Abstract
Enveloped viruses transfer their genomes into host cells by fusing their membrane to that of the cell. To visualize single-virus fusion in living cells, researchers take advantage of the proteolytic maturation of HIV, type 1 (HIV-1), which can generate free fluorescent proteins within the viral particle. Co-labeling viruses with a content marker and a fluorescently tagged Vpr (a viral core protein) enables detection of single-virus fusions, but a major limitation of this approach is that not all viral particles incorporate both markers. Here we designed a labeling strategy based on the bifunctional mCherry-2xCL-YFP-Vpr construct, in which 2xCL denotes a tandem cleavage site for the viral protease. This bifunctional marker was efficiently cleaved during virus maturation, producing free mCherry and the core-associated YFP-Vpr. A nearly perfect colocalization of these two markers in virions and their fixed 1:1 ratio enabled automated detection of single-particle fusion in both fixed and live cells based on loss of the mCherry signal. Furthermore, a drop in FRET efficiency between YFP and mCherry because of cleavage of the bifunctional marker, which manifested as a marked shift in the normalized YFP/mCherry fluorescence ratio, reliably predicted viral protease activity in single virions. This feature could discriminate between the particles containing free mCherry, and therefore likely representing mature viruses, and immature particles whose fusion cannot be detected. In summary, our new labeling strategy offers several advantages compared with previous approaches, including increased reliability and throughput of detection of viral fusion. We anticipate that our method will have significant utility for studying viral fusion and maturation.
Keywords: confocal microscopy, FRET, retrovirus, single particle analysis, virus entry, HIV maturation, live-cell imaging, fusion pore
Introduction
Enveloped viruses deposit their genomes into the cytoplasm by fusing their membrane to the cell membranes. The inefficient and asynchronous nature of this process, in which the majority of virions will fail to fuse, necessitates the use of functional assays that report productive entry events. Both population- and single particle–based functional assays have been implemented to examine entry, fusion, and infection of enveloped viruses (e.g. Refs. 1–9). Although bulk assays are generally easier to perform and have higher throughput than single-virus imaging, the latter technique has revealed a wealth of information regarding the dynamics and sites of viral fusion as well as the heterogeneity of virus entry pathways and fusion outcomes.
The most commonly used approach to visualize single-virus fusion relies on detection of lipid mixing between virions labeled with self-quenching concentrations of lipophilic fluorescent dyes and the plasma membrane or endosomes of target cells (e.g. Refs. 10–15). Lipid exchange between viral and the much larger cell membranes leads to readily detectable fluorescence dequenching. However, hemifusion (contacting monolayers of two membranes merge but not distal monolayers (16, 17)) is both necessary and sufficient for lipid mixing to occur, and thus the dequenching cannot be definitively interpreted as full fusion. Indeed, there are reports of hemifusion occurring in the absence of viral content release (e.g. Refs. 14, 15, 18). It is therefore important to monitor viral content release into a target cell.
Viral content mixing assays are limited by the ability to trap a fluid phase marker within intact virions. “Soaking” of the influenza viruses in a concentrated solution of sulforhodamine B has reportedly allowed entrapment of sufficient amounts of this dye to detect single-virus content release upon fusion with supported lipid bilayers (19). However, this strategy does not appear to be broadly applicable to introducing a content marker into virions or for virus–cell fusion experiments. In this regard, retroviruses offer a unique advantage, as they undergo maturation after budding off the infected cells. Maturation involves cleavage of the Gag and Pol polyproteins by the viral protease (20). We and others have taken advantage of this process to generate free fluorescent proteins within virions through appending those to the C terminus of the murine leukemia virus Gag (6, 8, 21) or inserting between the matrix and capsid subdomains of the HIV-1 Gag (5–7).
We have previously labeled HIV-1 particles with YFP-Vpr or GFP-Vpr, which incorporate into the viral core, and Gag containing an “internal” mCherry protein inserted between matrix and capsid subdomains and flanked by the protease cleavage sites (7, 22, 23). Free mCherry released from Gag upon maturation is trapped in the virions, serving as a fluid phase marker for sensitive detection of small fusion pores. This labeling strategy worked well for studies of fusion mediated by various viral glycoproteins (7, 22, 23). However, imperfect co-transfection of YFP-Vpr and Gag-imCherry plasmids into virus-producing cells and highly variable levels of their expression limit co-localization of these markers and tend to negatively affect virus maturation (22).
Here we introduce a novel approach to co-labeling of HIV-1 particles with the viral core and fluid phase markers for reliable visualization of single-virus fusion. This labeling strategy is based on tandem fluorescent proteins separated by the HIV-1 protease cleavage sequences and fused in-frame to the N terminus of the HIV-1 Vpr. This new strategy offers a number of advantages compared with the original labeling approach. First, introducing the two markers into virus particles at a fixed number ratio yields nearly perfect co-localization and enables robust automated identification of fused particles in the cytoplasm. Second, the use of a FRET pair of fluorescent proteins to tag Vpr provides a convenient reporter for the viral protease activity, which is manifested as a loss of FRET in single particles because of separation of the donor and acceptor proteins. We show that the FRET levels, as well as the relative intensities of the donor and acceptor signals associated with individual virions, can potentially discriminate between immature particles and mature virions capable of fusing with target cells.
Results
Design and characterization of a bifunctional content/core marker for near-perfect co-labeling of HIV-1 virions
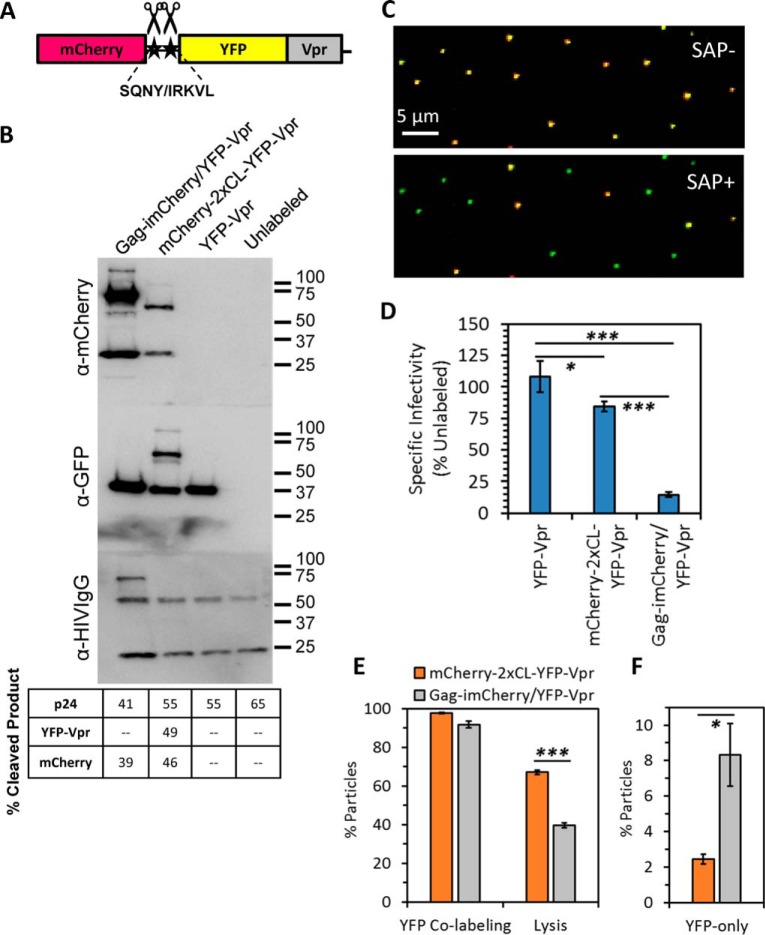
We have previously visualized single-virus fusion in living cells by co-labeling HIV-1 particles through co-transfection of virus-producing cells with two separate plasmids encoding for YFP-Vpr or GFP-Vpr (core markers (22, 24)) and Gag-imCherry (a virus maturation-dependent fluid phase marker) (5, 7). To improve virus labeling with the core and fluid-phase markers, we designed a novel bifunctional marker that consists of a tandem of fluorescent proteins fused in-frame to the N terminus of Vpr (referred to as mCherry-2xCL-YFP-Vpr, Fig. 1A). To generate free mCherry upon virus maturation, a linker containing two HIV-1 protease cleavage sites, SQNY and IRKVL, was inserted between mCherry and YFP. We found that the inclusion of a second protease site increased the cleavage efficiency (data not shown). This construct efficiently incorporated into the HIV-1 pseudoviruses and, upon processing by the viral protease, produced both releasable mCherry and core-associated YFP-Vpr (Fig. 1, B, C, and E).
Figure 1.
Design and characterization of the bifunctional content/core marker for visualization of pseudovirus fusion. A, schematic depicting the design of the Vpr-based bifunctional marker containing the mCherry/YFP-Vpr tandem separated by a linker made of two cleavage sites, SQNY and IRKVL, for HIV-1 protease. B, HXB2 pseudotyped virus stocks were produced unlabeled or labeled with YFP-Vpr, the bifunctional marker (mCherry-2xCL-YFP-Vpr), or Gag-imCherry/YFP-Vpr. Concentrated stocks were subjected to SDS-PAGE and immunoblotted for mCherry, GFP, or HIV proteins. Percent cleavage products report the fraction of cleaved product released after proteolytic processing found by densitometry measurements. C, HXB2 pseudoviruses labeled with the bifunctional marker were immobilized on coverslips and imaged at high signal-to-noise before and after the addition of saponin to mildly permeabilize the viral membrane and release cleaved mCherry. D, the specific infectivity of viruses in B was determined at 48 hours post-infection (h.p.i.) using TZM-bl cells. Error bars indicate standard error from three independent experiments. E and F, fractions of co-labeled YFP+ particles and co-labeled particles with releasable content (E) and occurrence of YFP-only particles (F) were assessed. Error bars indicate standard error from three independent experiments. *, p < 0.05; ***, p < 0.001.
We compared virus samples labeled with the bifunctional marker to samples containing only YFP-Vpr or co-labeled with Gag-imCherry/YFP-Vpr. Co-incorporation of Gag-imCherry and YFP-Vpr into the same virus particle and subsequent proteolytic processing of Gag-imCherry yielded releasable mCherry (Fig. 1B), as shown previously (5, 7). Although larger amounts of Gag-imCherry incorporated into virions compared with the Vpr-based bifunctional marker, the mCherry incorporated via the latter marker was readily detectable by single-virus particle imaging (Fig. 1C). Western blotting with anti-HIV serum showed a comparable Gag processing efficiency among virus samples labeled with any of the Vpr-based markers. However, incorporation of the Vpr-based markers somewhat reduced the Gag processing efficiency, and this efficiency was further diminished upon co-labeling with YFP-Vpr and Gag-imCherry (Fig. 1B, table). Viral Gag-imCherry and the bifunctional marker mCherry-2xCL-YFP-Vpr were partially cleaved, suggesting that these preparations contain a mixture of mature and immature viruses, similar to unlabeled virions. Next, we assayed the specific infectivity of virus samples. Although a bifunctional marker caused only a modest reduction in infectivity, co-incorporation of Gag-imCherry/YFP-Vpr more strongly impacted virus function (Fig. 1D), perhaps because of the reduced maturation efficiency (Fig. 1B).
Visualization of the coverslip-immobilized viruses under stringent imaging conditions to ensure an optimal signal-to-background ratio revealed that ∼98% and ∼91% of YFP-positive particles co-labeled with mCherry in bifunctional and Gag-imCherry/YFP-Vpr samples, respectively (Fig. 1, C and E, and data not shown). The novel labeling approach significantly (about 4-fold) reduced the number of YFP-Vpr–only particles that did not contain detectable amounts of mCherry (Fig. 1F). The imperfect apparent colocalization of YFP and mCherry in virions containing the bifunctional marker was somewhat surprising. We therefore asked whether the lack of mCherry fluorescence could reflect the presence of particles with a “leaky” membrane. In fact, even a transient small (>4 nm) pore in the viral membrane would quickly deplete the virus of the mCherry marker. To test whether the fluid phase marker could be lost because of virus handling, we compared YFP/mCherry co-localization in freshly produced unconcentrated virus versus preparations that were subjected to a freeze–thaw cycle, shear forces, prolonged incubation in the cold, or ultracentrifugation (supplemental Fig. S1). Because none of the treatments altered the apparent colocalization of the two markers, it appears that a small fraction of virions lost their content marker either through membrane defects forming at the time, or shortly after virus budding, or because of premature activation of the viral protease that causes the release of mCherry into the cytoplasm of producer cells.
We next assessed the extent of cleavage of fluorescent markers in single particles containing Gag-imCherry or bifunctional marker by subjecting these to mild saponin lysis. A greater fraction of particles incorporating a bifunctional marker contained cleaved mCherry than those co-labeled with Gag-imCherry/YFP-Vpr, as evidenced by the larger fraction of virions that lost mCherry upon addition of saponin (Fig. 1, C and E). Collectively, the above results demonstrate that the bifunctional labeling strategy improves the standard Gag-imCherry/YFP-Vpr two-marker labeling system.
Detection of single-particle fusion with the bifunctional marker
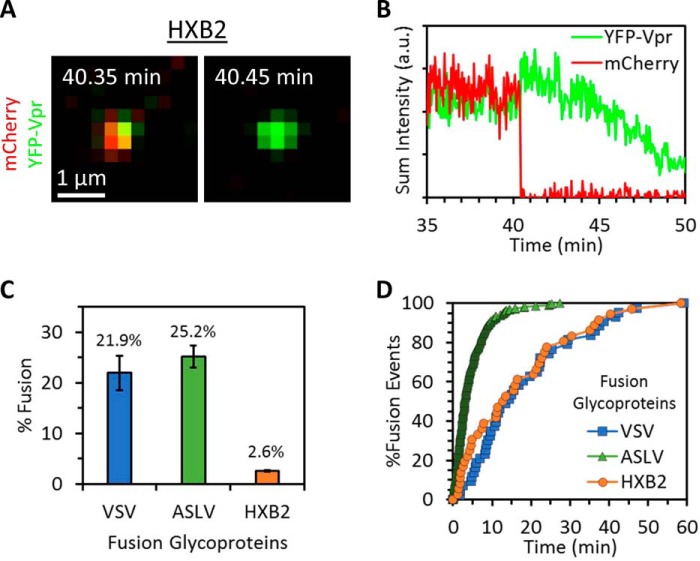
To demonstrate the utility of mCherry-2xCL-YFP-Vpr for detecting single-virus fusion, we pseudotyped HIV-1 cores labeled with the bifunctional marker with the Env glycoprotein of HIV-1 HXB2 strain. Live-cell imaging of single pseudovirus entry/fusion into CV1-derived cells expressing CD4 was performed. Viral fusion was detected as the near instantaneous loss of the fluid phase mCherry marker against the retention of the YFP-Vpr core-associated fiducial marker (Fig. 2, A and B), similar to events detected using the conventional Gag-imCherry/YFP-Vpr labeling approach (7, 14, 23). In agreement with our previous results (23), 2.6% of co-labeled HXB2 pseudoviruses fused with these cells in the course of 1 h (Fig. 2, C and D). We found no difference in the single-particle fusion kinetics when comparing HXB2 pseudoviruses labeled with the bifunctional marker and Gag-imCherry/YFP-Vpr (fusion half-times, ∼13 min; Fig. 2D and Ref. 23).
Figure 2.
The bifunctional content/core marker enables detection of fusion mediated by viral fusion proteins. Virus samples labeled with the bifunctional marker and pseudotyped with either VSV G, ASLV Env, or HIV HXB2 Env were prebound to permissive cells in the cold. Virus entry and fusion were initiated by addition of prewarmed buffer, and samples were imaged at 37 °C. A and B, visualization (A) and intensity traces from single-particle tracking (B) of an HXB2 pseudovirus labeled with the bifunctional marker shows a fusion event that manifests as loss of mCherry. a.u., arbitrary units. C and D, comparison of fusion kinetics (D) and extent (C) of viral particles labeled with the bifunctional marker and pseudotyped with HXB2, VSV, or ASLV fusion glycoproteins. In C, fusion extent is shown as fraction of co-labeled particles. Error bars indicate standard error from three or more independent experiments.
We next asked whether the new labeling strategy is broadly applicable to visualization of fusion induced by viral proteins other than HXB2 Env. HIV-1 particles labeled as above were pseudotyped with the Vesicular Stomatitis Virus (VSV)4 G or Env glycoprotein of the avian sarcoma and leukosis virus (ASLV), and single-particle imaging was performed in live CV1 cells expressing the TVA receptor for subtype A ASLV (25). Both proteins direct virus entry through endocytic pathways and mediate efficient low pH-dependent fusion with endosomes (26–28). In agreement with results published previously (22, 23, 25, 26, 29), we found that virus samples labeled with the bifunctional marker and pseudotyped with VSV G and ASLV Env fused much more efficiently than those pseudotyped with HXB2 Env (22.9% and 25.2%, respectively; Fig. 2C). ASLV Env mediated much faster fusion with CV1/TVA cells than VSV G or HXB2 Env (half-times, 3.0, 13.9, and 12.7 min, respectively; Fig. 2D), also in agreement with our previous studies (22, 23, 25, 26, 29). Thus, the bifunctional marker enables reliable measurements of the extent and kinetics of single virus fusion in living cells.
YFP-mCherry FRET in the bifunctional marker predicts virus protease activity and the presence of free mCherry in single virions
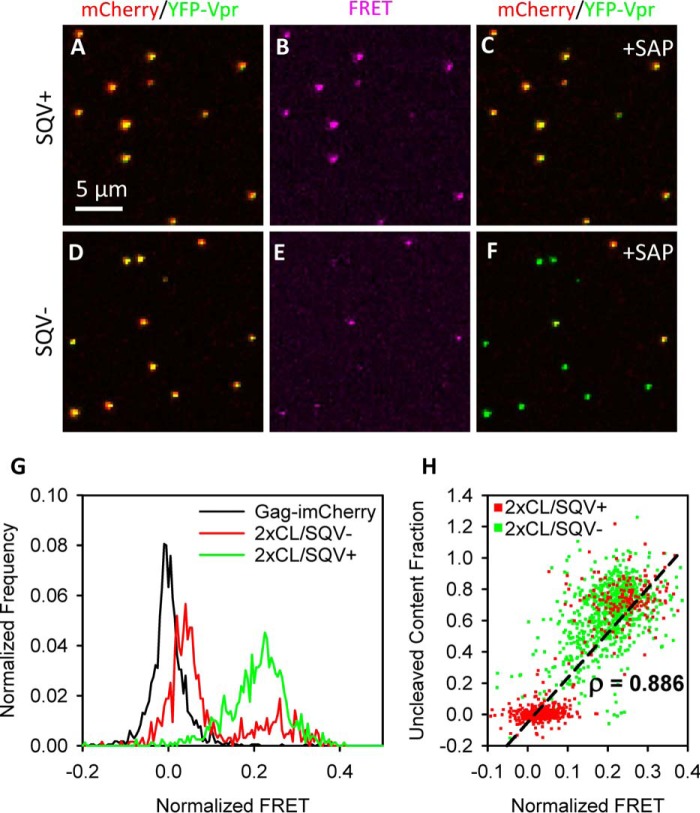
We used a FRET pair (YFP/mCherry) in the bifunctional marker to help identify immature virions and potentially exclude those from analysis of single virus fusion events. The short (∼3.5-nm) linker sequence containing the two HIV-1 viral protease cleavage sites between mCherry and YFP allows for an efficient FRET between this donor/acceptor pair with a Forster critical distance of 5.66 nm (30). As expected, we readily detected the YFP-mCherry FRET signal in viruses produced in the presence of the HIV-1 protease inhibitor saquinavir (SQV+; Fig. 3, A–C; and supplemental Fig. S2). By comparison, single-particle FRET in untreated samples (SQV−) ranged from high to zero or near-zero (Fig. 3, D–F). This broad distribution of FRET values is consistent with a heterogeneous population containing mature and immature viruses, as suggested by the Western blotting and virus lysis results (Fig. 1). Indeed, the single-particle acceptor-normalized FRET (see “Experimental procedures” and supplemental Fig. S2) (31, 32) for untreated samples was bimodally distributed (Fig. 3G). The high-FRET component overlapped the unimodal distribution for the saquinavir-treated sample, and the low-FRET mode was only slightly shifted from the control (no-FRET) viruses labeled with Gag-imCherry (Fig. 3G). These results suggest that the YFP/mCherry FRET reflects the extent of cleavage of the bifunctional marker upon virus maturation.
Figure 3.
Intrabifunctional marker FRET predicts HIV-1 protease activity. A–F, SQV-treated (SQV+, A–C) and untreated (SQV−, D–F) viruses labeled with mCherry-2xCL-YFP-Vpr were immobilized on coverslips and imaged by sequentially exciting with 488 nm and 594 nm light while detecting in both YFP and mCherry emission bands. YFP and mCherry intensity detected in 488-nm/YFP and 594-nm/mCherry channels, respectively, was measured before (A and D) and after (C and F) addition of saponin. Sensitized emission FRET intensity (B and E) was detected in the 488-nm/mCherry channel and corrected for cross-talk (see “Experimental procedures” and supplemental Fig. S2). G, single-particle distributions of acceptor-normalized FRET were measured for virus labeled with Gag-imCherry and for SQV+/− virus labeled with the bifunctional marker. H, scatterplot of normalized FRET versus uncleaved mCherry content fraction (determined as the mCherry signal remaining after addition of saponin) for SQV− virus labeled with the bifunctional marker. Single-particle normalized FRET strongly correlated with single-particle fraction of uncleaved mCherry content (Pearson correlation, ρ = 0.886). More than 600 particles were analyzed for each condition.
To further assess the link between single-virus FRET and cleavage of the bifunctional marker, viruses were imaged before and after mild lysis with saponin, which forms small pores in the viral membrane (33). The resulting loss of free mCherry (if any) provides a convenient means to verify the cleavage efficiency of the Vpr-based bifunctional marker. Comparison of the same image field before and after saponin lysis confirmed an inverse relationship between normalized FRET and protease activity. Single-particle FRET efficiency significantly correlated with the fraction of mCherry signal remaining after addition of saponin (Pearson correlation, ρ = 0.886) (Fig. 3H). Thus, YFP/mCherry FRET reliably predicts the HIV-1 protease activity on a single-particle level (Fig. 3H) and, thus, likely reports the particle maturation status.
The relative intensity of YFP and mCherry in single virions is an indicator of protease activity
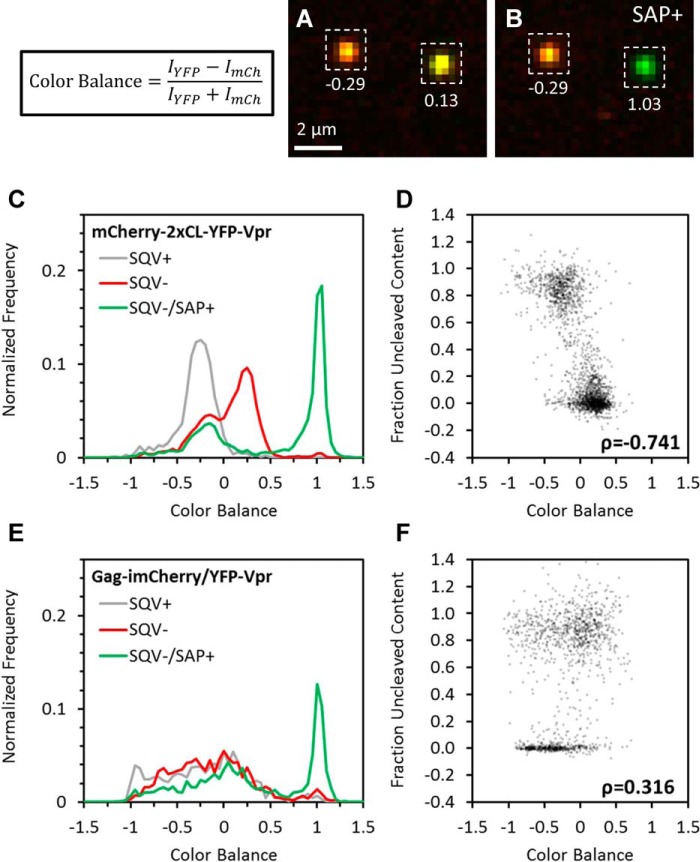
The distinct FRET levels found in the untreated samples (Fig. 3G) implied that single virions could be classified a priori as protease-active or -inactive. However, the cumbersome nature of measuring sensitized emission FRET inherently limited the utility of this approach, especially in a live-cell setting. We therefore used the relative intensities of YFP and mCherry as a proxy for FRET, which is manifested in quenching of the donor (YFP) fluorescence by the acceptor (mCherry). The relative intensities of the two fluorescent proteins in a single particle were characterized by “color balance,” defined as CB = (IYFP − ImCherry)/(IYFP + ImCherry). This alternative parameterization trades the dynamic range of the FRET measurement for ease of implementation. Color balance in the high-FRET, protease-inactive particles with large amounts of uncleaved mCherry should be lower than in the low-FRET particles because of rerouting of YFP emission to mCherry emission (particles appear more red; Fig. 4, A and B). In this sense, particles that appear as “mCherry-only” must contain uncleaved bifunctional marker in which the YFP fluorescence is quenched and are therefore unlikely to contain free mCherry that can report the fusion events. Moreover, HIV-1 Env–mediated fusion is dependent on virus maturation (34, 35) and is unlikely to occur with virions lacking protease activity.
Figure 4.
Single-particle color balance as an indicator of HIV-1 protease activity. HXB2 pseudoviruses labeled with either the bifunctional marker or Gag-imCherry/YFP-Vpr (data not shown) and treated with 500 nm SQV or left untreated were immobilized and imaged at high signal-to-noise before and after the addition of saponin. Single-particle color balance (see “Experimental procedures”) was measured using local background-corrected sum intensities of particles in the YFP and mCherry channels. A and B, representative images and measured color balance of particles labeled with bifunctional markers before (A) and after (B) addition of saponin. C and E, single-particle color balance distributions of HXB2 pseudovirus labeled with the bifunctional marker (C) or Gag-imCherry/YFP-Vpr (E) produced in the presence (gray) or absence of saquinavir before (red) and after (green) addition of saponin. D and F, scatterplot of single-particle color balance before saponin treatment and uncleaved mCherry content fraction of SQV− virus labeled with the bifunctional marker (D) or Gag-imCherry/YFP-Vpr (F). Color balance strongly (Pearson correlation, ρ = −0.741) and weakly (Pearson correlation, ρ = 0.316) correlated with uncleaved mCherry content for bifunctional labeling and Gag-imCherry/YFP-Vpr, respectively.
Importantly, color balance analysis can be applied to assess the protease activity in single virions containing the bifunctional marker, which produces a fixed (1:1) ratio of YFP and mCherry molecules in single virions. In agreement with the partial Gag cleavage in our preparations (Fig. 1B), we observed a bimodal distribution of CB values (Fig. 4C). The lower CB peak/shoulder roughly matched the values of SQV+ sample, whereas the larger intermediate peak likely corresponded to protease-active particles. Indeed, upon saponin lysis, the higher CB mode shifted to CB ∼1, whereas the lower CB mode was not affected by viral membrane permeabilization (Fig. 4C). Similar to the FRET experiments shown in Fig. 3, imaging before and after saponin lysis demonstrated a good correlation between the releasable mCherry pool and CB (Fig. 4, A–D). By contrast, there is no constraint on the numbers of YFP and mCherry incorporated into virions co-labeled with Gag-imCherry/YFP-Vpr so that the distribution of CB does not have any predictive value (Fig. 4, E and F).
Automated detection of post-fusion viral cores in fixed cells
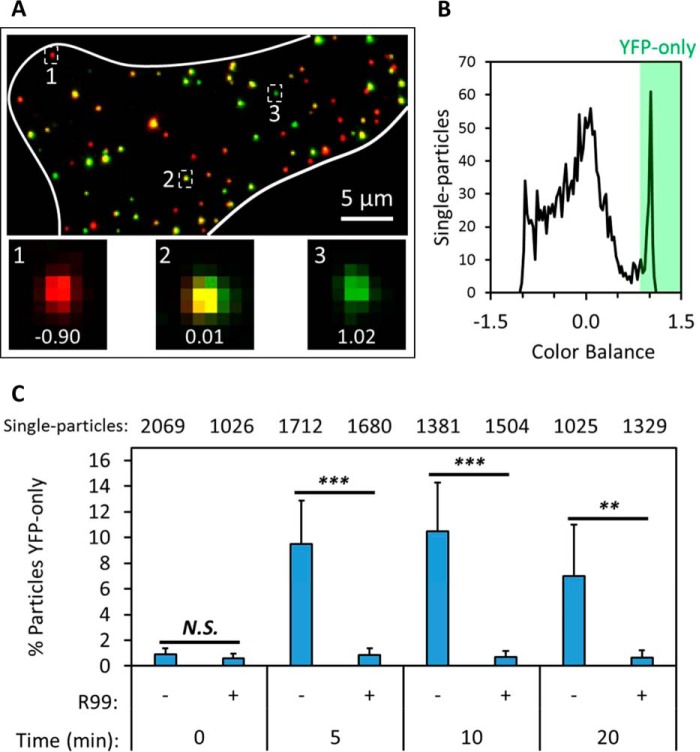
The marked shift in color balance after saponin lysis of single viruses labeled with the bifunctional marker (Fig. 4, A–C) prompted us to use this parameter to detect viral fusion, which manifested as a loss of mCherry (Fig. 2, A and B). We performed imaging of ASLV pseudovirus fusion with target cells using stringent, high signal-to-background imaging conditions (see “Experimental procedures”). The cells were fixed at different times post-infection, and the color balance parameter was calculated for every particle (Fig. 5, A and B). This analysis clearly identified the appearance of a sizeable population of YFP-only labeled spots with above-threshold CB values under conditions permissive for viral fusion. Approximately 10% of all cell-associated particles pseudotyped with the ASLV Env lost mCherry within the first 5 min of infection (Fig. 5C and supplemental Fig. S3 and Movie S1), in agreement with the fast kinetics of fusion mediated by this glycoprotein (Fig. 2D and Refs. 21, 25, 26, 36). By contrast, only a very low background number of YFP-only particles was detected at different times post-infection in the presence of the inhibitory peptide R99 (Fig. 5C).
Figure 5.
Detection of post-fusion viral cores in fixed cells. ASLV pseudoviruses labeled with the bifunctional marker were prebound to TVA950-expressing CV1 cells in the cold, and virus fusion was initiated by shifting to 37 °C for a short interval, after which cells were immediately fixed and imaged. A, maximum intensity projection of a fixed cell (white boundary) in which the virus was allowed to enter and fuse for 5 min. Insets highlight selected virus particles and their color balance, calculated from local background-corrected single-particle YFP and mCherry intensities. B, distribution of single-particle color balance at the 5-min time point. Post-fusion cores (YFP-only) are detected as puncta with color balance > 0.85. C, fraction of YFP-only particles per total particles in a field of view was measured for five to seven fields for each incubation interval. Parallel samples maintained in the presence of 50 μg/ml of the fusion inhibitor R99 were examined as negative controls. The total number of particles examined for each condition is listed on top. Error bars indicate standard deviation among the fields imaged for each condition. **, p < 0.01; ***, p < 0.001; N.S., not significant.
As expected from the fast fusion kinetics observed in live-cell settings (Fig. 2D), the fraction of high-color balance (YFP-only) particles automatically detected in fixed cells plateaued at ∼10 min post-infection (Fig. 5C). Interestingly, the number of these particles tended to decline at later times (Fig. 5C and data not shown) instead of remaining steady. This trend is in agreement with the previously reported loss of YFP-Vpr after fusion of viruses co-labeled with Gag-imCherry/YFP-Vpr (22). Indeed, we routinely observed YFP-Vpr intensity decay shortly after fusion of particles containing the bifunctional marker (Figs. 2B and Fig. 6B, bottom).
Figure 6.
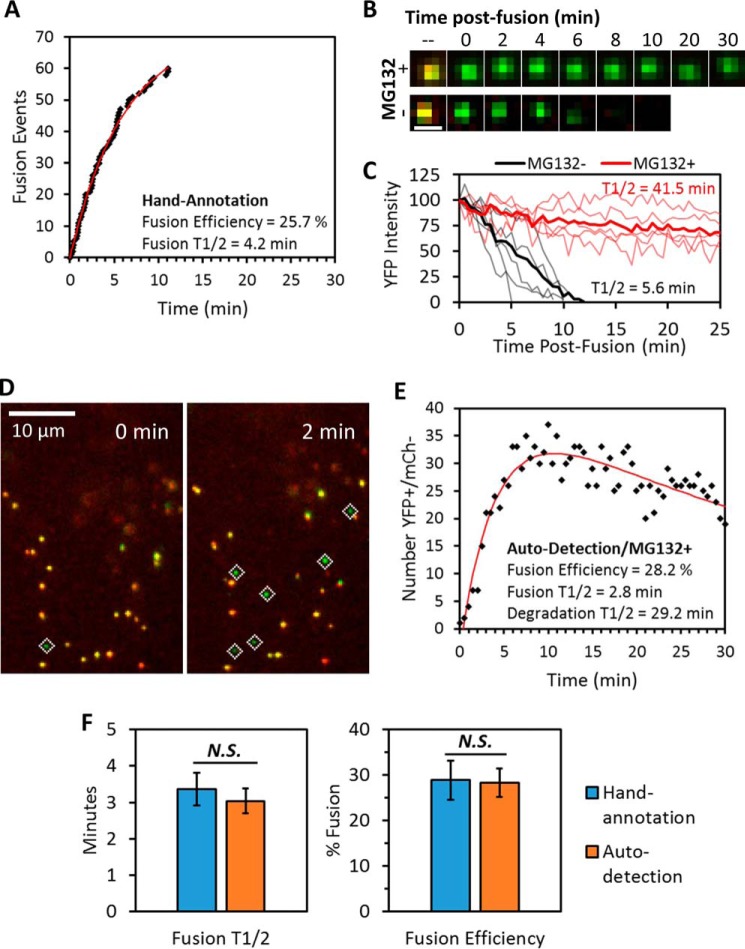
Comparison of live-cell single-particle fusion characterized by either visual or computer-automated annotation. ASLV pseudoviruses labeled with the bifunctional marker were prebound to TVA950-expressing CV1 cells in the cold, and the temperature was shifted to 37 °C by addition of prewarmed buffer immediately prior to imaging. A, representative fusion kinetics obtained by manual annotation of a live-cell experiment. Fusion efficiency and half-time (t½) were found by curve-fitting to a single exponential model. B and C, imaging (B) and single-particle tracking (C) of post-fusion cores revealed that pretreatment of cells with 20 μm proteasome inhibitor MG132 drastically prolonged detection of post-fusion YFP signal (see also supplemental Movie S3). Degradation half-times were estimated from the ensemble average (thick lines) of single-particle YFP intensity traces obtained immediately after fusion (thin lines). Scale bar in B is 1 μm. D and E, high signal-to-noise imaging (see “Experimental procedures”) and MG132 treatment enabled robust automated detection of post-fusion viral cores in live cells (D). Fusion efficiency and the rate (t½) of YFP signal degradation were found by fitting the time course of number post-fusion viral cores to a double-exponential model described under “Experimental procedures” (E). F, comparison of fusion efficiency and t½ from manual and computer-automated annotation of live-cell single-particle fusion experiments. Error bars indicate standard error of five or more independent experiments. Because the exponential model estimates steady-state solution at time → ∞, the efficiency reported for visual annotations in E is slightly larger than that reported for the 30-min imaging window in Fig. 2C. N.S., not significant.
Automated detection of viral fusion in live cells
The above results demonstrate the feasibility of automated detection of fusion of virions containing the bifunctional marker based on color balance analysis. However, the loss of the Vpr marker after fusion (Fig. 2B) confounds quantitative measurements of the fusion efficiency. Although the rate of the Vpr signal decay varied, on average, the YFP fluorescence decreased with a half-time of ∼6 min (Fig. 6C). The existence of two competing processes that lead to appearance (fusion) and loss (YFP-Vpr decay) of detectable post-fusion particles with high CB values is also likely to distort the measured fusion kinetics because these processes occur on a similar time scale (Figs. 2F and 6, A and B, and supplemental Movie S2). We therefore attempted to prevent the loss of a fiduciary marker from post-fusion cores. We asked whether the YFP-Vpr loss may be due to degradation of post-fusion cores in proteasomes. Cells were pretreated with an inhibitor, MG132, and the live-cell fusion experiments were carried out in the presence of the proteasome inhibitor. Strikingly, the loss of YFP-Vpr after fusion was dramatically delayed in treated cells compared with control cells (Fig. 6B and supplemental Fig. S4 and Movie S3), showing that post-fusion HIV-1 cores were targeted to proteasomal degradation in CV1-derived cells. We are currently pursuing the mechanism of MG132-mediated protection of post-fusion HIV-1 cores.5
The prevention of the quick loss of YFP-Vpr after fusion in MG132-treated cells allowed accumulation of YFP-only puncta under fusion-permissive conditions and greatly facilitated quantitative automated analysis of viral fusion (Fig. 6, C and D, and supplemental Fig. S5). For automated detection of ASLV fusion in living cells, we employed stringent imaging conditions to ensure a high signal-to-background ratio combined with less frequent image acquisition (every 30 s) to avoid excessive photobleaching. Curve-fitting the dependence of the number of high-CB (YFP-only) spots as a function of time using a simple equation (see “Experimental procedures”) yielded the kinetics and extent of ASLV fusion as well as the rate of core degradation in the presence of MG132 (Fig. 6D). This enabled comparison of the fusion parameters obtained by manual annotation and by automated analysis. Encouragingly, both the kinetics and the extent of ASLV fusion obtained through automated analysis were indistinguishable from those measured by conventional and much more tedious manual annotation of images (Fig. 6E).
The automated analysis was clearly augmented by the highly efficient and quick fusion mediated by ASLV Env compared with the slow loss of YFP-Vpr after fusion in MG132-treated CV1/TVA cells. We therefore tested whether this analysis could be applied to the much less efficient fusion driven by the HIV-1 Env. However, although a few HXB2 pseudotyped YFP-only particles seemed to appear in MG132-treated cells at relevant times post-infection (15–30 min), these numbers were comparable with the background level and did not allow for meaningful automated quantitative analysis (supplemental Fig. S6).
Discussion
A relatively small number of studies have visualized single-virus fusion in living cells, especially using a fluid phase marker, which necessitates virus co-labeling with a fiduciary core marker for reliable detection of fusion events (5–7, 21, 23, 37, 38). Most importantly, these experiments rely on specialized and rather expensive equipment and image processing software. Live cell imaging experiments also require a temperature/humidity/CO2-controlled microscope enclosure or a stage-top incubator and sensitive cameras suitable for time-resolved imaging of faint and often quickly moving viral particles to track their location in cells. Another major limitation of these experiments is that frequent image acquisition can cause excessive photobleaching of samples. Post-acquisition image processing requires single-particle tracking software, which, because of a poor signal-to-background ratio typical for live-cell imaging, does not generally allow for reliable automated detection of fusion events (e.g. Refs. 6, 23). These obstacles appear to have limited the use of real-time single-virus imaging techniques for studies of viral fusion. The novel virus labeling strategy introduced in this study offers a number of important advantages compared with virus co-labeling with YFP-Vpr and Gag-imCherry used in the previous single-virus fusion experiments. These improvements stem from the nearly perfect co-localization and fixed molecular ratio of the viral core and fluid phase markers in virions that enable automated detection of single-virus fusion and report the protease activity in single particles.
First, the negligible fraction of particles lacking the mCherry marker and the low variability of the YFP/mCherry signal ratio of the virion reduces the chances for falsely identifying YFP-only particles as post-fusion viral cores that released the mCherry marker. We demonstrated that the normalized ratio of YFP/mCherry intensities (color balance) can be used to automatically find fused particles in live-cell experiments or following cell fixation at different times after infection. The ability to automatically detect viral fusion in fixed cell experiments (Fig. 5A) and to deduce the kinetics and extent of fusion in a live-cell setting (Fig. 6E) increases the throughput of post-acquisition analysis. Quantification of the number of fusion events after cell fixation would greatly simplify the imaging experiment because environmental microscope enclosures or highly sensitive electron multiplying charge-coupled device cameras are not required for high signal-to-background imaging of viruses in fixed cells.
Second, the excellent co-localization of the viral core and content markers afforded by the bifunctional probe should augment the application of correlative light-electron microscopy approaches to study post-fusion particles. Fused (YFP-only) particles can be identified following fixation or plunge freezing and analyzed by electron microscopy.
Third, the level of YFP/mCherry FRET and, thus, the color balance are reliable indicators of the HIV-1 protease activity in single virions containing the bifunctional marker and, thus, likely report the virus maturation status. We have previously demonstrated a correlation between Gag-imCherry/Gag processing and the fraction of viruses losing mCherry upon saponin treatment (7). Of course, cleavage of the bifunctional Vpr-based marker does not directly reflect Gag cleavage. It is worth noting, however, that the extent of Gag cleavage in samples containing the bifunctional marker or Gag-imCherry/YFP-Vpr (Fig. 1B) tends to correlate with the fraction of virions releasing mCherry in response to saponin (Fig. 1E). Indeed, 55% of Gag cleaved in samples containing the bifunctional marker is in good agreement with the extent of saponin lysis (67%) of these particles. For virions containing Gag-imCherry/YFP-Vpr, these numbers are 41% and 39%, respectively. It is thus likely that YFP/mCherry FRET/color balance of single pseudoviruses reflects the extent of Gag cleavage and can thus discriminate between single mature and immature particles.
Despite the above advantages offered by the new labeling strategy, there are also limitations. A small uncertainty in identification of fused particles due to an imperfect co-localization of the core and fluid phase markers (∼2.5% of pre-existing YFP-only particles, Fig. 1F) diminishes the utility of automatic detection of the fusion mediated by less efficient viral glycoproteins, such as HIV-1 Env (supplemental Fig. S6). Under our conditions, HIV-1 Env mediates fusion of only 2.6% of double-labeled pseudoviruses with CV1-derived cells (Fig. 2C) and a similarly low fusion efficiency with other cell lines (6, 23, 39), so the number of fused particles is comparable with the background fraction of particles lacking mCherry (Fig. 1F). This problem could be alleviated by optimizing the fusion conditions and/or improving the Env incorporation into virions on one hand and further slowing down post-fusion decay of the YFP-Vpr signal (Fig. 6B) on the other hand. Of course, this labeling strategy is perfectly compatible with detection of fusion based on real-time particle tracking (Fig. 2). In conclusion, the experimental and analytical tools developed in this study should enable reliable measurements of robust viral fusion in fixed cells without the need for specialized equipment or single-particle tracking software.
Experimental procedures
Cell lines, reagents, and plasmids
HEK293T/17 cells were obtained from the ATCC (Manassas, VA). TZM-bl cells (donated by Drs. J. C. Kappes and X. Wu (40)) and the HIV-1 protease inhibitor saquinavir were from the National Institutes of Health AIDS Research and Reference Reagent Program. CV1/CD4/CXCR4 (CF3 clone) were a gift from Dr. David Kabat (Oregon State University). The CV1/TVA950 cell line has been described previously (26). All cell lines were cultured in DMEM supplemented with 10% FBS and 100 units/ml penicillin-streptomycin (Gemini Bio-Products, Sacramento, CA). The growth medium for HEK293T/17 cells was supplemented with 0.5 mg/ml G418 (Cellgro, Mediatech, Manassas, VA).
Heat-inactivated FBS and poly-l-lysine were from Sigma-Aldrich (St. Louis, MO). Live-cell imaging buffer and FluoroBriteTM DMEM were from Life Technologies. The pCAGGS plasmid encoding HXB2 Env was provided by Dr. J. Binley (Torrey Pines Institute). The pMDG-VSVG plasmid expressing VSV-G was a gift from J. Young (Roche). The subgroup A ASLV Env glycoprotein lacking the cytoplasmic domain (EnvAΔCT) has been described previously (9). The HIV-1-based packaging vector pR9ΔEnv was from Dr. C. Aiken (Vanderbilt University). The YFP-Vpr plasmid was a gift from Dr. T. Hope (Northwestern University). The HIV-1 Gag-imCherryΔEnv plasmid has been described previously (7).
mCherry-2xCL-YFP-Vpr was cloned into the pMM310 BlaM-Vpr construct (National Institutes of Health, AIDS Repository, catalog no. 11444) by excising BlaM using enzymes HindIII and XhoI. DNA encoding for mCherry-SQNY and IRKVL-eYFP-Vpr was PCR-amplified from the HIV-1 Gag-imCherry and eYFP-Vpr plasmids, respectively, using primers with overhangs that contained protease cleavage sites. The two DNA sequences were later digested with HindIII+NotI (mCherry-SQNY) and NotI+XhoI (IRKVL-eYFPVpr) and introduced into the pMM310ΔBlamVpr plasmid by three-way ligation. The identity of the insert was verified by sequencing.
Pseudovirus production and infectivity assays
Pseudoviruses were produced by transfection of HEK293T/17 cells, essentially as described in Ref. 7. Briefly, cells were transfected with the following amounts of plasmids per 60-mm dish: for bifunctional marker labeling, 0.8 μg of pR9ΔEnv, 0.16 μg of mCherry-2xCL-YFP-Vpr, 0.2 μg of pcRev, and 0.8 μg of HXB2 Env or ASLV EnvΔCT or 0.4 μg of VSV G; for conventional two-plasmid labeling, 0.4 μg of pR9ΔEnv, 0.2 μg of pcRev, 0.4 μg of HIV-1-Gag-imCherryΔEnv, 0.2 μg of YFP-Vpr, and 0.8 μg of HXB2 Env. Transfection was carried out using JetPrime transfection reagent. Twelve hours post-transfection, the medium was exchanged to fresh medium with or without 500 nm SQV, an HIV-1 protease inhibitor. Forty-eight hours post-transfection, the supernatant was collected, aliquoted, and stored at −80 °C. To obtain concentrated viral stocks, the viral supernatants were pelleted through a 20% sucrose cushion by centrifugation at 100,000 × g for 2 h at 4 °C. RT activity in the virus preparation was measured using the real-time quantitative PCR-based product-enhanced reverse transcription assay (41), and, where indicated, the p24 content was determined by ELISA (7). Viral supernatants with equal RT activity were used to determine specific infectivity on TZM-bl cells, as described previously (42, 43).
Western blotting
We loaded 400 pg of p24 onto a 10% polyacrylamide gel (Bio-Rad). Proteins were transferred onto a nitrocellulose membrane, blocked with 10% blotting-grade blocker (Bio-Rad) for 30 min at room temperature, and incubated with human anti-HIV serum (HIV IG) (1:2000 dilution), Living Colors monoclonal anti-eGFP mouse antibody (Invitrogen, 1:1000), or rabbit-anti-mCherry antibody (Abcam, 1:1000) for 1 h at 25 °C or overnight at 4 °C. Following three 10-min washes with PBST (PBS + 0.1% Tween), secondary antibodies, HRP goat anti-rabbit antibody (1:500 dilution, Santa Cruz Biotechnology, Dallas, TX), HRP goat anti-human (1:2000, Bio-Rad), or HRP rabbit anti-mouse (1:1000, Millipore) and a chemiluminescence reagent from GE Healthcare were used for protein detection. Precision Plus Protein Standards (KaleidoscopeTM, Bio-Rad) were used as molecular weight markers. Densitometry performed using ImageJ (National Institutes of Health) yielded percent of protein cleavage (the amount of cleaved product divided by the sum of cleaved and precursor products).
Single-virus imaging in live and fixed cells
Single-virus fusion experiments were performed with either CV1.CD4.CXCR4 cells (CF3 clone, for HXB2 pseudoviruses) or CV1.TVA950 cells (for ASLV or VSV G pseudoviruses). Cells were plated on collagen-coated glass-bottom dishes (MatTek, Ashland, MA) in FluoroBrite DMEM and grown to 70% confluency. Before imaging, the cells were chilled on ice, washed with ice-cold Ca2+/Mg2+-containing phosphate buffer (PBS2+), and spinoculated with freshly thawed pseudovirus for 20 min at 1500 × g and 4 °C. After spinoculation, cells were washed with ice-cold PBS2+ to remove unbound virus. Images were acquired with a Personal DeltaVision imaging system (Applied Precision, GE Healthcare) using an UPlanFluo ×60/1.4 numerical aperture (NA) oil objective (Olympus, Tokyo, Japan) and a YFP/mCherry standard filter set (Chroma, Bellows Falls, VT), and two-channel fluorescence emission was recorded in series by an EM-CCD camera (Photometrics, Tucson, AZ) for a single field of view. Live cells were imaged in a single axial plane for either 30 (ASLV) or 60 min (VSV G or HXB2). Immediately before imaging, virus entry was initiated by addition of prewarmed live-cell imaging buffer. For event detection by visual annotation, low signal-to-noise fluorescence emission was sampled every 3 s using low excitation intensities. For automated event detection, excitation intensities were increased 3-fold, image integration times were increased 2-fold, and high signal-to-noise fluorescence emission was sampled every 30 s. During time-lapse imaging, an environmental chamber was used to maintain samples at 37 °C and high humidity, and the UltimateFocus module (Applied Precision, GE Healthcare) was used to compensate for axial drift. For fixed cell imaging, samples were incubated at 37 °C for indicated times and immediately fixed by treatment with 4% paraformaldehyde for 30 min at room temperature. Image stacks with a Z interval of 0.2 μm were acquired, deconvolved, and collapsed by maximum intensity projection. Where indicated, cells were preincubated for 30 min with 20 μm MG-132, and cells were imaged in the presence of the same MG132 concentration.
Single-virus imaging for measuring co-labeled/lysed fraction, color balance, and FRET
Pseudoparticles were immobilized on poly-l-lysine–coated 8-well chamber coverslips (Lab-Tek, Nalge Nunc International, Penfield, NY) for 30 min at 4 °C, and wells were washed with cold PBS2+ to remove unbound virus. Virus samples were imaged before and after the addition of 0.1 mg/ml saponin to mildly permeabilize the viral membrane and release the mCherry content marker. For co-labeling/lysis fraction and color balance experiments, images were acquired in a single axial plane with a Personal DeltaVision imaging system (using the high signal-to-noise ratio protocol described above). Single-particle color balance was defined as CB = (IYFP − n × ImCh)/(IYFP + n × ImCh), where the integer n is a free parameter that is fixed to define the zero point of the CB distribution. CB is related to the “generalized polarization” parameter described in Ref. 44. Generalized polarization is calculated on a pixel-by-pixel basis, and CB is calculated for preidentified objects/single virus particles. For FRET experiments, virus samples were imaged for intensity of FRET with a Zeiss LSM780 confocal scanning microscope using a Plan-Apo ×63/1.4 NA oil immersion objective in a single Z plane by sequential excitation with 488 nm and 594 nm while detecting in both YFP (517–587 nm) and mCherry (613–692 nm) emission bands. YFP (IYFP/YFP) and mCherry (ImCh/mCh) intensities were detected in 488 nm/YFP and 594 nm/mCherry channels, respectively, and sensitized emission FRET intensity (IYFP/mCh) was detected in the 488 nm/mCherry channel. All analyses were performed on local background-corrected single-particle intensities. First-order cross-talk contributions were measured in parallel using single-labeled YFP-Vpr or mCherry virus as 2.24% for YFP emission bleedthrough and 3.67% for mCherry excitation bleedthrough (supplemental Fig. S2). The single-particle uncleaved content fraction was corrected for mCherry photobleaching, which was measured as 4.58% in a parallel experiment (supplemental Fig. S2). To find the normalized FRET, the single-particle FRET intensity was adjusted for cross-talk and normalized by acceptor intensity (31, 32) as follows: normalized FRET = (IYFP/mCh − 0.0224 × IYFP/YFP − 0.0367 × ImCh/mCh)/ImCh/mCh.
Event annotation, curve fitting, and single-particle tracking
For event detection by visual inspection, single-particle color change events were annotated using the ROI Manager in ImageJ (National Institutes of Health). Annotated particles were tracked using Volocity software (PerkinElmer Life Sciences) as described previously (14, 21, 23). For automated event detection, a Matlab (Mathworks, Natick, MA) script was designed to identify particles, calculate background-corrected single-particle CB, and classify particles as either co-labeled or YFP+/mCherry− (CB > 0.85) (supplemental Fig. S3). Manually annotated fusion kinetics were fit to a single exponential model, y(t) = A0 × (1 − exp{−k × t}), where A0 is the number of fusion-competent particles and k is the fusion rate. Curves from computer-automated detection of YFP-only particles were fit to a double-exponential model that solves the two-step reaction A→B→C: B(t) = (k1 × A0/(k2 − k1)) × exp{−k1 × t} + (B0 − (k1 × A0/(k2 − k1))) × exp{−k2 × t}. Here, A0 and B0 are the numbers of fusion-competent particles and post-fusion particles at time zero, respectively; k1 and k2 are rates of fusion (A→B) and degradation of post-fusion particles (B→C), respectively.
Statistical analysis
Unless stated otherwise, statistical analysis was performed using Student's t test or non-parametric Man-Whitney test, as appropriate. Triplicate data for each independent experiment (usually two to three experiments) performed under identical conditions were normalized to the internal control and pooled to calculate the mean and standard error. *, p < 0.05; **, p < 0.01; ***, p < 0.001; N.S., not significant.
Author contributions
A. C. F. created and optimized the bifunctional marker. C. S., A. C. F., and G. B. M. conceived and planned the experiments. C. S., A. C. F., and T. M. D. performed the experiments and analyzed the results. C. S. wrote software for automated detection of fusion. C. S., A. C. F., and G. B. M. wrote the manuscript. All authors read and approved the manuscript.
Supplementary Material
Acknowledgments
We thank Mariana Marin for help with Western blotting, critical reading of the manuscript, and helpful discussions. We thank the National Institutes of Health AIDS Reagent Program, David Kabat, and James Binley for cell lines and plasmids and Dr. E. Gratton for valuable suggestions regarding virus fusion analysis.
This work was supported by National Institutes of Health Grants R01 GM054787 and R01 AI053668 (to G. B. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S6 and Movies S1–S3.
C. Sood, A. C. Francis, T. M. Desai, and G. B. Melikyan, manuscript in preparation.
- VSV
- vesicular stomatitis virus
- ASLV
- avian sarcoma and leukosis virus
- SQV
- saquinavir
- CB
- color balance
- Env
- envelope glycoprotein
- eYFP
- enhanced YFP
- eGFP
- enhanced GFP
- HIV-1
- HIV, type 1
- TVA
- tumor virus A.
References
- 1. Cavrois M., De Noronha C., and Greene W. C. (2002) A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 2. Rosa A., Chande A., Ziglio S., De Sanctis V., Bertorelli R., Goh S. L., McCauley S. M., Nowosielska A., Antonarakis S. E., Luban J., Santoni F. A., and Pizzato M. (2015) HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526, 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Da Silva Santos C., Tartour K., and Cimarelli A. (2016) A novel entry/uncoating assay reveals the presence of at least two species of viral capsids during synchronized HIV-1 infection. PLoS Pathog. 12, e1005897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones D. M., and Padilla-Parra S. (2015) Imaging real-time HIV-1 virion fusion with FRET-based biosensors. Sci. Rep. 5, 13449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dale B. M., McNerney G. P., Thompson D. L., Hubner W., de Los Reyes K., Chuang F. Y., Huser T., and Chen B. K. (2011) Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host Microbe 10, 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miyauchi K., Kim Y., Latinovic O., Morozov V., and Melikyan G. B. (2009) HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137, 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Padilla-Parra S., Marin M., Gahlaut N., Suter R., Kondo N., and Melikyan G. B. (2013) Fusion of mature HIV-1 particles leads to complete release of a Gag-GFP-based content marker and raises the intraviral pH. PLoS ONE 8, e71002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Markosyan R. M., Cohen F. S., and Melikyan G. B. (2005) Time-resolved imaging of HIV-1 Env-mediated lipid and content mixing between a single virion and cell membrane. Mol. Biol. Cell 16, 5502–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melikyan G. B., Barnard R. J., Abrahamyan L. G., Mothes W., and Young J. A. (2005) Imaging individual retroviral fusion events: from hemifusion to pore formation and growth. Proc. Natl. Acad. Sci. U.S.A. 102, 8728–8733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lakadamyali M., Rust M. J., Babcock H. P., and Zhuang X. (2003) Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 100, 9280–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Blanc I., Luyet P. P., Pons V., Ferguson C., Emans N., Petiot A., Mayran N., Demaurex N., Fauré J., Sadoul R., Parton R. G., and Gruenberg J. (2005) Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 7, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banerjee I., Yamauchi Y., Helenius A., and Horvath P. (2013) High-content analysis of sequential events during the early phase of influenza A virus infection. PLoS ONE 8, e68450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spence J. S., Krause T. B., Mittler E., Jangra R. K., and Chandran K. (2016) Direct visualization of Ebola virus fusion triggering in the endocytic pathway. mBio 7, e01857–01815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desai T. M., Marin M., Chin C. R., Savidis G., Brass A. L., and Melikyan G. B. (2014) IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 10, e1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zaitseva E., Yang S.-T., Melikov K., Pourmal S., and Chernomordik L. V. (2010) Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 6, e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chernomordik L. V., and Kozlov M. M. (2005) Membrane hemifusion: crossing a chasm in two leaps. Cell 123, 375–382 [DOI] [PubMed] [Google Scholar]
- 17. Cohen F. S., and Melikyan G. B. (2004) The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J. Membr. Biol. 199, 1–14 [DOI] [PubMed] [Google Scholar]
- 18. Matos P. M., Marin M., Ahn B., Lam W., Santos N. C., and Melikyan G. B. (2013) Anionic lipids are required for vesicular stomatitis virus G protein-mediated single particle fusion with supported lipid bilayers. J. Biol. Chem. 288, 12416–12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Floyd D. L., Ragains J. R., Skehel J. J., Harrison S. C., and van Oijen A. M. (2008) Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 105, 15382–15387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ganser-Pornillos B. K., Yeager M., and Sundquist W. I. (2008) The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18, 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Padilla-Parra S., Matos P. M., Kondo N., Marin M., Santos N. C., and Melikyan G. B. (2012) Quantitative imaging of endosome acidification and single retrovirus fusion with distinct pools of early endosomes. Proc. Natl. Acad. Sci. U.S.A. 109, 17627–17632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Desai T. M., Marin M., Sood C., Shi J., Nawaz F., Aiken C., and Melikyan G. B. (2015) Fluorescent protein-tagged Vpr dissociates from HIV-1 core after viral fusion and rapidly enters the cell nucleus. Retrovirology 12, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sood C., Marin M., Mason C. S., and Melikyan G. B. (2016) Visualization of content release from cell surface-attached single HIV-1 particles carrying an extra-viral fluorescent pH-sensor. PLoS ONE 11, e0148944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McDonald D., Vodicka M. A., Lucero G., Svitkina T. M., Borisy G. G., Emerman M., and Hope T. J. (2002) Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Padilla-Parra S., Marin M., Kondo N., and Melikyan G. B. (2012) Synchronized retrovirus fusion in cells expressing alternative receptor isoforms releases the viral core into distinct sub-cellular compartments. PLoS Pathog. 8, e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jha N. K., Latinovic O., Martin E., Novitskiy G., Marin M., Miyauchi K., Naughton J., Young J. A., and Melikyan G. B. (2011) Imaging single retrovirus entry through alternative receptor isoforms and intermediates of virus-endosome fusion. PLoS Pathog. 7, e1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mothes W., Boerger A. L., Narayan S., Cunningham J. M., and Young J. A. (2000) Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103, 679–689 [DOI] [PubMed] [Google Scholar]
- 28. Johannsdottir H. K., Mancini R., Kartenbeck J., Amato L., and Helenius A. (2009) Host cell factors and functions involved in Vesicular stomatitis virus entry. J. Virol. 83, 440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oum Y. H., Desai T. M., Marin M., and Melikyan G. B. (2016) Click labeling of unnatural sugars metabolically incorporated into viral envelope glycoproteins enables visualization of single particle fusion. J. Virol. Methods 233, 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akrap N., Seidel T., and Barisas B. G. (2010) Forster distances for fluorescence resonant energy transfer between mCherry and other visible fluorescent proteins. Anal. Biochem. 402, 105–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jiang X., and Sorkin A. (2002) Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol. Biol. Cell 13, 1522–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zal T., and Gascoigne N. R. (2004) Photobleaching-corrected FRET efficiency imaging of live cells. Biophys. J. 86, 3923–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Francis G., Kerem Z., Makkar H. P., and Becker K. (2002) The biological action of saponins in animal systems: a review. Br. J. Nutr. 88, 587–605 [DOI] [PubMed] [Google Scholar]
- 34. Jiang J., and Aiken C. (2006) Maturation of the viral core enhances the fusion of HIV-1 particles with primary human T cells and monocyte-derived macrophages. Virology 346, 460–468 [DOI] [PubMed] [Google Scholar]
- 35. Wyma D. J., Jiang J., Shi J., Zhou J., Lineberger J. E., Miller M. D., and Aiken C. (2004) Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 78, 3429–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Padilla-Parra S., Marin M., Kondo N., and Melikyan G. B. (2014) Pinpointing retrovirus entry sites in cells expressing alternatively spliced receptor isoforms by single virus imaging. Retrovirology 11, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mamede J. I., Cianci G. C., Anderson M. R., and Hope T. J. (2017) Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc. Natl. Acad. Sci. U.S.A. 114, E7169–E7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simmons J. A., D'Souza R. S., Ruas M., Galione A., Casanova J. E., and White J. M. (2015) Ebolavirus glycoprotein directs fusion through NPC1+ endolysosomes. J. Virol. 90, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de la Vega M., Marin M., Kondo N., Miyauchi K., Kim Y., Epand R. F., Epand R. M., and Melikyan G. B. (2011) Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., and Kappes J. C. (2002) Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pizzato M., Erlwein O., Bonsall D., Kaye S., Muir D., and McClure M. O. (2009) A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J. Virol. Methods 156, 1–7 [DOI] [PubMed] [Google Scholar]
- 42. Francis A. C., Di Primio C., Quercioli V., Valentini P., Boll A., Girelli G., Demichelis F., Arosio D., and Cereseto A. (2014) Second generation imaging of nuclear/cytoplasmic HIV-1 complexes. AIDS Res. Hum. Retroviruses 30, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Francis A. C., Marin M., Shi J., Aiken C., and Melikyan G. B. (2016) Time-resolved imaging of single HIV-1 uncoating in vitro and in living cells. PLoS Pathog. 12, e1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu W., So P. T., French T., and Gratton E. (1996) Fluorescence generalized polarization of cell membranes: a two-photon scanning microscopy approach. Biophys. J. 70, 626–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.