Abstract
Background and Objectives:
Human milk is a continuous supply of Lactic Acid bacteria (LAB), including enterococci with probiotic potentials. The aim of this study was to analyze two Enterococcus species, isolated from human milk for their probiotic potential, bacteriocin producing ability and virulence traits.
Materials and Methods:
Enterococcus faecium TA0033 and E. faecalis TA102 were tested for acid and bile tolerance, survival in simulated gastric and intestinal conditions. The antibacterial spectrum of the isolates was tested by agar well diffusion assay. The antagonistic agent was characterized by physico-chemical methods. The enterocin structural genes, virulence determinants, vancomycin resistance and biogenic amine genes, such as hdc1, hdc2, tdc, ldc and odc were also determined.
Results:
The tested isolates survived acidic conditions, high bile salt (1%), simulated gastric and intestinal conditions. The culture supernatant fluids of the two isolates inhibited the growth of Escherichia coli, Listeria monocytogenes, Salmonella typhi, Staphylococcus aureus, Shigella dysenteriae and Streptococcus agalactiae. The antagonistic activity was lost in the presence of proteolytic enzymes but tolerated the action of catalase, lysozyme and lipase. In contrast to enterocin TA102, enterocin TA0033 possessed bactericidal mode of action. Bacteriocin structural genes, entA and entB were present in the genome of the two isolates, while E. faecalis TA102 additionally harboured entP and bac31 genes. The phenotypic and genotypic virulence assessment studies indicated hyaluronidase (hyl) production and vancomycin resistance in E. faecalis TA102 while, none of the isolates harboured the biogenic amine genes.
Conclusion:
The presence of virulence genes in E. faecalis TA102 calls for careful monitoring of Enterococcus isolates for their safety parameters.
Keywords: Enterococcus, Bacteriocins, Biogenic amines, Human milk, Vancomycin, Virulence genes
INTRODUCTION
Enterococci are Gram-positive cocci well known for their role in enhancing the shelf life, developing flavor compounds in food products, improving the intestinal microbial balance and treating gastroenteritis in humans and animals (1). Owing to the known health benefits of these bacteria they are used in a number of probiotic formulations. A significant character of Enterococcus species is their bacteriocin producing ability, which is a useful biotechnological trait (2). To date, a number of bacteriocin producing enterococci has been isolated from breast milk, and their bacteriocins (enterocins) been characterized (3). Enterocins are defined as small, ribosomally synthesized peptides displaying inhibitory actions, against the target bacteria by either dissipation of proton motive force by pore formation, cell lysis, or interference with degradation and metabolism of macromolecules (4). A number of reports have indicated the presence of virulence traits in this group of bacteria and their safety aspects are still under considerations. All this calls for strict scrutinzations before considering them as a suitable probiotic candidate. Some of the significant virulence genes reported to date are; ace (collagen binding protein), asa1 (aggregation substance), cpd (sex pheromone peptides), cylA (cytolysin activation), cylB (cytolysin transport), cylM (posttranslational modification of cytolysin), efaA (endocarditis antigen), esp (enterococcal surface protein), gelE (Gelatinase), and hyl (hyaluronidase) (5).
In the last few decades, vancomycin resistant Enterococci have emerged, causing major problems in infection control. Three phenotypic classes (VanA, VanB and VanC) are defined by the level of resistance to vancomycin and susceptibility or resistance to teicoplanin (6). Moreover, biogenic amines (BA) present in food are known to have deleterious health effects on humans, and thus selection of strains with no BA producing ability is highly recommended (7, 8). Hence, before we could exploit these bacteria in the food or biopharmaceutical industry, and or as a probiotic supplement, it is essential to evaluate their safety. In this study, we evaluated the bacteriocinogenic and virulence characteristics of two Enterococcus isolates by phenotypic and genotypic methods.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
E. faecium TA0033 and E. faecalis TA102, isolated from human milk samples of healthy young mothers in Tehran, capital of Iran, were identified by their carbohydrate fermentation pattern and 16S rRNA sequencing (9, 10). The isolates were cultured on KF agar, supplemented with 1% TTC (2, 3, 5-Triphenyl-Tetrazolium Chloride Solution, SIGMA, UK) and Kanamycin aesculin-Azide Agar (KAA, Oxoid, UK). All other Gram-positive and Gram-negative pathogens, used in the study were grown in BHI (Brain heart Infusion, Merck, USA) broth, at 37°C for 24 h in aerobic conditions. Stock-cultures were maintained in MRS broth supplemented with 20% glycerol and stored at −70°C.
Probiotic characterization of the isolates: Acid and bile tolerance.
The isolates were screened for their acid and bile resistance, by culturing in MRS broth with different set pH values (2, 2.5, 3, 4, 5, 6) and bile concentrations (0.1, 0.5, 0.7, 1%). The resistance of the isolates at the tested pH values was recorded, by determining their growth (cfu/ml) at different time intervals. While, bile tolerance was estimated, by calculating the Coefficient of inhibition (C inh ), according to the formula described by Gopal et al. (11).
Where, Δ T8-T0 represents the difference in absorbance at time zero (T0) and after 8 h (T8). C inh of less than 0.4 was considered significant, for the isolates to be considered as a suitable probiotic candidate.
Survival in simulated gastric and intestinal conditions.
The survival of the selected isolates under simulated gastric and intestinal contents was studied, by previously described method (12). The bacterial survival under the tested conditions was calculated as:
According to the formula, R =1 when no effect on the growth and survival of bacteria is seen, while a ratio of 0.5, indicated a loss of 50% of the viability. Ratios greater than 1 indicated bacterial growth.
Antibacterial spectrum.
The inhibitory spectrum of the selected isolates was studied, by determining the antagonistic action of their neutralized supernatant fluids (NSF), against a number of pathogens by agar well diffusion assay (13). Freshly grown cultures of a number of Gram-positive and Gram-negative pathogens, listed in Table 3, were used as sensitive indicator cells. Zone diameters, surrounding the wells were measured in millimeters, and based on these results the producer isolates were recorded as strong (≥20mm), moderate (≥16–19 mm) and weak (≤15 mm). The antibacterial activity demonstrated by the isolates was quantified, by slight modifications in the critical dilution method, described by Schillinger and Lucke (13). Two fold serial dilutions (100 μl) of NSF were poured in wells in agar plates seeded with the indicator strain, and incubated aerobically at 37ºC for 24 h. The diameters of the inhibition zones were measured in millimeters, by subtracting the well diameter from the zone diameter. The results were expressed in arbitrary units per millimeter (AU/ml), defined as the reciprocal of the highest dilution demonstrating zone of inhibition.
Table 3.
Effect of heat treatment on the enterocin activity (AU/ml) of the Enterococcus isolates, at different time intervals
| Time (min) | Enterocin TA0033 (AU/ml) | Enterocin TA102 (AU/ml) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 60°C | 80°C | 100°C | 120°C | 60°C | 80°C | 100°C | 120°C | ||||||
| 0 | 25600 | 25600 | 25600 | 25600 | 6400 | 6400 | 6400 | 6400 | |||||
| 15 | 25600 | 25600 | 0 | 0 | 6400 | 6400 | 6400 | 6400 | |||||
| 30 | 12800 | 6400 | 0 | ND | 6400 | 6400 | 6400 | 0 | |||||
| 60 | 6400 | 6400 | 0 | ND | 6400 | 6400 | 1600 | ND | |||||
| 90 | 6400 | 1600 | 0 | ND | 6400 | 6400 | 0 | ND | |||||
ND: not determined
Physicochemical characterization of the antagonistic agents.
The antagonistic agents produced by the two isolates were physico-chemically characterized. The effect of H 2 O 2 for the antimicrobial activity was excluded, by determining the antagonistic activity in the NSF after subjection to the enzyme catalase (1 mg/l). While, the chemical nature of the antagonistic agent was analyzed by treating the NSF of the producer strains with the enzymes like lipase, lysozyme, pepsin, pronase E, and proteinase K (Fluka, England), at a final concentration of 1 mg/l in phosphate buffer (10, 14).
Effect of variable pH (2.0, 4.0, 6.0, 8.0, and 10.0) and temperature ranges (60, 80, 100, 121°C) on the NSF of the two producer isolates, at different time intervals of 15, 30, 60, 90 min, was studied.
The bactericidal or bacteriostatic activity of the studied bacteriocins was determined by critical dilution assay using S. aureus as indicator strain. After 24 h, 5 μl of 10 mg/ml proteinase K solution (Sigma, USA) were spotted near each inhibition zone and the plates were again incubated at 37°C and observed for presence or absence of growth of indicator strain. Bactericidal mode of action of the bacteriocins was determined by the absence of indicator strain growth, after the destruction of the inhibitor by proteases. Absence of inhibition zones after enzymatic treatment indicated a bacteriostatic mode of action.
Enterocin structural genes.
The presence of bacteriocin structural genes in the producer isolates was studied, using a set of primers in a PCR assay. Multiple pairs of enterocin structural genes were used, as described previously (14, 15). DNA template was prepared by suspending a loop full of bacterial colony in 10μl of lysis buffer (0.25% SDS/ 0.05 % NaOH), heated at 95°C for 5 min and centrifuged at 15 000 × g for 5 min. The samples were diluted in 90 μl of sterile distilled water, centrifuged as above and the supernatant used as template DNA. Cycling parameters included a 2 min initial denaturation at 94°C, followed by 40 cycles of 45 s at 95°C, 30 s at 56°C for entP, bac31 and entL50A/B, 58°C in the case of entA and 60°C for entB, entQ, and cyl as annealing temperature, and 60 s at 72°C. Amplified PCR fragments were resolved on 1% agarose gels, using a 100 bp ladder for size verification.
Biochemical virulence traits.
The two isolates were screened for hemolytic activity, arginine hydrolysis, gelatinase, lipase, DNase, lecithinase and hyaluronidase production (16).
The ability of the mentioned Enterococcus isolates to produce biogenic amines (tyramine, histamine, putrescine, and cadaverine) was determined, using the decarboxylase broth and the method described by Bover-Cid and Holzapfel (17).
The phenotypic resistance of the selected isolates to vancomycin (30ug, Sigma, USA) was determined, by disc diffusion assay reported previously (18). According to the recommendation of the Clinical and Laboratory Standards Institute (CLSI), the strain was considered to be resistant to antibiotics, if the inhibition zone was ≤ 14 mm for the indicated antibiotic.
Genotypic virulence determinants.
Virulence in Enterococcus species was determined by PCR assay, using a set or primers targeting the virulence genes. Primer sequences and expected amplicons size are listed in Table 1. E. faecalis ATCC 29212; ATCC 51299 and E. faecium ATCC-BAA 2320; ATCC 19434 were used as positive control during this study. PCR parameters used, were according to the respective references (Table 1).
Table 1.
Virulence genes and their sequences, used in the study
| Genes | Primer Sequences (5-3) | Amplicon size (bp) | Reference |
|---|---|---|---|
| Virulence genes | |||
| agg-F | AAGAAAAAGAAGTAGAGACCAAC | 1553 | (1) |
| agg-R | AAACGGCAAGACAAGTAAATA | ||
| asa1-F | GCACGCTATTACGAACTATGA | 375 | (19) |
| asa1-R | TAAGAAAGAACATCACCACGA | ||
| cpd-F | TGGTGGGTTATTTTTCAATTC | 782 | (19) |
| cpd-R | TACGGACTCTGGCTTACTA | ||
| cylA-F | ACTCGGGGATTGATAGGC | 688 | (19) |
| cylA-R | GCTGCTAAAGCTGCGCTT | ||
| cylB-F | AAGTACACTAGTAGAACTAAGGGA | 2020 | (19) |
| cylB-R | ACAGTGAACGATATAACTCGCTATT | ||
| cylM-F | AAAAGGAGTGCTTACACTGGAAGAT | 2940 | (19) |
| cylM-R | CATAACCCACACCACTGATTCC | ||
| efaA-F | GACAGACCCTCACGAATA | 705 | (1) |
| efaA-R | AGTTCATCATGCTGTAGTA | ||
| esp-F | AGATTTCATCTTTGATTCTTGG | 510 | (19) |
| esp-R | AATTGATTCTTTAGCATCTGG | ||
| gelE-F | TATGACAATGCTTTTTGGGAT | 213 | (19) |
| gel-R | AGATGCACCCGAAATAATATA | ||
| hyln-F | ACAGAAGAGCTGCAGGAAATG | 276 | (19) |
| hyln-R | GACTGACGTCCAAGTTTCCAA | ||
| Vancomycin resistance genes | |||
| VanA-F | CGGGGAAGATGGCAGTAT | 732 | (22) |
| VanA-R | CGCAGGGACGGTGATTTT | ||
| VanB-F | CGGGGAAGATGGCAGTAT | 635 | (22) |
| VanB-R | CGCAGGGACGGTGATTTT | ||
| VanC-F | CGGGGAAGATGGCAGTAT | 484 | (22) |
| VanC-R | CGCAGGGACGGTGATTTT | ||
| Genes for biogenic amines | |||
| hdc1-F | AGATGGTATTGTTTCTTATG | 367 | (20) |
| hdc1-R | AGACCATACACCATAACCTT | ||
| hdc2-F | AAYTCNTTYGAYTTYGARAARGARG | 534 | (20) |
| hdc2-R | ATNGGNGANCCDATCATYTTRTGNCC | ||
| tdc-F | ACATAGTCAACCATRTTGAA | 1100 | (21) |
| tdc-R | CAAATGGAAGAAGAAGTAGG | ||
| ldc1-F | TTYGAYWCNGCNTGGGTNCCNTAAC | 1098 | (20) |
| ldc1-R | CCRTGDATRTCNGTYTCRAANCCN | ||
| odc-F | TGCACTTCCATATCCTCCAG | 127 | (23) |
| odc-R | GAATTTCTGGAGCAAATC |
In order to determine Vancomycin resistance genes in the test isolates, three sets of primers namely VanA, VanB and VanC were used in a PCR reaction, as described earlier (19).
The primer sequences targeting genes for amino acid decarboxylases, including hdc1 and hdc2 (both related to histidine decarboxylase), tdc (tyrosine decarboxylase), ldc (lysine decarboxylase) and odc (ornithine decarboxylase) were selected for studies. Primer sequences and PCR parameters were, as described previously (1, 6, 19–23).
Statistical Analyses.
Differences in the prevalence of virulence genes between the two Enterococcus species were compared, using the Chi-square test with a p-value < 0.05, indicating statistical significance.
RESULTS
E. faecium TA0033 and E. faecalis TA102 isolated from colostrum of healthy mothers in a previous study (NCBI Gene Bank with accession numbers KX158836.1 and KY009901.1, respectively), were evaluated for their probiotic potential and safety traits.
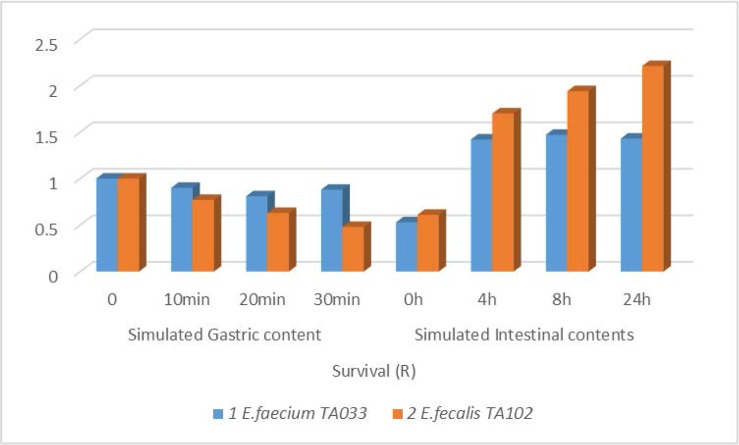
In vitro probiotic characterization of the isolates indicated the isolates to be resistant to acidic pH values of 2.5 and above, while none could survive lower pH values of 2. Enhanced survival rate of the tested Enterococcus isolates in the presence of 1% bile concentrations was observed, as their coefficient of inhibition (C inh ) values, appeared to be less than 0.4%. The survival rate of the selected isolates, in simulated gastric and intestinal content was indicated by their calculated R value. Fig. 1 shows survival of E. faecalis TA102 in simulated gastric conditions to be below 50 %, while in simulated intestinal conditions their survival percentage was significantly higher (R value > 2), compared to E. faecium TA0033.
Fig. 1.
The survival rate of Enterococcus isolates in stimulated gastric and upper intestine contents, at different time intervals
The cell free supernatant fluid (CFSF) of the Enterococcus isolates demonstrated wide antibacterial spectrum, against a range of Gram-positive and Gam-negative pathogens. The CFSF of both the isolates were inhibitory towards the growth of E. faecium, E. coli, L. monocytogenes, S. typhi, S. aureus, Sh. dysenteriae and S. agalactiae. However, none of the isolates could inhibit the growth of S. pyogenes (Table 2). The spectrum of activity of E. faecium TA0033 appeared significantly higher than E. faecalis TA102 (P≤0.05).
Table 2.
Antibacterial spectrum of cell free supernatant fluids of Enterococcus species, against pathogens
| Bacterial Pathogens | Reference Type | E. faecium TA0033 | E. faecalis TA102 |
|---|---|---|---|
| Bacillus cereus | PTCC 1015 | W | N |
| Bacillus subtilis | RTCC 1058 | S | M |
| Enterococcus faecium | ATCC 19434 | S | S |
| Enterococcus faecium | ATCC-BAA2320 | W | N |
| Enterococcus faecalis | ATCC 29212 | S | W |
| Enterococcus faecalis | ATCC 51299 | W | N |
| Escherichia coli | RTCC 1162 | S | S |
| Escherichia coli O157:H7 | ATCC 43888 | N | S |
| Escherichia coli K99 | Unknown | W | N |
| Klebsiella pneumoniaae | PTCC1290 | W | N |
| Listeria ivanovii | RTCC 13311 | N | M |
| Listeria monocytogenes | RTCC 1240 | M | S |
| Pasteurella multocida | ATCC 43137 | W | N |
| Pseudomonas aeruginosa | RTCC 1502 | S | M |
| Salmonella typhi | Local isolate | S | S |
| Shigella dysenteriae | PTCC 1188 | W | S |
| Staphylococcus aureus | ATCC 64542 | S | S |
| Staphylococcus epidermidis | ATCC 12228 | N | W |
| Streptococcus agalactiae | RTCC 2051 | S | S |
| Streptococcus pyogenes | ATCC 19615 | N | N |
ATCC: American type culture collection; PTCC: Persian type culture collection; RTCC: Razi type culture collection.
S: Strong anti-bacterial activity (zone diameter ≥20mm)
M: Moderate anti-bacterial activity (zone diameter ≥16–19mm)
W: Weak anti-bacterial activity (zone diameter ≤15mm)
N: No anti-bacterial activity (absence of zone of inhibition)
All experiments performed in triplicate.
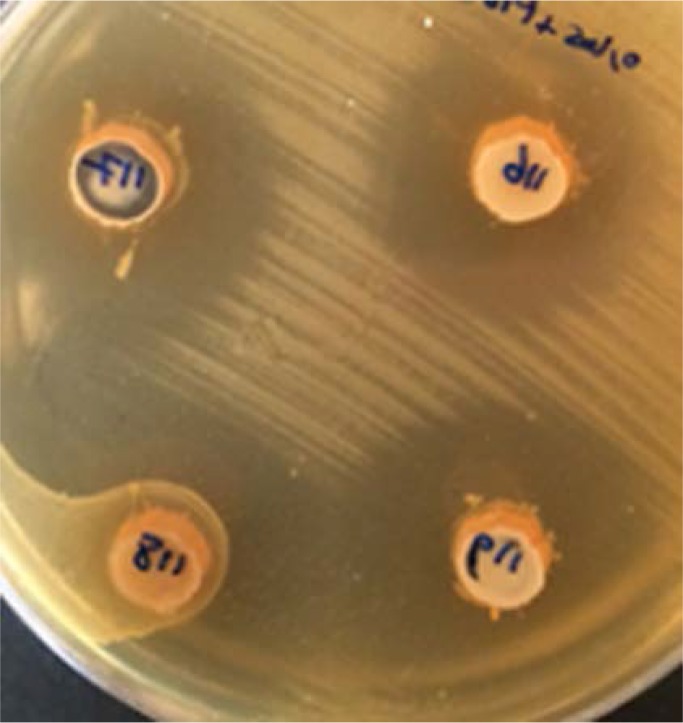
The inhibitory activity demonstrated by the isolates, appeared unaffected by neutralization of their supernatant fluid and the action of enzyme catalase. These results indicated the absence of acid and H 2 O 2 for the anti-bacterial activity ( Fig. 2). The proteolytic nature of the antagonistic agent produced by both the isolates was confirmed, by their sensitivity to the tested proteolytic enzymes. In contrast, lipase and lysozyme had no effect on the inhibitory actions of the two isolates.
Fig. 2.
Effect of neutral pH, catalase, lipase and lysozyme on the enterocin produced by E. faecium TA0033
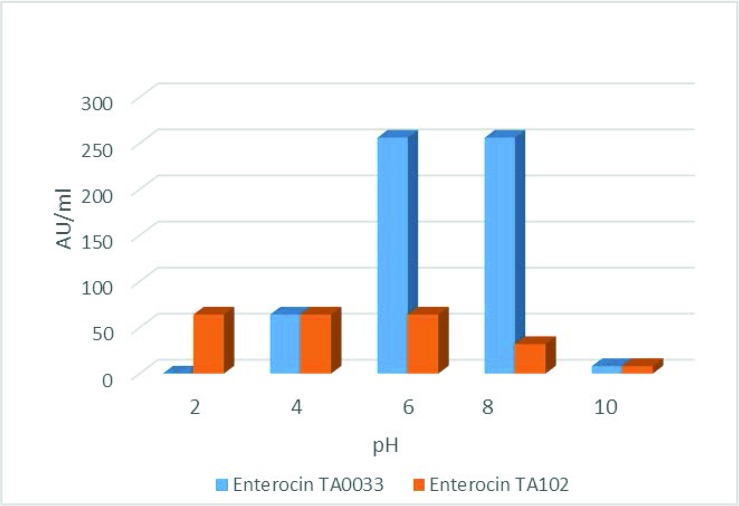
Effect of variable pH on the proteinaceous antagonistic compound produced, by the Enterococcus isolates in study is illustrated in Fig. 3 Significant differences were recorded in the pH stability of the studied enterocins (P≤0.05). According to the obtained results, enterocins TA0033 was not able to resist the extreme acidic (pH 2.0) and alkaline (pH 10) conditions, whereas, enterocin TA102 retained its stability at pH 2.0 for 24 h. Both the enterocins lost their activity completely, at an alkaline pH 10 and above. Maximum activity (AU/ml) was recorded at pH values of 2.0, 6.0 and 8.0 in both the isolates.
Fig. 3.
Acid resistance of enterocin TA0033 and enterocin TA102 at different pH within 24 h.
Thermal stability of enterocins TA0033 and TA102 is depicted in Table 3. Significant differences (p≤0.05) were recorded, in the thermal stability of the two enterocins. Enterocin TA102 appeared to be highly temperature resistant, compared to enterocins TA0033, as it was able to resist 100°C for 60 min and 121°C for 15 minutes.
In contrast, enterocin TA0033 was unstable at 100°C and autoclaving temperatures and lost its activity after exposure to the indicated temperatures. Partial loss in activity was seen with enterocin TA0033 at 60 and 80°C after 60 min of exposure with 75% reduction in activity within 90 min. In contrast, no loss of activity of enterocin TA102 was seen at these temperatures, during the tested time intervals and no significant difference (p>0.05), in activity between treated and untreated supernatant were observed.
Enterocin TA0033 displayed bactericidal mode of action at 6, 400 AU/ml, whereas bacteriostatic activity was detected at lower concentrations (200 AU/ml). Enterocin TA102 demonstrated bacteriostatic mode of action, as seen by the absence of inhibition zones after enzymatic treatment.
Multiple enterocin genes were detected in the tested isolates. The isolates possessed entA and entB, as indicated by a visible band of 126 and 159 bp, respectively. Whereas, entP and bac31, corresponding to DNA bond size of 121 bp and 248 bp, respectively, were detected only in E. faecalis TA102.
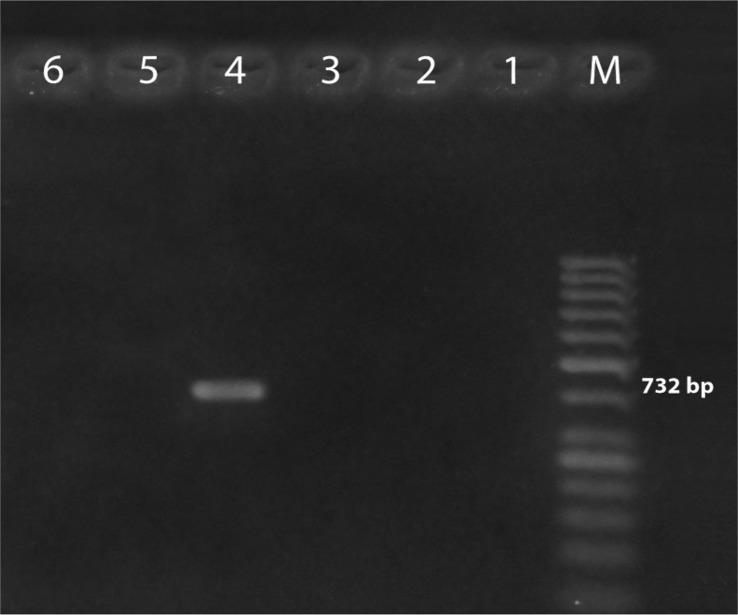
The phenotypic and genotypic virulence traits of the isolates are indicated in Table 4. During virulence trait characterizations, E. faecium TA0033 appeared non virulent as none of the tested virulence factors were observed in this isolate during biochemical and molecular genetic analysis. However, E. faecalis TA102 showed the presence of cylA (688 bp, cytolysin), esp (510bp, Enterococcal surface protein) and vanA (732 bp, Vancomycin) (Fig. 4).
Table 4.
Phenotypic and genotypic virulent characters in Enterococcus isolates
| Virulence determinants | E. faecium TA0033 | E. faecalis TA102 |
|---|---|---|
| Phenotypic virulence traits | ||
| Hemolytic activity | α | γ |
| Arginine hydrolysis | −ve | −ve |
| Gelatinase production | −ve | −ve |
| Lipase production | −ve | −ve |
| DNase production | −ve | −ve |
| Lecithinase production | −ve | −ve |
| Hyaluronidase production | −ve | −ve |
| Vancomycin (30ug) | S | R |
| Genotypic Virulence traits | ||
| agg | −ve | −ve |
| asa | −ve | −ve |
| cpd | −ve | −ve |
| cylA | −ve | +ve |
| cylB | −ve | −ve |
| cylM | −ve | −ve |
| efa | −ve | −ve |
| esp | −ve | +ve |
| gelE | −ve | −ve |
| hyl | −ve | −ve |
| Genes for Vancomycin Resistance | ||
| vanA | −ve | +ve |
| vanB | −ve | −ve |
| vanC | −ve | −ve |
| Genes for Biogenic Amines | ||
| hdc1 | −ve | −ve |
| hdc2 | −ve | −ve |
| tdc | −ve | −ve |
| ldc | −ve | −ve |
| odc | −ve | −ve |
Fig. 4.
Vancomycin resistance genes in E. faecium TA033 and E. faecalis TA102. Lanes M: Molecular weight marker; Lane 1, 2, 3: vanA, vanB and vanC in E. faecium TA0033; Lanes 4, 5, 6: vanA, vanB and vanC in E. faecalis TA102
The genomic DNA of the two Enterococcal isolates was subjected to the genes coding for the enzymes involved in biogenic amines (BA) production, including hdc1, hdc2, tdc, ldc, and odc. None of the respective genes appeared to be present in the genome of the tested Enterococcus species and the two isolates were considered BA negative.
DISCUSSION
Human milk is a potential source of probiotic bacteria, including lactobacilli, streptococci, bifidobacteria and enterococci to the infant gut, affecting the overall composition of the neonate gut microbiota. Although enterococci are widely recognized as probiotic bacteria, but opposed to other LAB genus they have yet not been assigned the GRAS (generally recognized as safe) status. E. faecium is considered a suitable probiotic candidate for the modulation of immune responses against pathogens (24). In this study, we observed the probiotic properties of the two Enterococcus species, including their resistance in stress conditions like acidic environment, high bile salt concentrations, and simulated gastric and intestinal conditions. Another important characteristic of the isolates in the study was their wide antibacterial spectrum against a number of Gram-positive and Gram-negative pathogens. In agreement with our findings, Ghrairi and colleagues (25), reported E. faecium MMT21 bacteriocin ability to inhibit not only closely related LAB, but also L. monocytogenes and S. aureus. Different spectrum of inhibitory action may be based on the bacteriocin producing strain, the indicator strain, and also the method used for bacteriocin detection. During physicochemical characterizations of the antagonistic agent produced by the two Enterococcus species in study, it was observed that the antibacterial actions exerted by the isolates were not due to acids or hydrogen peroxide. Similar to other reports (26), enterocin TA0033 and TA102 appeared to be simple proteins rather than conjugated proteins linked to lipid or carbohydrate moiety. Temperature and pH stability of enterocins is considered an essential aspect, which may compromise their use in dairy products (27). Comparable to enterocin SE-K4, enterocin CRL 1826, mundticin KS and enterocin QU2 (28), Enterocin TA102 showed significant pH and thermal stability. The mentioned enterocin TA102, showed significantly higher resistance in these conditions, compared to other reported enterocins produced by E. faecium D081821, and E. faecium D081833.
Presence of more than one bacteriocin gene in Enterococcus species has been reported previously ( 4, 10, 12, 14). In agreement with these reports, we observed multiple enterocins genes in both of the producer isolates. Nevertheless, the incidence of numerous enterocin genes in enterococci is not necessarily associated with an enhanced bacteriocin activity, and not all enterocin genes should be expressed at the same time (29).
A microorganism considered, as a probiotic is essentially either of GRAS status or their safety parameters are required to be investigated, before they could be considered a probiotic. Enterococcus species have been known to be responsible for nosocomial infections, especially in neonates, and those who suffer from underlying diseases (30). Therefore, the enterococcal strain of clinical or industrial interest should be carefully and individually evaluated for their safety and associated risk factors. In this study, we could demonstrate the safety of E. faceium TA0033, as none of the tested phenotypic or genotypic virulent traits were observed in this isolate. In contrast, E. faecalis TA102 showed the presence of few pathogenic genes like cylA and esp. Our results are in accordance with the previous findings, stating that pathogenic virulence factors are more common in E. faecalis strains compared to E. faecium strains (31).
Antibiotic resistance is one of the major safety concerns in Enterococci strains, particularly vancomycin resistance, as it is the drug of choice efficient against clinical infections by multidrug resistance pathogens. In our studies, E. faecalis TA102 appeared vancomycin resistant, during phenotypic and genotypic analysis. According to the reports, if the strain has either of the vancomycin resistance genes such as vanA, vanB and vanC, it is considered unsafe and could not be applied in the food or feeds (32). Another important safety concerns, are biogenic amine production by some probiotic isolates to be used in the dairy industry. Biogenic amines contained foods are known to have toxicological properties and are known to trigger health problems such as allergies, hypertension, hypotension, headaches, depressions, schizophrenia and Parkinson disorder (7) and thus should be avoided in food (33, 34). Absence of BA genes highlights the importance of a strain for use in starter and adjunct cultures.
In conclusion, results of this study determine the possible advantage of the indicated enterocins in dairy or biotechnology industry. The presence of couple of virulence genes in E. faecalis TA102 calls for careful monitoring of Enterococcus isolates, for their safety parameters.
ACKNOWLEDGEMENTS
A section of this work was financially supported by the Iran National Science Foundation (INSF), project No 92044469, during the year 2015–2017.
The authors declare no conflict of interest.
REFERENCES
- 1.Eaton TJ, Gasson MJ. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol 2001; 67: 1628–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franz CM, Huch M, Abriouel H, Holzapfel WH, Galvez A. Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 2011; 151: 125–140. [DOI] [PubMed] [Google Scholar]
- 3.Kozak K, Charbonneau D, Sanozky-Dawes R, Klaehammer T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes 2015; 6: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishibashi N, Himeno K, Fujita K, Masuda Y, Perez RH, Zendo T, et al. Purification and characterization of multiple bacteriocins and an inducing peptide produced by E. faecium NKR-5-3 from Thai fermented fish. Biosci Biotechnol Biochem 2012; 76: 947–953. [DOI] [PubMed] [Google Scholar]
- 5.Carlos A, Semedo-Lemsaddek T, Barreto-Crespo M, Tenreiro R. Transcriptional analysis of virulence-related genes in enterococci from distinct origins. J Appl Microbiol 2010; 108: 1563–1575. [DOI] [PubMed] [Google Scholar]
- 6.Miele A, Bandera M, Goldstien BH. Use of primers selective for vancomycin resistance genes to determine van genotype in enterococci and to study gene organization in VanA isolates. Antimicrob Agents Chemother 1995; 39: 1772–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elsanhoty RM, Ramadan MW. Genetic screening of biogenic amines production capacity from some lactic acid bacteria strains. Food Control 2016; 68: 220–228. [Google Scholar]
- 8.Laukova A, Szaboova R, Pleva P, Bunkova L, Chrastinova L. Decarboxylase-positive Enterococcus faecium strains isolated from rabbit meat and their sensitivity to enterocins. Food Sci Nutr 2017; 5: 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalkhali S, Mojgani N. Characterization of candidate probionts isolated from human breast milk. Cell Mol Biol 2017; 82–88. [DOI] [PubMed] [Google Scholar]
- 10.Mojgani N, Vaseji N, Khalkhali S, Naz Baloch M. Biochemical and molecular analysis of the antilisterial peptides produced by Enterococcus hirae strains isolated from raw ewe milk. J Adv Biol Biotech 2017;11: 1–11. [Google Scholar]
- 11.Gopal PK, Prasad J, Smart J, Gill HS. In vitro adherence properties of L. rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int J Food Microbiol 2001; 67:207– 216. [DOI] [PubMed] [Google Scholar]
- 12.Mojgani N, Hussaini F, Vaseji N. Characterization of indigenous Lactobacillus strains for probiotic properties. Jundishapur J Microbiol 2015; 8(2): e17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schillinger U, Lücke FK. Antibacterial activity of L. sake isolated from meat. Appl Environ Microbiol 1989; 55: 1901–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aymerich T, Holo H, Håvarstein LS, Hugas M, Garriga M, Nes IF. Biochemical and genetic characterization of enterocin A from E. faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol 1996; 62: 1676–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nami Y, Haghshenas B, Haghshenas M, Khosroushahi AD, Rosli R, Khosroushahi Y. Antimicrobial activity and the presence of virulence factors and bacteriocin structural genes in E. faecium CM33 isolated from ewe colostrum. Frontiers Microbiol 2015; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Azeredo LA, Leite SG, Freire DM, Benchetrit LC, Coelho R. Proteases from actinomycetes interfere in solid media plate assays of hyaluronidase activity. J Microbiol Methods 2001; 45: 207–212. [DOI] [PubMed] [Google Scholar]
- 17.Bover-Cid S, Holzapfel WH. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int J Food Microbiol 1999; 53: 33–41. [DOI] [PubMed] [Google Scholar]
- 18.Bauer A, Kirby WM, Sheris JKT. Antimicrobial susceptibility testing by a standard single disc method. Am J Clin Pathol 1966; 45: 493–496. [PubMed] [Google Scholar]
- 19.Vankerckhoven V, Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of E. faecium. J Clin Microbiol 2004; 42: 4473–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reviriego C, Eaton T, Martín R, Jiménez E, Fernández L, Gasson MJ, Rodríguez JM. Screening of virulence determinants in E. faecium strains isolated from breast milk. J Hum Lact 2005; 21: 131–138 [DOI] [PubMed] [Google Scholar]
- 21.Semedo MA, Santos P, Martins MFS, Lopes JJ, Figueiredo Marques R, Tenreiro MT, et al. Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J Clin Microbiol 2003; 41: 2569–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satake S, Clark N, Rimland D, Nolte FS, Tenover FC. Detection of vancomycin-resistant Enterococci in fecal samples by PCR. J Clin Microbiol 1997; 35:2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landete JM, de Las Rivas B, Marcobal A, Muñoz R. Molecular methods for the detection of biogenic amine-producing bacteria on foods. Int J Food Microbiol 2007; 117(3):258–269. [DOI] [PubMed] [Google Scholar]
- 24.Khalkhali S, Mojgani N. Enterococcus faecium; a suitable probiotic candidate for modulation of immune responses against pathogens. Int J Basic Sci Med 2017;2:77–82. [Google Scholar]
- 25.Ghrairi T, Frere J, Berjeaud JM, Manai M. Purification and characterization of bacteriocins produced by E. faecium from Tunisian Rigouta cheese. Food Control 2008; 19:162–169. [Google Scholar]
- 26.Borzenkov V, Surovtsev V, Dyatlov I. Obtaining bacteriocins by chromatographic methods. Adv Biosci Biotechnol 2014; 5: 446–451. [Google Scholar]
- 27.Chen YS, Yanagida F, Srionnual S. Characteristics of bacteriocin-like inhibitory substances from dochi-isolated E. faecium D081821 and D081833. Lett Appl Microbiol 2007; 44: 320–325. [DOI] [PubMed] [Google Scholar]
- 28.Zendo T, Eungruttanagorn N, Fujioka S, Tashiro Y, Nomura K, et al. Identification and production of a bacteriocin from E. mundtii QU 2 isolated from soybean. J Appl Microbiol 2005; 99: 1181–1190. [DOI] [PubMed] [Google Scholar]
- 29.Montel Mendoza G, Ale CE, Nader-Macías MEF, Pasteris SE. Characterization of a bacteriocin produced by E. gallinarum CRL 1826 isolated from captive bullfrog: evaluation of its mode of action against L. monocytogenes and Gram-Negatives. J Bioprocess Biotechnol 2015; 5: 250–256. [Google Scholar]
- 30.Casaus P, Nilsen T, Cintas L, Nes IF, Hernández PE, Holo H. Enterocin B, a new bacteriocin from E. faecium T136 which can act synergistically with enterocin A. Microbiology 1997; 143: 2287–2294. [DOI] [PubMed] [Google Scholar]
- 31.Trivedi K, Radmila S, Renata K. Bacteriocin activity of enterococci and presence of genes related to pathogenesis. Chez J food Scis 2012; 30: 330–335. [Google Scholar]
- 32.Garbutt JM, Ventrapragada M, Littenberg B, Mundy LM. Association between resistance to vancomycin and death in cases of E. faecium bacteremia. Clin Infect Dis 2000; 30: 466–472. [DOI] [PubMed] [Google Scholar]
- 33.Ogier JC, Serror P. Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol 2008; 126: 291–301. [DOI] [PubMed] [Google Scholar]
- 34.Linares DM, del Rio B, Ladero V, Martinez N, Fernandez M, Martin MC. Factors influencing biogenic amines accumulation in dairy products. Front Microbiol 2012; 3: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]