Abstract
Aspergillus flavus and Fusarium verticillioides infect maize kernels and contaminate them with the mycotoxins aflatoxin, and fumonisin, respectively. Genetic resistance in maize to these fungi and to mycotoxin contamination has been difficult to achieve due to lack of identified resistance genes. The objective of this study was to identify new candidate resistance genes by characterizing their temporal expression in response to infection and comparing expression of these genes with genes known to be associated with plant defense. Fungal colonization and transcriptional changes in kernels inoculated with each fungus were monitored at 4, 12, 24, 48, and 72 h post inoculation (hpi). Maize kernels responded by differential gene expression to each fungus within 4 hpi, before the fungi could be observed visually, but more genes were differentially expressed between 48 and 72 hpi, when fungal colonization was more extensive. Two-way hierarchal clustering analysis grouped the temporal expression profiles of the 5,863 differentially expressed maize genes over all time points into 12 clusters. Many clusters were enriched for genes previously associated with defense responses to either A. flavus or F. verticillioides. Also within these expression clusters were genes that lacked either annotation or assignment to functional categories. This study provided a comprehensive analysis of gene expression of each A. flavus and F. verticillioides during infection of maize kernels, it identified genes expressed early and late in the infection process, and it provided a grouping of genes of unknown function with similarly expressed defense related genes that could inform selection of new genes as targets in breeding strategies.
Keywords: maize kernel, Aspergillus flavus, Fusarium verticillioides, histology, RNA-sequencing
Introduction
Aspergillus flavus is an opportunistic fungal pathogen that can grow either as a saprophyte in the soil or as a pathogen of many plant species. Hosts of A. flavus include maize kernels, peanuts, cottonseeds and tree nuts (Payne, 1992; Payne and Yu, 2010; Scheidegger and Payne, 2003; St. Leger et al., 2000). Unlike A. flavus, Fusarium verticillioides can colonize maize seeds as an endophyte or necrotroph. Under certain conditions, often related to plant stress, F. verticillioides can cause seedling blight, ear rot, and stalk rot (Bacon et al., 1992; Munkvold, 2003; Pei-Bao et al., 2010).
Aspergillus flavus and F. verticillioides are capable of invading kernels in the field by several routes. F. verticillioides can grow from infected seeds, colonize plant stalks, and grow into ears and infect kernels (Bacon et al., 1992; Duncan and Howard, 2010). While this route of kernel infection is important, kernels are infected more commonly in the field from airborne inoculum, which infects silks and grows down the silk channel into the ear (Munkvold, 2003). A. flavus also colonizes maize silks and grows down the silk channel into the ear (Marsh and Payne, 1984; Smart et al., 1990). Both fungi can colonize and produce mycotoxins in kernels without visible damage (Munkvold et al., 1997; Widstrom et al., 1981) but insect damage and other wounds provide sites for infection and lead to higher levels of mycotoxin contamination (Koehler, 1942; Lillehoj et al., 1975; Sobek and Munkvold, 1999). Effective management of these two diseases must account for the role of insects in the disease.
Effective resistance to either of these two fungi in commercial maize lines has been difficult to achieve, due in part to the quantitative nature of resistance, large environment effects on the diseases, and the lack of characterized genes for host resistance (Payne et al., 1986; Munkvold, 2003; Dolezal et al., 2014; Shu, 2014; Warburton and Williams, 2014; Lanubile et al., 2015). Further complicating the identification of resistance genes is a poor understanding of the temporal pattern of gene expression during the colonization of multiple tissue types within seeds, each of which may differ in their expression profiles. Better characterization of resistance gene expression at stages in the infection process could indicate optimum times for resistance evaluation.
This study was directed at the identification of defense-related genes expressed in maize kernels at several stages during colonization by A. flavus and F. verticillioides. By observing fungal colonization and measuring gene expression in maize seeds from 4 to 72 h post inoculation (hpi) we were able to identify genes expressed early in pathogenesis as well as those expressed during the colonization of specific tissue types of the kernel. The study also allowed analysis of similarities and differences in response of maize seeds to these two pathogens with different tropic behaviors. We are unaware of any study that compares both histological examination and gene profiling of these two fungi under the same growing conditions or one that has examined very early gene expression in maize in response to these fungi.
Large-scale RT-qPCR, RNA-seq, microarray and proteomic studies have been conducted to dissect maize transcriptional changes during infection by A. flavus (Chen et al., 2002; Dolezal, 2010; Luo et al., 2011; Pechanova et al., 2011; Kelley et al., 2012; Brown et al., 2013; Dolezal et al., 2013; Asters et al., 2014; Musungu et al., 2016). Results from these studies have shown that maize seeds respond to infection by the expression of characterized defense related genes. As an example, a recent study by Musungu et al. (2016) reported that maize JA, ET, and ROS pathways are associated with defense response to A. flavus infection. Similar classes of maize genes show elevated gene expression during infection by F. verticillioides (Lanubile et al., 2010, 2012, 2013, 2014; Kim et al., 2015; Wang et al., 2016). Wang et al. (2016) reported that maize ABA, JA and SA signaling pathways are associated with triggering immunity against F. verticillioides.
Fusarium verticillioides also has been shown to induce more drastic gene expression changes in the susceptible maize lines than in the resistant lines (Lanubile et al., 2010, 2013, 2014; Campos-Bermudez et al., 2013). The pathogenesis-related genes are also transcribed at higher levels in kernels of the F. verticillioides-resistant lines before infection (Lanubile et al., 2010, 2014; Campos-Bermudez et al., 2013). By using RNA-seq technology, Lanubile et al. (2014) found that the resistant maize line showed higher activation of genes involved in JA and ET signaling pathways and shikimate biosynthesis pathway. Lanubile et al. (2015) reported that higher gene expression and enzymatic activities were detected in maize kernels that are resistant to A. flavus as well as other two Fusarium spp., F. proliferatum and F. subglutinans. They also found that expression levels of PR genes remained higher in the resistant lines after inoculation (Lanubile et al., 2015).
The consistent observation that infection by A. flavus and F. verticillioides causes a change in expression of these upstream signaling molecules suggest that they may be good markers for a host resistance response to these fungi in developing maize seeds. The overall objectives of this study were to provide fundamental information on the colonization of maize seeds by these two fungi, determine the temporal profile of maize gene expression during infection, and identify genes with similar expression profiles to resistance associated genes.
Materials and Methods
Plant Material, Fungal Inoculation, and Sampling
Fungal strains (A. flavus NRRL 3357, F. verticillioides n16) were grown on potato dextrose agar (PDA) plates at 28°C for 5 days. Conidial suspensions were harvested by adding sterile distilled water containing 0.05% (v/v) Triton X-100 (Fisher) and scraping the plates using a glass spreader. The concentration of conidia was quantified using a hemocytometer and diluted to 1 × 106 conidia/ml for inoculation. Maize inbred line B73 was grown at the Central Crops Research Station near Clayton, NC plots as described by Shu et al. (2015). Maize ears were hand-pollinated and covered with paper bags until inoculation at 21–22 days after pollination. Inoculation was conducted by wounding the kernel with a 3 mm needle bearing approximately 13 conidia (Dolezal et al., 2014; Figure 1a). Kernels for the mock treatment were inoculated with sterile distilled water containing 0.05% (v/v) Triton X-100.
FIGURE 1.

Aspergillus flavus (Af) and Fusarium verticillioides (Fv) colonization of maize kernels developing in the field. Kernels in early dough stage (20–22 days after pollination) were inoculated by inserting a pin dipped into either an Af or Fv conidial suspension into the crown of exposed kernels, introducing approximately 13 spores into the endosperm (En) tissue (a). The percent of fungal RNA-seq reads in the total reads of Af or Fv infected kernels at 4, 12, 24, 48, and 72 h post inoculation (hpi) indicated that fungal tissues increased over time in the kernel, and A. flavus more extensively colonized the kernel than F. verticillioides at 24, 48, and 72 hpi (b). Kernel sections collected at 72 hpi were stained with safranin and fast green (c–h). No fungal colonization was observed in the aleurone (Al), En or scutellum (Sc) of a mock inoculated kernel (c,f). Af colonized and destroyed the Al (d), whereas Fv colonized around the intact Al (e). Af colonized the EnSc interface forming a biofilm-like structure (g). But the biofilm like structure was not observed in Fv inoculated kernels (h). Arrows denote fungal colonization. Scale bars: 30 μm.
For each treatment three ears randomly selected from the 70 plots were harvested at 4, 12, 24, 48, and 72 hpi. Each of the three ears was treated as a biological replication. At harvest kernels from ears collected at each time point were divided into two subsets. One subset of kernels at each of the time points was examined for tissue specific colonization by the fungi as described by Shu et al. (2015). The other subset of kernels at each time point was frozen in liquid nitrogen immediately and stored at -80°C until RNA extraction and sequencing by Illumina HiSeq.
Tissue Fixation, Embedding, and Microscopy
Six kernels from each ear replicate were fixed and dehydrated using the protocol modified from Livingston et al. (2009, 2013) and Shu et al. (2015). Briefly, we used a modified FAA fixative consisting of 45% methanol, 10% formaldehyde and 5% glacial acetic acid. The paraffin blocks were sectioned with a RM2255 microtome (Leica) and mounted on slides (Gold Seal). Slides were dried on a hot plate overnight and stored at room temperature. Paraffin was removed by dipping the slides in 100% xylene. Sections were then rehydrated with an ethanol series. Safranin and fast green staining were applied to differentiate tissue structure of maize kernels and the fungus grown in the kernel (Figures 1c–h). The rehydrated sections were stained with safranin, dehydrated with an ethanol series, and counter stained with fast green (Fisher) (Shu et al., 2015). Stained sections were mounted in permount mounting medium (Fisher) and covered with coverslips. Images of stained tissues were collected on an Eclipse E600 light microscope (Nikon). Images were captured on an Infinity1-3C digital camera, and analyzed with the software Infinity Analyze (Lumenera).
RNA-Isolation, Illumina Library Preparation, and Sequencing
Eight frozen kernels from each ear replicate were pooled and ground in liquid nitrogen with a mortar and pestle. About 100 mg of ground tissue was added to 0.75 ml of saturated phenol, pH 6.6 (Fisher), and homogenized for 2 min. Samples were then dissolved in Tris EDTA buffer, pH 8.0 (ACROS Organics), extracted with 5:1 acid phenol: chloroform, pH 4.5 (Fisher), and precipitated with ice-cold 100% ethanol overnight. Total RNA was further purified with an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The quality and concentration of RNA was analyzed using an RNA Pico chip on an Agilent Bioanalyzer. The cDNA library construction and sequencing runs on an Illumina HiSeq were done by the Genomic Sciences Laboratory, North Carolina State University. Multiple samples with different barcodes were loaded in three lanes and sequenced to obtain 100 bp single-end reads.
RNA-Seq Data Analysis
Illumina reads were sorted by barcodes, and adapter sequences were trimmed. The raw sequencing reads were then analyzed using the iPlant Collaborative Discovery Environment (Matasci and McKay, 2013). Reads of the same individual from multiple lanes were concatenated using the software named ‘Concatenate Multiple Files.’ The quality of the reads was checked using FastQC 0.10.1 (data not shown) and then aligned to the maize genome (Ensembl 14) by using TopHat2-SE (Trapnell et al., 2012). Maize transcripts were assembled using Cufflinks2. All transcripts from Cufflinks output were merged into a single transcriptome annotation file using Cuffmerge2 and used for Cuffdiff2. The TopHat2-SE output files were sorted using SAMtools sort BAM file before conducting Cuffdiff2 analysis. Gene expression levels were analyzed using Cuffdiff2 (Trapnell et al., 2012).
We assigned the differentially expressed maize genes (q-value < 0.05 and fold change > 2) into functional categories using MapMan (Usadel et al., 2009). Then we used the MaizeSequence database to obtain gene annotation (Schnable et al., 2009). For the genes without annotation in MaizeSequence database, we combined a few databases, including MaizeGDB, ProFITS, GRASSIUS, CoGePedia, Maize Protein Atlas databases (Lawrence et al., 2004; Lyons and Freeling, 2008; Yilmaz et al., 2009; Ling et al., 2010; Castellana et al., 2013) and the R-like gene database published by Song et al. (2015). Differentially expressed maize genes were also placed into biosynthetic and regulatory pathways using the MaizeCyc database and the bioinformatics software Biocyc (Caspi et al., 2012). Clustering was performed using JMP, 11 (SAS Institute Inc., Cary, NC, United States).
Data Deposition
RNA-seq data generated in this study were deposited in the National Center for Biotechnology Information (NCBI) sequence read archive (SRA) collection, accession number PRJNA418364. The de novo assemblies of transcriptomes and assembled sequences together with sequence annotation were posted on figshare1.
Results
Maize kernels developing in the field were inoculated with either A. flavus or F. verticillioides (Figure 1a) and colonization followed by histological examination of stained kernel sections at 4, 12, 24, 48, and 72 hpi (Figures 1c–h). RNA was isolated from a subsample of kernels taken at each of these time points and subjected to RNA-seq analysis. This paired analysis on the same tissue samples permitted a direct association of tissue colonization with global gene expression in maize kernels in response to infection by these two fungi (Figures 2, 3).
FIGURE 2.
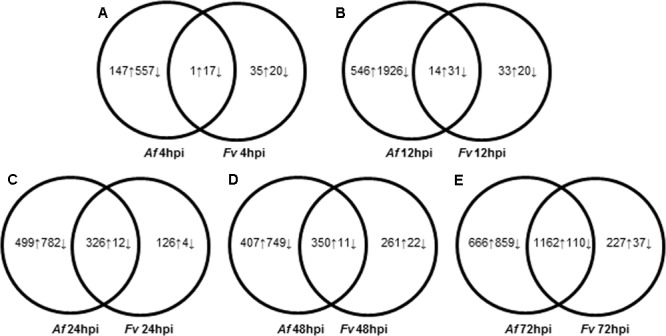
Dynamic changes of kernel transcriptome during A. flavus (Af) and F. verticillioides (Fv) infection. (A–E) Show total numbers of maize genes differentially expressed in response to A. flavus and F. verticillioides infection at 4 (A), 12 (B), 24 (C), 48 (D), and 72 (E) hours post inoculation (hpi). Total numbers of up (↑) and down (↓)-regulated genes of treatment-specific and shared between treatments are displayed in Venn diagrams.
FIGURE 3.

Functional categories of maize genes differentially regulated upon infection by A. flavus (Af) and F. verticillioides (Fv) at different time points. Colored bars represent the numbers of regulated genes with annotated function. Positive bars denote numbers of up-regulated genes. Negative bars denote numbers of down-regulated genes.
Colonization of Maize Kernels by A. flavus and F. verticillioides
Histological sections from kernels collected at 4, 12, or 24 hpi did not reveal fungal mycelia. In contrast, extensive colonization by A. flavus and F. verticillioides was observed at 48 hpi in the aleurone and the outermost layer of the endosperm (Figures 1d,e). Disruption of the aleurone was associated with the presence of A. flavus mycelium (Figure 1d), whereas colonization of the aleurone tissue by F. verticillioides at 48 hpi was less extensive and the cells appeared intact (Figure 1e). By 72 hpi, both pathogens had colonized the aleurone and endosperm. Mycelia of A. flavus, but not that of F. verticillioides, was found in the embryo at 72 hpi. A. flavus formed a dense mat of mycelium at the interface of the endosperm and scutellum of the embryo (Figure 1g). The scutellum was rarely colonized prior to the formation of the mat. No such structure was observed in the F. verticillioides colonized kernels. Visual observation as well as transcript abundance from RNA-seq analysis indicated more extensive colonization of kernels by A. flavus than F. verticillioides at each sample time (Figure 1b).
Temporal Expression of Maize Genes in Response to Fungal Colonization
Pathogenesis by A. flavus or F. verticillioides resulted in broad transcriptional and presumably large metabolic changes in maize kernels during infection. Expression analysis identified 5,863 maize genes differentially expressed at one or more of the time points during pathogenesis by these two fungi. The greatest number of maize genes differentially expressed in response to the two fungi occurred at 72 hpi. At each time point, more genes were differently expressed in response to A. flavus than to F. verticillioides. While A. flavus did colonize seeds more extensively than F. verticillioides (Figure 1), it is unlikely that differences in colonization alone resulted in greater transcriptional changes in maize kernels. Fungal biomass accumulation by A. flavus in maize seeds 48 hpi was similar to that of F. verticillioides biomass accumulation at 72 hpi. At these time points, 757 genes were upregulated and 760 genes were downregulated in response to A. flavus infection and 1389 genes were upregulated and 147 genes were downregulated in response to F. verticillioides infection. Furthermore, colonization of maize seed by F. verticillioides 72 hpi resulted in greater expression of more chitinase genes, stress-related genes, hormone metabolism and signaling genes, transcription and RNA processing, secondary metabolism and primary metabolism genes than colonization by A. flavus 48 hpi (Figure 3 and Supplementary Table S1).
Monitoring maize gene expression at five time points allowed association of gene expression profiles with stages of fungal colonization. Unique and overlapping sets of maize genes were differentially expressed in maize kernels in response to the two pathogens over the entire infection process (Figure 2 and Supplementary Table S1). Maize kernels recognized infection by A. flavus within 4 hpi and increased expression of 148 genes and decreased expression of 574 genes. Fewer maize genes were differentially expressed in response to F. verticillioides infection, with 36 maize genes up regulated and 37 down regulated at 4 hpi (Figure 2). At this sample point, both pathogens repressed the expression of chitinase2 (chn2), heat shock protein26, a 22.0 kDa class IV heat shock protein, and a cell wall modification related gene. Chitinase 2 (chn2) was elevated in response to either fungus 12, 24, 48, and 72 hpi. By 24 hpi, 231 genes with known function were more highly expressed in response to either fungus including a set of PR genes. Within 24 hpi, these fungi induced expression of maize genes assigned to all seven functional categories (Figure 3 and Supplementary Table S1), including genes associated with stress, hormone metabolism and signaling, transcription and RNA processing, redox regulation, as well as primary and secondary metabolism. Continued colonization of maize seeds resulted elevated expression of more genes with known function. At 72 hpi, 724 genes with known function are more highly expressed in response to either fungus, including defense-related genes. Notably, PR-4, PR-5, PR-10 and PR-10.1 were consistently more highly expressed in response to either fungus at 24, 48, and 72 hpi. At 72 hpi, 63 genes with known function are more highly expressed in response to either fungus, including a PR protein (GRMZM2G178199).
Ward’s Two-way Hierarchal Clustering Analysis allowed us to cluster expression levels of the 5,863 differentially expressed genes by time and by genes. When clustered by time, early time points (4–12 hpi) clustered together and later time points (24–72 hpi) clustered together. RNA samples collected at 4, 12, and 24 hpi provided transcriptional data at stages of colonization before the fungi were observed by histological examination, whereas 48 and 72 hpi RNA samples provided data from more advanced stages of fungal colonization (Figures 1b–h).
Among the genes expressed early during infection were genes that have been used as markers for activation of host defenses in maize and other plants (Supplementary Table S1). In maize kernels infected with A. flavus, more PR-like genes were down regulated than up regulated at 4 and 12 hpi. There was no significant increase in expression of any PR-like genes in maize seed at 4 hpi in response to infection by either fungus. In contrast, expression of genes for PR-1, PR-10, three chitinases, and a glucan endo-1,3-beta-glucosidase 7 significantly decreased after infection with A. flavus, and expression of chitinase 2 (chn2) decreased in F. verticillioides infected kernels (Supplementary Table S1). By 12 hpi, expression of PR-5 and chitinase 2 (chn2) were elevated in response to either fungus. By 24 hpi several PR-like genes were more highly expressed after infection by the two fungi, including PR-10. Overall, infection of maize by A. flavus resulted in a larger number of PR-like and R-like genes with decreased expression compared to F. verticillioides at 4 and 12 hpi.
In plants, both coiled-coil nucleotide-binding site leucine-rich repeat (CC-NBS-LRR) and leucine-rich repeat receptor-like kinase (LRR-RLK) receptors are R-like proteins important in the resistance response to pathogens. Of the 378 R-like genes predicted in the maize genome (Ling et al., 2010; Song et al., 2015), 40 of 151 predicted CC-NBS-LRR genes were differentially expressed during infection by A. flavus and/or F. verticillioides. Of the 227 predicted LRR-RLK genes in maize, 36 were differentially expressed during one or more time points. Expression of three of these genes decreased at 4 hpi upon infection by A. flavus. None of these 36 differentially expressed LRR-RLK genes showed decreased expression in kernels infected with F. verticillioides until 72 hpi. Some of the LRR-RLKs were differentially expressed at 24, 48, and 72 hpi during infection of either A. flavus or F. verticillioides.
Plant defense to pathogen colonization is mediated by hormone signaling pathways. SA, JA, and ET govern the best described signaling pathways through their regulation of resistance gene expression, including PR-like genes. Because these two fungi are capable of different tropic interactions with the host, we predicted that maize seed would differ between the two fungi in the type or timing of gene expression. Expression of genes for SA biosynthesis increased from 24 to 72 hpi in response to A. flavus infection, but not until 72 hpi in response to F. verticillioides. Cluster analysis of gene expression data collected at the five time points in response to A. flavus infection placed the SA biosynthetic gene encoding the 3-phosphoshikimate 1-carboxyvinyltransferase gene into a cluster with a set of PR-like genes, including PR-5 (Figure 4 and Supplementary Table S1).
FIGURE 4.

Two way Clustering of differentially expressed genes. FPKM values of differentially expressed genes were Ln transformed and Ward’s Hierarchical Clustering was performed. The number of clusters was set at 12 and clusters were color coded. The color map indicates relative gene expression over time with red indicating high expression and blue indicating low expression. (A) A. flavus; (B) F. verticillioides.
Several genes encoding maize 12-oxo-phytodienoic acid reductases (OPRs) and lipoxygenases (LOXs) were differentially expressed in maize seeds in response to either fungus (Supplementary Table S1). Expression of both LOX4 and LOX13 increased in response to A. flavus at 24 hpi and in response to F. verticillioides at 72 hpi. Two other LOX genes, LOX10 and LOX11, were down regulated at 12 hpi in response to A. flavus infection, and LOX10 was up regulated only at 24 hpi (Supplementary Table S1). These two genes were not differentially expressed in response to infection by F. verticillioides. Infection by A. flavus increased expression of OPR2 at 24 and 48 hpi, and expression of ORP2, ORP3, and ORP5 was up regulated in response to A. flavus at 48 hpi and to F. verticillioides at 72 hpi.
Hierarchical Clustering of Genes Expression Profiles
Many of the maize genes differentially expressed in response to infection by A. flavus or F. verticillioides lack annotation or known biological function (Supplementary Table S1). To gain additional insight into possible involvement of these genes in resistance, we performed Ward’s Two-way Hierarchical Clustering (Figure 4) to group genes based on their profile of expression over time. This analysis allowed the clustering of genes with known or putative function with genes of unknown function. Clustering of unknown genes with resistance genes does not indicate a direct role for them in recognition and defense, but a similar profile of expression may indicate a role for these genes during different stages of disease development. This information may also guide future studies on sampling times to further characterize these genes.
Figure 4 shows the clustering of all 5,863 genes into 12 clusters. The respective cluster membership for each gene is listed in Supplementary Table S1. The temporal expression of maize genes in response to these two fungi differed. This difference could be due to rate of growth or to differences in host response to the two species. For this reason, an independent cluster analysis of gene transcription was performed for each fungus (Figures 4A,B). Some clusters were enriched in defense related genes (Figure 4 and Supplementary Table S1), for example clusters 6A and 10A associated with A. flavus infection (‘A’ denotes A. flavus-associated cluster). A. flavus Cluster 6A (Supplementary Table S1 and Figure 4A) had as members several genes encoding pathogenesis related proteins. These included PR-5, PR-10, PR-10.1, Protein P21, and seven chitinases, four of which were identified by Hawkins et al. (2015) as associated with host resistance to A. flavus (Supplementary Table S1). Also in this cluster were several biotic and abiotic stress related genes, including six GSTs. Overall, there were 183-annotated and 165 unannotated maize genes in Cluster 6 associated with A. flavus infection. The greatest number of genes associated with host resistance expressed in response to F. verticillioides clustered in 1F (‘F’ denotes F. verticillioides-associated cluster; Figure 4B and Supplementary Table S1). This cluster included PR-5, PR-10, PR-10.1, Protein P21, and four chitinase genes.
Other clusters contained different defense associated genes. Cluster 1A contained the JA biosynthesis genes OPR3 and LOX13, while cluster 11A contained OPR2, OPR5 and LOX4 (Supplementary Table S1). LOX10 and LOX11 were clustered in Cluster 2A and 12A in response to A. flavus, respectively (Supplementary Table S1). In F. verticillioides infected kernels, the expression profile of two LOX genes, LOX and LOX4, clustered in Cluster 2F, while LOX10 and LOX13 clustered in Cluster 7F and 9F, respectively (Supplementary Table S1). OPR2, OPR3 and OPR5 clustered in Cluster 9F, 10F and 3F, respectively, in response to F. verticillioides infection (Supplementary Table S1).
Discussion
Aspergillus flavus and F. verticillioides present worldwide health concerns because of their production of the mycotoxins, aflatoxin, and fumonisin, respectively (Wild et al., 2015). Genetic resistance, which has been successful for the control of many maize diseases, has not been achieved for the diseases caused by these two fungi (Warburton and Williams, 2014; Wild et al., 2015). The inability to identify and move resistance genes into agronomically desirable genotypes is a recognized gap in the development of control strategies for these diseases (Wild et al., 2015).
This study sought to characterize the transcriptional response of developing maize seed to infection by A. flavus and F. verticillioides, fungi capable of different nutrient associations with maize. In agreement with previous histological observations (Dolezal et al., 2013; Shu et al., 2015), A. flavus formed a dense mat of mycelium at the interface of the endosperm and scutellum of the embryo. This mat had the appearance of the type of structure referred to as a biofilm in Aspergillus species (Wuren et al., 2014; Reichhardt et al., 2015). We found that F. verticillioides did not form this mat. Transcriptional profiling alongside with the histological observations allowed quantification of gene transcription during early recognition events, and during the colonization of different maize seed tissue types. Overall, A. flavus colonized seeds much more rapidly and induced gene expression changes in maize seed earlier than did F. verticillioides (Figures 1, 2 and Supplementary Table S1). Differential gene expression was observed in kernels inoculated with either fungus at 4 hpi; however, most genes were more highly expressed at later time points; 24–48 hpi for A. flavus and 48–72 hpi for F. verticillioides. Infection by A. flavus also changed expression of a greater number of maize genes than infection by F. verticillioides. In both fungi, more genes were down regulated than up regulated at 4 hpi, including PR-1 and PR-10 by A. flavus. These genes were in clusters 2A and 6A, respectively (Supplementary Table S1). During infection with F. verticillioides a similar number of genes were down regulated as were up regulated at 4 and 12 hpi (Figure 2). In contrast, infection by A. flavus resulted in nearly 3.5 times more down regulated than up regulated genes at these two time points. These observations suggest that one of the early events in infection by these two fungi is suppression of host gene expression, especially by A. flavus.
Differences in observed gene expression in maize in response to the two fungi may have been the result of more extensive kernel colonization by A. flavus than by F. verticillioides, differences in the colonization of specific kernel tissue types, or the differential response of maize kernels to the two fungi. Examination of histological sections and transcripts reads showed more extensive colonization by A. flavus. As reported earlier (Shu et al., 2015) and as shown in this study (Figure 1), the two differ in their colonization of maize kernels. F. verticillioides appears to cause less initial cell damage in the aleurone than A. flavus and A. flavus produces an extensive fungal mat adjacent to the scutellum that has not be observed for F. verticillioides. The scutellum is a metabolically active tissue known to secrete inhibitory compounds, including PRms (Guo et al., 1999; Murillo et al., 1999). In their studies following timing of PRms expression by in situ hybridization, Shu et al. (2015) observed PRms transcripts in cells of the aleurone of kernels infected by either A. flavus or F. verticillioides by 48 hpi, but not in the scutellum of A. flavus infected kernels until 72 hpi. Differences in colonization patterns and their adaptation of different strategies for nutrient acquisition could account for dissimilarities in the transcriptional response to the two fungi. Targeted gene expression to change either the type or quantity of fungal inhibitory compounds in the scutellum, for example, could reduce infection by these fungi. Strategies targeted to this tissue may be particularly important for control of aflatoxin contamination as A. flavus preferentially colonizes germ tissues and produces more aflatoxin in this tissue than other tissues of the kernel (Smart et al., 1990; Keller et al., 1994). In contrast, F. verticillioides produces more fumonisin in the endosperm than in the germ (Bluhm et al., 2008).
The analysis of gene transcripts from infected kernels at five time points, ranging from no visible fungal colonization to extensive colonization, allowed us to determine both temporal and quantitative changes in maize gene expression in response to infection by the two fungi. With this information, we were able to cluster genes of unknown function, which could include putative resistance genes, with characterized defense related genes. Several genes in maize have been associated with resistance to these two fungi by transcriptional profiling studies (Christensen et al., 2014; Dolezal et al., 2014; Lanubile et al., 2014, 2015; Warburton and Williams, 2014; Hawkins et al., 2015; Shu et al., 2015), genome-wide association analysis (Tang et al., 2015), molecular and proteomic analysis (Murillo et al., 1999; Baker et al., 2009a,b), or marker assisted selection (Zhang et al., 2016) (Table 1).
Table 1.
Comparison of maize genes and/or proteins that have been observed in this study and in previous studies.
Af, associated with defense response to Aspergillus flavus; Fv, associated with defense response to Fusarium verticillioides.
Pathogenesis related proteins, which are often expressed early during infection, are associated with disease resistance of several host species including maize. PR-4 transcripts are expressed in maize plants upon infection by F. verticillioides and Ustilago maydis (Bravo et al., 2003; Doehlemann et al., 2008). Maize PR-5 expression is associated with defense responses to F. proliferatum, F. subglutinans, and A. flavus (Lanubile et al., 2015). PR-10, which has been associated with resistance to A. flavus (Chen et al., 2010), was expressed 24 hpi after inoculation. In maize plants, chitinases and β-1, 3- glucanases were reported to be involved in resistance to A. flavus and F. verticillioides (Cordero et al., 1993; Lozovaya et al., 1998; Ji et al., 2000; Moore et al., 2004; Warburton and Williams, 2014; Hawkins et al., 2015).
Other defense related proteins associated with resistance include plant chitinases and endoglucanases, which may have cell wall-degrading activities (Huynh et al., 1992; Wu et al., 1994a,b; Bravo et al., 2003), that function downstream of SA and JA/ET signaling pathways (Vidal et al., 1998). Sánchez-Rangel et al. (2012) found that Fumonisin B1 targeted beta-1, 3-glucanase of the germinating maize embryo. Holmes et al. (2008) also identified the maize seed chitinase A and thaumatin-like proteins as inhibitors of A. flavus fungal growth and aflatoxin biosynthesis. We observed transcriptional changes of thaumatin-like proteins after A. flavus or F. verticillioides inoculation (Supplementary Table S1). Members of the thaumatin-like proteins were characterized to be PR proteins in A. thaliana, barley, wheat and apple (Hejgaard et al., 1991; Hu and Reddy, 1997; Krebitz et al., 2003; Wang et al., 2010). The PR-like genes that we identified in this study, including the thaumatin-like proteins, are putative PR genes that might play important roles in maize resistance to a broad spectrum of pathogens.
We also observed elevated expression of several CC-NBS-LRR and LRR-RLK genes, which are important in the resistance response to pathogens. Over 378 of these R-like genes are predicted in the maize genome. One of the 40 members of the CC-NBS-LRR genes induced by A. flavus and F. verticillioides was GRMZM2G032602 (Collins et al., 1998). Elevated expression of this gene has been observed in response to infection by the maize pathogen Cochliobolus heterostrophus and by treatment with SA (Cheng et al., 2012), suggesting that the gene is involved in SA-mediated defense responses.
Several studies have shown transcriptional changes in expression of plant genes involved in the biosynthesis and regulation of the plant hormones JA, SA, and ET to be associated with host defense (Nemchenko et al., 2006; Menkir et al., 2008; Pré et al., 2008; Mengiste, 2012; Christensen et al., 2014; Tang et al., 2015). A recent genome-wide association analysis showed the JA biosynthesis pathway to be the most significant metabolic pathway involved in resistance to aflatoxin accumulation (Tang et al., 2015). Christensen et al. (2014) also found JA to be the major defense hormone against F. verticillioides, with LOX12, OPR7, and OPR8 playing key roles in this interaction. In our study we observed different LOX and OPR genes to be differentially expressed in response to A. flavus and F. verticillioides in maize kernels (Supplementary Table S1).
In some cases, it may be difficult to associate changes in expression of specific hormone biosynthetic genes with host resistance due to cross talk among the JA-, SA-, and ET-mediated pathways (Derksen et al., 2013). For example, NPR1 (non-expresser of PR genes 1), a positive regulator of SA biosynthesis, is also a negative regulator of JA biosynthesis (Mengiste, 2012), We did not find this gene differently expressed in our study even though some SA biosynthesis genes were more highly expressed during infection by A. flavus. We also observed infection by either fungus to elevate expression of members of the MYB and ERF families, which act downstream of the of the JA/ET signaling pathways (Supplementary Table S1). Additionally, we found elevated expression of PR-5 and several WRKYs suggesting that the SA signaling pathway may be involved in maize defense to either fungus. Previous studies showed that the maize ethylene responsive factor 1 (ERF1), a key transcription factor involved in ET and JA signaling, was more highly expressed in the resistant maize inbred TZAR101 than the susceptible B73 in response to A. flavus infection (Menkir et al., 2008; Fountain et al., 2013). We also found AP2-EREBP, a gene known to integrate ET and JA signaling pathways, and to regulate defense-related gene expression in Arabidopsis thaliana (Pré et al., 2008) to be down regulated in response to infection by these two fungi prior to 72 hpi (Supplementary Table S1). In this study, we observed LOX4, LOX10, LOX11, and LOX13 to be differentially expressed in response to both fungi. Nemchenko et al. (2006) found that maize LOX10 was induced by both biotic and abiotic stresses, such as wounding, cold, Cochliobolus carbonum, JA, SA, and ABA treatment. But they reported that LOX11 was induced only by ABA.
Other phytohormone signaling pathways, such as the ABA, auxin, CKs, BRs, and GA pathways, interact with SA, JA and ET signaling pathways and form a regulatory network which is important in defense response to external stimulus (Angra-Sharma and Sharma, 1999; Siemens et al., 2006; Walters and McRoberts, 2006; Pieterse et al., 2009; Zhang et al., 2010; Iglesias et al., 2011; Behr et al., 2012; Denancé et al., 2013; Derksen et al., 2013; Yang et al., 2013; Zhu et al., 2015). We detected transcriptional changes of genes that are associated with ABA, auxin, and GA pathways that affect the three innate defense pathways (Supplementary Table S1), suggesting a complex regulatory network triggered by these fungi. Our results indicate that upregulation of ABA/auxin signaling pathways may contribute to the inhibition of SA signaling. The inhibition of SA signaling would attenuate host resistance and promote fungal colonization. ABA and auxin signaling pathways suppress SA signaling in plants (Wang et al., 2007; Pieterse et al., 2009). Elevated auxin levels and transcriptional induction of auxin-responsive genes were also observed in maize during U. maydis infection (Turian and Hamilton, 1960; Doehlemann et al., 2008). Our data indicate that both GA signaling and GA degradation pathways changed during infection.
Downregulation of the GA signaling pathway may release the inhibition of JA/ET signaling in the kernel. In A. thaliana, GA produced by Gibberella fujikuroi inhibits JA-dependent necrotroph resistance via GA-mediated degradation of DELLA proteins (Navarro et al., 2008). The role of GA in maize kernels may be more complex as it is also involved in seed development by stimulating the production of proteases and α-amylases necessary for the hydrolysis of endosperm starch and proteins, a process that ABA inhibits (Harvey and Oaks, 1974). We also observed elevated expression of BR and CK signaling pathways in response to infection by either fungus. BRs have been shown to contribute to disease resistance in A. thaliana, tobacco and rice plants (Nakashita et al., 2003; Belkhadir et al., 2012). Collectively, our data support the conclusion that an interacting hormone network contributes to maize resistance to A. flavus and F. verticillioides in developing maize kernels.
Several studies have shown that glutathiones produced by GSTs to be important in resistance to abiotic and biotic stress through their action as soluble antioxidants (Mauch and Dudler, 1993; Ogawa, 2005; Sytykiewicz, 2011; Wisser et al., 2011). As an example, elevated GST levels were found during infection of wheat by Erysiphe graminis (Dudler et al., 1991) and A. thaliana by Alternaria brassicicola (Mukherjee et al., 2010). We also observed elevated transcription of GST genes during infection by either A. flavus or F. verticillioides, suggesting a role of glutathiones in this host–pathogen interaction.
In this study, we directly compared transcriptional changes in maize seeds during colonization by the two major mycotoxin-producing fungi on maize. By pairing histological and transcriptional examinations during disease development in the field, we were able to associate transcriptional changes with tissue colonization by the two fungi under conditions conducive for their growth. The study revealed similarities and differences in the response of maize to two fungi differing in tropic relationships with the host. We were able to construct a comprehensive gene expression database for those interested in further characterization of these two diseases and those interested in breeding for resistance.
Using hierarchal clustering analysis, we were able to place genes into 12 clusters representing genes with similar expression profiles (Supplementary Table S1 and Figure 4). The placement of genes in clusters based on the timing and magnitude of their expression along with genes of unknown function promises to aid in future understanding of maize resistance regulatory circuits, as well as guide the selection of candidate genes for further studies on their role in host resistance. While expression of known defense related genes has been associated with host resistance to these two diseases in maize, their expression alone has not been shown to be sufficient for adequate resistance. This is presumably due to lack of sufficient expression of these genes or to improper timing of expression of downstream regulatory genes. The gene profile analysis provided in this manuscript will serve as a genetic resource and aid in the identification of candidate resistance genes.
Author Contributions
XS and CW designed the experiments. XS and DL performed the experimental work. XS performed the data analysis. GP supervised the research and manuscript preparation. All authors wrote and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Mr. Gregory R. OBrian for his invaluable technical help and advice on the manuscript.
Funding. This work was supported by the Agriculture and Food Research Initiative Competitive Grants Program of the United States Department of Agriculture, National Institute of Food and Agriculture (Nos. 2010-65108-20496 and 2013-68004-20359) and USDA REE Cooperative Agreement 58-6435-1-61.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02075/full#supplementary-material
References
- Angra-Sharma R., Sharma D. K. (1999). Cytokinins in pathogenesis and disease resistance of Pyrenophora teres-barley and Dreschslera maydis-maize interactions during early stages of infection. Mycopathologia 148 87–95. 10.1023/A:1007126025955 [DOI] [PubMed] [Google Scholar]
- Asters M. C., Williams W. P., Perkins A. D., Mylroie J. E., Windham G. L., Shan X. (2014). Relating significance and relations of differentially expressed genes in response to Aspergillus flavus infection in maize. Sci. Rep. 4:4815. 10.1038/srep04815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon C. W., Bennett R. M., Hinton D. M., Voss K. A. (1992). Scanning electron microscopy of Fusarium moniliforme within asymptomatic corn kernels and kernels associated with equine leukoencephalomalacia. Plant Dis. 76 144–148. 10.1094/PD-76-0144 [DOI] [Google Scholar]
- Baker R. L., Brown R. L., Chen Z. Y., Cleveland T. E., Fakhoury A. M. (2009a). A maize lectin-like protein with antifungal activity against Aspergillus flavus. J. Food Prot. 72 120–127. [DOI] [PubMed] [Google Scholar]
- Baker R. L., Brown R. L., Chen Z. Y., Cleveland T. E., Fakhoury A. M. (2009b). A maize trypsin inhibitor (ZmTIp) with limited activity against Aspergillus flavus. J. Food Prot. 72 185–188. [DOI] [PubMed] [Google Scholar]
- Behr M., Motyka V., Weihmann F., Malbeck J., Deising H. B., Wirsel S. G. (2012). Remodeling of cytokinin metabolism at infection sites of Colletotrichum graminicola on maize leaves. Mol. Plant Microbe Interact. 25 1073–1082. 10.1094/MPMI-01-12-0012-R [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Jaillais Y., Epple P., Balsemão-Pires E., Dangl J. L., Chory J. (2012). Brassinosteroids modulate the efficiency of plant immune responses to microbe-associated molecular patterns. Proc. Natl. Acad. Sci. U.S.A. 109 297–302. 10.1073/pnas.1112840108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluhm B. H., Kim H., Butchko R. A. E., Woloshuk C. P. (2008). Involvement of ZFR1 of Fusarium verticillioides in kernel colonization and the regulation of FST1 a putative sugar transporter gene required for fumonisin biosynthesis on maize kernels. Mol. Plant Pathol. 9 203–211. 10.1111/j.1364-3703.2007.00458.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J. M., Campo S., Murillo I., Coca M., San Segundo B. (2003). Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol. Biol. 52 745–759. 10.1023/A:1025016416951 [DOI] [PubMed] [Google Scholar]
- Brown R. L., Menkir A., Chen Z. Y., Bhatnagar D., Yu J., Yao H., et al. (2013). Breeding aflatoxin-resistant maize lines using recent advances in technologies - a review. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 30 1382–1391. 10.1080/19440049.2013.812808 [DOI] [PubMed] [Google Scholar]
- Campos-Bermudez V. A., Fauguel C. M., Tronconi M. A., Casati P., Presello D. A., Andreo C. S. (2013). Transcriptional and metabolic changes associated to the infection by Fusarium verticillioides in maize inbreds with contrasting ear rot resistance. PLOS ONE 8:e61580. 10.1371/journal.pone.0061580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R., Altman T., Dreher K., Fulcher C. A., Subhraveti P., Keseler I. M., et al. (2012). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 40 D742–D753. 10.1093/nar/gkv1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellana N. E., Shen Z., He Y., Walley J. W., Cassidy C. J., Briggs S. P., et al. (2013). An automated proteogenomic method utilizes mass spectrometry to reveal novel genes in Zea mays. Mol. Cell. Proteomics 13 157–167. 10.1074/mcp.M113.031260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Cleveland T. E., Damann K. E., Russin J. S. (2001). Comparison of constitutive and inducible maize kernel proteins of genotypes resistant or susceptible to aflatoxin production. J. Food Prot. 64 1785–1792. 10.4315/0362-028X-64.11.1785 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Damann K. E., Cleveland T. E. (2002). Identification of unique or elevated levels of kernel proteins in aflatoxin-resistant maize genotypes through proteome analysis. Phytopathology 92 1084–1094. 10.1094/PHYTO.2002.92.10.1084 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Damann K. E., Cleveland T. E. (2007). Identification of maize kernel endosperm proteins associated with resistance to aflatoxin contamination by Aspergillus flavus. Phytopathology 97 1094–1103. 10.1094/PHYTO-97-9-1094 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Damann K. E., Cleveland T. E. (2010). PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 11 69–81. 10.1111/j.1364-3703.2009.00574.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Lax A. R., Cleveland T. E., Russin J. S. (1999a). Inhibition of plant-pathogenic fungi by a corn trypsin inhibitor overexpressed in Escherichia coli. Appl. Environ. Microbiol. 65 1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Lax A. R., Guo B. Z., Cleveland T. E., Russin J. S. (1998). Resistance to Aspergillus flavus in corn kernels is associated with a 14-kDa protein. Phytopathology 88 276–281. 10.1094/PHYTO.1998.88.4.276 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Menkir A., Damann K., Cleveland T. E. (2005). Proteome analysis of near isogenic maize lines differing in the level of resistance against Aspergillus flavus infection/aflatoxin production. Phytopathology 95:S19. [Google Scholar]
- Chen Z. Y., Brown R. L., Rajasekaran K., Damann K. E., Cleveland T. E. (2006). Identification of a maize kernel pathogenesis-related protein and evidence for its involvement in resistance to Aspergillus flavus infection and aflatoxin production. Phytopathology 96 87–95. 10.1094/PHYTO-96-0087 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., Brown R. L., Russin J. S., Lax A. R., Cleveland T. E. (1999b). A corn trypsin inhibitor with antifungal activity inhibits Aspergillus flavus alpha-amylase. Phytopathology 89 902–907. 10.1094/PHYTO.1999.89.10.902 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Li X., Jiang H., Ma W., Miao W., Yamada T., et al. (2012). Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J. 279 2431–2443. 10.1111/j.1742-4658.2012.08621.x [DOI] [PubMed] [Google Scholar]
- Christensen S. A., Nemchenko A., Park Y. S., Borrego E., Huang P. C., Schmelz E. A., et al. (2014). The novel monocot-specific 9-lipoxygenase ZmLOX12 is required to mount an effective jasmonate-mediated defense against Fusarium verticillioides in maize. Mol. Plant Microbe Interact. 27 1263–1276. 10.1094/MPMI-06-13-0184-R [DOI] [PubMed] [Google Scholar]
- Collins N. C., Webb C. A., Seah S., Ellis J. G., Hulbert S. H., Pryor A. (1998). The isolation and mapping of disease resistance gene analogs in maize. Mol. Plant Microbe Interact. 11 968–978. 10.1094/MPMI.1998.11.10.968 [DOI] [PubMed] [Google Scholar]
- Cordero M. J., Raventos D., San Segundo B. (1993). Differential expression and induction of chitinases and β-13-glucanases in response to fungal infection during germination of maize seeds. Mol. Plant Microbe Interact. 7 23–31. 10.1094/MPMI-7-0023 [DOI] [Google Scholar]
- Denancé N., Sánchez-Vallet A., Goffner D., Molina A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. 10.3389/fpls.2013.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C., Daayf F. (2013). Signaling cross-talk in plant disease resistance. Plant Sci. 207 79–87. 10.1016/j.plantsci.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., Wahl R., Horst R. J., Voll L. M., Usadel B., Poree F., et al. (2008). Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56 181–195. 10.1111/j.1365-313X.2008.03590.x [DOI] [PubMed] [Google Scholar]
- Dolezal A. L. (2010). Interactions between Aspergillus flavus and the Developing Maize Kernel. Doctoral dissertation, North Carolina State University; Raleigh, NC. [Google Scholar]
- Dolezal A. L., Obrian G. R., Nielsen D. M., Woloshuk C. P., Boston R. S., Payne G. A. (2013). Localization, morphology and transcriptional profile of Aspergillus flavus during seed colonization. Mol. Plant Pathol. 14 898–909. 10.1111/mpp.12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal A. L., Shu X., OBrian G. R., Nielsen D. M., Woloshuk C. P., Boston R. S., et al. (2014). Aspergillus flavus infection induces transcriptional and physical changes in developing maize kernels. Front. Microbiol. 5:384. 10.3389/fmicb.2014.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd P. F., Johnson E. T. (2016). Maize peroxidase Px5 has a highly conserved sequence in inbreds fungal resistant to mycotoxin producing fungi which enhances resistance insect. J. Plant Res. 129 13–20. 10.1007/s10265-015-0770-3 [DOI] [PubMed] [Google Scholar]
- Dudler R., Hertig C., Rebmann G., Bull J., Mauch F. (1991). A pathogen-induced wheat gene encodes a protein homologous to glutathione-S-transferases. Mol. Plant Microbe Interact. 4 14–18. 10.1094/MPMI-4-014 [DOI] [PubMed] [Google Scholar]
- Duncan K. E., Howard R. J. (2010). Biology of maize kernel infection by Fusarium verticillioides. Mol. Plant Microbe Interact. 23 6–16. 10.1094/MPMI-23-1-0006 [DOI] [PubMed] [Google Scholar]
- Fountain J. C., Raruang Y., Luo M., Brown R. L., Chen Z. Y. (2013). Maize WRKY transcription factors and their potential roles in regulating defense gene expression during Aspergillus flavus infection. Phytopathology 103(Suppl. 1):S1.4. [Google Scholar]
- Fountain J. C., Raruang Y., Luo M., Brown R. L., Guo B., Chen Z.-Y. (2015). Potential roles of WRKY transcription factors in regulating host defense responses during Aspergillus flavus infection of immature maize kernels. Physiol. Mol. Plant Pathol. 89 31–40. 10.1016/j.pmpp.2014.11.005 [DOI] [Google Scholar]
- Gao X., Brodhagen M., Isakeit T., Brown S. H., Gobel C., Betran J., et al. (2009). Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Mol. Plant Microbe Interact. 22 222–231. 10.1094/MPMI-22-2-0222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Stumpe M., Feussner I., Kolomiets M. (2008). A novel plastidial lipoxygenase of maize (Zea mays) ZmLOX6 encodes for a fatty acid hydroperoxide lyase and is uniquely regulated by phytohormones and pathogen infection. Planta 227 491–503. 10.1007/s00425-007-0634-8 [DOI] [PubMed] [Google Scholar]
- Guo B. Z., Cleveland T. E., Brown R. L., Widstrom N. W., Lynch R. E., Russin J. S. (1999). Distribution of antifungal proteins in maize kernel tissues using immunochemistry. J. Food Prot. 62 295–299. 10.4315/0362-028X-62.3.295 [DOI] [PubMed] [Google Scholar]
- Harvey B. M. R., Oaks A. (1974). The role of gibberellic acid in the hydrolysis of endosperm reserves in Zea mays. Planta 121 67–74. 10.1007/BF00384007 [DOI] [PubMed] [Google Scholar]
- Hawkins L. K., Mylroie J. E., Oliveira D. A., Smith J. S., Ozkan S., Windham G. L., et al. (2015). Characterization of the maize chitinase genes and their effect on Aspergillus flavus and aflatoxin accumulation resistance. PLOS ONE 10:e0126185. 10.1371/journal.pone.0126185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejgaard J., Jacobsen S., Svendsen I. (1991). Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 291 127–131. 10.1016/0014-5793(91)81119-S [DOI] [PubMed] [Google Scholar]
- Holmes R. A., Boston R. S., Payne G. A. (2008). Diverse inhibitors of aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 78 559–572. 10.1007/s00253-008-1362-0 [DOI] [PubMed] [Google Scholar]
- Hu X., Reddy A. S. (1997). Cloning and expression of a PR5-like protein from Arabidopsis: inhibition of fungal growth by bacterially expressed protein. Plant Mol. Biol. 34 949–959. 10.1023/A:1005893119263 [DOI] [PubMed] [Google Scholar]
- Huynh Q. K., Hironaka C. M., Levine E. B., Smith C. E., Borgmeyer J. R., Shah D. M. (1992). Antifungal proteins from plants. Purification, molecular cloning, and antifungal properties of chitinases from maize seed. J. Biol. Chem. 267 6635–6640. [PubMed] [Google Scholar]
- Iglesias M. J., Terrile M. C., Casalongue C. A. (2011). Auxin and salicylic acid signalings counteract the regulation of adaptive responses to stress. Plant Signal. Behav. 6 452–454. 10.4161/psb.6.3.14676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C., Norton R. A., Wicklow D. T., Dowd P. F. (2000). Isoform patterns of chitinase and beta-1,3-glucanase in maturing corn kernels (Zea mays L.) associated with Aspergillus flavus milk stage infection. J. Agric. Food Chem. 48 507–511. 10.1021/jf9905119 [DOI] [PubMed] [Google Scholar]
- Jiang T., Zhou B., Luo M., Abbas H. K., Kemerait R., Lee R. D., et al. (2011). Expression analysis of stress-related genes in kernels of different maize (Zea mays L.) inbred lines with different resistance to aflatoxin contamination. Toxins 3 538–550. 10.3390/toxins3060538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N. P., Butchko R., Sarr B., Phillips T. D. (1994). A visual pattern of mycotoxin production in maize kernels by Aspergillus spp. Phytopathology 84 483–488. 10.1094/Phyto-84-483 [DOI] [Google Scholar]
- Kelley R. Y., Williams W. P., Mylroie J. E., Boykin D. L., Harper J. W., Windham G. L., et al. (2012). Identification of maize genes associated with host plant resistance or susceptibility to Aspergillus flavus infection and aflatoxin accumulation. PLOS ONE 7:e36892. 10.1371/journal.pone.0036892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Zhang H., Woloshuk C., Shim W. B., Yoon B. J. (2015). Computational identification of genetic subnetwork modules associated with maize defense response to Fusarium verticillioides. BMC Bioinformatics 13:S12. 10.1186/1471-2105-16-S13-S12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler B. (1942). Natural mode of entrance of fungi into corn ears and some symptoms that indicate infection. J. Agric. Res. 64 421–442. [Google Scholar]
- Krebitz M., Wagner B., Ferreira F., Peterbauer C., Campillo N., Witty M., et al. (2003). Plant-based heterologous expression of Mal d 2, a thaumatin-like protein and allergen of apple (Malus domestica), and its characterization as an antifungal protein. J. Mol. Biol. 329 721–730. 10.1016/S0022-2836(03)00403-0 [DOI] [PubMed] [Google Scholar]
- Lanubile A., Bernardi J., Marocco A., Logrieco A., Paciolla C. (2012). Differential activation of defense genes and enzymes in maize genotypes with contrasting levels of resistance to Fusarium verticillioides. Environ. Exp. Bot. 78 39–46. 10.1016/j.envexpbot.2011.12.006 [DOI] [Google Scholar]
- Lanubile A., Ferrarini A., Maschietto V., Delledonne M., Marocco A., Bellin D. (2014). Functional genomic analysis of constitutive and inducible defense responses to Fusarium verticillioides infection in maize genotypes with contrasting ear rot resistance. BMC Genomics 15:710. 10.1186/1471-2164-15-710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanubile A., Logrieco A., Battilani P., Proctor R. H., Marocco A. (2013). Transcriptional changes in developing maize kernels in response to fumonisin-producing and nonproducing strains of Fusarium verticillioides. Plant Sci. 210 183–192. 10.1016/j.plantsci.2013.05.020 [DOI] [PubMed] [Google Scholar]
- Lanubile A., Maschietto V., De Leonardis S., Battilani P., Paciolla C., Marocco A. (2015). Defense responses to mycotoxin-producing fungi Fusarium proliferatum, F. subglutinans, and Aspergillus flavus in kernels of susceptible and resistant maize genotypes. Mol. Plant Microbe Interact. 28 546–557. 10.1094/MPMI-09-14-0269-R [DOI] [PubMed] [Google Scholar]
- Lanubile A., Pasini L., Marocco A. (2010). Differential gene expression in kernels and silks of maize lines with contrasting levels of ear rot resistance after Fusarium verticillioides infection. J. Plant Physiol. 167 1398–1406. 10.1016/j.jplph.2010.05.015 [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., Dong Q., Polacco M. L., Seigfried T. E., Brendel V. (2004). MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Res. 32 D393–D397. 10.1093/nar/gkh011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj E. B., Kwolek W. F., Fennell D. I., Milburn M. S. (1975). Aflatoxin incidence and association with bright greenish-yellow fluorescence and insect damage in a limited survey of freshly harvested high-moisture corn. Cereal Chem. 52 403–411. [Google Scholar]
- Ling Y., Du Z., Zhang Z., Su Z. (2010). ProFITS of maize: a database of protein families involved in the transduction of signaling in the maize genome. BMC Genomics 11:580. 10.1186/1471-2164-11-580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston D. P., Henson C. A., Tuong T. D., Wise M. L., Tallury S. P., Duke S. H. (2013). Histological analysis and 3D reconstruction of winter cereal crowns recovering from freezing: a unique response in oat (Avena sativa L.). PLOS ONE 8:e53468. 10.1371/journal.pone.0053468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston D. P., Tuong T. D., Haigler C. H., Avci U., Tallury S. P. (2009). Rapid microwave processing of winter cereals for histology allows identification of separate zones of freezing injury in the crown. Crop Sci. 49 1837–1842. 10.2135/cropsci2009.02.0077 [DOI] [Google Scholar]
- Lozovaya V. V., Waranyuwat A., Widholm J. M. (1998). Beta-1,3-Glucanase and resistance to Aspergillus flavus infection in maize. Crop Sci. 38 1255–1260. 10.2135/cropsci1998.0011183X003800050024x [DOI] [Google Scholar]
- Luo M., Brown R. L., Chen Z. Y., Menkir A., Yu J., Bhatnagar D. (2011). Transcriptional profiles uncover Aspergillus flavus-induced resistance in maize kernels. Toxins 3 766–786. 10.3390/toxins3070766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E., Freeling M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 53 661–673. 10.1111/j.1365-313X.2007.03326.x [DOI] [PubMed] [Google Scholar]
- Marsh S., Payne G. (1984). Preharvest infection of corn silks and kernels by Aspergillus flavus. Phytopathology 74 1284–1289. 10.1094/Phyto-74-1284 [DOI] [Google Scholar]
- Maschietto V., Lanubile A., Leonardis S., Marocco A., Paciolla C. (2016). Constitutive expression of pathogenesis-related proteins and antioxydant enzyme activities triggers maize resistance towards Fusarium verticillioides. J. Plant Physiol. 200 53–61. 10.1016/j.jplph.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Maschietto V., Marocco A., Malachova A., Lanubile A. (2015). Resistance to Fusarium verticillioides and fumonisin accumulation in maize inbred lines involves an earlier and enhanced expression of lipoxygenase (LOX) genes. J. Plant Physiol. 188 9–18. 10.1016/j.jplph.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Matasci N., McKay S. (2013). Phylogenetic analysis with the iPlant discovery environment. Curr. Protoc. Bioinformatics 42 6.13.1–6.13.13. [DOI] [PubMed] [Google Scholar]
- Mauch F., Dudler R. (1993). Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol. 102 1193–1201. 10.1104/pp.102.4.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50 267–294. 10.1146/annurev-phyto-081211-172955 [DOI] [PubMed] [Google Scholar]
- Menkir A., Brown R. L., Bandyopadhyay R., Cleveland T. E. (2008). Registration of six tropical maize germplasm lines with resistance to aflatoxin contamination. J. Plant Regist. 2 246–250. 10.3198/jpr2008.01.0028crg [DOI] [Google Scholar]
- Moore K. G., Price M. S., Boston R. S., Weissinger A. K., Payne G. A. (2004). A chitinase from Tex6 maize kernels inhibits growth of Aspergillus flavus. Phytopathology 94 82–87. 10.1094/PHYTO.2004.94.1.82 [DOI] [PubMed] [Google Scholar]
- Mukherjee A. K., Carpm M. J., Zuchman R., Ziv T., Horwitz B. A., Gepstein S. (2010). Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J. Proteomics 73 709–720. 10.1016/j.jprot.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Munkvold G. P. (2003). Cultural and genetic approaches managing mycotoxins in maize. Annu. Rev. Phytopathol. 41 99–116. 10.1146/annurev.phyto.41.052002.095510 [DOI] [PubMed] [Google Scholar]
- Munkvold G. P., McGee D. C., Carlton W. M. (1997). Importance of different pathways for maize kernel infection by Fusarium moniliforme. Phytopathology 87 209–217. 10.1094/PHYTO.1997.87.2.209 [DOI] [PubMed] [Google Scholar]
- Murillo I., Cavallarin L., Segundo B. S. (1999). Cytology of infection of maize seedlings by Fusarium moniliforme and immunolocalization of the pathogenesis-related PRms protein. Phytopathology 89 737–747. 10.1094/PHYTO.1999.89.9.737 [DOI] [PubMed] [Google Scholar]
- Musungu B. M., Bhatnagar D., Brown R. L., Payne G. A., OBrian G., Fakhoury A. M., et al. (2016). A network approach of gene co-expression in the Zea mays/Aspergillus flavus pathosystem to map host/pathogen interaction pathways. Front. Genet. 7:206. 10.3389/fgene.2016.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashita H., Yasuda M., Nitta T., Asami T., Fujioka S., Arai Y., et al. (2003). Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33 887–898. 10.1046/j.1365-313X.2003.01675.x [DOI] [PubMed] [Google Scholar]
- Navarro L., Bari R., Achard P., Lisón P., Nemri A., Harberd N. P., et al. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18 650–655. 10.1016/j.cub.2008.03.060 [DOI] [PubMed] [Google Scholar]
- Nemchenko A., Kunze S., Feussner I., Kolomiets M. (2006). Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J. Exp. Bot. 57 3767–3779. 10.1093/jxb/erl137 [DOI] [PubMed] [Google Scholar]
- Ogawa K. (2005). Glutathione-associated regulation of plant growth and stress responses. Antioxid. Redox Signal. 7 973–981. 10.1089/ars.2005.7.973 [DOI] [PubMed] [Google Scholar]
- Payne A. G., Yu J. (2010). “Ecology, development and gene regulation in Aspergillus flavus,” in Aspergillus: Molecular Biology and Genomics eds Machida M., Gomi K. (Norfolk, VA: Caister Academic Press; ) 157–171. [Google Scholar]
- Payne G. A. (1992). Aflatoxin in maize. Crit. Rev. Plant Sci. 10 423–440. 10.1080/07352689209382320 [DOI] [Google Scholar]
- Payne G. A., Cassel D. K., Adkins C. R. (1986). Reduction of aflatoxin contamination in corn by irrigation and tillage. Phytopathology 76 679–684. 10.1094/Phyto-76-679 [DOI] [Google Scholar]
- Pechanova O., Pechan T., Williams W. P., Luthe D. S. (2011). Proteomic analysis of the maize rachis: potential roles of constitutive and induced proteins in resistance to Aspergillus flavus infection and aflatoxin accumulation. Proteomics 11 114–127. 10.1002/pmic.201000368 [DOI] [PubMed] [Google Scholar]
- Peethambaran B., Hawkins L., Windham G. L., Williams W. P., Luthe D. S. (2010). Anti-fungal activity of maize silk proteins and role of chitinases in Aspergillus flavus resistance. Toxin Rev. 29 27–39. 10.3109/15569540903402874 [DOI] [Google Scholar]
- Pei-Bao Z., Ren A. Z., Xu H. J., Li D. C. (2010). The gene fpk1 encoding a cAMP-dependent protein kinase catalytic subunit homolog, is required for hyphal growth, spore germination, and plant infection in Fusarium verticillioides. J. Microbiol. Biotechnol. 20 208–216. [PubMed] [Google Scholar]
- Pieterse C. M., Leon-Reyes A., Van der Ent S., Van Wees S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5 308–316. 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C. M., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147 1347–1357. 10.1104/pp.108.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichhardt C., Ferreira J. A., Joubert L. M., Clemons K. V., Stevens D. A., Cegelski L. (2015). Analysis of the Aspergillus fumigatus biofilm extracellular matrix by solid-state nuclear magnetic resonance spectroscopy. Eukaryot. Cell 14 1064–1072. 10.1128/EC.00050-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rangel D., Sánchez-Nieto S., Plasencia J. (2012). Fumonisin B1, a toxin produced by Fusarium verticillioides, modulates maize beta-1,3-glucanase activities involved in defense response. Planta 235 965–978. 10.1007/s00425-011-1555-0 [DOI] [PubMed] [Google Scholar]
- Scarpari M., Punelli M., Scala V., Zaccaria M., Nobili C., Ludovici M., et al. (2014). Lipids in Aspergillus flavus-maize interaction. Front. Microbiol. 5:74. 10.3389/fmicb.2014.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidegger K. A., Payne G. A. (2003). Unlocking the secrets behind secondary metabolism: a review of Aspergillus flavus from pathogenicity to functional genomics. Toxin Rev. 22 423–459. 10.1081/TXR-120024100 [DOI] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Shu X. (2014). Pathogenesis and Host Response during Infection of Maize Kernels by Aspergillus flavus and Fusarium verticillioides. Doctoral dissertation, North Carolina State University; Raleigh, NC. [Google Scholar]
- Shu X., Livingston D. P., Franks R. G., Boston R. S., Woloshuk C. P., Payne G. A. (2015). Tissue specific gene expression in maize seeds during colonization by Aspergillus flavus and Fusarium verticillioides. Mol. Plant Pathol. 16 662–674. 10.1111/mpp.12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens J., Keller I., Sarx J., Kunz S., Schuller A., Nagel W., et al. (2006). Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol. Plant Microbe Interact. 19 480–494. 10.1094/MPMI-19-0480 [DOI] [PubMed] [Google Scholar]
- Smart M. G., Wicklow D. T., Caldwell R. W. (1990). Pathogenesis in Aspergillus ear rot of maize: light microscopy of fungal spread from wounds. Phytopathology 80 1287–1294. 10.1094/Phyto-80-1287 [DOI] [Google Scholar]
- Sobek E. A., Munkvold G. P. (1999). European corn borer (Lepidoptera: Pyralidae) larvae as vectors of Fusarium moniliforme, causing kernel rot and symptomless infection of maize kernels. J. Econ. Entomol. 92 503–509. 10.1093/jee/92.3.503 [DOI] [Google Scholar]
- Song W., Wang B., Li X., Wei J., Chen L., Zhang D., et al. (2015). Identification of immune related LRR-containing genes in maize (Zea mays L.) by genome-wide sequence analysis. Int. J. Genomics 2015:231358. 10.1155/2015/231358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Leger R. J., Screen S. E., Shams-Pirzadeh B. (2000). Lack of host specialization in Aspergillus flavus. Appl. Environ. Microbiol. 66 320–324. 10.1128/AEM.66.1.320-324.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytykiewicz H. (2011). Expression patterns of glutathione transferase gene (GstI) in maize seedlings under juglone-induced oxidative stress. Int. J. Mol. Sci. 12 7982–7995. 10.3390/ijms12117982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. D., Perkins A., Williams W. P., Warburton M. L. (2015). Using genome-wide associations to identify metabolic pathways involved in maize aflatoxin accumulation resistance. BMC Genomics 16:673. 10.1186/s12864-015-1874-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubajika K. M., Damann K. E. (2001). Sources of resistance to aflatoxin production in maize. J. Agric. Food Chem. 49 2652–2656. 10.1021/jf001333i [DOI] [PubMed] [Google Scholar]
- Turian G., Hamilton R. H. (1960). Chemical detection of 3-indolylacetic acid in Ustilago zeae tumors. Biochim. Biophys. Acta 41 148–150. 10.1016/0006-3002(60)90381-4 [DOI] [PubMed] [Google Scholar]
- Usadel B., Poree F., Nagel A., Lohse M., Czedik-Eysenberg A., Stitt M. (2009). A guide to using MapMan to visualize and compare omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 32 1211–1229. 10.1111/j.1365-3040.2009.01978.x [DOI] [PubMed] [Google Scholar]
- Vidal S., Eriksson A. R. B., Montesano M., Denecke J., Palva E. T. (1998). Cell wall-degrading enzymes from Erwinia carotovora cooperate in the salicylic acid-independent induction of a plant defense response. Mol. Plant Microbe Interact. 11 23–32. 10.1094/MPMI.1998.11.1.23 [DOI] [Google Scholar]
- Walters D. R., McRoberts N. (2006). Plants and biotrophs: a pivotal role for cytokinins? Trends Plant Sci. 11 581–586. [DOI] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A. H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17 1784–1790. 10.1016/j.cub.2007.09.025 [DOI] [PubMed] [Google Scholar]
- Wang X., Tang C., Deng L., Cai G., Liu X., Liu B., et al. (2010). Characterization of a pathogenesis-related thaumatin-like protein gene TaPR5 from wheat induced by stripe rust fungus. Plant Physiol. 139 27–38. 10.1111/j.1399-3054.2009.01338.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhou Z., Gao J., Wu Y., Xia Z., Zhang H., et al. (2016). The mechanisms of maize resistance to Fusarium verticillioides by comprehensive analysis of RNA-seq data. Front. Plant Sci. 7:1654. 10.3389/fpls.2016.01654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton M. L., Williams W. P. (2014). Aflatoxin resistance in maize: what have we learned lately? Adv. Bot. 2014:352831 10.1155/2014/352831 [DOI] [Google Scholar]
- Widstrom N. W., Wilson D. M., McMillian W. W. (1981). Aflatoxin contamination of preharvest corn as influenced by timing and method of inoculation. Appl. Environ. Microbiol. 42 249–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild C. P., Miller J. D., Groopman J. D. (eds). (2015). Mycotoxin Control in Low- and Middle Income Countries. IARC Working Group Report No. 9 Lyon: International Agency for Research on Cancer. [PubMed] [Google Scholar]
- Wilson R. A., Gardner H. W., Keller N. P. (2001). Cultivar-dependent expression of a maize lipoxygenase responsive to seed infesting fungi. Mol. Plant Microbe Interact. 14 980–987. 10.1094/MPMI.2001.14.8.980 [DOI] [PubMed] [Google Scholar]
- Wisser R. J., Kolkman J. M., Patzoldt M. E., Holland J. B., Yu J., Krakowsky M., et al. (2011). Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proc. Natl. Acad. Sci. U.S.A. 108 7339–7344. 10.1073/pnas.1011739108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Kriz A. L., Widholm J. M. (1994a). Molecular analysis of two cDNA clones encoding acidic class I chitinase in maize. Plant Physiol. 105 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Kriz A. L., Widholm J. M. (1994b). Nucleotide sequence of a maize cDNA for a class II, acidic beta-13-glucanase. Plant Physiol. 106 1709–1710. 10.1104/pp.106.4.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuren T., Toyotome T., Yamaguchi M., Takahashi-Nakaguchi A., Muraosa Y., Yahiro M., et al. (2014). Effect of serum components on biofilm formation by Aspergillus fumigatus and other Aspergillus species. Jpn. J. Infect. Dis. 67 172–179. 10.7883/yoken.67.172 [DOI] [PubMed] [Google Scholar]
- Xie Y. R., Chen Z. Y., Brown R. L., Bhatnagar D. (2010). Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J. Plant Physiol. 167 121–130. 10.1016/j.jplph.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Yang D. L., Yang Y., He Z. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant 6 675–685. 10.1093/mp/sst056 [DOI] [PubMed] [Google Scholar]
- Yilmaz A., Nishiyama M. Y., Jr., Fuentes B. G., Souza G. M., Janies D., Gray J., et al. (2009). GRASSIUS: a platform for comparative regulatory genomics across the grasses. Plant Physiol. 149 171–180. 10.1104/pp.108.128579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Zhang J., Ye N., Cao J., Tan M., Zhang J., et al. (2010). ZmMPK5 is required for the NADPH oxidase-mediated self-propagation of apoplastic H2O2 in brassinosteroid-induced antioxidant defence in leaves of maize. J. Exp. Bot. 61 4399–4411. 10.1093/jxb/erq243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Simmons C., Yalpani N., Crane V., Wilkinson H., Kolomiets M. (2005). Genomic analysis of the 12-oxo-phytodienoic acid reductase gene family of Zea mays. Plant Mol. Biol. 59 323–343. 10.1007/s11103-005-8883-z [DOI] [PubMed] [Google Scholar]
- Zhang Y., Cui M., Zhang J., Zhang L., Li C., Kan X., et al. (2016). Confirmation and fine mapping of a major QTL for aflatoxin resistance in maize using a combination of linkage and association mapping. Toxins 8:E258. 10.3390/toxins8090258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Liu W., Sheng Y., Zhang J., Chiu T., Yan J., et al. (2015). ABA affects brassinosteroid-induced antioxidant defense via ZmMAP65-1a in maize plants. Plant Cell Physiol. 56 1442–1455. 10.1093/pcp/pcv061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


