knockout samples differed by at least 5%.
A similar strategy was applied to identify ADAR2 target sites in 5 mouse tissues by comparing wild-type and Adar2−/−Gria2R/R mice21. We required (1) reproducible editing levels between replicates in wild-type and knockout samples (s.d. <10%), (2) a significant difference between wild-type and knockout replicates (P <0.1, Student’s t-test), and (3) a difference of >5% between wild-type and Adar2−/− average editing levels. These criteria were also applied to identify ADAR1 targets from human 2fTGH cells and mouse embryonic fibroblasts treated or untreated with IFNα.
To understand the regulation of editing by ADARs further, we used the Adar1E861A/E861AIfih1−/− and Adar2−/−Gria2R/R mouse tissue data to identify editing sites that were preferentially edited by ADAR1 and ADAR2, respectively, in different contexts. We calculated the ratio between Adar1E861a/E861A and wild-type as well as the ratio between Adar2−/− and wild-type editing levels (knockout/ wild-type ratio). A knockout/wild-type ratio close to 0 signified low editing in mutant mice but higher editing in wild-type mice and therefore a dependence of editing on the corresponding enzyme. Vice versa, a knockout/wild-type ratio close to 1 suggested similar levels of editing in mutant and wild-type mice, and therefore not a dependence of editing on the corresponding enzyme. In some cases, the knockout/wild-type ratio was higher than 1, suggesting the inhibiting role of editing of the corresponding enzyme.
Identification of sites affected by FMRP and PIN1 from wild-type and knockout mice comparisons
To identify sites that differed in their editing levels between wild-type and Fmrp−/− mice, we required that (1) their editing levels were reproducible within biological replicates of wild-type samples and Fmrp−/− samples (s.d. <10%), (2) their editing levels differed significantly between wild-type and Fmrp−/− replicates (P <0.1, Student’s t-test), and (3) the difference between average editing levels was >5% between wild-type and Fmrp−/− samples. For brain tissues, we used four replicates for wild-type and knockout mice each. For non-brain tissues, two biological replicates were available for both wild-type and knockout mice.
We applied the same strategy to discover sites that differed significantly in editing between wildtype and Pin1−/− mice. For that purpose, three biological replicates were available in each tissue for wild-type and knockout mice.
Identification of editing regulator candidates and functional enrichment analysis
To identify genes in which the expression level positively or negatively correlated with overall editing level of different subsets of sites, we applied robust linear regression model to fit the expression levels of every gene in each of the major tissue types of GTEx data to the overall editing levels of all sites, Alu sites, non-Alu repetitive sites, non-repetitive sites and coding sites, respectively. Before fitting the linear models, we required the expression level of each gene to be higher than 1 in at least 60% of the samples, and normalized the expression levels by gene so that the mean =0 and variance =1. Subsequently, we built the linear model between gene expression levels and overall editing levels, taking into consideration the expression level of ADAR1 as an additional variable. We chose the cut-off P value for positive and negative regulator candidates within each tissue using Bonferroni correction (α=0.01). To identify regulators with broader effect on editing, we further required that in at least 8 tissues the candidate’s expression level was significantly correlated with editing levels and no conflicts were found between the tissues. After repeating this test for different categories of editing sites, we combined the results to generate a comprehensive list of 144 positive and 147 negative regulator candidates of RNA editing (Supplementary File 5).
We obtained GO annotation for each of the regulator candidates using BioMart54. To find functional enrichment, we used topGO54 to perform the enrichment test on positive and negative regulators based on gene counts (topNodes =20, nodeSize =5, P <0.01, Fisher’s exact test).
Co-immunoprecipitation
HEK293T cells were co-transfected using JETPrime with Myc-tagged ADAR1 or ADAR2 and Flag-tagged AIMP2 fragments for 48 h. Cells were lysed in RIPA buffer containing 150 mM NaCl, 25 mM Tris-HCl, pH7.2, 1% NP-40, 0.25% sodium dodecyl sulphate (SDS), and 1 mM dithiothreitol (DTT). Phenylmethylsulfonyl fluoride (PMSF) (1 mM final concentration) and protease inhibitor cocktail (Roche) were added freshly before lysis. Anti-Flag M2 beads (Sigma) were washed twice with RIPA buffer and equal amount of cell lysates were added to the beads and incubated overnight at 4 °C. The bound proteins were washed away from unspecific proteins by high-salt buffer containing 250 mM NaCl, 25 mM Tris-HCl, pH 7.2, 0.5% NP-40, 0.1% SDS and 1 mM DTT. The samples were run on 12% SDS–PAGE and transferred onto a nitrocellulose membrane using TurboBlot (Biorad). The blots were probed with anti-Myc (Santa Cruz) and anti-Flag (Sigma) for specific interaction of ADAR1/ADAR2 and AIMP2 respectively.
Cycloheximide chase
To study protein degradation rate of ADAR1, HEK293T cells were split into a 6-well plate at a seeding density of 300,000 cells per well and grown overnight. The cells were then either transfected with an empty p3×-Flag vector or Flag-tagged AIMP2 vector and grown for another 48 h before the addition of cycloheximide. After the addition of cycloheximide (100 μg ml−1), the cells were collected at different times points as indicated and lysed in RIPA buffer (25 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, and 1 mM PMSF). Equal amounts of protein were separated on a 12% SDS–PAGE gel and transferred to a nitrocellulose membrane. The blots were blocked in 5% milk overnight at 4 °C and incubated with anti-ADAR1 (Abcam) and anti-Flag primary antibodies followed by secondary antibody incubation. The blots were developed using Advanta chemiluminescence western blotting solution (Advanta). Anti-β-actin was used as loading control. Images were captured in Biorad Image station and analysed using ImageJ.
Quantitative PCR
Total RNA was extracted using Quick-RNA MiniPrep (Zymo Research) kit following the manufacturer’s protocol. For each sample, 1 μg of total RNA was taken for reverse transcription using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher) using standard protocol. The reverse transcription products were later used to run qPCR using KAPA SYBR FAST qPCR Master Mix (KAPA Biosystems) for specific genes. All assays were run in triplicates and normalized to Gapdh control. Four independent biological replicates were used to calculate the mean and s.e.m.
The following mouse primers were used in real-time PCR experiments: Gapdh forward, ACCACAGTCCATGCCATCAC; Gapdh reverse, TCCACCAC CCTGTTGCTGTA; Adar1 forward, GGGTCTTGATCGGGGAGA; Adar1 reverse, GCTGCCAGAGAGAGGAAGTG; Aimp2 forward, CGTGCTTGGAAA GGACTATG; Aimp2 reverse, ATTCTCGGGCACATTCTTG; Myod forward, CCAGGCACAGGAAGATTG; MYOD_R, CAGCACTCCATGCATATCTC; Myog forward, TTATCATAATATGCCTCG; Myog reverse, GAAGAGACTA GAACAGAT; Myh3 forward, ATCGAAGCTCAGAACCAG; Myh3 reverse, CCCTTGACATATTCTTCCTTTG.
Knockdowns of AIMP2 and ADAR1 in C2C12
We first predicted potential shRNA targets in silico using the following website: http://projects.insilico.us/SpliceCenter/siRNACheck. Subsequently, the following oligonucleotides were ordered from IDT: AIMP2_shRNA1 forward, CCGGCACACACATTCGTCTGTCAAGCTCGAGCTTGACAGACGAATGTGTGTGTTTTTG; AIMP2_ shRNA1 reverse, AATTCAAAAACACACACATTCGTCTGTCAAGCTCGAGCTTGACAGACGAATGTGTGTG; AIMP2_shRNA2 forward, CCGGCAGATGCAGACTTGGACGTAACTCGAGTTACGTCCAAGTCTGCATCTGTTTTTG; AIMP2_shRNA2 reverse, AATTCAAAAACAGATGCAGACTTGGACGTAACTCGAGTTACGTCCAAGTCTGCATCTG; AIMP2_shRNA3 forward, CCGGTAGCCACAAACACATTGGACTCTCGAGAGTCCAATGTGTTTGTGGCTATTTTTG; AIMP2_shRNA3 reverse, AATTCAAAAATAGCCACAAACACATTGGACTCTCGAGAGTCCAATGTGTTTGTGGCTA; ADAR1_ shRNA1 forward, CCGGGCCAAGAACTACTTCAAGAAACTCGAGTTTCTTGAAGTAGTTCTTGGCTTTTTG; ADAR1_shRNA1 reverse, AATTCAAAAAGCCAAGAACTACTTCAAGAAACTCGAGTTTCTTGAAGTAGTTCTTGGC; ADAR1_shRNA2 forward, CCGGGAAGAGCCCAGTTACTACACTCTCGAGAGTGTAGTAACTGGGCTCTTCTTTTTG; ADAR1_shRNA2 reverse, AATTCAAAAAGAAGAGCCCAGTTACTACACTCTCGAGAGTGTAGTAACTGGGCTCTTC. The underlined sequences were the predicted shRNA targets. Next, the forward and reverse oligonucleotides were annealed and cloned into the pLKO.1 lentiviral vector, which was predigested with EcoRI and AgeI. The final constructs were verified by sequencing. For the knockdown experiments, C2C12 cells were grown until 50% confluency in 6-well plates and then infected with the relevant shRNA lentiviruses (with 8 ng ml−1 polybrene). The infected cells were passaged the next day and selected in puromycin for 3 days.
Data availability
Sequencing data have been deposited in the NCBI Sequence Read Archive under accession number SRP039090 and the NCBI Gene Expression Omnibus (GEO) under accession codes GSE87068 and GSE87198. Scripts used to analyse the data and plot the figures are available upon request.
Extended Data
Extended Data Figure 1. Analysis of GTEx RNA-seq data.
a, PCA was applied to the editing levels of all sites in every GTEx body part. The brain tissues were separated from other non-brain tissues. b, A focused PCA of editing in individual brain tissues highlighted that the cerebellum was distinct from other brain regions. c, Correlation between the first editing principal component (PC1) and the expression level of ADAR2 in various brain tissues. d, Co-editing network analysis of 2,094 sites that exhibited high variation across tissues (coefficient of variance >0.8) detected 8 regulatory modules (coloured in grey, turquoise, green, black, yellow, red, brown and blue). e, Heat map of editing levels from sites that are specifically edited in a single human tissue. The editing levels are normalized across samples for each site.
Extended Data Figure 2. Analysis of adult human tissues by mmPCR–seq.
a, Comparisons between mmPCR–seq editing level measurements and RNA-seq data from the GTEx project for different human tissues. R2 values were calculated by simple linear regression. b, Pearson correlations between the editing profiles of different adult human tissues from a single individual (N37), as measured by mmPCR–seq. c, PCA of editing levels in different tissues from N37 revealed that the brain samples were separated from non-brain samples. d, Scatterplot between the loading of PC1 and the average editing level for each N37 tissue. PC1, which explained over 30% of the editing differences between tissues, corresponded to average editing levels of the tissues. Editing activity was lowest in the skeletal muscle of N37, similar to what was observed in the GTEx data. e, PCA of editing in various brain tissues from a single individual (N6) revealed that the cerebellum was distinct from other brain anatomical regions. Cer, cerebellum; Corpus, corpus callosum; Di, diencephalon; FL, frontal lobe; TL, temporal lobe.
Extended Data Figure 3. Analysis of adult mouse tissues by mmPCR–seq.
a, Average editing levels of sites at coding and untranslated region (UTR) positions in 12 mouse tissues from a single individual (129S1 strain). b, Correlations between ADAR expression levels (quantified as the number of RNA-seq fragments per kilobase of transcript per million mapped reads (FPKM)) and overall editing levels in different mouse tissues. The overall editing level is defined as the percentage of edited nucleotides at all known editing sites. c, Pearson correlations for the editing levels of individual sites between various adult mouse tissues (129S1 strain). d, Numbers of significantly differentially edited sites between various brain parts from 129S1 adult mice (n =2 biological replicates). e, Editing levels of two exemplary sites that are differentially edited between various brain parts from 129S1 adult mice (n =2 biological replicates). f, Pearson correlations for the editing levels of individual sites between various adult mouse tissues (FVB strain). g, Editing levels of two exemplary sites that are differentially edited between various brain parts from FVB adult mice (n =4 biological replicates). h, Comparison of editing levels in the cerebellum and frontal lobe between mice of two different genetic backgrounds (129S1 and FVB). The editing levels of sites that are marked in red differ by more than 10% between the two mouse strains in both cerebellum and frontal lobe. Editing levels were calculated as the average between technical replicates at reproducible sites (P >0.05, Fisher’s exact test, for the comparison of edited and unedited nucleotide counts between technical replicates). i, Predicted RNA secondary structure for part of the NT5DC3 3′UTR that contains an SNP (blue) and an editing site (orange). The editing site in the FVB strain (edited at 63%) is located in a more stable dsRNA stem than the same site in the 129S1 strain (edited at 15%). j, Changes in RNA editing levels during a four-day period of liver regeneration after carbon tetrachloride (CCl4)-induced injury in the mouse. A total of 262 editing sites were significantly variable from day 0 to day 4 after injury (P <0.2, ANOVA). k-means clustering revealed that the 262 sites can be divided into five distinct groups with different patterns of editing level changes. For each cluster, an exemplary editing site was shown on the right. k, GO analysis of the genes in which editing was dynamically regulated during liver regeneration. During liver injury, hepatocytes undergo necrosis and the surviving hepatocytes proliferate. The enriched GO terms suggest that RNA editing may have an important role during the reparative process of the liver.
Extended Data Figure 4. Analysis of mouse development by mmPCR–seq.
a, Comparison of average editing levels between mouse brain and liver at mid-embryogenesis stage E12.0–E13.0 (n =4 biological replicates). b, Comparison of RNA editing between mouse brain and liver. At mid-embryogenesis (E12.0–E13.0), most sites are edited at higher levels in the liver than in the brain. However, as development progresses over time (postnatal 2 days and 6 months), the brain becomes the dominant tissue of editing activity instead. c, Heat map of editing levels in mouse liver and brain during development. We observed an overall trend of increased editing over development in brain. d, Sanger validation of two editing sites in the mouse Cacna1d gene that show an increase in editing levels over development. e, A total of 30 sites, in which the editing levels remained stable over development, including the Gria2 Q/R site. These sites were required to have an average editing within the 75th percentile and no significant increase or decrease in editing over development (P >0.02, F-test, and slope <0.01, linear regression). f, Sanger validation of one site in the Copa gene that showed constant editing levels over mouse brain development. g, Average editing levels in different mouse tissues over development. h, ADAR expression levels in different mouse tissues over development.
Extended Data Figure 5. Comparison of human and mouse editing landscapes.
a, Workflow for the identification of 215 editing sites that are targeted in mmPCR–seq and conserved between and edited in human and mouse. b, Heat map showing editing levels of the 215 conserved sites for various human and mouse adult tissues. The tissues (columns) were clustered hierarchically based on correlations of editing levels between them. The dendrogram on top represents the distances between tissue samples. Sites (rows) were clustered into positions that either differed significantly in editing between human and mouse (group 1) (P <0.01, Wilcoxon rank sum test) or were similarly edited between the two species (groups 2A, 2B and 2C). Group 2A: highest editing level <0.04 in both human and mouse; group 2B: 0.04 ≤highest editing level <0.2; group 2C: highest editing level ≥0.2. c, Heat map showing editing levels of the 215 conserved sites for various human and mouse developmental stages. Clustering was performed in a similar manner to that in b, and the same groupings were used. d, RNA duplex free energies for human and mouse sites with differential (group 1) or similar (groups 2A, 2B and 2C) levels of editing. The secondary structures in human displayed significantly lower free energy than those in mouse (P <0.001, Wilcoxon rank sum test) for group 1 sites, which were generally edited at higher levels in human and primarily responsible for the separation of human and mouse in the clustering. e, Distance from nearest Alu element for differentially edited sites (group 1) and similarly edited sites (groups 2A, 2B and 2C). In human, group 1 sites were significantly closer to Alu repeats than group 2 sites (P <0.05, Wilcoxon rank sum test).
Extended Data Figure 6. Comparison of editing landscapes across different primates.
a, Workflow for the identification of 46,344 editing sites that are conserved between and edited in human and non-human primates. b, PCA of editing profiles in various tissues from different chimpanzee individuals. The samples are largely separated by tissue type. c, PCA of editing profiles in various tissues from four human subjects who participated in the GTEx project. We selected the top four individuals with RNA-seq data from the most number of tissue types. d, ADAR1 expression levels in various tissues of human and four non-human primates. e, Distribution of editing variance with sites binned according to the extent to which their surrounding sequences are conserved between different primates. Sites that are more highly conserved between species (high phastCons scores) showed lower variation in editing (low coefficient of variance). PhastCons scores were calculated using 500 bp flanking each editing site. Association test was performed using ANOVA.
Extended Data Figure 7. Identification of ADAR1 and ADAR2 targets in human.
a, Editing levels for human 2fTGH cells that were either untreated or treated with IFNα. Sites that differ in editing by more than 10% between untreated and treated samples are marked in red. GO analysis of the differentially edited sites revealed a functional enrichment for genes involved in viral response or cytokine production, fatty acid metabolism, and intracellular transport. b, Comparison of editing levels between HEK293T cells with ADAR1 overexpression and control cells. P values were calculated using the Fisher’s exact test. c, Comparison of editing levels between HEK293T cells with ADAR2 overexpression and control cells. P values were calculated using the Fisher’s exact test. d, Venn diagram showing number of ADAR1 targets identified from different ADAR1 knockdown cell lines (see Supplementary Note 5 for details).
Extended Data Figure 8. Identification of ADAR1 and ADAR2 targets in mouse.
a, Editing levels for mouse embryonic fibroblasts that were either untreated or treated with IFNα. Sites that differ in editing by more than 10% between untreated and treated samples are marked in red. b, Average editing levels for wild-type, Adar1+/− and Adar1−/− E12.0 mouse embryos. Error bars represent s.d. of two (wild type), seven (Adar1+/−), or five (Adar1−/−) biological replicates. c, Comparison of editing levels between wild-type and Adar1−/− E12.0 mouse embryos. Sites that differ in editing by more than 10% between wild-type and knockout mice are marked in red. d, Average editing levels of sites in different tissues from wild-type and Adar1E861A/E861A mice. Error bars represent s.d. of two biological replicates. e, Average editing levels of sites in different tissues from wild-type and Adar2−/− mice. Error bars represent s.d. of two (heart), four (spleen and thymus), or six (brain and liver) biological replicates. f, Normalized expression levels of Adar2 in various tissues from wild-type and Adar1E861A/E861A mice. Error bars represent s.d. of two biological replicates. g, Normalized expression levels of Adar1 in various tissues from wild-type and Adar2−/− mice. Error bars represent s.d. of two (heart), four (spleen and thymus), or six (brain and liver) biological replicates. h, Chromatograms from Sanger sequencing of two clustered sites on chromosome X at positions 160415964 and 160415965 in the Car5b gene (reverse strand) are shown as examples for different modes of regulation across tissues.
Extended Data Figure 9. Analysis of FMRP, PIN1 and other potential regulators of RNA editing.
a, Comparison of average editing levels in 10 tissues and neural stem cells of wild-type and Fmrp−/− mice at reproducible sites (s.d. <10% in wild-type and Fmrp−/− replicates). Sites that differ by more than 10% in editing levels between wild-type and Fmrp−/− mice are marked in red. b, Comparison of average editing levels in 9 tissues of wild-type and Pin1−/− mice at reproducible sites (s.d. <10% in wild-type and Pin1−/− replicates). Sites that differ by more than 10% in editing levels between wild-type and Pin1−/− mice are marked in red. c, Correlation of the expression levels of the top negative (FASTKD5 and MRPL15) or positive (CLK1, N4BP2L1 and CDKN1B) candidate regulators with overall editing of all sites in the GTEx samples. R2 values were calculated by robust linear regressions on overall editing levels and logarithmic transformed RPKM values. d, GO analysis of the 144 putative positive regulators and 147 putative negative regulators of editing. The top three biological processes that are reported by both DAVID and Panther are given for each set of regulators. e, Both ADAR1 and ADAR2 co-immunoprecipitates with FASTKD5, MRPL15 and N4BP2L1. HEK293T cell lysates were incubated with anti-Flag M2 beads to immunoprecipitate each regulator and concurrently pull down the ADAR enzymes.
Extended Data Figure 10. Characterization of AIMP2 as a negative regulator of RNA editing.
a, Deletion mapping of AIMP2. The schematic diagram depicts the wild-type AIMP2 gene and various fragments (F1–F7) of AIMP2 that were tested for interaction with the ADAR enzymes. The first and last numbers of each construct indicate the amino acid residues that were included in that particular fragment. b, c, Co-immunoprecipitation experiments using anti-Flag M2 beads revealed that only fragments F5 and F6 failed to interact biochemically with ADAR1 (b) and ADAR2 (c), thereby suggesting that the TP53 interaction domain (in pink) is required for AIMP2 to bind with ADAR1 and ADAR2. Additionally, the PARK2 interaction domain (in orange) seems to hinder the interaction of AIMP2 with ADAR1 because its absence in fragment F3 led to an increase in the amount of ADAR1 that was pulled down together with the regulator. d, Western blot analysis showed that overexpression of AIMP2 in MCF7 cells reduced the protein levels of both the p150 and p110 isoforms of ADAR1. e, Expression levels of ADAR1 and ADAR2 in HEK293T cells with or without AIMP2 overexpression, as assayed by RNA-seq. f, Expression levels of AIMP2 in various human tissues from the GTEx RNA-seq datasets. g, Expression levels of AIMP2 in various non-human primate tissues from the NHPRTR RNA-seq datasets. h, Replications of Fig. 4i with independent shRNAs.
Supplementary Material
Acknowledgments
We thank C. Mason and L. Pipes for help with non- human primate RNA-seq data, Y. Hu, P. Sahbaie, A. Chang, K. McGowan and R. Hannibal for technical assistance, and J. Baker and H. H. Ng for use of laboratory resources. We also thank A. Fire, Y. Wan, K. W. K. Sung, W. Zhai, S. Prabhakar, and members of the Li and Tan laboratories for discussions and critical reading of the manuscript. This work is supported by National Institutes of Health (NIH) grants R01GM102484, R01GM124215 and U01HG007593 (J.B.L.), R01GM040536 (K.N.), R01CA175058 (K.N.), and R01AI012520 (C.E.S.), Ellison Medical Foundation (J.B.L. and K.N.), Stanford University Department of Genetics (J.B.L.), Genome Institute of Singapore (M.H.T.), Nanyang Technological University School of Chemical and Biomedical Engineering (M.H.T.), National Medical Research Council OFIRG15nov151 (M.H.T.), the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health (K.N.), MRC and European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement No 621368 (M.A.O’C.), NHMRC project grant 1102006 (C.W. and J.B.L.), Italian Health Ministry (RF-2011-02346976) and the Italian Association for Cancer Research (AIRC) Special Program Molecular Clinical Oncology ‘5 per mille’ (grant no. 10016), AIRC IG (grant no. 17659) (G.D.S.), the Cariplo Foundation (grant no. 2014-0812) (G.D.S.), Stanford Graduate Fellowship (G.R.), German Academic Exchange Service research fellowship (R.P.) and Stanford University School of Medicine Dean’s Fellowship (Q.L., R.P. and R.Z.). The Genotype-Tissue Expression (GTEx) project was supported by the Common Fund of the Office of the Director of the NIH. Additional funds were provided by the NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS. Donors were enrolled at Biospecimen Source Sites funded by NCI\SAIC-Frederick, Inc. (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). The Brain Bank was supported by a supplement to University of Miami grants DA006227 and DA033684 and to contract N01MH000028. Statistical Methods development grants were made to the University of Geneva (MH090941 and MH101814), the University of Chicago (MH090951, MH090937, MH101820 and MH101825), the University of North Carolina-Chapel Hill (MH090936 and MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University St Louis (MH101810), and the University of Pennsylvania (MH101822).
GTEx Consortium
Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group François Aguet1, Kristin G. Ardlie1, Beryl B. Cummings1,2, Ellen T. Gelfand1, Gad Getz1,3, Kane Hadley1, Robert E. Handsaker1,4, Katherine H. Huang1, Seva Kashin1,4, Konrad J. Karczewski1,2, Monkol Lek1,2, Xiao Li1, Daniel G. MacArthur1,2, Jared L. Nedzel1, Duyen T. Nguyen1, Michael S. Noble1, Ayellet V. Segrè1, Casandra A. Trowbridge1, Taru Tukiainen1,2
Statistical Methods groups—Analysis Working Group Nathan S. Abell5,6, Brunilda Balliu6, Ruth Barshir7, Omer Basha7, Alexis Battle8, Gireesh K. Bogu9,10, Andrew Brown11,12,13, Christopher D. Brown14, Stephane E. Castel15,16, Lin S. Chen17, Colby Chiang18, Donald F. Conrad19,20, Nancy J. Cox21, Farhan N. Damani8, Joe R. Davis5,6, Olivier Delaneau11,12,13, Emmanouil T. Dermitzakis11,12,13, Barbara E. Engelhardt22, Eleazar Eskin23,24, Pedro G. Ferreira25,26, Laure Frésard5,6, Eric R. Gamazon21,27,28, Diego Garrido-Martín9,10, Ariel D.H. Gewirtz29, Genna Gliner30, Michael J. Gloudemans5,6,31, Roderic Guigo9,10,32, Ira M. Hall18,19,33, Buhm Han34, Yuan He35, Farhad Hormozdiari23, Cedric Howald11,12,13, Hae Kyung Im36, Brian Jo29, Eun Yong Kang23, Yungil Kim8, Sarah Kim-Hellmuth15,16, Tuuli Lappalainen15,16, Gen Li37, Xin Li6, Boxiang Liu5,6,38, Serghei Mangul23, Mark I. McCarthy39,40,41, Ian C. McDowell42, Pejman Mohammadi15,16, Jean Monlong9,10,43, Stephen B. Montgomery5,6, Manuel Muñoz-Aguirre9,10,44, Anne W. Ndungu39, Dan L. Nicolae36,45,46, Andrew B. Nobel47,48, Meritxell Oliva36,49, Halit Ongen11,12,13, John J. Palowitch47, Nikolaos Panousis11,12,13, Panagiotis Papasaikas9,10, YoSon Park14, Princy Parsana8, Anthony J. Payne39, Christine B. Peterson50, Jie Quan51, Ferran Reverter9,10,52, Chiara Sabatti53,54, Ashis Saha8, Michael Sammeth55, Alexandra J. Scott18, Andrey A. Shabalin56, Reza Sodaei9,10, Matthew Stephens45,46, Barbara E. Stranger36,49,57, Benjamin J. Strober35, Jae Hoon Sul58, Emily K. Tsang6,31, Sarah Urbut46, Martijn van de Bunt39,40, Gao Wang46, Xiaoquan Wen59, Fred A. Wright60, Hualin S. Xi51, Esti Yeger-Lotem7,61, Zachary Zappala5,6, Judith B. Zaugg62, Yi-Hui Zhou60
Enhancing GTEx (eGTEx) groups Joshua M. Akey29,63, Daniel Bates64, Joanne Chan5, Lin S. Chen17, Melina Claussnitzer1,65,66, Kathryn Demanelis17, Morgan Diegel64, Jennifer A. Doherty67, Andrew P. Feinberg35,68,69,70, Marian S. Fernando36,49, Jessica Halow64, Kasper D. Hansen68,71,72, Eric Haugen64, Peter F. Hickey72, Lei Hou1,73, Farzana Jasmine17, Ruiqi Jian5, Lihua Jiang5, Audra Johnson64, Rajinder Kaul64, Manolis Kellis1,73, Muhammad G. Kibriya17, Kristen Lee64, Jin Billy Li5, Qin Li5, Xiao Li5, Jessica Lin5,74, Shin Lin5,75, Sandra Linder5,6, Caroline Linke36,49, Yaping Liu1,73, Matthew T. Maurano76, Benoit Molinie1, Stephen B. Montgomery5,6, Jemma Nelson64, Fidencio J. Neri64, Meritxell Oliva36,49, Yongjin Park1,73, Brandon L. Pierce17, Nicola J. Rinaldi1,73, Lindsay F. Rizzardi68, Richard Sandstrom64, Andrew Skol36,49,57, Kevin S. Smith5,6, Michael P. Snyder5, John Stamatoyannopoulos64,74,77, Barbara E. Stranger36,49,57, Hua Tang5, Emily K. Tsang6,31, Li Wang1, Meng Wang5, Nicholas Van Wittenberghe1, Fan Wu36,49, Rui Zhang5
NIH Common Fund Concepcion R. Nierras78
NIH/NCI Philip A. Branton79, Latarsha J. Carithers79,80, Ping Guan79, Helen M. Moore79, Abhi Rao79, Jimmie B. Vaught79
NIH/NHGRI Sarah E. Gould81, Nicole C. Lockart81, Casey Martin81, Jeffery P. Struewing81, Simona Volpi81
NIH/NIMH Anjene M. Addington82, Susan E. Koester82
NIH/NIDA A. Roger Little83
Biospecimen Collection Source Site—NDRI Lori E. Brigham84, Richard Hasz85, Marcus Hunter86, Christopher Johns87, Mark Johnson88, Gene Kopen89, William F. Leinweber89, John T. Lonsdale89, Alisa McDonald89, Bernadette Mestichelli89, Kevin Myer86, Brian Roe86, Michael Salvatore89, Saboor Shad89, Jeffrey A. Thomas89, Gary Walters88, Michael Washington88, Joseph Wheeler87
Biospecimen Collection Source Site—RPCI Jason Bridge90, Barbara A. Foster91, Bryan M. Gillard91, Ellen Karasik91, Rachna Kumar91, Mark Miklos90, Michael T. Moser91
Biospecimen Core Resource—VARI Scott D. Jewell92, Robert G. Montroy92, Daniel C. Rohrer92, Dana R. Valley92
Brain Bank Repository—University of Miami Brain Endowment Bank David A. Davis93, Deborah C. Mash93
Leidos Biomedical—Project Management Anita H. Undale94, Anna M. Smith95, David E. Tabor95, Nancy V. Roche95, Jeffrey A. McLean95, Negin Vatanian95, Karna L. Robinson95, Leslie Sobin95, Mary E. Barcus96, Kimberly M. Valentino95, Liqun Qi95, Steven Hunter95, Pushpa Hariharan95, Shilpi Singh95, Ki Sung Um95, Takunda Matose95, Maria M. Tomaszewski95
ELSI Study Laura K. Barker97, Maghboeba Mosavel98, Laura A. Siminoff97, Heather M. Traino97
Genome Browser Data Integration & Visualization—EBI Paul Flicek99, Thomas Juettemann99, Magali Ruffier99, Dan Sheppard99, Kieron Taylor99, Stephen J. Trevanion99, Daniel R. Zerbino99
Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California Santa Cruz Brian Craft100, Mary Goldman100, Maximilian Haeussler100, W. James Kent100, Christopher M. Lee100, Benedict Paten100, Kate R. Rosenbloom100, John Vivian100, Jingchun Zhu100
Footnotes
The Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, Massachusetts 02142, USA.
Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
Massachusetts General Hospital Cancer Center and Department of Pathology, Massachusetts General Hospital, Boston, Massachusetts 02114, USA.
Department of Genetics, Harvard Medical School, Boston, Massachusetts 02114, USA.
Department of Genetics, Stanford University, Stanford, California 94305, USA.
Department of Pathology, Stanford University, Stanford, California 94305, USA.
Department of Clinical Biochemistry and Pharmacology, Faculty of Health Sciences, Ben-Gurion University of the Negev, Beer-Sheva 84105, Israel.
Department of Computer Science, Johns Hopkins University, Baltimore, Maryland 21218, USA.
Centre for Genomic Regulation (CRG), The Barcelona Institute for Science and Technology, 08003 Barcelona, Spain.
Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain.
Department of Genetic Medicine and Development, University of Geneva Medical School, 1211 Geneva, Switzerland.
Institute for Genetics and Genomics in Geneva (iG3), University of Geneva, 1211 Geneva, Switzerland.
Swiss Institute of Bioinformatics, 1211 Geneva, Switzerland.
Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA.
New York Genome Center, New York, New York 10013, USA.
Department of Systems Biology, Columbia University Medical Center, New York, New York 10032, USA.
Department of Public Health Sciences, The University of Chicago, Chicago, Illinois 60637, USA.
McDonnell Genome Institute, Washington University School of Medicine, St. Louis, Missouri 63108, USA.
Department of Genetics, Washington University School of Medicine, St. Louis, Missouri 63108, USA.
Department of Pathology & Immunology, Washington University School of Medicine, St. Louis, Missouri 63108, USA.
Division of Genetic Medicine, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.
Department of Computer Science, Center for Statistics and Machine Learning, Princeton University, Princeton, New Jersey 08540, USA.
Department of Computer Science, University of California, Los Angeles, California 90095, USA.
Department of Human Genetics, University of California, Los Angeles, California 90095, USA.
Instituto de Investigação e Inovação em Saúde (i3S), Universidade do Porto, 4200-135 Porto, Portugal.
Institute of Molecular Pathology and Immunology (IPATIMUP), University of Porto, 4200-625 Porto, Portugal.
Department of Clinical Epidemiology, Biostatistics and Bioinformatics, Academic Medical Center, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands.
Department of Psychiatry, Academic Medical Center, University of Amsterdam, 1105 AZ Amsterdam, The Netherlands.
Lewis Sigler Institute, Princeton University, Princeton, New Jersey 08540, USA.
Department of Operations Research and Financial Engineering, Princeton University, Princeton, New Jersey 08540, USA.
Biomedical Informatics Program, Stanford University, Stanford, California 94305, USA.
Institut Hospital del Mar d’Investigacions Mèdiques (IMIM), 08003 Barcelona, Spain.
Department of Medicine, Washington University School of Medicine, St. Louis, Missouri 63108, USA.
Department of Convergence Medicine, University of Ulsan College of Medicine, Asan Medical Center, Seoul 138–736, South Korea.
Department of Biomedical Engineering, Johns Hopkins University, Baltimore, Maryland 21218, USA.
Section of Genetic Medicine, Department of Medicine, The University of Chicago, Chicago, Illinois 60637, USA.
Department of Biostatistics, Mailman School of Public Health, Columbia University, New York, New York 10032, USA.
Department of Biology, Stanford University, Stanford, California 94305, USA.
Wellcome Trust Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford OX3 7BN, UK.
Oxford Centre for Diabetes, Endocrinology and Metabolism, University of Oxford, Churchill Hospital, Oxford OX3 7LE, UK.
Oxford NIHR Biomedical Research Centre, Churchill Hospital, Oxford OX3 7LJ, UK.
Computational Biology & Bioinformatics Graduate Program, Duke University, Durham, North Carolina 27708, USA.
Human Genetics Department, McGill University, Montreal, Quebec H3A 0G1, Canada.
Departament d’Estadística i Investigació Operativa, Universitat Politècnica de Catalunya, 08034 Barcelona, Spain.
Department of Statistics, The University of Chicago, Chicago, Illinois 60637, USA.
Department of Human Genetics, The University of Chicago, Chicago, Illinois 60637, USA.
Department of Statistics and Operations Research, University of North Carolina, Chapel Hill, North Carolina 27599, USA.
Department of Biostatistics, University of North Carolina, Chapel Hill, North Carolina 27599, USA.
Institute for Genomics and Systems Biology, The University of Chicago, Chicago, Illinois 60637, USA.
Department of Biostatistics, The University of Texas MD Anderson Cancer Center, Houston, Texas 77030, USA.
Computational Sciences, Pfizer Inc, Cambridge, Massachusetts 02139, USA.
Universitat de Barcelona, 08028 Barcelona, Spain.
Department of Biomedical Data Science, Stanford University, Stanford, California 94305, USA.
Department of Statistics, Stanford University, Stanford, California 94305, USA.
Institute of Biophysics Carlos Chagas Filho (IBCCF), Federal University of Rio de Janeiro (UFRJ), 21941902 Rio de Janeiro, Brazil.
Department of Psychiatry, University of Utah, Salt Lake City, Utah 84108, USA.
Center for Data Intensive Science, The University of Chicago, Chicago, Illinois 60637, USA.
Department of Psychiatry and Biobehavioral Sciences, University of California, Los Angeles, California 90095, USA.
Department of Biostatistics, University of Michigan, Ann Arbor, Michigan 48109, USA.
Bioinformatics Research Center and Departments of Statistics and Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
National Institute for Biotechnology in the Negev, Beer-Sheva, 84105, Israel.
European Molecular Biology Laboratory, 69117 Heidelberg, Germany.
Department of Ecology and Evolutionary Biology, Princeton University, Princeton, New Jersey 08540, USA.
Altius Institute for Biomedical Sciences, Seattle, Washington 98121, USA.
Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts 02215, USA.
University of Hohenheim, 70599 Stuttgart, Germany.
Huntsman Cancer Institute, Department of Population Health Sciences, University of Utah, Salt Lake City, Utah 84112, USA.
Center for Epigenetics, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
Department of Mental Health, Johns Hopkins University School of Public Health, Baltimore, Maryland 21205, USA.
McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins School of Medicine, Baltimore, Maryland 21205, USA.
Department of Biostatistics, Johns Hopkins University, Baltimore, Maryland 21205, USA.
Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.
Department of Medicine, University of Washington, Seattle, Washington 98195, USA.
Division of Cardiology, University of Washington, Seattle, Washington 98195, USA.
Institute for Systems Genetics, New York University Langone Medical Center, New York, New York 10016, USA.
Department of Genome Sciences, University of Washington, Seattle, Washington 98195, USA.
Office of Strategic Coordination, Division of Program Coordination, Planning and Strategic Initiatives, Office of the Director, NIH, Rockville, Maryland 20852, USA.
Biorepositories and Biospecimen Research Branch, Division of Cancer Treatment and Diagnosis, National Cancer Institute, Bethesda, Maryland 20892, USA.
National Institute of Dental and Craniofacial Research, Bethesda, Maryland 20892, USA.
Division of Genomic Medicine, National Human Genome Research Institute, Rockville, Maryland 20852, USA.
Division of Neuroscience and Basic Behavioral Science, National Institute of Mental Health, NIH, Bethesda, Maryland 20892, USA.
Division of Neuroscience and Behavior, National Institute on Drug Abuse, NIH, Bethesda, Maryland 20892, USA.
Washington Regional Transplant Community, Falls Church, Virginia 22003, USA.
Gift of Life Donor Program, Philadelphia, Pennsylvania 19103, USA.
LifeGift, Houston, Texas 77055, USA.
Center for Organ Recovery and Education, Pittsburgh, Pennsylvania 15238, USA.
LifeNet Health, Virginia Beach, Virginia 23453, USA.
National Disease Research Interchange, Philadelphia, Pennsylvania 19103, USA.
Unyts, Buffalo, New York 14203, USA.
Pharmacology and Therapeutics, Roswell Park Cancer Institute, Buffalo, New York 14263, USA.
Van Andel Research Institute, Grand Rapids, Michigan 49503, USA.
Brain Endowment Bank, Miller School of Medicine, University of Miami, Miami, Florida 33136, USA.
National Institute of Allergy and Infectious Diseases, NIH, Rockville, Maryland 20852, USA.
Biospecimen Research Group, Clinical Research Directorate, Leidos Biomedical Research, Inc., Rockville, Maryland 20852, USA.
Leidos Biomedical Research, Inc., Frederick, Maryland 21701, USA.
Temple University, Philadelphia, Pennsylvania 19122, USA.
Department of Health Behavior and Policy, School of Medicine, Virginia Commonwealth University, Richmond, Virginia 23298, USA.
European Molecular Biology Laboratory, European Bioinformatics Institute, Hinxton CB10 1SD, UK.
UCSC Genomics Institute, University of California Santa Cruz, Santa Cruz, California 95064, USA.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations
Reviewer Information Nature thanks M. Warnefors and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions M.H.T. and J.B.L. conceived the study and provided overall supervision for the project. M.H.T. designed mmPCR–seq primers, with inputs from R.Z. and G.R. M.H.T., J.K., A.N.Y. and J.B.L. performed exome-seq, RNA-seq, and mmPCR–seq experiments. M.H.T. performed validation experiments to verify newly identified editing sites and editing level measurements from RNA-seq and mmPCR–seq. R.S. performed gene perturbation experiments, co-immunoprecipitation experiments, cycloheximide-chase experiments, western blots, muscle cell differentiation experiments, cell proliferation assays, and real-time PCR experiments for the newly identified editing regulators, with help from K.I.L. and M.H.T. M.H.T., Q.L., R.P. and J.B.L. analysed the exome-seq, RNA-seq, and mmPCR–seq data. Q.L. and R.P. led the analysis of the GTEx data and performed cross-species comparisons, with active participation from M.H.T. and J.B.L. K.A., A.G., L.P.K., C.X.G., A.R., N.H., E.A.P., D.L., A.R., Y.P.S.G., A.C., G.D.S., G.P., A.B., D.F.C., C.E.S., M.A.O’C., C.R.W., K.N. and the GTEx Consortium provided samples and data. M.H.T., Q.L. and J.B.L. wrote the manuscript with inputs from the other authors.
References
- 1.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahn JH, et al. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012;22:142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danecek P, et al. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012;13:R26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 5.Peng Z, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 6.Ramaswami G, et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswami G, et al. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behm M, Öhman M. RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet. 2016;32:165–175. doi: 10.1016/j.tig.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Rosenthal JJC, Seeburg PH. A-to-I RNA editing: effects on proteins key to neural excitability. Neuron. 2012;74:432–439. doi: 10.1016/j.neuron.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci. 2013;16:1518–1522. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014;42:D109–D113. doi: 10.1093/nar/gkt996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramaswami G, Li JB. Identification of human RNA editing sites: A historical perspective. Methods. 2016;107:42–47. doi: 10.1016/j.ymeth.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahlstedt H, Daniel C, Ensterö M, Öhman M. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009;19:978–986. doi: 10.1101/gr.089409.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stulić M, Jantsch MF. Spatio-temporal profiling of Filamin A RNA-editing reveals ADAR preferences and high editing levels outside neuronal tissues. RNA Biol. 2013;10:1611–1617. doi: 10.4161/rna.26216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, Xiao X. Genome sequence-independent identification of RNA editing sites. Nat Methods. 2015;12:347–350. doi: 10.1038/nmeth.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picardi E, et al. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci Rep. 2015;5:14941. doi: 10.1038/srep14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang T, et al. Dynamic regulation of RNA editing in human brain development and disease. Nat Neurosci. 2016;19:1093–1099. doi: 10.1038/nn.4337. [DOI] [PubMed] [Google Scholar]
- 18.Stellos K, et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat Med. 2016;22:1140–1150. doi: 10.1038/nm.4172. [DOI] [PubMed] [Google Scholar]
- 19.Zhang R, et al. Quantifying RNA allelic ratios by microfluidic multiplex PCR and sequencing. Nat Methods. 2014;11:51–54. doi: 10.1038/nmeth.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oakes E, Anderson A, Cohen-Gadol A, Hundley HA. Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit b pre-mRNA inhibits RNA editing in glioblastoma. J Biol Chem. 2017;292:4326–4335. doi: 10.1074/jbc.M117.779868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liddicoat BJ, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daniel C, Silberberg G, Behm M, Öhman M. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol. 2014;15:R28. doi: 10.1186/gb-2014-15-2-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto Y, Cohen HY, Levanon EY. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 2014;15:R5. doi: 10.1186/gb-2014-15-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sapiro AL, Deng P, Zhang R, Li JB. Cis regulatory effects on A-to-I RNA editing in related Drosophila species. Cell Reports. 2015;11:697–703. doi: 10.1016/j.celrep.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramaswami G, et al. Genetic mapping uncovers cis-regulatory landscape of RNA editing. Nat Commun. 2015;6:8194. doi: 10.1038/ncomms9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 27.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhogal B, et al. Modulation of dADAR-dependent RNA editing by the Drosophila fragile X mental retardation protein. Nat Neurosci. 2011;14:1517–1524. doi: 10.1038/nn.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcucci R, et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MJ, et al. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet. 2003;34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh CL, et al. ADAR1 deaminase contributes to scheduled skeletal myogenesis progression via stage-specific functions. Cell Death Differ. 2014;21:707–719. doi: 10.1038/cdd.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tariq A, et al. RNA-interacting proteins act as site-specific repressors of ADAR2-mediated RNA editing and fluctuate upon neuronal stimulation. Nucleic Acids Res. 2013;41:2581–2593. doi: 10.1093/nar/gks1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 2013;10:192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pipes L, et al. The non-human primate reference transcriptome resource (NHPRTR) for comparative functional genomics. Nucleic Acids Res. 2013;41:D906–D914. doi: 10.1093/nar/gks1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 37.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 38.Tan MH, et al. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23:201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yalcin B, et al. Sequence-based characterization of structural variation in the mouse genome. Nature. 2011;477:326–329. doi: 10.1038/nature10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stekhoven DJ, Bühlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–118. doi: 10.1093/bioinformatics/btr597. [DOI] [PubMed] [Google Scholar]
- 49.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadota K, Ye J, Nakai Y, Terada T, Shimizu K. ROKU: a novel method for identification of tissue-specific genes. BMC Bioinformatics. 2006;7:294. doi: 10.1186/1471-2105-7-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes A, et al. Drift and conservation of differential exon usage across tissues in primate species. Proc Natl Acad Sci USA. 2013;110:15377–15382. doi: 10.1073/pnas.1307202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato K, Kato Y, Hamada M, Akutsu T, Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27:i85–i93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited in the NCBI Sequence Read Archive under accession number SRP039090 and the NCBI Gene Expression Omnibus (GEO) under accession codes GSE87068 and GSE87198. Scripts used to analyse the data and plot the figures are available upon request.