Abstract
Population diversity was examined in individual and natural mixed infections of Cowpea chlorotic mottle virus (CCMV) and Cucumber mosaic virus (CMV) isolates in two systemic hosts, cowpea and Nicotiana benthamiana. Isolates of CCMV and CMV obtained from a cowpea field in Arkansas were separated biologically in cowpea and tobacco plants, respectively. After separation, individual and mixed cultures of both viruses were serially passaged ten times by mechanical inoculation in cowpea and N. benthamiana. High-fidelity reverse transcriptase-polymerase chain reaction (HiFi RT-PCR) of RNA 3, followed by cDNA cloning and sequence analysis was used to assess the quasispecies cloud size of CCMV and CMV populations in passages zero and ten of each host species. The levels of population variation were generally higher in individual infections of CCMV-Car1 and-Car2 isolates, and the CMV-Car2 isolate compared with mixed infections, in both host species, although the significance of the differences varied depending on how mutations were counted. There were no significant differences in the levels of population variation in individual and mixed infections of the CMV-Car1 isolate. Partially fixed mutations were observed in both individual and mixed infections of the CCMV-Car2 isolate in N. benthamiana and CMV-Car1 and-Car2 isolates in both cowpea and N. benthamiana.
Keywords: virus populations, plant viruses, RNA viruses
1. Introduction
The study of the epidemiology and evolution of plant virus populations is important for understanding the emergence of new diseases and may also help us to develop strategies for the control of viral diseases. The existence of viral strains in the field that cause different kinds of diseases is an interesting and important aspect of plant virology. A single virus isolate does not contain a single uniform population, but has a genetically diverse population referred to as quasispecies (reviewed in Domingo et al. 2012). The concept of quasispecies is fundamental to the understanding of virus variation and evolution. In previous experimental evolution studies, the initial infection produced from cDNA clones for various viruses in a controlled environment showed that the level of genetic diversity in individual quasispecies varied among related viruses [Cowpea chlorotic mottle virus (CCMV), Cucumber mosaic virus (CMV), and Tobacco mosaic virus (TMV)] (Schneider and Roossinck 2000) and among hosts infected with the same virus (Schneider and Roossinck 2001). Variation also was seen comparing strains of CMV (Pita and Roossinck 2013). However, no studies have been done on viruses in mixed infections, a condition that is common in field isolates of plant viruses.
We obtained field samples of cowpea plants that contained natural mixed infections of two viruses: CCMV and CMV. Here, we report the quasispecies variation of CCMV and CMV field isolates from mixed infections. We separated the two viruses from each other biologically, passaged them ten times as individual and mixed infections in two host species, and determined the quasispecies cloud size of these viruses in passage zero and ten of cowpea and Nicotiana benthamiana.
2. Materials and methods
2.1 Plants and culture of viruses
Young seedlings of cowpea (Vigna unguiculata cv Red Chinese) and N. benthamiana were used in this study. Seeds of cowpea were provided by George Bruening, University of California. All plants were grown in a greenhouse at 25 °C and a 16-h photoperiod with supplementary light. Two field samples of cowpea plants that contained natural mixed infections of CCMV and CMV were provided by Rose Gergerich, University of Arkansas. These two mixed samples were designated Car1 (Cowpea Arkansas 1) and Car2 (Cowpea Arkansas 2). Mixed infections were designated as Car1-mix and Car2-mix cultures, respectively. CCMV and CMV were successfully separated from these samples as described previously (Ali and Roossinck 2008) and individual cultures were designated as CCMV-Car1, CCMV-Car2, CMV-Car1, and CMV-Car2 isolates.
2.2 Plant inoculations and serial passaging
A total of six virus cultures that included two mixed cultures of CCMV and CMV (Car1-mix and Car2-mix), two individual cultures of CCMV (CCMV-Car1 and CCMV-Car2), and two individual cultures of CMV (CMV-Car1 and CMV-Car2) were used in serial passage experiments.
For mixed cultures, sap was extracted directly in inoculation buffer (50 mM NaH2PO4, pH 7.0) from original leaves infected with Car1-mix and Car2-mix cultures and inoculated to young cowpea and N. benthamiana seedlings to constitute passage zero. Individual cultures of CCMV-Car1 and CCMV-Car2 isolates were separated on cowpea and total nucleic acids from that passage constituted passage zero of CCMV isolates. CMV was separated from CCMV in tobacco and total nucleic acid obtained from tobacco was inoculated to cowpea to constitute passage zero of CMV isolates.
Plants infected with the above six virus cultures were used as a source for the subsequent passages (one through ten) at 14-day intervals, using sap extracted from the serially-passaged systemic leaves of each infected plant. Two plants per passage of both species for each treatment were inoculated for the serial passaged experiments. Two systemically infected leaves from each plant were combined and ground in inoculation buffer (50 mM Na2HPO4, pH 7.0), and the combined sap was used to inoculate plants in the next passage.
2.3 Extraction of total RNA and HiFi RT-PCR
High-fidelity reverse transcriptase-polymerase chain reaction (HiFi RT-PCR) of RNA 3 (including the 3' portion of the movement protein gene, the coat protein gene, and a portion of the 3' untranslated region) (Fig. 1) followed by cDNA cloning and sequence analysis were used to assess the quasispecies cloud size of CCMV and CMV populations in passage zero and passage ten. Fourteen days post-inoculation total RNA from 100 mg of systemically infected leaves of each host species at passages zero and ten was isolated by using Tri-Reagent, according to the manufacturer’s protocol (Molecular Research Centre, Inc). One fifth of the total RNA obtained was used as template for HiFi RT-PCR using Superscript reverse transcriptase as prescribed by the manufacturer (Invitrogen). The reverse transcription reaction was primed with the RNA 3 primers (covering the coat protein gene and flanking regions) of both CCMV and CMV as described previously (Schneider and Roossinck 2001). PCR was carried out in capillary tubes using an Idaho Technology Rapid Cycler for a maximum of 10 cycles (94 °C denaturation for 6 s, 50 °C annealing for 6 s, and 72 °C extension for 20 s) and included a DNA polymerase with proofreading capability (Pfu; Stratagene) that was mixed at a ratio of 1:8 with Taq DNA polymerase (Roche).
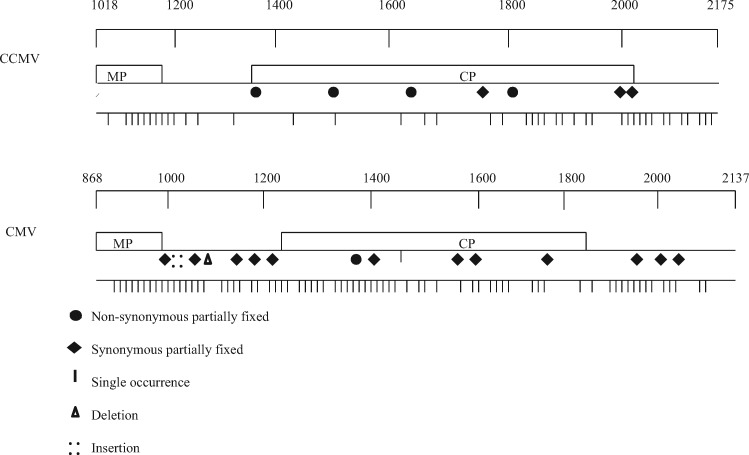
Figure 1.
Distribution of mutations in analyzed regions of CCMV and CMV. Analyzed regions of RNAs 3, including the 3' portion of the movement protein (MP), complete coat protein gene (CP) and a portion of the 3'-untranslated region of CCMV and CMV are shown, with nucleotides positions marked above. The locations of single mutations (|), partially fixed non-synonymous (•) and synonymous mutations (♦), point deletions (Δ) and insertions (::) are shown below the maps.
2.4 Cloning, sequencing, and analysis of viral clones
The HiFi PCR products were purified using a PCR purification kit (Qiagen) and cloned into pGEM-T easy vector (Promega, USA). The sequence of the inserts in the recombinant plasmid was determined by a DNA analyzer ABI 3730. Sequencing was performed with the flanking plasmid primers. From ten to twenty-nine clones were sequenced for each population (a minimum of 11,000 bases/population). The consensus sequence for each population was determined, the sequence of each clone was compared to the consensus sequence and changes were recorded. The mutation frequency (total number of changes observed in all clones for a given viral population/total number of bases sequenced for the viral population) and the percentage of mutated clones were used as indicators of genetic diversity. Partially fixed mutations observed on a number of clones on the same positions were considered either as a single mutation event (Method 1) or as individual mutation events (Method 2) as described previously (Pita and Roossinck 2013). Differences in mutation frequencies between viral populations from single versus mixed infections were tested for statistical significance using one-way analysis of variance (ANOVA) using StatPlus Mac LE.
3. Results
3.1 Mutation frequency in control reactions
The level of error created by the experimental procedure was measured using control reactions with plasmid clones of the Fny-CMV strain as a template for in vitro transcripts to estimate the level of variability introduced by transcription, reverse transcription, and thermal cycling. One of the eighteen control clones (22,860 nucleotides) derived from in vitro transcripts contained a single mutation, which established the background level of the experimental procedure at a mutation frequency at 4.3 × 10−5 mutations per nucleotide. Mutation frequency from these control experiments indicated that the level of sequence variability introduced by the experimental method was significantly lower than what was observed in the viral populations.
3.2 Analysis of CCMV populations
In cowpea, CCMV-Car1 produced severe mosaic symptoms and CCMV-Car2 produced mild symptoms; however, in N. benthamiana, the infection was symptomless. Car1-mix and Car2-mix cultures that contained both CCMV and CMV isolates together produced severe and mild symptoms respectively in cowpea while both cultures produced severe symptoms in N. benthamiana. Symptoms did not change in any case over the ten passages.
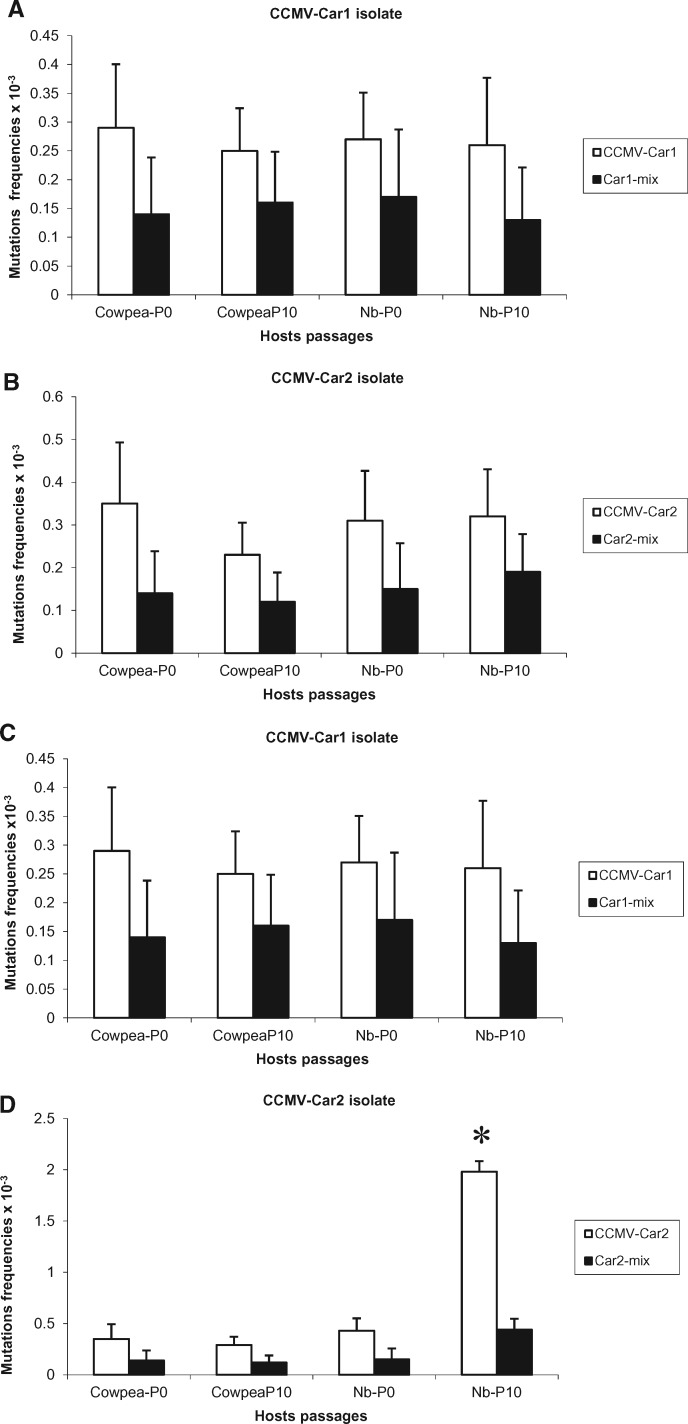
Sequence analysis of the RNAs 3 of CCMV-Car1 and CCMV-Car2 isolates showed that they have 95.2 per cent and 94.4 per cent nucleotide sequence identity respectively, with the CCMV-type strain. The size of CCMV amplified product used for population analysis was 1,157 nucleotides (Fig. 1). However, a stretch of seventeen nucleotides containing only poly As in CCMV sequence was excluded for the analysis of quasispecies variation as the number of As is highly variable. Thus, the total length of the cloned PCR product considered for quasispecies variation was 1,140 nucleotides. Ten to twenty-nine clones (1,140 nucleotides/clone) of CCMV isolates were sequenced. Virus populations from passages zero and ten were analyzed for all six individual and mixed infections. The level of population variation remained consistent for CCMV-Car1 and CCMV-Car2 isolates in passages of both cowpea and N. benthamiana and did not fluctuate significantly. Similarly, in mixed infection, the population variation between the hosts and passages did not significantly rise or fall for CCMV isolates. The overall trend of population variation in CCMV was higher in individual infections compared to mixtures, but most of these were not statistically significant at P < 0.05 when the partially fixed mutations were calculated as single events. However, when the partially fixed mutations were counted as individual events the level of population variation was significantly higher in individual infections of CCMV-Car2 isolates compared to mixed infections in passages zero and ten for both host species (Fig. 2). The majority of mutations in the populations of CCMV isolates occurred in the movement protein gene, and 3' untranslated regions while very few occurred in the coat protein gene (Fig. 1). Interestingly, all of the fixed mutations in CCMV were found in the CP gene, and several of these were nonsynonomous, indicating that the CP gene is the analyzed portion of the genome that is evolving.
Figure 2.
Quasispecies variation in CCMV population in different hosts. Mutation frequency of individual CCMV-Car1 isolate and Car1-mix culture (A, C) and CCMV-Car2 isolate and Car2-mix culture (B, D) in passages zero and ten of cowpea and Nicotiana benthamiana (Nb) plants. Frequencies were calculated by counting partially fixed mutations as single events (A, B) or as multiple independent events (C, D). Levels of variation were compared for single and mixed infections, and those with significantly higher levels are marked with an asterisk. Error bars correspond to standard errors.
3.3 Analysis of CMV populations
CMV-Car1 and CMV-Car2 isolates produced mild symptoms in both cowpea and N. benthamiana seedlings, and symptoms did not change over passages. Sequence analysis of RNAs 3 of CMV-Car1 and CMV-Car2 isolates showed that they have 88.6 per cent and 88.0 per cent nucleotide sequence identity with the CMV-Fny strain used in previous experimental evolution studies (Schneider and Roossinck 2000; Pita and Roossinck 2013).
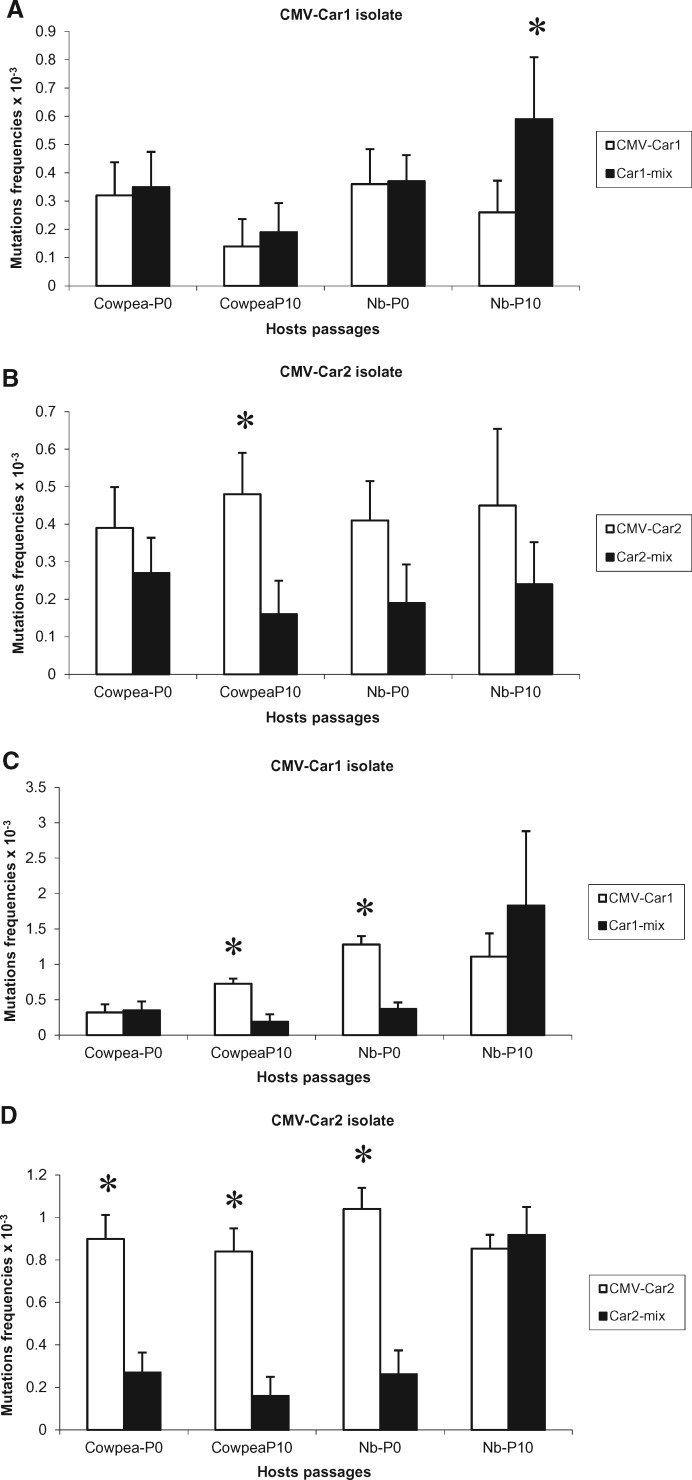
The size of the CMV amplified product was 1,270 nucleotide (Fig. 1) covering part of the movement protein gene, the intergenic region, the complete coat protein gene and the 3' untranslated region (UTR). All 1,270 nucleotides of the cloned PCR product were used for the analysis of quasispecies variation. From eleven to nineteen clones of CMV were sequenced for each population. Levels of population variation of CMV-Car1 were generally higher in individual infections versus mixed infections in both host species except in passage ten in N. benthamiana where the mutation frequency of CMV-Car1 isolate was significantly higher in mixed infection than individual infection (Fig. 3). In the case of the CMV-Car2 isolate, the trend was similar to that of CCMV-Car1 and -Car2 isolates. The population variation of CMV-Car2 was higher in individual infections than in mixed infections with CMV in both host species; the significance was greater when partially fixed mutations were considered as independent events. Most of the mutations in CMV populations occurred throughout the MP, CP and 3' UTR regions, but very few were found in the 3' end of CP gene.
Figure 3.
Quasispecies variation in CMV population in different hosts. Mutation frequency of individual CMV-Car1 isolate and Car1-mix culture (A, C) and CMV-Car2 isolate and Car2-mix culture (B, D) in passages zero and ten of cowpea and Nicotiana benthamiana (Nb) plants. Frequencies were calculated by counting partially fixed mutations as single events (A, B) or as multiple independent events (C, D). Levels of variation were compared for single and mixed infections, and those with significantly higher levels are marked with an asterisk. Error bars correspond to standard errors.
3.4 Distribution of partially fixed mutations
Partially fixed mutations (i.e. those that appeared in the same site in more than one individual clone) were observed in some populations of both individual and mixed infections of CCMV and CMV. Out of 120 clones (136,800 nucleotides) sequenced for CCMV-Car1 in both hosts species, no partially fixed mutations were observed in cowpea or N. benthamiana (Table 1). However, in CCMV-Car2, seven partially fixed mutations were observed out of 128 clones (145,920 nucleotides) sequenced (Table 1). All seven partially fixed mutations were in N. benthamiana populations, except one that was observed in passage ten of cowpea. All partially fixed mutations were observed in the CP gene (Fig. 1). Out of seven partially fixed mutations four were non-synonymous and three were synonymous substitutions. There were no deletions or insertions detected in CCMV populations. Partially fixed mutations detected in passage zero of N. benthamiana were never detected in passage ten for CCMV-Car2.
Table 1.
Partially fixed mutations observed in CCMV populations.
| Virus | Host | Passage 0 | Passage 10 |
|---|---|---|---|
| CCMV-Car1 | Cowpea | ||
| Car1-mix | Cowpea | ||
| CCMV-Car1 | Nbc | ||
| Car1-mix | Nb | ||
| CCMV-Car2 | Cowpea | A1530a → G (24: 3)b | |
| Car2-mix | Cowpea | ||
| CCMV-Car2 | Nb | T1388→G (11: 3) | C1718 → T (3: 12) |
| C1744 → T (12: 3) | |||
| T1895 → C (5: 10) | |||
| A1905 → G (10: 5) | |||
| Car2 mix | Nb | G1632 → T (12: 6) |
Position of nucleotide in RNA 3.
Number of clones that have particular mutation.
Nicotiana benthamiana.
In the case of CMV, a total of twenty-two partially fixed mutations were observed in both host species for CMV-Car1 and CMV-Car2 isolates. Four of these mutations were in cowpea while the remaining eighteen were observed in N. benthamiana. (Table 1) Most of these mutation (68%) occurred in individual infections of CMV, compared to 32 per cent in the mixed infections (Table 2). In addition, 60 per cent of the mutations for CMV were observed in passage ten. The partially fixed mutations detected in CMV isolates were observed in the MP, CP and 3' UTR regions. The majority of partially fixed mutations in CMV isolates were synonymous substitutions. However, one mutation was an insertion in passage ten of N. benthamiana for CMV-Car1 mixed infection while three mutations were deletions in passage zero of cowpea and N. benthamiana for CMV-Car2 in both individual and mixed infections. These were all found in the intergenic region of the virus, and hence did not effect aa sequences. Out of twenty-two partially fixed mutations, nine were detected in passage zero of both host species populations for CMV-Car1 and CMV-Car2 isolates, and most were maintained in passage ten. (Table 2). Since it is not possible to determine if partially fixed mutations are the result of a single event or multiple events in a mutational hot spot, mutations frequencies were calculated in two ways, as shown in Figs 2 and 3, parts A and B, versus C and D.
Table 2.
Partially fixed mutations observed in CMV populations.
| Virus | Host | Passage 0 | Passage 10 |
| CMV-Car1 | Cowpea | A1163a→ G (9: 2)b | |
| C1404 → T (4: 7) | |||
| Car1-mix | Cowpea | ||
| CMV-Car1 | Nbc | A1163 → G (8: 3) | A1163 → G (10: 2) |
| G1173 → A, T (0: 10: 1) | G1173 → A, T (0: 2: 10) | ||
| Car1-mix | Nb | T1035 → :: d (6: 6) | |
| A1163 → G (6: 6) | |||
| G1777 → T (7: 5) | |||
| C2083 → T (9: 3) | |||
| CMV-Car2 | Cowpea | Δe1149 → C (4: 10) | A1490 → G (6: 7) |
| Car2-mix | Cowpea | ||
| CMV-Car2 | Nb | Δ1149 → C (10: 5) | T1607 → C (7: 5) |
| A1163 → G (13: 2) | G2000 → A (10: 2) | ||
| T1190 → C (13: 2) | |||
| A1637 → G (13: 2) | |||
| A2014 → G (13: 2) | |||
| Car2-mix | Nb | Δ1149 → C 10: 2 | T990 → C (3: 9) |
| A1163 → G (10: 2) |
Position of nucleotide in RNA 3.
Number of clones that have particular mutation.
Nicotiana benthamiana.
Insertion.
Deletion.
4. Discussion
In this study, we present quasispecies variation data obtained from individual and mixed infection of field isolates of CCMV and CMV that were used in a controlled environment by passaging them in cowpea and N. benthamiana. According to experimental evolution studies conducted previously (Schneider and Roossinck 2000), the mean mutation frequencies of the CMV-Fny strain and CCMV-type strains in N. benthamiana were 0.66 × 10−3 and zero, respectively. In this study, the mean mutation frequencies of CMV-Car1 and CMV-Car2 isolates in N. benthamiana were 0.31 × 10−3 and 0.43 × 10−3, respectively (Fig. 2) and that of CCMV-Car1 and CCMV-Car2 isolates were 0.26 × 10−3 and 0.30 × 10−3, respectively (Fig. 3). Hence, these field isolates of CMV have slightly lower mutation frequencies compared to the CMV-Fny strain, while the CCMV isolates have higher mutation frequencies than CCMV-type strain (Schneider and Roossinck 2000, 2001), indicating that different strains of the same virus can have different levels of population variation. This was shown recently for two strains of CMV from different subgroups, and variation control was mapped to RNAs 1 and 2 (Pita and Roossinck 2013). However, the source of infection in the experimental evolution studies was a uniform population of CMV obtained from RNA transcripts generated in vitro from cDNA clones, while in the case of this study the starting populations of the field isolates of CCMV and CMV were not uniform. We found evidence of evolution in the genomes during passages, with a number of partially fixed mutations, including nonsynonymous changes. However, no changes in pathology occurred over passages, and hence these changes may be affecting other aspects of the virus.
In most cases, the levels of variation did not change over passage in either host, although the level of CCMV-Car2 variation was higher in passage 10 of N. benthamiana when partially fixed mutations were considered as individual events. Our results are similar to that observed for field populations of other viruses (Rodríguez-Cerezo and García-Arenal 1989, Rodríguez-Cerezo et al. 1989; Moya et al. 1993; Fraile et al. 1996) supporting the view of a significant genetic stability for field populations of RNA plant viruses, that do not appear to change much over time. Interestingly, the level of population variation was generally higher in individual infections of both CCMV and CMV isolates, although the significance varied depending on the method of calculation. These results were unexpected as one might predict that during mixed infections, the two viruses will be competing for resources and may generate more mutations due to a lower pool of nucleotides in the host cells. We do not know if these viruses actually infect the same cells, but in plants there is a great deal of resource flow between adjoining cells, with thousands of plasmadesmatal connections between mesophyll cells where most virus replication occurs. However, in the case of CMV-Car1 the mutation frequency in individual infections was lower than in mixed infections implying that there was some competition between CMV-Car1 and CCMV-Car1 isolates. This illustrates the complicated relationships that may occur during mixed viral infections that may influence the evolutionary trajectory of viruses.
Host–virus interactions can have dramatic effects on viral populations as shown previously for CMV-Fny strain (Schneider and Roossinck 2001), and TMV populations (Kearney et al. 1999). The fidelity of RNA virus replication also can be affected by different host environments (Pita et al. 2007). However, we did not see that trend in our current study using these field isolates of CCMV and CMV. In most cases, mutation frequencies of CCMV-Car1 and CCMV-Car2 isolates as well as CMV-Car1 and CMV-Car2 isolates did not change significantly during passages in either cowpea or N. benthamiana, with the exception of Passage 10 of CCMV-Car2 in N. benthamiana noted above.
In summary, we report here that quasispecies variation in field isolates of CCMV and CMV can be stable and did not seem to be particularly affected by the host. However, this might be different for different viruses and virus strains in other hosts. Most significantly, with one exception population variations were generally lower in mixed viral infection compared to individual viral infection. This suggests that mixed infections of viruses may significantly alter the rate of viral evolution by increasing or decreasing high levels of diversity of viral populations, and emphasizes the point that general conclusions about RNA virus populations dynamics cannot be drawn based on specific interactions, each virus-host combination will have its own population dynamics and evolutionary trajectory.
Acknowledgements
We thank George Bruening, University of California for providing cowpea seed, Rose Gergerich, University of Arkansas for providing field samples of CCMV and CMV, and Luciano Nunes, Universidade Federal de Uberlândia for help with statistical analyses. This work was supported by the Samuel Roberts Noble Foundation and by Pennsylvania State University.
Conflict of interest: None declared.
References
- Ali A., Roossinck M. J. (2008) ‘A simple technique for separation of Cowpea chlorotic mottle virus from Cucumber mosaic virus in natural mixed infections’, Journal of Virological Methods, 153/2: 163–7. [DOI] [PubMed] [Google Scholar]
- Domingo E. et al. (2012) ‘Viral quasispecies evolution’, Microbiology and Molecular Biology Reviews, 76/2: 159–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile A. et al. (1996) ‘Genetic diversity in tobacco mild green mosaic tobamovirus infecting the wild plant Nicotiana glauca’, Virology, 223/1: 148–55. [DOI] [PubMed] [Google Scholar]
- Kearney C. M. et al. (1999) ‘Genome evolution of tobacco mosaic virus populations during long-term passaging in a diverse range of hosts’, Archives of Virology, 144/8: 1513–26. [DOI] [PubMed] [Google Scholar]
- Moya A. et al. (1993) ‘Genetic structure of natural populations of the plant RNA virus tobacco mild green mosaic virus’, Molecular Biology and Evolution, 10/2: 449–56. [Google Scholar]
- Pita J. S. et al. (2007) ‘Environment determines fidelity for an RNA virus replicase’, Journal of Virology, 81/17: 9072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita J. S., Roossinck M. J. (2013) ‘Mapping viral functional domains for genetic diversity in plants’, Journal of Virology, 87/2: 790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cerezo E., García-Arenal F. (1989) ‘Genetic heterogeneity of the RNA genome population of the plant virus U5-TMV’, Virology, 170/2: 418–23. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Cerezo E. et al. (1989) ‘Variability and evolution of the plant RNA virus pepper mild mottle virus’, Journal of Virology, 63/5: 2198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W. L., Roossinck M. J. (2000) ‘Evolutionarily related sindbis-like plant viruses maintain different levels of population diversity in a common host’, Journal of Virology, 74/7: 3130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W. L., Roossinck M. J. (2001) ‘Genetic diversity in RNA viral quasispecies is controlled by host-virus interactions’, Journal of Virology, 75/14: 6566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]