Abstract
Δ9-Desaturases are central enzymes in unsaturated fatty acid synthesis regulated at the transcriptional and mRNA levels and by proteasomal degradation. A new study by Murakami et al. uncovers a novel regulatory pathway in which an N-terminal di-proline motif in the Drosophila Δ9-desaturase mediates protein degradation by a calcium-dependent cysteine protease in response to unsaturated fatty acids. This study provides new details of desaturase regulation with therapeutic implications for the treatment of metabolic syndrome.
Introduction
Δ9-Desaturases are critical enzymes involved in unsaturated fatty acid metabolism in all living cells. They catalyze the synthesis of monounsaturated fatty acids (MUFAs),2 mainly palmitoleate and oleate, from the corresponding saturated fatty acid substrates. MUFAs are important substrates for the synthesis of complex lipids including triglycerides, cholesterol esters, wax esters, and phospholipids (1). Moreover, the fatty acid composition of phospholipids in membranes modulates membrane fluidity, affecting a broad range of cellular and physiological functions. Given these important and wide-ranging roles for MUFAs, Δ9-desaturases are regulated at multiple levels. However, none of the known regulatory mechanisms are directly responsive to changes in the levels of fatty acid desaturation among cellular lipids, raising the question as to how cells coordinate desaturase activity in response to changing environmental conditions. A new study by Murakami et al. (2) provides one answer with the discovery of a fatty acid saturation–sensitive pathway for protein degradation in Drosophila that surprisingly depends on a di-proline motif in the N-terminal region of the desaturase.
The mechanisms underlying the regulation of fatty acid desaturase expression have been explored previously for both mammalian and yeast enzymes. For example, the mammalian desaturase, stearoyl-CoA desaturase 1 (SCD1), is regulated at the transcriptional level and by proteasome-mediated degradation while the yeast desaturase Ole1 is regulated at transcriptional and mRNA levels (3–5). While some of these mechanisms indirectly report on cellular lipid composition, such as in the transcriptional regulation of SCD1 by a master regulator of lipid biosynthesis (5), it is unknown whether lipid changes, monitored as either the lipid structures themselves or as influencing membrane properties more generally, might allow faster or otherwise complementary strategies to regulate desaturase activity. This is particularly relevant in Drosophila, which cannot synthesize sterols and contain only trace amounts of polyunsaturated fatty acids in membrane phospholipids (6), meaning small changes in unsaturated fatty acids could substantially alter the biophysical properties (i.e. fluidity) of membranes.
In the study by Murakami et al. (2), the authors focus on the Drosophila enzyme DESAT1, the only desaturase expressed in the S2 cell line. Using a polyclonal antibody raised against a C-terminal peptide from the enzyme, they measured DESAT1 levels upon treatment of cells with saturated, monounsaturated, or polyunsaturated fatty acids. Unsaturated but not saturated fatty acids decreased the protein abundance of DESAT1 without affecting mRNA expression. In contrast, incubation with known SCD1 inhibitors that were confirmed to inhibit DESAT1 resulted in protein levels being markedly increased without changes in mRNA levels. Repetition of these experiments in the presence of the protein synthesis inhibitor cycloheximide enhanced the changes seen with each treatment. These results suggest that the Drosophila Δ9-desaturase is regulated at the protein level by a mechanism coupled to cellular levels of unsaturated fatty acids.
To explore the molecular basis for this new pathway, the authors tested several chimeric DESAT1 proteins with FLAG insertions, finding that the N terminus was the key domain involved. Additional constructs with truncations at the N terminus or alanine mutations identified residues 1–6, and specifically Pro2 and Pro3, as necessary and sufficient for the MUFA-dependent degradation of DESAT1 (Fig. 1). Interestingly, this “di-proline motif” is not present in most other desaturases; for example, the mouse SCD1 contains one proline residue at position 2 (7) and is not responsive to unsaturated fatty acid–mediated degradation. However, splicing the mouse SCD1 with the N terminus of Drosophila DESAT1 or introducing a second Pro into the mouse sequence sensitized the mouse enzyme to MUFA-mediated degradation, confirming the importance of this simple motif (Fig. 1).
Figure 1.
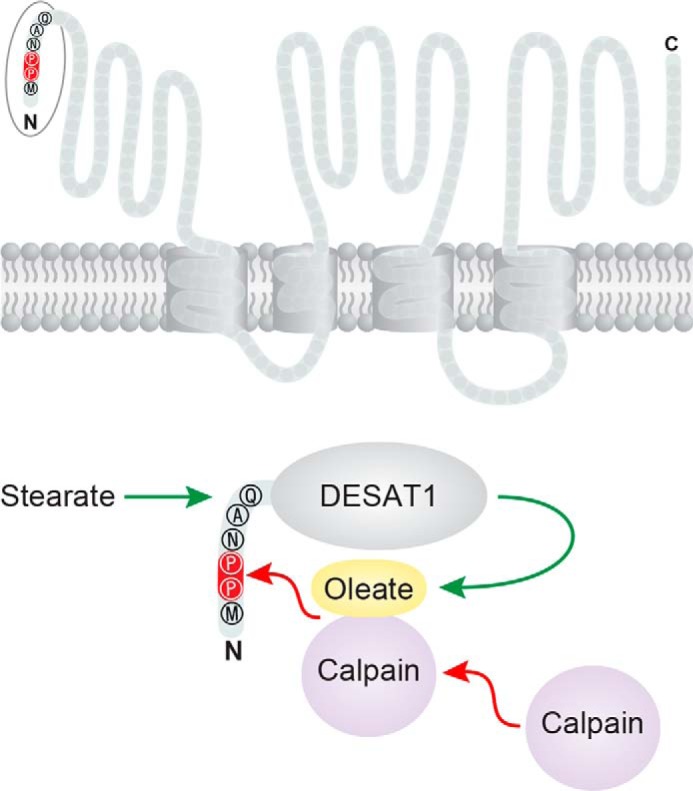
Model for MUFA-dependent degradation of a Δ9- desaturase. Top, putative location of the N-terminal di-proline motif in DESAT1, a Δ9-desaturase expressed in Drosophila, identified by Murakami et al. (2). Topology shown is modeled on the mouse enzyme (7). Bottom, prolines 2 and 3, highlighted in red, mediate degradation by calpain in MUFA-replete conditions. One possible mechanism is that calpain that exists in the cytosol as an inactive enzyme translocates to membranes where it is activated in the presence of Ca2+ and phospholipids containing oleate or unsaturated fatty acids. N and C represent N and C termini.
Finally, the authors sought to identify the protease responsible for DESAT1 degradation. Proteasomal and lysosomal inhibitors did not impact DESAT1 processing, but an inhibitor of calpain, a calcium-dependent cysteine protease, was effective in blocking degradation of DESAT1. Knockdown of the different isoforms of calpain expressed in Drosophila indicated that calpains A and B were the most important, and analysis of DESAT1 mutants confirmed that calpain-mediated degradation is dependent on the di-proline motif (Fig. 1).
These results establish a new pathway by which cells can respond to changes in unsaturated fatty acid levels to directly regulate desaturase degradation. The question that remains unanswered is how the pathway works. For example, how do the unsaturated fatty acids in either membrane phospholipids or other lipids collaborate with the di-proline motif to facilitate this process? Might the unsaturated fatty acids be ligand activators of a calpain, or do they induce a conformational change in DESAT1 that affects trafficking and degradation? Calpain exists in the cytosol as an inactive enzyme and translocates to membranes in response to increases in the cellular Ca2+ level (8). Is it possible that the activation of calpain occurs at the membrane when phospholipids containing the unsaturated fatty acids bind to its lipid-binding domain?
Beyond mechanistic considerations, this finding could also point in new directions for therapeutic interventions. Much of what we know regarding the physiological function of SCD comes from studies conducted in mice with a targeted deletion of the Scd1 gene. These mice are characterized by a lean hypermetabolic phenotype, which includes resistance to diet-induced and genetically-induced obesity, hepatic steatosis, and insulin resistance (1). Furthermore, tissue-specific deletion of SCD1 gene in skin and liver both recapitulate some of the metabolic benefits of the global SCD1 knockout mouse (9). Interestingly, calpain activators have also previously been suggested for the treatment of metabolic diseases such as type II diabetes in which calpain activity is markedly decreased (10). Thus, searching for mammalian regulatory pathways similar to the calpain-mediated degradation of Δ9-desaturase could potentially lead to new therapeutic opportunities for decreasing SCD1 activity and help in the treatment of diseases of the metabolic syndrome.
Acknowledgments
Critical reading and helpful comments on the manuscript by the members of the Ntambi laboratory are greatly appreciated. The stearoyl-CoA desaturase work in the Ntambi laboratory has been supported by National Institutes of Health Grant R01 DK062388, American Diabetes Association Grant DA 7–13-BS-540 118, and U.S. Department of Agriculture Hatch W2005 grants (to J. M. N.).
The authors declare that they have no conflicts of interests with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, American Diabetes Association, or U.S. Department of Agriculture.
- MUFA
- monounsaturated fatty acid
- SCD
- stearoyl-CoA desaturase.
References
- 1. Ntambi J. M., Miyazaki M., Stoehr J. P., Lan H., Kendziorski C. M., Yandell B. S., Song Y., Cohen P., Friedman J. M., and Attie A. D. (2002) Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. U.S.A. 99, 11482–11486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Murakami A., Nagao K., Juni N., Hara Y., and Umeda M. (2017) An N-terminal di-proline motif is essential for fatty acid-dependent degradation of Δ9-desaturase in Drosophila. J. Biol. Chem. 292, 19976–19986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kato H., Sakaki K., and Mihara K. (2006) Ubiquitin-proteasome-dependent degradation of mammalian ER stearoyl-CoA desaturase. J. Cell Sci. 119, 2342–2353 [DOI] [PubMed] [Google Scholar]
- 4. Gonzalez C. I., and Martin C. E. (1996) Fatty acid-responsive control of mRNA stability. Unsaturated fatty acid-induced degradation of the Saccharomyces OLE1 transcript. J. Biol. Chem. 271, 25801–25809 [DOI] [PubMed] [Google Scholar]
- 5. Miyazaki M., Flowers M. T., Sampath H., Chu K., Otzelberger C., Liu X., and Ntambi J. M. (2007) Hepatic stearoyl-CoA desaturase-1 deficiency protects mice from carbohydrate-induced adiposity and hepatic steatosis. Cell Metab. 6, 484–496 [DOI] [PubMed] [Google Scholar]
- 6. Shen L. R., Lai C. Q., Feng X., Parnell L. D., Wan J. B., Wang J. D., Li D., Ordovas J. M., and Kang J. X. (2010) Drosophila lacks C20 and C22 PUFAs. J. Lipid Res. 51, 2985–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Man W. C., Miyazaki M., Chu K., and Ntambi J. M. (2006) Membrane topology of mouse stearoyl-CoA desaturase 1. J. Biol. Chem. 281, 1251–1260 [DOI] [PubMed] [Google Scholar]
- 8. Suzuki K., Hata S., Kawabata Y., and Sorimachi H. (2004) Structure, activation, and biology of calpain. Diabetes 53, Suppl. 1, S12–S18 [DOI] [PubMed] [Google Scholar]
- 9. Sampath H., Flowers M. T., Liu X., Paton C. M., Sullivan R., Chu K., Zhao M., and Ntambi J. M. (2009) Skin-specific deletion of stearoyl-CoA desaturase-1 alters skin lipid composition and protects mice from high fat diet-induced obesity. J. Biol. Chem. 284, 19961–19973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Saez M. E., Ramirez-Lorca R., Moron F. J., and Ruiz A. (2006) The therapeutic potential of the calpain family: New aspects. Drug Discovery Today 11, 917–923 [DOI] [PubMed] [Google Scholar]


