Abstract
RNA methylation is an abundant modification identified in various RNA species in both prokaryotic and eukaryotic organisms. However, the functional roles for the majority of these methylations remain largely unclear. In eukaryotes, many RNA methylations have been suggested to participate in fundamental cellular processes. Mutations in eukaryotic RNA methylating enzymes, and a consequent change in methylation, often lead to the development of diseases and disorders. In contrast, loss of RNA methylation in prokaryotes can be beneficial to microorganisms, especially under antibiotic pressure. Here we discuss several recent advances in understanding mutational landscape of both eukaryotic and prokaryotic RNA methylating enzymes and their relevance to disease and antibiotic resistance.
Graphical abstract
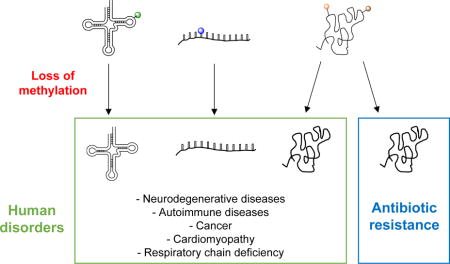
Introduction
RNA methylation is one of the simplest and most abundant post-transcriptional modifications (Figure 1). Methylation is present in all types of RNA and in all kingdoms of life; however, the distribution of various methylation types and their abundance varies among archea, bacteria and eukaryotes [1]. With recent advances in whole-genome sequencing, novel methylation sites are being identified at an increasing rate [2*,3*,4]. Methylations are introduced by RNA methylating enzymes, which are mechanistically diverse and can be divided into three broad groups based on the electronic demand of the substrate. Nucleobase heteroatoms and ribose hydroxyl groups are methylated by enzymes utilizing an SN2 displacement mechanism between the nucleophilic heteroatom and electrophilic methyl group of S-adenosyl-L-methionine (SAM) (e.g. ref [5–7]). Methylation of C5 carbon atoms of cytosine and uridine is accomplished by enzyme-mediated conjugate addition, which builds nucleophilic character at the substrate carbon (e.g. ref [8,9]). Unique among RNA methylating enzymes are those that methylate C2 and C8 carbons of adenosines via a distinctive radical mechanism [10–14].
Figure 1.
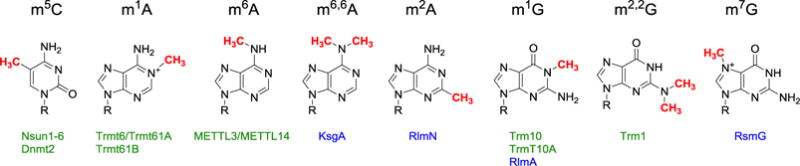
Structures of methylated nucleotides with their corresponding RNA methylating enzymes discussed in this review. Eukaryotic RNA methylating enzymes are noted in green and prokaryotic enzymes in blue. R represents RNA.
Functional roles of numerous RNA methylations are diverse and depend on the location and type of RNA molecule being modified. RNA methylations have been implicated in the regulation of RNA stability, RNA quality control, antibiotic susceptibility, mRNA reading frame maintenance, and immune response, among others. In eukaryotes, nucleotide methylations regulate a wide range of fundamental cellular processes, and mutations in these methylating enzymes have been associated with human diseases, including cancer and neurological disorders [15,16,17*,18–31]. In prokaryotes, many rRNA and tRNA methylations are not essential, and not a single rRNA methylation is critical for cell survival [32*]. Consequently, loss-of-function mutations in RNA methylating enzymes are common and under specific conditions beneficial. In this short review, we will focus on several nucleobase methylations where either gain- or loss-of-function mutations in respective RNA methylating enzymes have been directly linked to human health.
Loss of RNA methylation in prokaryotic ribosomal RNA
The most common rRNA modification in bacteria is methylation of nucleobases. Various methylations are tightly clustered in functional regions of the ribosome: peptidyl-transferase center, decoding centers, and ribosomal subunit interfaces. None of these base methylations are essential; however, they are thought to improve the efficiency and fidelity of mRNA decoding by the ribosome. As the bacterial ribosome represents a major antibiotic target, modulation of rRNA methylation has also emerged as a mechanism of antibiotic resistance. While hypermethylation of antibiotic site has long been recognized as a common way to antagonize antibiotic binding, several instances of antibiotic resistance as a consequence of a loss of physiological methylation of the nucleobase have emerged (Table 1) [33,34]. The resulting aberrant RNA methylation alters the antibiotic binding site, affecting the antibiotic’s ability to inhibit translation. Additionally, lack of most physiological RNA methylations has a minor effect on the cell fitness, enabling bacteria to easily adapt to antibiotic-rich environments. This loss of methylation is conferred by loss-of-function mutations in physiological RNA methylating enzymes, and often results in low-level antibiotic resistance that can support emergence of high-level resistance mechanisms [35**–39]. For example, inactivating mutations in the physiological RNA methyalting enzyme KsgA were first detected in the E. coli isolate resistant to kasugamycin [40]. The kasugamycin resistance was caused by the lack of KsgA-mediated dimethylation at A1518 and A1519 of 16S rRNA [35**]. Though both nucleotides are located far from the drug-binding site, the X-ray crystal structure of the 30S ribosomal subunit from a T. thermophilus KsgA mutant indicates that lack of dimethylation of these two consecutive nucleotides causes a conformational change that likely affects the kasugamycin binding site (Figure 2) [41]. Inactivation of another physiological RNA methylating enzyme RsmG, which methylates m7G527 in 16S rRNA, leads to low-level streptomycin resistance [36,38]. Interestingly, RsmG loss-of-function mutations, detected in the streptomycin-resistant clinical isolates of M. tuberculosis, emerge spontaneously at a high frequency, and support the rapid appearance of other mechanisms that confer high-level resistance to streptomycin [38].
Table 1.
Ribosomal RNA methylating enzymes associated with antibiotic resistance in bacteria, where resistance is conferred by the absence of specific methylations of RNA nucleobases.
| Gene encoding methylating enzyme | Methylated nucleotides (E. coli numbering) | Position of methylation | Antibiotic resistance phenotype |
|---|---|---|---|
| rlmN | A2503, 23S rRNA | C2 | Linezolid |
| Tiamulin | |||
| Virginiamycin M1 | |||
|
| |||
| rlmA1 | G745, 23S rRNA | N1 | Viomycin |
|
| |||
| rlmA11 | G748, 23S rRNA | N1 | Telithromycin |
|
| |||
| ksgA | A1518 & A1519, 16S rRNA | N6, N6 | Kasugamycin |
|
| |||
| rsmG | G527, 16S rRNA | N7 | Streptomycin |
Figure 2.
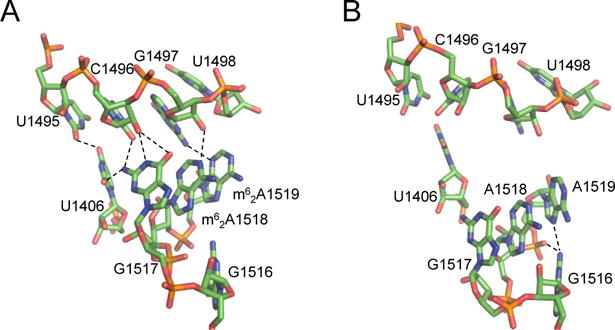
Structural changes induced by the lack of KsgA-mediated methylation in T. thermophilus 30S ribosomal subunit. Methylation of A1518 and A1519 is necessary for the formation of a hydrogen-bonding network between two helices, and stabilization of kasugamycin’s binding site. (A) Crystal structure of the wild-type, fully methylated 30S subunit (PDB 1J5E). Resolution of the structure (3.05 Å) prevented modeling of methylation. (B) Crystal structure of the 30S subunit from ΔksgA strain (PDB 3OTO).
Methylation of G745 in 23S rRNA (m1G745) is a conserved modification in all gram-negative bacteria and is installed by RlmAI. Inactivation of RlmAI, detected in an E. coli isolate, leads to a decrease in the translational efficiency and cell growth, and confers moderate resistance to viomycin [6]. Though viomycin binds to the interface of ribosomal subunits distal from G745, it has been suggested that the presence of m1G at nucleotide 745 could have an allosteric effect on antibiotic binding [42]. Similarly, inactivation of RlmAII, a homolog of RlmAI that methylates G748 (m1G748), leads to low-level resistance to telithromycin and is a common mechanism of resistance detected in clinical isolates of S. pneumoniae [39]. Contrary to viomycin, telithromycin binds in the close proximity to G748, where the methylation of G748 contributes to stabilization of telithromycin binding to the ribosome. Importantly, loss-of-function mutations in RlmAII were detected in all telithromycin-resistant S. pneumoniae clinical isolates reported in a study by Takaya et al., however the extent of the telithromycin resistance depended on the presence of other resistance mechanisms [39].
Finally, inactivating mutation in RlmN, an enzyme that methylates C2 position of A2503 (m2A) in the peptidyltransferase center region of the ribosome, has been shown to cause linezolid resistance in a methicillin-resistant S. aureus (MRSA) clinical isolate [43,44]. Similar loss-of-function mutations accompanied by mutations in 23S rRNA and ribosomal proteins have been observed in clinical isolates of linezolid resistant S. capitis [45]. Recently, we have shown that absence of m2A at A2503 leads to tiamulin resistance [37]. Using directed evolution under antibiotic selection, we have evolved variants of RlmN that confer resistance to tiamulin. These variants act as dominant negative proteins that prevent methylation of 23S rRNA by endogenous RlmN in WT E. coli. Though lack of endogenous m2A leads to low-level antibiotic resistance, it has minimal effect on the cell fitness, and as a result loss-of-function mutations in RlmN can easily emerge [37,43,45]. Furthermore, in combination with other modes of resistance, absence of methylation at A2503 can result in highly resistant pathogens [45]. These findings indicate the importance of monitoring the mutation status of endogenous rRNA methylating enzymes in antibiotic-resistant pathogens.
Alterations in RNA methylation in eukaryotic non-coding RNA
5-methylcytosine
5-Methylcytosine (m5C) is a widespread mark in the transcriptome of eukaryotes [2], and its various biological roles in non-coding RNA (ncRNA) have been recently reviewed in detail [24]. In multicellular organisms, m5C modifications in ncRNAs are installed by DNA methyltransferase homolog Dnmt2 and NSun-domain RNA methyltransferase family members, both of which exhibit broad substrate specificity. For example, Dnmt2, NSun2 and NSun6 methylate cytosolic tRNA. While Dnmt2 methylates position 38 in several tRNAs, NSun2 targets multiple cytosines in the variable loop of most tRNAs [46]. NSun6 methylates cytosine 72 at the 3′ end of the tRNA acceptor stem of cysteine and threonine tRNAs [47]. Lack of m5C in tRNAs leads to endonucleolytic cleavage resulting in tRNA fragments that cause translational defects by interfering with efficient transpeptidation or by inducing codon-specific mistranslation [48,49]. The precise role of m5C mark and its respective RNA methylating enzymes in human health is currently unclear. Both gain-of-function and loss-of-function mutations in RNA methylating enzymes have been detected and linked to development of cancer, autoimmune diseases, and variety of intellectual disability syndromes [15,22,23,25,28,50]. Specifically, loss-of-function mutations in NSun2 have been linked to autosomal-recessive intellectual disability (ARID) and a Dubowitz-like syndrome (Table 2) [15,22,23,28]. Studies in patients were further supported by knockdown and rescue experiments in Drosophila, and additional functional studies in mice showed that these mutations prevent NSun2 localization to the correct cellular organelles [15,28]. On the other hand, both upregulation and gain-of-function mutations in Dnmt2 have been detected in different cancer tissues, where increased activity of Dnmt2 increases the metabolic activity of human cancer cell lines [19,50]. This is the only example of gain-of-function mutation within the scope of this review that we have identified in the literature. A common gain-of-function mutation is a substitution of Glu63 by a Lys. Interestingly, this residue is located on the back-side of Dnmt2, far from the active site. It was suggested that the Glu to Lys substitution and subsequent change in the charge of the residue 63 may either increase the binding of the tRNA substrate or lead to allosteric activation of the enzyme [50]. These possibilities remain to be tested.
Table 2.
Human diseases associated with inactivation of enzymes that modify RNA nucleobases.
| RNA methylation | Type of RNA | Methyltransferase gene | Human disease |
|---|---|---|---|
| m5C | 25 rRNA | nsun1 | Cri-du-chat syndrome [27] |
| tRNA, mRNA | nsun2 | Autosomal recessive intellectual disability [15,22,28] | |
| Dubowitz-like syndrome [23] | |||
| mt tRNA | nsun3 | Cardiomyopathy [51] | |
| Respiratory chain deficiency [51] | |||
| tRNA | dnmt2 | Cancer [19,50] | |
|
| |||
| m1A | mt tRNA | trmt61B | Alzheimer’s disease [26] |
| mt 16S rRNA | ER-negative breast cancer [17] | ||
|
| |||
| m6A | mRNA | mettl3 | Colon cancer [19] |
| mettl14 | Endometrial cancer [16,19] | ||
|
| |||
| m1G | tRNA | trmt10A | Young onset diabetes [20,21] |
| Microcephaly [20,21] | |||
| Epilepsy [21] | |||
| Intellectual disability [20,21] | |||
|
| |||
| m2,2G | tRNA | trmt1 | Autosomal recessive Intellectual disability [29,60] |
Another member of NSun family that methylates tRNA is NSun3. Recently, it was discovered that NSun3-mediated methylation at the wobble cytosine position in mitochondrial tRNAMet is necessary for efficient mitochondrial protein synthesis [51**]. Thus, loss-of-function mutations which abrogate the production of full-length NSun3 lead to the development of mitochondrial respiratory chain disorder (Table 2) [51**].
Recently, several other NSun family members, namely NSun1, 4 and 5, have been described to methylate cytoplasmic and mitochondrial rRNA. The 5-methylcytosine modification in rRNA is crucial for ribosome biogenesis [52]. Loss of NSun1- and NSun4-mediated methylation affects ribosomal subunit and polysome assemblies, leading to inhibition of translation [24,52]. On the other hand, NSun5-mediated methylation of cytoplasmic rRNA does not play a role in polysome assembly but alters translational fidelity under certain conditions [53]. While loss-of-function mutations in these enzymes have not been identified nor associated with any diseases, deletion of NSun1 has been linked to the development of Cri-du-Chat syndrome (Table 2) [27].
N1-methyladenosine
Another prominent and frequently methylated nucleotide in ncRNA is N1-methyladenosine (m1A). Typically found at position 58 in select tRNAs, m1A is installed by the Trmt6/Trmt61A complex in cytoplasmic tRNAs and by Trmt61B in mitochondrial tRNAs. The presence of m1A in tRNA is critical for stabilizing the initiator tRNAMet [54]. N1-methyladenosine is also present at A947 in mitochondrial 16S rRNA and is installed by Trmt61B in all vertebrates [55]. Since this nucleotide is located near one of the intersubunit bridges, it is possible that this Trmt61B-mediated methylation plays a role in mitochondrial translation. Decreased expression of Trmt61B has been observed in Alzheimer’s disease and ER-negative breast cancer (Table 2) [17,26]; however, it is unclear if the global reduction of Trmt61B-mediated methylation or rather hypomethylation of a specific Trmt61B substrate is responsible for disease development.
N1-methylguanosine
One of the less studied ncRNA methylations is 1-methylguanosine (m1G). In yeast, Trm10 is an RNA methylating enzyme responsible for methylation of nucleotide G9 in several tRNAs. Three Trm10 orthologs have been identified in humans, one mitochondrial and two cytoplasmic. Recently identified nonsense mutations in one of the cytoplasmic orthologs, TRMT10A, have been associated with microcephaly, intellectual disability, epilepsy, and adolescent onset diabetes (Table 2) [20,21]. These nonsense mutations lead to either reduction in TRMT10A expression through the introduction of an early stop codon or to a complete loss of TRMT10A activity via a Gly206Arg mutation. Interestingly, loss of catalytic activity is caused by deficient binding of the methyl donor, SAM, while binding of RNA substrate is not affected [20].
N2,N2-dimethylguanosine
Post-transcriptional N2,N2-dimethylguanosine (m2,2G) modification is one of the first identified RNA modifications [56]. It is present at the position 26 in most nuclear and mitochondrial tRNAs, where together with other tRNA modifications assists in proper folding and stability of tRNA [57–59]. In mammals, enzyme responsible for placing this mark is TrmT1. Recently, it was shown that lack of functional TrmT1 leads to alteration in global protein synthesis and perturbation in redox homeostasis, including hypersensitivity to oxidation agents [30]. Inactivation of TrmT1 has been identified as the cause of ARID (Table2) [29,60]. Mutations detected in patients with ARID led to expression of a truncated protein that cannot bind tRNA substrate [30].
Loss of RNA methylation in messenger RNA
The most abundant modified nucleotide in eukaryotic mRNA is N6-methyladenosine (m6A) [61*]. The majority of m6A modifications are localized in 5′-UTRs, around the stop codons, and in 3′-UTRs adjacent to stop codons [4,62]. In accordance with its localization within UTRs, the presence of m6A has been shown to affect mRNA stability and translation [63–65]. This modification has also been found in introns; however, the exact role of m6A in splicing is not understood. In mammals, the deposition of m6A is carried out by a multicomponent methyltransferase complex, which consists of methyltransferase-like 3 protein (METTL3), METTL14, Wilms tumor 1-associated protein (WTAP) and KIAA1429. It was recently demonstrated that METTL3 is the sole methyltransferase, while METTL14 positions RNA substrate for methylation by METTL3 [66*].
The Tyr406Cys loss-of-function mutation in METTL3 has been identified in the patients with colon cancer (Table 2) [19]. This mutation appears to prevent proper interaction between METTL3 and its RNA substrate, thus affecting methylation. Additionally, loss-of-function METTL14 mutations have been identified in patients with endometrial cancer [16,19]. A common mutation is Arg298Pro, where residue 298 is located within a basic patch on the interface between METTL3 and METTL14 and is also adjacent to the METLL3 active site. This mutation seems to affect both the rate of methylation and RNA substrate specificity through allosteric changes [66*].
Two additional methylation marks identified in eukaryotic mRNA are m1A and m5C. While m5C is a well-established eukaryotic mRNA modification [67], the presence of m1A in mRNA has only been recently identified [3*,68]. The biological roles of both of these modifications are unknown, but it has been suggested that m5C is a dynamic mark with a potential regulatory role [68]. Additionally, transcriptome-wide mapping revealed that m1A peaks are highly enriched within 5′-UTRs and in the vicinity of start codons [3*], suggesting that m1A may promote translation. The RNA methylating enzyme responsible for installing m1A mark is currently unknown, and future work will be essential to identify key mediators of m1A methylation and how their misregulation affects human health. The enzyme identified to methylate cytosine at the C5 position in mRNA is a well-known tRNA methyltransferase, NSun2. Presence of inactivating mutations in NSun2 has been linked to several human diseases. Since NSun2 additionally methylates several ncRNAs, it is unclear whether lack of specific Nsun2-mediated methylation or lack of all NSun2-mediated methylations is linked to the development of certain intellectual disorders [15,22,23].
Finally, it is interesting to draw parallels between prokaryotic and eukaryotic mRNA methylation. Recently, it was reported that m6A is also a widespread mark in bacterial mRNA [69]; however, the RNA methylating enzyme responsible for installing this modification is still unknown and no METTL3 or METTL14 homologs have been identified. Importantly, unlike in eukaryotes where m6A is a dynamic modification [70,71], m6A seems to be a stable mark during bacterial growth, suggesting that prokaryotes and eukaryotes regulate this modification by different mechanisms [69]. Additionally, neither m1A nor m5C have been identified in prokaryotic mRNA; however, with the recent discovery of m6A in prokaryotic mRNA, it is possible that m1A and m5C modifications are present in all kingdoms of life as well [69].
Conclusions and future directions
In recent years, our understanding regarding the biological roles of RNA methylations has increased significantly largely due to the identification of RNA methylating enzymes responsible for installing these marks. Changes in activities of RNA methylating enzymes, usually through loss-of-function mutations, have been linked to many diseases. Loss-of-function mutations are more common than the gain-of-function mutations, and interestingly, loss of catalytic activity can be achieved through just a few mutations, often far from the active site. In certain instances, such inactivated enzymes can still retain their ability to bind RNA substrate and act as dominant negative proteins, as is the case for RlmN and TRMT10A. Further structural and biochemical studies are necessary to reveal whether and how these mutations affect SAM binding or catalysis.
Additionally, little is known about the mechanism by which loss of methylation leads to a specific phenotype. For example, with exceptions of the KsgA- and RlmAII-mediated rRNA methylations, it is unknown how loss of rRNA methylation leads to antibiotic resistance. Unfortunately, this mechanism is often understudied, and low-level antibiotic resistance is usually not detected by standard susceptibility testing nor is screened for in clinical settings. Additional studies are necessary to understand how lack of modifications in rRNAs affects the structure and function of the prokaryotic ribosome, as these mechanisms represent a gateway to high-level, clinically relevant resistance. Similarly, it remains largely unknown how changes in RNA methylation in eukaryotes contribute to disease pathology and whether these changes are causative of the disease or simply a result of disease-induced cellular alterations.
Lastly, new advances are needed to further our understanding of RNA methylation. These include new transcriptome-wide methods to detect RNA methylations, especially in low abundant RNAs, and new strategies to identify RNA methylating enzymes since several methylation sites have yet undiscovered cognate enzymes responsible for placing these marks. Improvements of recently developed techniques, such as RNA-immunoprecipitation methods followed by next generation sequencing, are likely to accelerate this area of research [2,4,62,72]. Additionally, identification of the regulatory pathways that control the activity of RNA methylating enzymes as well as identification of RNA demethylases for dynamic RNA methylation marks will be necessary to truly understand the biological function of RNA methylations. Ultimately, this knowledge will aid the development of small-molecule inhibitors for pharmacological modulation of RNA methylation in disease.
Highlights.
-
-
Methylations of nucleobases regulate a wide range of fundamental cellular processes.
-
-
Distribution of RNA methylation is not uniform across different kingdoms of life.
-
-
Mutations that modulate the activity of eukaryotic RNA methylating enzymes are linked to human diseases.
-
-
Loss-of-function mutations in prokaryotic RNA methylating enzymes can be beneficial in the presence of antibiotics.
Acknowledgments
We thank James Longbotham and Kaitlyn Tsai for comments on the review, and the National Institute of Health (R01AI095393) for research support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 2*.Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. This study developed a novel method to globally map m5C in RNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3*.Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. This study developed a novel method to map m6A in the mammalian transcriptome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 5.Dunkle JA, Vinal K, Desai PM, Zelinskaya N, Savic M, West DM, Conn GL, Dunham CM. Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA. Proc Natl Acad Sci U S A. 2014;111:6275–6280. doi: 10.1073/pnas.1402789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gustafsson C, Persson BC. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen LH, Kirpekar F, Douthwaite S. Recognition of nucleotide G745 in 23S ribosomal RNA by the rrmA methyltransferase. J Mol Biol. 2001;310:1001–1010. doi: 10.1006/jmbi.2001.4836. [DOI] [PubMed] [Google Scholar]
- 8.Agarwalla S, Stroud RM, Gaffney BJ. Redox reactions of the iron-sulfur cluster in a ribosomal RNA methyltransferase, RumA: optical and EPR studies. J Biol Chem. 2004;279:34123–34129. doi: 10.1074/jbc.M405702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–11225. doi: 10.1021/bi026055q. [DOI] [PubMed] [Google Scholar]
- 10.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 11.Grove TL, Livada J, Schwalm EL, Green MT, Booker SJ, Silakov A. A substrate radical intermediate in catalysis by the antibiotic resistance protein Cfr. Nat Chem Biol. 2013;9:422–427. doi: 10.1038/nchembio.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCusker KP, Medzihradszky KF, Shiver AL, Nichols RJ, Yan F, Maltby DA, Gross CA, Fujimori DG. Covalent intermediate in the catalytic mechanism of the radical S-adenosyl-L-methionine methyl synthase RlmN trapped by mutagenesis. J Am Chem Soc. 2012;134:18074–18081. doi: 10.1021/ja307855d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan F, Fujimori DG. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc Natl Acad Sci U S A. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli-Nieh S, Cremer K, et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Hum Genet. 2012;90:847–855. doi: 10.1016/j.ajhg.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell DW. Novel genetic targets in endometrial cancer. Expert Opin Ther Targets. 2014;18:725–730. doi: 10.1517/14728222.2014.909414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Couch FJ, Kuchenbaecker KB, Michailidou K, Mendoza-Fandino GA, Nord S, Lilyquist J, Olswold C, Hallberg E, Agata S, Ahsan H, et al. Identification of four novel susceptibility loci for oestrogen receptor negative breast cancer. Nat Commun. 2016;7:11375. doi: 10.1038/ncomms11375. This study implicated TRMT16B in ER-negative breast cancer aetiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dezi V, Ivanov C, Haussmann IU, Soller M. Nucleotide modifications in messenger RNA and their role in development and disease. Biochem Soc Trans. 2016;44:1385–1393. doi: 10.1042/BST20160110. [DOI] [PubMed] [Google Scholar]
- 19.Forbes SA, Beare D, Gunasekaran P, Leung P, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillis D, Krishnamohan A, Yaacov B, Shaag A, Jackman JE, Elpeleg O. TRMT10A dysfunction is associated with abnormalities in glucose homeostasis, short stature and microcephaly. J Med Genet. 2014;51:581–586. doi: 10.1136/jmedgenet-2014-102282. [DOI] [PubMed] [Google Scholar]
- 21.Igoillo-Esteve M, Genin A, Lambert N, Desir J, Pirson I, Abdulkarim B, Simonis N, Drielsma A, Marselli L, Marchetti P, et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 2013;9:e1003888. doi: 10.1371/journal.pgen.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komara M, Al-Shamsi AM, Ben-Salem S, Ali BR, Al-Gazali L. A Novel Single-Nucleotide Deletion (c.1020delA) in NSUN2 Causes Intellectual Disability in an Emirati Child. J Mol Neurosci. 2015;57:393–399. doi: 10.1007/s12031-015-0592-8. [DOI] [PubMed] [Google Scholar]
- 23.Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, Frye M, Al-Gazali L, Gleeson JG. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet. 2012;49:380–385. doi: 10.1136/jmedgenet-2011-100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popis MC, Blanco S, Frye M. Posttranscriptional methylation of transfer and ribosomal RNA in stress response pathways, cell differentiation, and cancer. Curr Opin Oncol. 2016;28:65–71. doi: 10.1097/CCO.0000000000000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross KA. Coherent somatic mutation in autoimmune disease. PLoS One. 2014;9:e101093. doi: 10.1371/journal.pone.0101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekar S, McDonald J, Cuyugan L, Aldrich J, Kurdoglu A, Adkins J, Serrano G, Beach TG, Craig DW, Valla J, et al. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging. 2015;36:583–591. doi: 10.1016/j.neurobiolaging.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Q, Niebuhr E, Yang H, Hansen L. Determination of the ‘critical region’ for cat-like cry of Cri-du-chat syndrome and analysis of candidate genes by quantitative PCR. Eur J Hum Genet. 2005;13:475–485. doi: 10.1038/sj.ejhg.5201345. [DOI] [PubMed] [Google Scholar]
- 28.Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, et al. Mutation in NSUN2, which Encodes an RNA Methyltransferase, Causes Autosomal-Recessive Intellectual Disability. American Journal of Human Genetics. 2012;90:856–863. doi: 10.1016/j.ajhg.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davarniya B, Hu H, Kahrizi K, Musante L, Fattahi Z, Hosseini M, Maqsoud F, Farajollahi R, Wienker TF, Ropers HH, et al. The Role of a Novel TRMT1 Gene Mutation and Rare GRM1 Gene Defect in Intellectual Disability in Two Azeri Families. PLoS One. 2015;10:e0129631. doi: 10.1371/journal.pone.0129631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dewe JM, Fuller BL, Lentini JM, Kellner SM, Fu D. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol Cell Biol. 2017:e00214–00217. doi: 10.1128/MCB.00214-17. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA. 2017 doi: 10.1261/rna.063503.117. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32*.Sergeeva OV, Bogdanov AA, Sergiev PV. What do we know about ribosomal RNA methylation in Escherichia coli? Biochimie. 2015;117:110–118. doi: 10.1016/j.biochi.2014.11.019. This review summarizes our knowledge regarding RNA modifications present in E. coli rRNA. [DOI] [PubMed] [Google Scholar]
- 33.Vester B, Long KS. Antibiotic Resistance in Bacteria Caused by Modified Nucleosides in 23S Ribosomal RNA. In: Grosjean H, editor. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Landes Bioscience; 2009. pp. 537–549. [Google Scholar]
- 34.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 35.Poldermans B, Goosen N, Van Knippenberg PH. Studies on the function of two adjacent N6, N6-dimethyladenosines near the3′ end of 16 S ribosomal RNA of Escherichia coli. I. The effect of kasugamycin on initiation of protein synthesis. J Biol Chem. 1979;254:9085–9089. [PubMed] [Google Scholar]
- 36.Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HI, Barry CE., 3rd Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55:2515–2522. doi: 10.1128/AAC.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37**.Stojkovic V, Noda-Garcia L, Tawfik DS, Fujimori DG. Antibiotic resistance evolved via inactivation of a ribosomal RNA methylating journal. Nucleic Acids Res. 2016;44:8897–8907. doi: 10.1093/nar/gkw699. In this study, authors used directed evolution under antibiotic selection to obtain RlmN variants that mediate low-level antibiotic resistance by acting as dominant negative proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol. 2007;63:1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x. [DOI] [PubMed] [Google Scholar]
- 39.Takaya A, Sato Y, Shoji T, Yamamoto T. Methylation of 23S rRNA nucleotide G748 by RlmAII methyltransferase renders Streptococcus pneumoniae telithromycin susceptible. Antimicrob Agents Chemother. 2013;57:3789–3796. doi: 10.1128/AAC.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helser TL, Davies JE, Dahlberg JE. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat New Biol. 1971;233:12–14. doi: 10.1038/newbio233012a0. [DOI] [PubMed] [Google Scholar]
- 41.Demirci H, Murphy Ft, Belardinelli R, Kelley AC, Ramakrishnan V, Gregory ST, Dahlberg AE, Jogl G. Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA. 2010;16:2319–2324. doi: 10.1261/rna.2357210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 43.Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, Holmes NE, Rhee HW, Hong JI, Hartland EL, et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaMarre JM, Howden BP, Mankin AS. Inactivation of the indigenous methyltransferase RlmN in Staphylococcus aureus increases linezolid resistance. Antimicrob Agents Chemother. 2011;55:2989–2991. doi: 10.1128/AAC.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takaya A, Kimura A, Sato Y, Ishiwada N, Watanabe M, Matsui M, Shibayama K, Yamamoto T. Molecular characterization of linezolid-resistant CoNS isolates in Japan. J Antimicrob Chemother. 2015;70:658–663. doi: 10.1093/jac/dku443. [DOI] [PubMed] [Google Scholar]
- 46.Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 47.Haag S, Warda AS, Kretschmer J, Günnigmann MA, Höbartner C, Bohnsack MT. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA. 2015;21:1532–1543. doi: 10.1261/rna.051524.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G, Reitter S, Liebers R, Stoecklin G, Grone HJ, et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. Embo Journal. 2015;34:2350–2362. doi: 10.15252/embj.201591382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elhardt W, Shanmugam R, Jurkowski TP, Jeltsch A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie. 2015;112:66–72. doi: 10.1016/j.biochi.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 51**.Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun. 2016;7:12039. doi: 10.1038/ncomms12039. This study identified RNA target of NSun3 and linked m5C in RNA with energy metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Metodiev MD, Spahr H, Polosa PL, Meharg C, Becker C, Altmueller J, Habermann B, Larsson NG, Ruzzenente B. NSUN4 Is a Dual Function Mitochondrial Protein Required for Both Methylation of 12S rRNA and Coordination of Mitoribosomal Assembly. Plos Genetics. 2014;10 doi: 10.1371/journal.pgen.1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schosserer M, Minois N, Angerer TB, Amring M, Dellago H, Harreither E, Calle-Perez A, Pircher A, Gerstl MP, Pfeifenberger S, et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nature Communications. 2015;6 doi: 10.1038/ncomms7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281–1290. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, Blumberg A, Schlesinger O, Bieri P, Greber B, et al. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 2016;14:e1002557. doi: 10.1371/journal.pbio.1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith JD, Dunn DB. The occurrence of methylated guanines in ribonucleic acids from several sources. Biochem J. 1959;72:294–301. doi: 10.1042/bj0720294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bavi RS, Kamble AD, Kumbhar NM, Kumbhar BV, Sonawane KD. Conformational Preferences of Modified Nucleoside N-2-methylguanosine (m(2)G) and Its Derivative N-2, N-2-dimethylguanosine (m (2) (2) G) Occur at 26th Position (Hinge Region) in tRNA. Cell Biochemistry and Biophysics. 2011;61:507–521. doi: 10.1007/s12013-011-9233-1. [DOI] [PubMed] [Google Scholar]
- 59.Pallan PS, Kreutz C, Bosio S, Micura R, Egli M. Effects of N2, N2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m2 2G:A pairs. RNA. 2008;14:2125–2135. doi: 10.1261/rna.1078508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Najmabadi H, Hu H, Garshasbi M, Zemojtel T, Abedini SS, Chen W, Hosseini M, Behjati F, Haas S, Jamali P, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 61*.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. Exhaustive review on current knowledge about m6A modification in mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66*.Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. This study described how Mettl3 and Mettl14 cooperate to catalyze m6A in mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dubin DT, Taylor RH. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975;2:1653–1668. doi: 10.1093/nar/2.10.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 69.Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015;43:6557–6567. doi: 10.1093/nar/gkv596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]


