Abstract
Impaired mitochondrial oxidative phosphorylation (OXPHOS) capacity, accompanied by enhanced glycolysis, is a key metabolic feature of cancer cells, but its underlying mechanism remains unclear. Previously, we reported that human hepatoma cells that harbor OXPHOS defects exhibit high tumor cell invasiveness via elevated claudin-1 (CLN1). In the present study, we show that OXPHOS-defective hepatoma cells (SNU354 and SNU423 cell lines) exhibit reduced expression of mitochondrial ribosomal protein L13 (MRPL13), a mitochondrial ribosome (mitoribosome) subunit, suggesting a ribosomal defect. Specific inhibition of mitoribosomal translation by doxycycline, chloramphenicol, or siRNA-mediated MRPL13 knockdown decreased mitochondrial protein expression, reduced oxygen consumption rate, and increased CLN1-mediated tumor cell invasiveness in SNU387 cells, which have active mitochondria. Interestingly, we also found that exogenous lactate treatment suppressed MRPL13 expression and oxygen consumption rate and induced CLN1 expression. A bioinformatic analysis of the open RNA-Seq database from The Cancer Genome Atlas (TCGA) liver hepatocellular carcinoma (LIHC) cohort revealed a significant negative correlation between MRPL13 and CLN1 expression. Moreover, in patients with low MRPL13 expression, two oxidative metabolic indicators, pyruvate dehydrogenase B expression and the ratio of lactate dehydrogenase type B to type A, significantly and negatively correlated with CLN1 expression, indicating that the combination of elevated glycolysis and deficient MRPL13 activity was closely linked to CLN1-mediated tumor activity in LIHC. These results suggest that OXPHOS defects may be initiated and propagated by lactate-mediated mitoribosomal deficiencies and that these deficiencies are critically involved in LIHC development.
Keywords: cell invasion, hepatocellular carcinoma, mitochondrial metabolism, mitochondrial respiratory chain complex, ribosome
Introduction
The mitochondrion is a semi-autonomous organelle with distinct machinery for transferring information from its own DNA into proteins. This machinery achieves replication, transcription, and translation. However, more than a thousand nuclear-encoded proteins are required for maintaining the integrity and function of mitochondria (1). Human mitochondrial DNA (mtDNA)3 comprises a double-stranded circular DNA with 16,569 base pairs. It carries 2 ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs), and it produces 13 proteins (2). Recently, small peptides encoded as short open reading frames that possess various biological roles have also been identified in mtDNA (3). All 13 proteins are essential components of the mitochondrial oxidative phosphorylation (OXPHOS) system for aerobic ATP generation. The 2 rRNAs and the 22 tRNAs are only required for synthesizing these 13 proteins (4). Therefore, ATP synthesis through the OXPHOS system represents the major function of the mitochondrion, despite its diverse molecular and biochemical activities.
Many solid tumor cells harbor impaired OXPHOS activity and acquire a dependence on glycolytic ATP production (5). This reprogrammed bioenergetic state is often regarded as a hallmark of cancer cells. It has long been accepted that defective OXPHOS is merely a cancer epiphenomenon, because many tumor cells experience intermittent hypoxia in the process of forming a nodular mass. However, recent accumulating evidence has supported the hypothesis that OXPHOS impairments are involved in tumor progression (6, 7). Mitochondrial dysfunction promotes migratory and invasive tumor cell activities in many cancer types through reactive oxygen species generation or hypoxia-inducible factor 1α accumulation (6, 8–10). These properties explain the causative role of mitochondria in tumors that gain metastatic properties. In addition, mitochondrial defects are involved in tumor cell growth (11) and oncogenesis (12). Tumoral OXPHOS defects are primarily associated with mtDNA deletions and/or point mutations, which are often related to altered mitochondrial biogenesis or degradation, as described in many types of cancers (10, 13–16). However, it remains unclear how OXPHOS defects are triggered during cancer development, particularly in normoxic conditions.
The mitochondrial translation machinery or the mitochondrial ribosome (mitoribosome) is the final executor in the synthesis of the 13 essential OXPHOS proteins encoded by mtDNA. This machinery includes 2 structurally incorporated mitochondrial rRNAs and 22 functionally involved mitochondrial tRNAs. The human mitoribosome comprises small 28S and large 39S subunits (17). The small subunit (MRPS) is composed of 30 mitoribosomal proteins (MRPs) and the large subunit (MRPL) is composed of 52 MRPs (18). All MRPs are encoded by nuclear genes. Because of this complexity, the complete structure of the mammalian mitoribosome was only recently revealed (19). The importance of OXPHOS impairments in cancer development corresponds well with differential MRP gene expression and MRP gene alterations; thus, these alterations have often been investigated in diverse cancers (19). For example, low expression of MRPS29, also called DAP3, was significantly associated with local recurrence, distant metastasis, and mortality in breast cancer (20). MRPS23 gene amplification was observed in cervical and breast cancer (21). A single nucleotide polymorphism in the genomic region of MRPS30 was associated with postmenopausal breast cancer risk (22). However, the underlying mechanisms that link MRP alterations to cancer cell activity remain to be elucidated.
In the present study, we show that reduced MRPL13 expression is a key factor in mitoribosome regulation and subsequent OXPHOS defects. We demonstrate that this mechanism can regulate hepatoma cell invasion activity and that lactate can act as an upstream regulator of MRPL13 suppression. Finally, a bioinformatic analysis of The Cancer Genome Atlas (TCGA) liver hepatocellular carcinoma (LIHC) open database revealed that claudin-1 (CLN1), a key regulator of hematoma cell invasion activity, is strongly negatively correlated with oxidative metabolic phenotypes in low MRPL13-expressing LIHC tumors. These results indicate that mitoribosomal defects are key regulators of mitochondrial OXPHOS and that these defects may be initiated and propagated by lactate released from neighboring glycolytic tumor cells in liver cancer development.
Results
Mitoribosomal defect is linked to OXPHOS dysfunction in SNU hepatoma cells
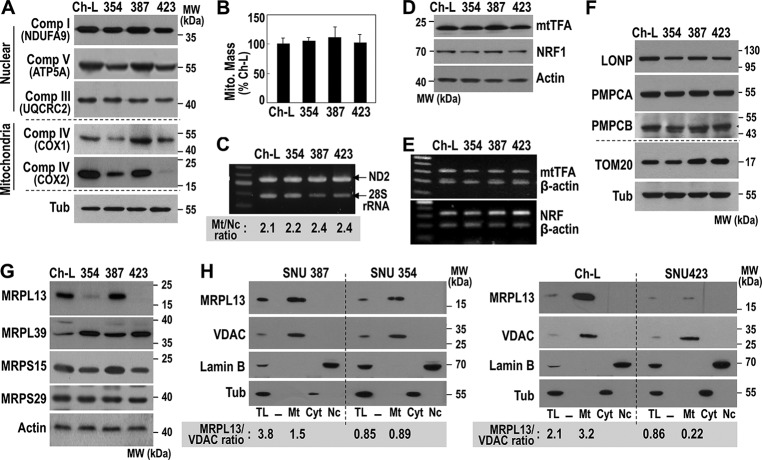
We previously characterized some metabolic features of SNU hepatoma cells; we showed that SNU354 and SNU423 cells had mitochondrial respiratory defects and high cell invasiveness compared with hepatoma cells with active mitochondria, Chang-liver (Ch-L), and SNU387 cells (10, 23). Interestingly, in SNU354 and SNU423 cells, the expression levels of mtDNA-encoded OXPHOS subunits, such as COX1 and COX2, were reduced more severely than the nuclear encoded subunits (Fig. 1A). To understand the mechanism underlying this low expression of mtDNA-encoded genes, we first monitored mitochondrial mass in these cells with MitoTracker Red, a mitochondria-specific fluorescence dye. Then we performed PCR-based evaluations of genomic mtDNA levels. However, no significant differences were observed among the different cell lines (Fig. 1, B and C). We detected similar expression levels of mitochondrial transcription factor A (mtTFA), a key transcription factor in mitochondrial replication and transcription, and its upstream regulator, nuclear respiratory factor 1 (NRF1) (Fig. 1. D and E). Similar levels of some mitochondrial proteases, Lon protease (LONP), mitochondrial-processing peptidase subunit α (PMPCA), and mitochondrial-processing peptidase subunit β (PMPCB) were also observed (Fig. 1F). However, a clear difference was found in the expression levels of some mitoribosomal subunits. We found a minor reduction in MRPS15, a minor increase in MRPL39, and a severe reduction in MRPL13 levels in SNU354 and SNU423 cells (Fig. 1G). These results implied that imbalanced subunit expression might mediate a mitoribosomal defect that may be linked to the OXPHOS defect. Among the mitoribosomal subunits, the reduction in MRPL13 expression was most evident. As expected, MRPL13 was exclusively localized in the mitochondria of all cells, and the levels were clearly reduced in the mitochondria of SNU354 and SNU423 (Fig. 1H). This result suggested that down-regulation of MRPL13 expression may be a critical factor in the OXPHOS defect.
Figure 1.
Mitochondria-defective hepatoma cells possess MRPL13-mediated mitoribosomal defect. SNU hepatoma cells (SNU354, SNU387, and SNU423) and Ch-L clone were cultured to maintain an exponentially growing state. A, Western blot analysis for OXPHOS subunits. B, intracellular mitochondria mass was estimated by flow cytometric analysis after staining cells with MitoTracker Red as described under “Experimental procedures.” C, intracellular mtDNA levels were analyzed by genomic PCR with a primer set for mtDNA ND2 and a primer set for 28S rRNA as an indicator for nuclear DNA. The ratio of ND2 versus 28S rRNA was presented in the bottom panel. D, Western blot analysis. E, RT-PCR for mtTFA (upper panel) and NRF1 (lower panel). F and G, Western blot analysis for some mitochondrial proteases (F) and some mitoribosomal subunits (G). H, cellular compartments, such as mitochondria (Mt), cytoplasm (Cyt), and nuclei (Nc), were isolated and subjected to Western blot analysis. voltage-dependent anion-selective channel (VDAC) (mitochondria), Lamin B (nuclei), and tubulin (Tub, cytoplasm) were used as the markers of the compartments. MRPL13 expression levels in total lysate (TL) and mitochondria fraction were presented as the ratio of MRPL13 versus VDAC in the bottom panel. Comp, complex; MW, molecular mass.
Mitoribosomal defect enhances hepatoma cell invasiveness
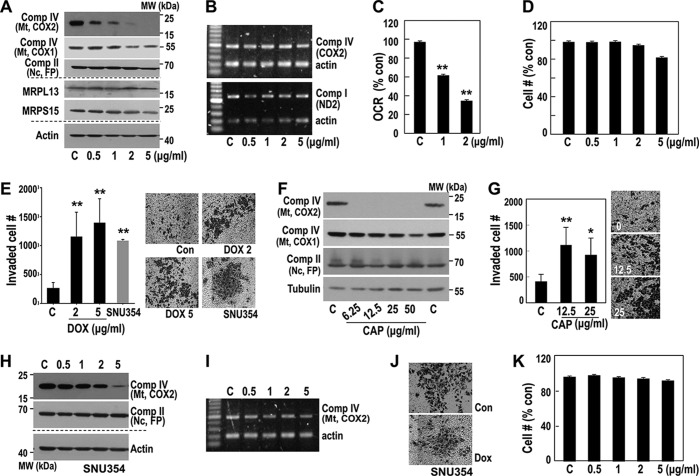
Because mitochondria-defective hepatoma cells (SNU354 and SNU423) showed high cell invasion activity (10, 23), we examined whether mitoribosomal inhibition could regulate hepatoma cell invasiveness. We inhibited mitoribosomes with mitochondria-specific translation inhibitors doxycycline (DOX) and chloramphenicol (CAP). In SNU387 cells, which have active mitochondria and low invasiveness, mitochondrial COX2 protein expression was highly sensitive to DOX treatment without affecting its mRNA level (Fig. 2, A and B, upper panel). The COX2 mRNA transcript levels were further confirmed by detecting ND2 transcript, which was located in a different region of the same heavy strand polycistronic transcript (Fig. 2B, lower panel). COX1 expression showed a delayed response (Fig. 2, A and B). As a result, DOX treatment significantly reduced the mitochondrial oxygen consumption rate (OCR) without affecting the cell growth rate (Fig. 2, C and D), and cell invasion activity increased (Fig. 2E). Similar results were obtained with CAP, a blocker of the peptidyl transfer step in the mitochondrial 50S ribosomal subunit (Fig. 2, F and G). In contrast, DOX had only minor effects on mitochondrial expression and cell invasion activity at high concentrations in OXPHOS defective, highly invasive SNU354 cells (Fig. 2, H–K). These results supported our hypothesis that the OXPHOS defect in this hepatoma cell line might be due to a mitoribosomal defect.
Figure 2.
Pharmacological inhibition of mitochondrial translation activity increases invasion activity. A–D, SNU387 cell was exposed to the indicated concentrations of DOX for 3 days and then subjected to individual analysis. A, Western blot analysis for some OXPHOS subunits and mitochondrial ribosomal subunits. B, RT-PCR to monitor some OXPHOS subunit mRNA levels. C, cellular OCR. **, p < 0.01 versus control by Student's t test. D, cell growth rates were monitored by counting live cell number. No significant amount of dead cells was found. E, cell invasion activity was performed using MatrigelTM-coated TranswellTM as described under “Experimental procedures.” Invaded cell numbers were counted. Representative images for invaded cells are shown in the right panel. **, p < 0.01 versus control by Student's t test. F and G, SNU387 cell was exposed to the indicated concentrations of CAP for 3 days, and the cells were subjected to individual analysis. F, Western blot analysis. G, cell invasion activity. *, p < 0.05 and **, p < 0.01 versus control by Student's t test. H–K, SNU354 cell was exposed to the indicated concentrations of DOX for 3 days, and then cells were subjected to Western blot analysis (H), RT-PCR (I), cell invasion activity (J), and cell growth rate (K). Comp, complex; MW, molecular mass; Mt, mitochondria; Nc, nuclei; Con, control.
MRPL13 knockdown-mediated mitoribosomal defect promotes hepatoma cell invasiveness via claudin-1 induction
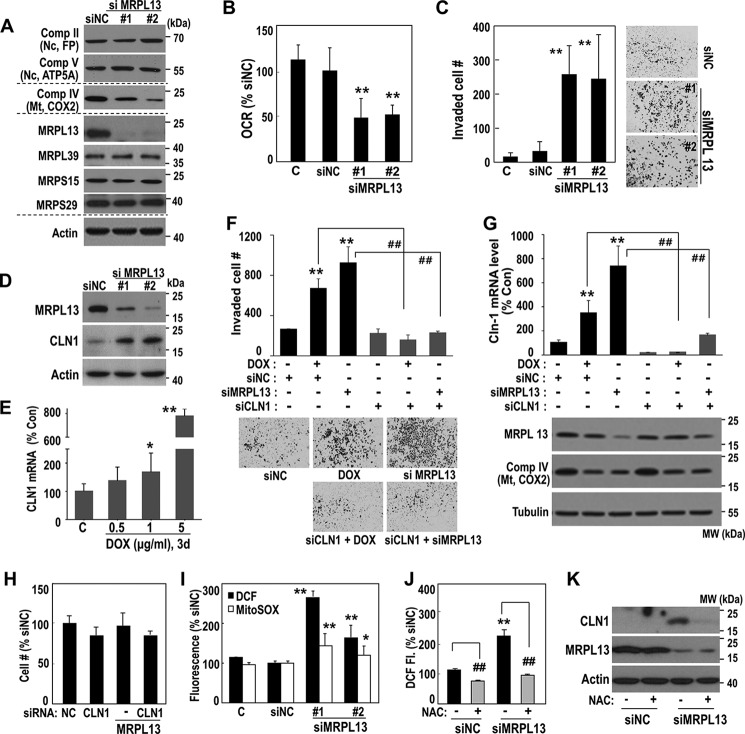
Next, we examined whether MRPL13 suppression could induce mitoribosomal defects and regulate hepatoma cell invasiveness. A MRPL13 knockdown in SNU387 cells effectively decreased mitochondrial COX2 expression without affecting the other mitoribosomal subunits or the nuclear-encoded OXPHOS subunits (flavoprotein (FP) of complex II and ATP5A of complex V). The MRPL13 knockdown also significantly decreased OCR and enhanced cell invasion activity (Fig. 3, A–C). We previously demonstrated that CLN1 is a key regulator of hepatoma cell invasion activity in response to OXPHOS defect (23). Similarly, mitoribosomal defects, caused by siRNA-mediated MRPL13 knockdown or DOX treatment, clearly induced CLN1 expression (Fig. 3, D and E). We further confirmed that SNU387 invasiveness, induced by DOX or MRPL13 suppression, was effectively obviated by a CLN1 knockdown (Fig. 3, F and G) without significantly affecting cell growth (Fig. 3H). Interestingly, MRPL13 knockdown augmented both mitochondrial and overall intracellular reactive oxygen species (ROS) levels (Fig. 3I). Removal of the intracellular ROS by N-acetylcysteine, an antioxidant, blocked the CLN1 induction (Fig. 3, J and K). These results suggest that CLN1 is involved in the invasiveness mediated by mitoribosomal defects, and its expression is regulated by ROS-mediated retrograde signals.
Figure 3.
Mitoribosomal inhibition regulates hepatoma cell invasiveness via CLN1. A–D, SNU387 cell was transfected with siRNAs for MRPL13 for 3 days. A, Western blot analysis. B, cellular O2 consumption rate. **, p < 0.01 versus siNC by Student's t test. C, cell invasion activity. **, p < 0.01 versus siNC by Student's t test. Representative images for invaded cells are shown in the right panel. D, Western blot analysis. E, SNU387 cell was exposed to the indicated concentrations of DOX for 3 days and then subjected to qPCR for CLN1 mRNA. *, p < 0.05 and **, p < 0.01 versus control by Student's t test. F–H, SNU387 cell was transfected with siMRPL13 or exposed to DOX (5 μg/ml) with or without combination of siCLN1 transfection for 2 days. F, cell invasion activity. Representative images for invaded cells are shown in the bottom panel. G, qPCR for CLN1 mRNA levels. Western blot analyses are also shown in the bottom panel. **, p < 0.01 versus siNC by Student's t test; ##, p < 0.01 versus siNC+DOX or siMRPL13 by Student's t test. H, cell growth was monitored by counting cell number. I, SNU387 cell was transfected with siMRPL13 for 2 days, and then intracellular (■) and mitochondrial (□) ROS levels were monitored as described in “Experimental procedures.” *, p < 0.05 and **, p < 0.01 versus siNC by Student's t test. J and K, SNU387 cell was transfected with siMRPL13 for 2 days and subsequently exposure to 10 mm NAC for 6 h. J, intracellular ROS levels. **, p < 0.01 versus siNC without NAC; ##, p < 0.01 versus without NAC in the same siRNA-treated condition by Student's t test. K, Western blot analysis. Comp, complex; MW, molecular mass; Mt, mitochondria; Nc, nuclei; Con, control.
Exogenous lactate regulates MRPL13 expression
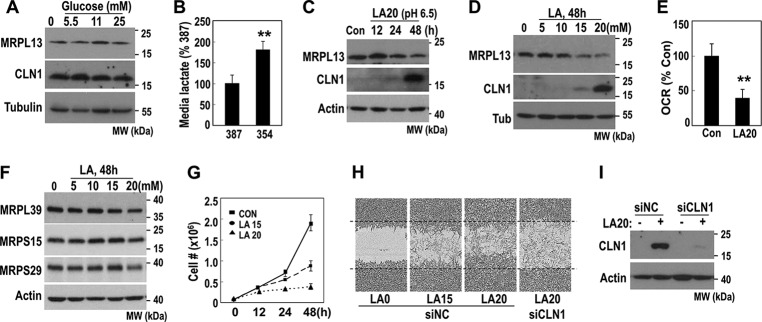
We then investigated how MRPL13 expression was suppressed in hepatoma cells. Yeast MRPL13, a homologue of mammalian MRPL50, was reported to be repressed in the presence of glucose (24). Therefore, we tested whether MRPL13 was regulated by glucose. Unexpectedly, manipulating extracellular glucose concentrations did not affect MRPL13 expression in SNU387 cells (Fig. 4A). Alternatively, OXPHOS-defective SNU354 cells produced and released high amounts of lactate into media (Fig. 4B). Therefore, we tested whether lactate could regulate MRPL13 expression. When we exposed SNU387 to 20 mm lactate at pH 6.5, the physiological level in tumor microenvironments (25, 26), MRPL13 expression progressively decreased in parallel with CLN1 induction (Fig. 4C). This response was obtained with over 15 mm lactate (Fig. 4D). We also confirmed that 20 mm lactate treatment significantly decreased OCR (Fig. 4E). Interestingly, the lactate treatment not only decreased MRPL13 expression but also slightly suppressed the expression of other MRP at 20 mm (Fig. 4F). This result is consistent with a previous report by Tang et al. (27) in which a microarray analysis showed that lactate treatment suppressed the expression of several MRPs in MCF7 cells. Despite its delayed effect on cell growth (Fig. 4G), lactate promoted cell invasion activity, and this invasiveness was inhibited by CLN1 knockdown (Fig. 4, H and I). These results implied that exogenous lactate suppressed mitochondrial OXPHOS by down-regulating MRPs, especially MRPL13, and promoted tumor cell invasiveness via CLN1. Thus, extracellular lactate released from glycolytic tumor cells may induce mitoribosomal defects in an autocrine and/or paracrine manner and thereby trigger mitochondrial OXPHOS defects and hepatoma cell invasiveness.
Figure 4.
Exogenous lactate suppresses MRPL13 expression. A, SNU387 cell was treated with the indicated glucose concentration for 2 days and then subjected to Western blot analysis. B, extracellular lactate concentration was measured in SNU387 cell and SNU354 cell as described under “Experimental procedures.” **, p < 0.01 versus SNU387 by Student's t test. C, SNU387 cell was treated with 20 mm lactic acid (LA), adjusted to pH 6.5, for the indicated times, and subjected to Western blot analysis. D–G, SNU387 cell was treated with the indicated concentration of LA for the indicated time periods. D, Western blot analysis. E, cellular O2 consumption rate. **, p < 0.01 versus control by Student's t test. F, Western blot analysis. G, cell growth. H and I, SNU387 cell was transfected with siRNAs for CLN1 and exposed to LA for 48 h. Cell invasion activity was monitored by IncuCyteTM as described under “Experimental procedures.” Representative images are shown (H), and protein levels were validated by Western blot analysis (I). MW, molecular mass; Con, control; Tub, tubulin.
Mitoribosomal defect mediated by the combination of elevated glycolysis and reduced MRPL13 expression is correlated with CLN1 expression in LIHC tumors
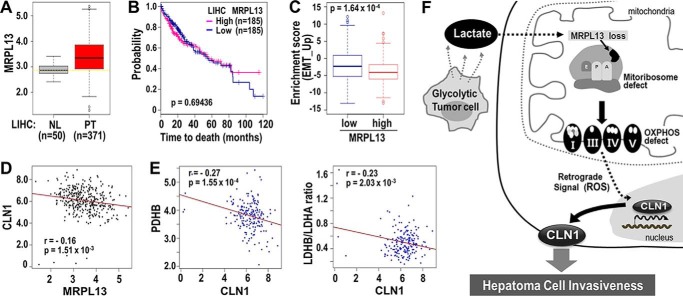
Finally, to address the clinical significance of the MRPL13 gene, we analyzed its expression levels in hepatocellular carcinoma with RNA-Seq data from the TCGA-LIHC cohort. Unexpectedly, we found that, in primary tumors (PT), MRPL13 expression levels were higher but much more dispersed than normal liver tissues (NL) (Fig. 5A). The broad range of MRPL13 expression levels in PTs reflected the heterogeneity of liver cancer. To elucidate the clinical significance of MRPL13, we performed a Kaplan–Meier survival analysis to compare clinical outcomes between MRPL13-high and MRPL13-low expression groups, where high and low expression levels were defined, respectively, as above and below the median MRPL13 expression level. However, overall survival was not significantly different between groups, except that the MRPL13-low group showed slightly worse survival after 80 months (Fig. 5B). These results implied that MRPL13 alone was not a good prognostic indicator of liver cancer.
Figure 5.
Validation of clinical significance of MRPL13 in TCGA-LIHC. A, the expression level of MRPL13 in different tissue type was plotted. NL and PT indicate tissues from normal liver and primary tumor, respectively. **, p < 0.01 PT versus NL by Welch two-sample t test. B, Kaplan–Meier survival analysis was performed between MRPL13-high and MRPL13-low groups that were divided from 371 patients, based on the median value of MRPL13 expression level. The statistical p value was generated by the Cox–Mantel log-rank test. C, enrichment scores of EMT_UP genes were compared between MRPL13-low and -high groups. p value from Kolmogorov–Smirnov test of ssGSEA was transformed in log scale and used in the plot. D, association of MRP13 with CLN1 among the 371 patients was assessed by Pearson's product moment correlation test. E, association of CLN1 with PDHB (left panel) and the ratio of LDHB to LDHA (right panel) among the MRPL13 low group (n = 185) was assessed by Pearson's product moment correlation test. The ratio of LDHB to LDHA was calculated at individual sample level. Pearson correlation estimate, r, and p values were depicted. F, schematic diagram for our hypothesis explaining the underlying mechanisms of OXPHOS defect in hepatoma cell invasiveness.
Next, we further evaluated whether MRPL13 level is associated with epithelial-mesenchymal transition (EMT) activity by analyzing core EMT interactome genes that were previously identified by Taube et al. (28). When we performed the single sample gene set enrichment analysis (ssGSEA) in the TCGA-LIHC cohort, EMT core gene signature (EMT_UP; n = 91) was significantly more enriched in the MRPL13-low group than MRPL-high group (Fig. 5C), indicating a strong relevance of MRPL13 to EMT-related activity. Moreover, MRPL13 expression was significantly negatively correlated with CLN1 expression in the cohort (Fig. 5D). Not surprisingly, in the MRPL13-low group, CLN1 expression showed strongly negative correlation with two indicators of oxidative mitochondrial metabolism, pyruvate dehydrogenase B (PDHB) expression and the ratio of LDH type B to LDH type A (LDHB/LDHA), which indicated a metabolic shift to glycolysis (Fig. 5E). These results implied that a combined phenotype of elevated glycolysis and MRPL13-mediated mitoribosomal defects may be strongly associated with CLN1-mediated tumor activity, especially EMT, in patients with liver cancer. Collectively, our results indicated that OXPHOS defects may be initiated and propagated by lactate-mediated mitoribosomal defects and critically involved in hepatoma cell invasiveness (Fig. 5F).
Discussion
The human mitoribosome is composed of 82 MRPs, all encoded by nuclear DNA and imported into the mitochondria. The complete high-resolution structure of the mitochondrion was recently determined with cryo-electron microscopy (29, 30). Protein synthesis inside the mitochondria produces only 13 mtDNA-encoded OXPHOS subunits; this implies that the key role of mitochondria is to regulate OXPHOS activity at the protein level. Interestingly, a recent study revealed that MRPS39, also called PTCD3, played a critical role in Myc-driven lymphoma (31). In addition, MRPL44 expression was identified as a representative marker of a metabolic phenotype, and it was predictive of lymph node metastasis in papillary thyroid carcinoma (32). Those results suggested that detailed studies on the link between the mitoribosome and cancer activity may extend our understanding of novel molecular and metabolic mechanisms involved in tumor development. In the present study, we observed that suppressed MRPL13 expression was critical in the manifestation of mitoribosomal defect-mediated OXPHOS dysfunctions and subsequently enhanced hepatoma cell invasiveness. The importance of mitoribosomal activity in regulating hepatoma cell invasiveness was further demonstrated with the pharmacological mitoribosomal inhibitors DOX and CAP. To the best of our knowledge, this study was the first to show that mitoribosomal defects could cause cancerous OXPHOS damage and identify a link between mitoribosomal activity and tumor cell invasiveness.
Many non-hypoxic cancers commonly display an OXPHOS defect (33, 34), and thus, OXPHOS defects represent a metabolic hallmark of cancer. However, the detailed mechanism underlying the OXPHOS defect, and its maintenance in normoxic conditions has remained elusive. Oncogenic Kras activation was shown to reduce OXPHOS by decreasing complex I or autophagic degradation (12, 35). Myc and hypoxia-inducible factor 1 can suppress OXPHOS activity via PDK1-mediated PDH inhibition (36). However, non-oncogenic regulation of tumoral OXPHOS activity is not clearly understood. Thus, an important observation of this study was that exogenous lactate effectively suppressed OXPHOS activity by reducing MRPL13 expression. These results support our hypothesis that an OXPHOS defect may be propagated and sustained in normoxic conditions by elevated extracellular lactate, which is released from surrounding glycolytic tumor cells. This mechanism emphasizes the role of lactate as a tumor microenvironmental propagator of metabolic alterations.
In tumor cells with OXPHOS defects, cellular energy is compensated by glycolytic ATP production. To facilitate and maintain glycolytic activity, two metabolic events are required: activation of LDH, which converts pyruvate to lactate, and suppression of PDH, which converts pyruvate to acetyl-CoA for aerobic mitochondrial metabolism. LDH is a tetramer composed of two different proteins encoded by the LDHA and LDHB genes. Different combinations of these two gene products give rise to five different LDH isozymes: LDH1 (B4), LDH2 (AB3), LDH3 (A2B2), LDH4 (A3B), and LDH5 (A4). LDH1 prefers to convert lactate to pyruvate, and LDH5 effectively catalyzes the conversion of pyruvate to lactate (37). Therefore, the ratio of LDHB to LDHA expression governs the direction of LDH action. PDH is a multienzyme complex composed of three enzymatic components: PDH (E1), dihydrolipoamide acetyltransferase (E2), and lipoamide dehydrogenase (E3). E1 is the key enzyme; it is a heterotetramer composed of two α and two β subunits. The PDHB gene encodes the E1 β subunit, and mutations are associated with pyruvate dehydrogenase E1-β deficiencies (38). That finding implied that PDHB plays an essential role in and might serve as a representative marker of PDH activity. Here, we found that PDHB and the LDHB/LDHA ratio were strongly negatively correlated with CLN1 expression in patients in the MRPL13-low LIHC group. However, liver cancer cells showed high heterogeneity in MRPL13 expression, which suggests that they represent a mixture of hepatoma cells with high and low mitoribosomal activity.
Here, we have demonstrated that MRPL13-mediated OXPHOS defects enhanced hepatoma cell invasion activity via CLN1 expression. Finally, we want to emphasize that this mitoribosomal defect is a key metabolic alteration, which is triggered and sustained by microenvironmental lactate released by glycolytic hepatoma cells, even in normoxic conditions.
Experimental procedures
Cell condition and cell growth rate
Human hepatoma cells SNU354, SNU387, and SNU423 were purchased from Korean Cell Line Bank (Seoul, Korea) and were cultured in GIBCO® RPMI 1640 medium (Invitrogen) supplemented with 10% GIBCO® FBS (Invitrogen) and GIBCO® antibiotics (Invitrogen) at 37 °C in a humidified incubator with 5% CO2. Chang cells were obtained from the American Tissue Culture Collection (Rockville, MD), and a Chang cell clone, denoted as Ch-L, with higher hepatic characteristics (albumin production and liver-specific carbamoyl-phosphate synthase-1 expression), was isolated by single cell dilution and expansion (23). Ch-L clone was cultured in GIBCO® DMEM (Invitrogen) supplemented with 10% GIBCO® FBS. Doxycycline (D9891), chloramphenicol (C1919), lactic acid solution (L1875), and glucose (G7021) were purchased from Sigma.
The cell growth rate was monitored by counting the trypan blue-negative viable cells. At the end of each experiment, the cells were harvested by trypsinization and counted using the CountessTM automated cell counter (Invitrogen) after staining with 0.4% (w/v) trypan blue (Invitrogen) to exclude dead cells.
Estimation of mitochondrial mass
To estimate mitochondrial mass, cells were stained with 100 nm Mitotracker Red CMXRos (M7512l Molecular Probes, Eugene, OR), a membrane potential-independent mitochondria-specific fluorescence dye, for 30 min at 37 °C. After harvesting cells in PBS, their fluorescence intensities were analyzed by flow cytometry (FACSCantoTM II; Becton Dickinson, San Jose, CA). Intracellular mitochondria mass was represented as a percentage of Ch-L clone using mean values of arbitrary fluorescence unit of 10,000 cells.
Estimation of intracellular mtDNA level
To estimate mtDNA level, total genomic DNA including mtDNA was isolated as described previously (39). Two primer sets, a set for ND2 gene for mitochondrial DNA and a set of 28S rRNA gene for nuclear DNA, were simultaneously used to PCR analysis with the isolated total genomic DNA as template. The final PCR products were electrophoresed on 1.5% agarose gel, stained with SYBR® Safe DNA Gel Stain (Invitrogen), and visualized on UV transilluminator (Gel Doc 2000; 4Bio-Rad, Milan, Italy). The ratio of ND2 level to 28S rRNA was obtained by scanning and comparing the band intensities. The sequences of the primer sets were as follows: for ND2, 5′-AGGTTACCCAAGGCACCCCT and 5′-AGTAGATTAGGCGTAGGTAG; for 28S rRNA, 5′-TAGCAGCCGACTTAGAACTGG, and 5′-CTCCCACTTATTCTACACCTC.
Subcellular fractionation
The nuclear, mitochondrial and cytoplasmic fractions were obtained from cells cultured in 90% confluency on 100-mm culture dishes as described previously with slight modification (40). Briefly, the cells were harvested by trypsinization and resuspended in medium A (250 mm sucrose, 0.1 mm EDTA, 2 mm HEPES, pH 7.4). The cell slurry was homogenized in a Dounce homogenizer (StedFastTM stirrer; Fisher Scientific), followed by centrifugation at 500 × g for 10 min to precipitate nuclei. The nuclei pellets were washed three times with buffer A (0.1 mm EDTA, 10 mm KCl, 10 mm HEPES, pH 7.9) containing 1% Nonidet P-40, and the final pellets were collected for nuclei fraction. The supernatant obtained after the first centrifugation at 500 × g was further centrifuged at 7,000 × g for 10 min. The supernatant (cytoplasmic fraction) and the pellet (crude mitochondrial fraction) were collected. The fractions were lysed in radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris, pH 8.0) for Western blot analysis.
Introduction of siRNAs into cells
To introduce siRNAs into cells, the cells were transfected with target siRNA duplexes using OligofectamineTM reagent (Invitrogen), according to the manufacturer's instructions. All the siRNA for targets, CLN1 (5′-UCACUGAACAAAACCUACA and 5′-UGUAGGUUUUGUUCAGUGA), MRPL13(5′-GUCUAGAUGAGUACACACA and 5′-UGUGUGUACUCAUCUAGAC), and negative control (5′-CCUACGCCACCAAUUUCGU and 5′-ACGAAAUUGGUGGCGUAGG) were generated from Bioneer (Daejeon, Korea).
Endogenous cellular oxygen consumption rate
Endogenous cellular OCR was measured in situ with cultured cells using an XF-24 extracellular flux analyzer (Seahorse Bioscience, Santa Clara, CA) according to the protocol provided. Briefly, the cells were seeded on XF24 cell culture microplates (Seahorse Bioscience) at a density of 10,000 cells/well and preincubated with XF base medium (Seahorse Bioscience) containing 1 mm sodium pyruvate (Invitrogen), 2 mm GlutaMAX (Invitrogen), and 11 mm glucose. Its mitochondrial specificity was confirmed by adding 5 mm KCN.
Cell invasion assay
Cell invasion assay was performed with TranswellTM permeable supports (Corning, Acton, MA) as described previously with slight modifications (23). Briefly, cells (2 × 104) prestarved with serum-free RPMI for 16 h were placed into the upper chamber with 100 μl of serum-free RPMI. RPMI (800 μl) supplemented with 10% FBS was placed in the lower chamber as a chemo-attractant. Uncoated TranswellTM insert membranes (8-μm pore size) were precoated with 7% growth factor reduced BD MatrigelTM Matrix (Becton Dickinson Labware, Franklin Lakes, NJ) for the cell invasion assay. After 2 days, the cells that invaded the lower surface were fixed with 100% methanol, stained with hematoxylin solution (Sigma) and Eosin Y Solution (Sigma), and counted by CELLCOUNTER (41). All experiments were performed in at least three independent experiments.
To acquire time-lapse images of invaded cells, cell invasion activity was assessed by IncuCyteTM (Essen Bioscience, Ann Arbor, MI) according to the protocol provided. 96-well ImageLockTM plates (Essen Bioscience) were precoated with 100 μg/ml MatrigelTM matrix (Becton Dickinson Labware) overnight. After cells (35,000/well) were seeded on each matrix-coated well with serum-free RPMI for overnight, the monolayered confluent cells were scratched using an Essen Wound Maker (Essen Bioscience). Then Matrigel matrix (50 μl of 2 mg/ml) was again added to each well and covered with 100 μl of medium containing 20 mm lactic acid (pH 6.5). Invaded cell images were automatically acquired by the IncuCyte imaging system.
Measurement of intracellular and mitochondrial ROS levels
To monitor intracellular and mitochondrial ROS levels, dichlorofluorescin diacetate (Molecular Probes) and MitoSOXTM(Molecular Probes) fluorogenic probes were used, respectively. Briefly, the cells were incubated in media containing 20 μm dichlorofluorescin diacetate or 2.5 μm MitoSOX for 15 min at 37 °C. Stained cells were washed and resuspended in PBS and analyzed by flow cytometry (FACSCantoTM II). Mean values of arbitrary fluorescence unit of 10,000 cells were obtained and expressed as percentages of control.
RT-PCR
Total RNA was isolated using TRIzol (Invitrogen), and cDNA was prepared using avian myeloblastosis virus reverse transcriptase (Promega). PCR was performed simultaneously using two primer sets: a set for target gene and a set for β-actin gene as an internal control and in an optimized conventional method. The PCR primer sets were produced by Bioneer to detect COX2 (5′-TGCCCTTTTCCTAACACTCAC and 5′-GGTTTGCTCCACAGATTTCAG), ND2 (5′-AGGTTACCCAAGGCACCCCT and 5′-AGTAGATTAGGCGTAGGTAG), mtTFA (5′-AAAAAGGAAAGCTATGAC and 5′-AGCACCATATTTTCGTTG), NRF1 (5′-GCAAACGCAAACACAGGCC and 5′-CTGCATCTCCCTGAGAAGC), and β-actin (5′-CCTTCCTGGGCATGGAGTCCTGT and 5′-GGAGCAATGATCTTGATCTTC).
Quantitative real-time RT-PCR (qPCR) was performed using THUNDERBIRDTM SYBRTM qPCR Mix (Toyobo, Osaka, Japan) following the manufacturer's protocol. PCR was performed with 45 cycles of the reaction at 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 20 s. The PCR primer sets were produced by Bioneer for CLN1 (5′-TGCCCCAGTGGAGGATTTAC and 5′-AGGATGCCAACCACCATCAA) and β-actin (5′-CCTTCCTGGGCATGGAGTCCTGT and 5′-GGAGCAATGATCTTGATCTTC). Expression levels of target mRNAs were normalized by β-actin mRNA level.
Western blot analysis
Western blotting was performed using standard procedures. Antibody for mtTFA was kindly provided by Youngmi Kim Pak from Kyung Hee University (Seoul, South Korea). Antibodies against MRPL13 (ab103801), MRPS15 (ab137070), NRF1 (ab55744), complex III (UQCRC2, ab14745), and complex IV (COX2, ab110258) were purchased from Abcam (Cambridge, MA). PMPCA antibody was obtained from Sigma (HPA021648). Antibodies against β-actin (sc-1616) and PMPCB (sc-160672) were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). Antibodies against α-tubulin (05–829), LONP (H00009361), and Complex II (FP, MS204) were obtained from Millipore (Billerica, MA), Abnova (Taipei City, Taiwan), and Mitoscience (Eugene, OR), respectively. Antibodies for MRPL39 (GTX87886) and MRPL29 (GTX113864) were obtained from Genetex Inc. (Irvine, CA). Claudin-1 antibody (717800) was purchased from Invitrogen. Antibodies against NDUFA9 of complex I (A21344), COX1 of complex IV (A6403), and ATP5A1 of complex V (A21350) were purchased from Molecular Probes.
Measurement of extracellular lactate concentration
Concentration of extracellular lactate was measured using a YSI 7100 bioanalyzer (Yellow Springs, OH). Briefly, cell was seeded in 12-well plates and cultured for 24 h. After replenishing with the fresh medium, the conditioned medium was harvested 2 days later, and the cell numbers were counted to normalize the values.
Validation in the open liver cancer cohort
To validate the clinical significance of MRPL13, we used liver cancer open database, TCGA-LIHC. Level 3 RNA-SeqV2 data for LIHC were downloaded from TCGA data portal (https://portal.gdc.cancer.gov/). RNASeqV2 expression data (n = 421) including 371 primary tumors and 50 normal livers from TCGA-LIHC cohort were used in analysis. All expression data were log2 transformed and quantile normalized using R. LIHC patients (n = 371) were divided into high and low groups based on the median value of MRPL13 expression level to perform the Kaplan–Meier survival analysis. Statistical significance for Kaplan–Meier survival was estimated by Cox–Mantel log-rank test. To elucidate the association between two genes, correlation estimation and p value calculated from Pearson's correlation test were used. The ratio of LDHB to LDHA was calculated individually.
To evaluate the relevance of EMT related genes in MRPL13 expression level, ssGSEA was performed using the EMT core gene signature (EMT_UP; n = 91), which were previously identified by Taube et al. (28). p value from the Kolmogorov–Smirnov test was transformed in log scale and used to compare enrichment score between MRPL13-high and MRPL13-low groups. All statistics were performed using R (R version 3.3.2).
Author contributions
Y.-K. L., J. J. L., U.-W. J., S. M., and E.-B. L. performed experiments. S. M. K. performed bioinformatics analysis. C. L. analyzed and interpreted data. G. Y. designed and supervised the project and prepared the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Note added in proof
There were two errors in the version of this paper that was published as a Paper in Press on October 4, 2017. The wrong MRPL13 immunoblot was used in Fig. 3A, and the wrong cell image was used in the siMRPL13 knockdown shown in Fig. 3C. These errors have now been corrected and do not affect the results or conclusions of this work.
This work was supported by National Research Foundation of Korea Grant 2015R1A2A1A10055038 from the Korean Ministry of Science, ICT and Future Planning. The authors declare that they have no conflicts of interest with the contents of this article.
- mtDNA
- mitochondrial DNA
- OCR
- oxygen consumption rate
- TCGA
- The Cancer Genome Atlas
- LIHC
- liver hepatocellular carcinoma
- LDH
- lactate dehydrogenase
- OXPHOS
- oxidative phosphorylation
- rRNA
- ribosomal RNA
- tRNA
- transfer RNA
- MRP
- mitoribosomal protein
- CLN1
- claudin-1
- Ch-L
- Chang-liver
- mtTFA
- mitochondrial transcription factor A
- NRF1
- nuclear respiratory factor 1
- DOX
- doxycycline
- CAP
- chloramphenicol
- ROS
- reactive oxygen species
- PT
- primary tumor
- NL
- normal liver tissue
- EMT
- epithelial-mesenchymal transition
- ssGSEA
- single sample gene set enrichment analysis
- qPCR
- quantitative real-time RT-PCR
- VDAC
- voltage-dependent anion-selective channel.
References
- 1. Gustafsson C. M., Falkenberg M., and Larsson N. G. (2016) Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 85, 133–160 [DOI] [PubMed] [Google Scholar]
- 2. Falkenberg M., Larsson N. G., and Gustafsson C. M. (2007) DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 76, 679–699 [DOI] [PubMed] [Google Scholar]
- 3. Lee C., Kim K. H., and Cohen P. (2016) MOTS-c: A novel mitochondrial-derived peptide regulating muscle and fat metabolism. Free Radic. Biol. Med. 100, 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Attardi G., and Schatz G. (1988) Biogenesis of mitochondria. Annu. Rev. Cell Biol. 4, 289–333 [DOI] [PubMed] [Google Scholar]
- 5. Gatenby R. A., and Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 6. He X., Zhou A., Lu H., Chen Y., Huang G., Yue X., Zhao P., and Wu Y. (2013) Suppression of mitochondrial complex I influences cell metastatic properties. PLoS One 8, e61677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandra D., and Singh K. K. (2011) Genetic insights into OXPHOS defect and its role in cancer. Biochim. Biophys. Acta 1807, 620–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hung W. Y., Huang K. H., Wu C. W., Chi C. W., Kao H. L., Li A. F., Yin P. H., and Lee H. C. (2012) Mitochondrial dysfunction promotes cell migration via reactive oxygen species-enhanced β5-integrin expression in human gastric cancer SC-M1 cells. Biochim. Biophys. Acta 1820, 1102–1110 [DOI] [PubMed] [Google Scholar]
- 9. Ma J., Zhang Q., Chen S., Fang B., Yang Q., Chen C., Miele L., Sarkar F. H., Xia J., and Wang Z. (2013) Mitochondrial dysfunction promotes breast cancer cell migration and invasion through HIF1α accumulation via increased production of reactive oxygen species. PLoS One 8, e69485. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Lee Y. K., Jee B. A., Kwon S. M., Yoon Y. S., Xu W. G., Wang H. J., Wang X. W., Thorgeirsson S. S., Lee J. S., Woo H. G., and Yoon G. (2015) Identification of a mitochondrial defect gene signature reveals NUPR1 as a key regulator of liver cancer progression. Hepatology 62, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee Y. K., Youn H. G., Wang H. J., and Yoon G. (2013) Decreased mitochondrial OGG1 expression is linked to mitochondrial defects and delayed hepatoma cell growth. Mol. Cells 35, 489–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J. H., Kim H. Y., Lee Y. K., Yoon Y. S., Xu W. G., Yoon J. K., Choi S. E., Ko Y. G., Kim M. J., Lee S. J., Wang H. J., and Yoon G. (2011) Involvement of mitophagy in oncogenic K-Ras-induced transformation: overcoming a cellular energy deficit from glucose deficiency. Autophagy 7, 1187–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chatterjee A., Mambo E., and Sidransky D. (2006) Mitochondrial DNA mutations in human cancer. Oncogene 25, 4663–4674 [DOI] [PubMed] [Google Scholar]
- 14. Nishikawa M., Nishiguchi S., Shiomi S., Tamori A., Koh N., Takeda T., Kubo S., Hirohashi K., Kinoshita H., Sato E., and Inoue M. (2001) Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res. 61, 1843–1845 [PubMed] [Google Scholar]
- 15. Petros J. A., Baumann A. K., Ruiz-Pesini E., Amin M. B., Sun C. Q., Hall J., Lim S., Issa M. M., Flanders W. D., Hosseini S. H., Marshall F. F., and Wallace D. C. (2005) mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 102, 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sotgia F., Whitaker-Menezes D., Martinez-Outschoorn U. E., Salem A. F., Tsirigos A., Lamb R., Sneddon S., Hulit J., Howell A., and Lisanti M. P. (2012) Mitochondria “fuel” breast cancer metabolism: fifteen markers of mitochondrial biogenesis label epithelial cancer cells, but are excluded from adjacent stromal cells. Cell Cycle 11, 4390–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pietromonaco S. F., Denslow N. D., and O'Brien T. W. (1991) Proteins of mammalian mitochondrial ribosomes. Biochimie 73, 827–835 [DOI] [PubMed] [Google Scholar]
- 18. Greber B. J., and Ban N. (2016) Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem. 85, 103–132 [DOI] [PubMed] [Google Scholar]
- 19. Gopisetty G., and Thangarajan R. (2016) Mammalian mitochondrial ribosomal small subunit (MRPS) genes: a putative role in human disease. Gene 589, 27–35 [DOI] [PubMed] [Google Scholar]
- 20. Wazir U., Jiang W. G., Sharma A. K., and Mokbel K. (2012) The mRNA expression of DAP3 in human breast cancer: correlation with clinicopathological parameters. Anticancer Res. 32, 671–674 [PubMed] [Google Scholar]
- 21. Gatza M. L., Silva G. O., Parker J. S., Fan C., and Perou C. M. (2014) An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat. Genet. 46, 1051–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng S., Lü B., Ruan W., Zhu Y., Sheng H., and Lai M. (2011) Genetic polymorphisms and breast cancer risk: evidence from meta-analyses, pooled analyses, and genome-wide association studies. Breast Cancer Res. Treat. 127, 309–324 [DOI] [PubMed] [Google Scholar]
- 23. Kim J. H., Kim E. L., Lee Y. K., Park C. B., Kim B. W., Wang H. J., Yoon C. H., Lee S. J., and Yoon G. (2011) Decreased lactate dehydrogenase B expression enhances claudin 1-mediated hepatoma cell invasiveness via mitochondrial defects. Exp.. Cell Res. 317, 1108–1118 [DOI] [PubMed] [Google Scholar]
- 24. Grohmann L., Kitakawa M., Isono K., Goldschmidt-Reisin S., and Graack H. R. (1994) The yeast nuclear gene MRP-L13 codes for a protein of the large subunit of the mitochondrial ribosome. Curr. Genet. 26, 8–14 [DOI] [PubMed] [Google Scholar]
- 25. Rattigan Y. I., Patel B. B., Ackerstaff E., Sukenick G., Koutcher J. A., Glod J. W., and Banerjee D. (2012) Lactate is a mediator of metabolic cooperation between stromal carcinoma associated fibroblasts and glycolytic tumor cells in the tumor microenvironment. Exp. Cell Res. 318, 326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rudrabhatla S. R., Mahaffey C. L., and Mummert M. E. (2006) Tumor microenvironment modulates hyaluronan expression: the lactate effect. J. Invest. Dermatol. 126, 1378–1387 [DOI] [PubMed] [Google Scholar]
- 27. Tang X., Lucas J. E., Chen J. L., LaMonte G., Wu J., Wang M. C., Koumenis C., and Chi J. T. (2012) Functional interaction between responses to lactic acidosis and hypoxia regulates genomic transcriptional outputs. Cancer Res. 72, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taube J. H., Herschkowitz J. I., Komurov K., Zhou A. Y., Gupta S., Yang J., Hartwell K., Onder T. T., Gupta P. B., Evans K. W., Hollier B. G., Ram P. T., Lander E. S., Rosen J. M., Weinberg R. A., et al. (2010) Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U.S.A. 107, 15449–15454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Greber B. J., Boehringer D., Leibundgut M., Bieri P., Leitner A., Schmitz N., Aebersold R., and Ban N. (2014) The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515, 283–286 [DOI] [PubMed] [Google Scholar]
- 30. Greber B. J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., and Ban N. (2015) Ribosome: the complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 [DOI] [PubMed] [Google Scholar]
- 31. D'Andrea A., Gritti I., Nicoli P., Giorgio M., Doni M., Conti A., Bianchi V., Casoli L., Sabò A., Mironov A., Beznoussenko G. V., and Amati B. (2016) The mitochondrial translation machinery as a therapeutic target in Myc-driven lymphomas. Oncotarget 7, 72415–72430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J., Seol M. Y., Jeong S., Lee C. R., Ku C. R., Kang S. W., Jeong J. J., Shin D. Y., Nam K. H., Lee E. J., Chung W. Y., and Jo Y. S. (2015) A metabolic phenotype based on mitochondrial ribosomal protein expression as a predictor of lymph node metastasis in papillary thyroid carcinoma. Medicine 94, e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Modica-Napolitano J. S., and Singh K. K. (2004) Mitochondrial dysfunction in cancer. Mitochondrion 4, 755–762 [DOI] [PubMed] [Google Scholar]
- 34. Singh A. K., Pandey P., Tewari M., Pandey H. P., and Shukla H. S. (2014) Human mitochondrial genome flaws and risk of cancer. Mitochondrial DNA 25, 329–334 [DOI] [PubMed] [Google Scholar]
- 35. Baracca A., Chiaradonna F., Sgarbi G., Solaini G., Alberghina L., and Lenaz G. (2010) Mitochondrial complex I decrease is responsible for bioenergetic dysfunction in K-ras transformed cells. Biochim. Biophys. Acta 1797, 314–323 [DOI] [PubMed] [Google Scholar]
- 36. Dang C. V., Kim J. W., Gao P., and Yustein J. (2008) The interplay between MYC and HIF in cancer. Nat. Rev. Cancer 8, 51–56 [DOI] [PubMed] [Google Scholar]
- 37. Drent M., Cobben N. A., Henderson R. F., Wouters E. F., and van Dieijen-Visser M. (1996) Usefulness of lactate dehydrogenase and its isoenzymes as indicators of lung damage or inflammation. Eur. Respir. J. 9, 1736–1742 [DOI] [PubMed] [Google Scholar]
- 38. Koike K., Urata Y., and Koike M. (1990) Molecular cloning and characterization of human pyruvate dehydrogenase β subunit gene. Proc. Natl. Acad. Sci. U.S.A. 87, 5594–5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoon Y. S., Yoon D. S., Lim I. K., Yoon S. H., Chung H. Y., Rojo M., Malka F., Jou M. J., Martinou J. C., and Yoon G. (2006) Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J. Cell. Physiol. 209, 468–480 [DOI] [PubMed] [Google Scholar]
- 40. Byun H. O., Jung H. J., Seo Y. H., Lee Y. K., Hwang S. C., Hwang E. S., and Yoon G. (2012) GSK3 inactivation is involved in mitochondrial complex IV defect in transforming growth factor (TGF) β1-induced senescence. Exp. Cell Res. 318, 1808–1819 [DOI] [PubMed] [Google Scholar]
- 41. Li X., Yang H., Huang H., and Zhu T. (2014) CELLCOUNTER: novel open-source software for counting cell migration and invasion in vitro. BioMed Res. Int. 2014, 863564. [DOI] [PMC free article] [PubMed] [Google Scholar]