SUMMARY
Ataxia-Telangiectasia Mutated (ATM) regulates the DNA damage response as well as DNA double-strand break repair through homologous recombination. Here we show that ATM is hyperactive when the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is chemically inhibited or when the DNA-PKcs gene is deleted in human cells. Pre-incubation of ATM protein with active DNA-PKcs also significantly reduces ATM activity in vitro. We characterize several phosphorylation sites in ATM that are targets of DNA-PKcs and show that phospho-mimetic mutations at these residues significantly inhibit ATM activity and impair ATM signaling upon DNA damage. In contrast, phospho-blocking mutations at one cluster of sites increase the frequency of apoptosis during normal cell growth. DNA-PKcs, which is integral to the non-homologous end joining pathway, thus negatively regulates ATM activity through phosphorylation of ATM. These observations illuminate an important regulatory mechanism for ATM that also controls DNA repair pathway choice.
Keywords: ATM, DNA-PKcs, phosphorylation, protein kinase, DNA repair, cell cycle checkpoint
INTRODUCTION
DNA double-strand breaks (DSBs) are the most deleterious genetic lesions as failure of repair may lead to cell death, genomic instability or tumorigenesis (Ciccia and Elledge, 2010). Eukaryotic cells utilize two major DSB repair pathways, non-homologous end joining (NHEJ) and homologous recombination (HR). DSBs also activate a complex signaling network called the DNA damage response, which coordinates cell cycle checkpoint activation and repair of DSBs (Shiloh and Ziv, 2013).
ATM is a serine/threonine protein kinase belonging to the phosphatidylinositol-3-kinase like kinase (PIKK) family and is a master regulator of the DNA damage response (Paull, 2015). Another member of the PIKK family, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), associates with the Ku70/80 heterodimer (Ku) to form the DNA-PK holoenzyme (Jette and Lees-Miller, 2015). DNA-PKcs and ATM are both actively involved in DSB repair: DNA-PKcs plays a key role in NHEJ, the predominant pathway for repair of DSBs in human cells, while ATM promotes the HR pathway (Shrivastav et al., 2008). The Mre11-Rad5-Nbs1 (MRN) complex acts as a DSB sensor and is essential for ATM recruitment to DSB ends and for ATM activation (Paull, 2015). Similarly, DNA-PKcs is recruited to DSBs and activated by Ku (Jette and Lees-Miller, 2015). However, it is not well understood how cells make choices between the ATM/MRN-dependent HR pathway and the DNA-PKcs/Ku-dependent NHEJ pathway in S/G2 phase when both pathways are active.
Recent evidence shows that cross-talk occurs between DNA-PKcs and ATM and that they may cooperatively initiate DSB repair signaling and regulate DSB repair pathway choice. DNA-PKcs is known to undergo autophosphorylation that regulates the maintenance of its association with Ku-bound DNA ends and is required for DNA end processing and end joining (Jette and Lees-Miller, 2015). DNA-PKcs is also phosphorylated by ATM at residues T2609 and T2647 in response to DNA damage, which is essential for the repair of DSBs by NHEJ (Chen et al., 2007). ATM phosphorylation of DNA-PKcs has been shown to overcome Ku/DNA-PKcs inhibition of resection in vitro (Zhou and Paull, 2013) and plays an important role in end-processing mediated by DNA-PKcs and Artemis endonucleases (Jette and Lees-Miller, 2015). Lastly, the expression of ATM is regulated by DNA-PKcs such that ATM protein levels are decreased in DNA-PKcs-deficient human cells (Peng et al., 2005).
Autophosphorylation of ATM has been shown to be essential for ATM activation and function in response to DNA damage in human cells (Bakkenist and Kastan, 2003; Kozlov et al., 2011; Kozlov et al., 2006). However, phospho-blocking mutations at these four autophosphorylation sites do not affect ATM kinase activity in vitro or in mouse models (Daniel et al., 2008; Guo et al., 2010; Lee and Paull, 2005; Pellegrini et al., 2006), suggesting that there are other mechanisms for regulation of ATM activity.
In this study, we show that DNA-PKcs negatively regulates ATM activity through phosphorylation of ATM at multiple sites. We characterize DNA-PKcs phosphorylation sites that repress ATM activation both in vitro and in cells. Overexpression of ATM with phospho-mimetic mutations at the identified sites in ATM-deficient cells fails to restore cell survival, DSB end resection, or intra-S-phase checkpoint activation in response to DNA damage. Our data suggests that the NHEJ factor DNA-PKcs suppresses the catalytic activity of ATM through phosphorylation and that these modifications negatively regulate ATM signaling and DNA repair upon DNA damage. Since ATM is known to promote the HR pathway, this may provide an important mechanism for DNA repair pathway choice upon DNA damage as well as for ATM inactivation after DNA repair.
RESULTS
ATM is hyperactive in the absence of DNA-PKcs kinase activity in human cells
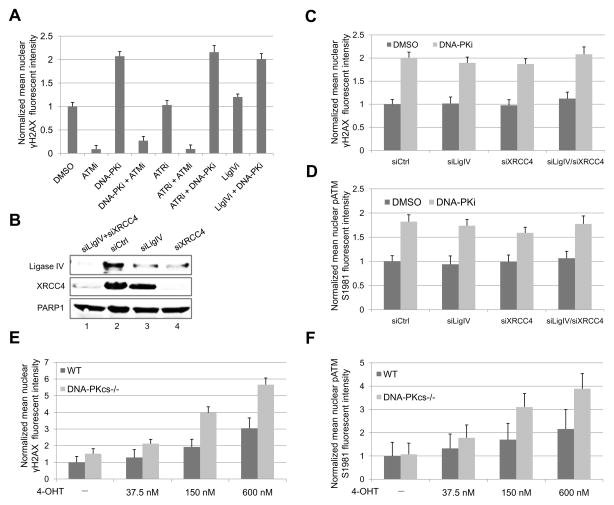
To assess the effect of PIKK enzymes on DSB signaling, we used the ER-AsiSI system in which DSBs can be induced by the AsiSI restriction enzyme that is induced to translocate to the nucleus upon hydroxytamoxifen (4-OHT) treatment (Iacovoni et al., 2010). We found that the formation of γH2AX foci at AsiSI-induced DSBs is dependent on ATM but not on DNA-PKcs or ATR in human U2OS cells. As shown in Figure 1A, while γH2AX foci formation is blocked by the ATM inhibitor KU-55933 (ATMi), it is not affected by the ATR inhibitor AZ20 (ATRi). Interestingly, γH2AX foci signal is significantly enhanced by the DNA-PK inhibitor NU7026 (DNA-PKi) (Figure 1A and Figure S1A). A combination of DNA-PKi and ATRi also leads to elevated γH2AX signal, similar to the effect observed with DNA-PKi alone. These data suggest that ATM, but not DNA-PKcs or ATR, mediates γH2AX foci formation at AsiSI-induced DSBs. This conclusion is further confirmed by combinational treatments of cells with DNA-PKi/ATMi or ATRi/ATMi, which both lead to attenuated γH2AX foci signal.
Figure 1. ATM is Hyperactive When DNA-PKcs Kinase Activity Is Blocked by DNA-PK Inhibitor or When the DNA-PKcs Gene Is Deleted in Human Cells.
(A) ER-AsiSI U2OS cells were pre-treated with 10 μM ATM inhibitor KU55933 (ATMi), 10 μM DNA-PK inhibitor NU7026 (DNA-PKi), 10 μM ATR inhibitor AZ20 (ATRi), and 100 μM DNA Ligase IV inhibitor SCR7 (LigIVi) as indicated for 1 h, followed by 4 h of 4-OHT (600 nM) treatment. Cells were fixed and stained with γH2AX antibody. The intensity of nuclear γH2AX foci signal per cell was quantitated from at least 200 cells for each cell line with the value of DMSO-treated cells being normalized to 1. Error bars: S.E.M. (B) ER-AsiSI U2OS cells were transfected with control siRNA (siCtrl), siRNA against DNA Ligase IV (siLigIV) or XRCC4 (siXRCC4), or siRNAs against both Ligase IV and XRCC4 (siLigIV/siXRCC4). After 48 h, cells were lysed and subjected to western blotting analysis for DNA Ligase IV and XRCC4. PARP1 was used as a loading control. (C, D) Cells from (B) were pre-treated or mock treated with 10 μM DNA-PKi for 1 h. After 4 h of 4-OHT induction, cells were fixed and stained with γH2AX and phospho-ATM S1981 antibodies. The intensities of nuclear γH2AX foci signal (C) and phospho-ATM S1981 foci signal (D) per cell were quantitated from at least 200 cells for each cell line with the value of control cells (siCtrl + DMSO) being normalized to 1. Error bars: S.E.M. (E, F) WT and DNA-PKcs−/− ER-AsiSI HCT116 cells (Zhou et al., 2014) were treated with various concentrations of 4-OHT for 4 h as indicated in the presence or absence of 10 μM DNA-PKi. Cells were fixed and stained with phospho-ATM S1981 antibody and γH2AX antibody. The intensities of nuclear γH2AX foci signal (E) and phospho-ATM S1981 foci signal (F) per cell were quantitated from at least 200 cells for each cell line. The value for WT cells without 4-OHT treatment was normalized to 1. Error bars: S.E.M.
The intriguing observation that DNA-PKcs inhibition leads to ATM hyperactivation upon DNA damage may be explained by DNA-PKi-mediated inhibition of the NHEJ DSB repair pathway, which may lead to elevated accumulation of DSBs and thus increased ATM activation. However, we noticed that inhibition of NHEJ with a DNA Ligase IV specific inhibitor SCR7 (LigIVi) fails to stimulates ATM activation as DNA-PKi does, and that a combination of DNA-PKi and LigIVi does not cause an additive effect (Figure 1A), suggesting that the stimulatory effect of DNA-PKi on ATM activation is not simply due to inhibition of the NHEJ pathway. In addition, we did not observe altered accumulation of DSBs after 4-OHT induction in the presence of DNA-PKi (data not shown), likely due to the fact that the AsiSI enzyme stays in the nucleus during 4-OHT induction, where it can induce DSBs constitutively. Because of this characteristic of the system, we are not measuring repair but simply the signaling efficiency at sites of DSBs. We further depleted DNA Ligase IV and XRCC4 by siRNA and examined γH2AX foci and phospho-ATM S1981foci formation at AsiSI-induced DSBs in ER-AsiSI U2OS cells. Consistent with inhibition of Ligase IV (Figure 1A), depletion of Ligase IV/XRCC4 alone does not affect ATM activity, while further treatment with DNA-PKi again leads to ATM hyperactivation, as shown by elevated γH2AX and phospho-ATM S1981foci signal (Figure 1B–D).
In agreement with these findings, higher levels of ATM phospho-S1981 signal and γH2AX signal are also observed at AsiSI-induced DSBs in DNA-PKcs−/− HCT116 cells compared with wild-type (WT) cells (Figure 1E, F). The phosphorylation levels of ATM targets, including p53, Chk2 and KAP1, are also increased in the absence of DNA-PKcs when the cells are treated with ionizing radiation (IR) (Figure S1B). In mouse cells, loss of DNA-PKcs also increases IR-induced ATM autophosphorylation, phosphorylation of ATM substrates KAP1, RPA, and H2AX, as well as the phosphorylation of Akt at S473 which has been shown to be dependent on ATM activity (Viniegra et al., 2005) (Figure S1C). Consistent with our observations, increased global phosphorylation of H2AX as well as higher enrichment of γH2AX at AsiSI-induced DSBs have been observed in U2OS cells upon DNA-PKi treatment (Caron et al., 2015). Hyperactivation of ATM upon DNA-PKcs inhibition is also reported in A549 and MCF10A cells after IR treatment (Finzel et al., 2016). Notably, this effect was not observed when the NHEJ pathway was inhibited by DNA Ligase IV inhibitor, suggesting that an increase in DSBs alone is not sufficient for ATM hyperactivation upon DNA-PKcs inhibition (Finzel et al., 2016). Taken together, these results suggest that DNA-PKcs negatively regulates ATM activation upon DNA damage in mammalian cells.
DNA-PKcs Phosphorylates ATM and MRN in Vitro
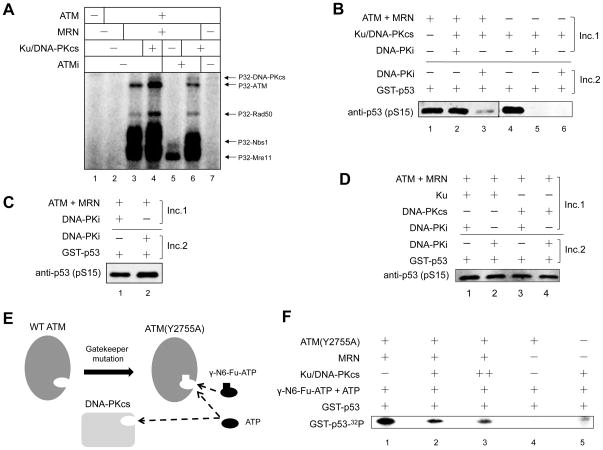
To test the hypothesis that DNA-PKcs may regulate ATM kinase activity through phosphorylation, we performed an in vitro kinase assay with [γ-32P]-ATP. As shown in Figure 2A, ATM is autophosphorylated in the presence of MRN/DNA, which is increased by the addition of Ku/DNA-PKcs (compare lanes 3 and 4). Addition of ATM inhibitor to the reaction completely blocks ATM phosphorylation, while DNA-PKcs can restore the phosphorylation of ATM (lanes 5 and 6). However, ATM is poorly phosphorylated by DNA-PKcs in the absence of MRN, likely due to the low DNA-binding affinity of ATM (Lee and Paull, 2005). In addition, the MRN complex is also strongly phosphorylated by both ATM and DNA-PKcs in vitro. These results suggest that ATM is phosphorylated by DNA-PKcs in vitro and that MRN promotes this phosphorylation event, presumably by recruiting ATM to DNA ends where DNA-PKcs is located.
Figure 2. DNA-PKcs Inhibits ATM Kinase Activity in Vitro.
(A) Purified recombinant ATM protein was incubated with Ku/DNA-PKcs, MRN, linearized DNA, [γ-32P]-ATP at 37 °C for 1 h in the presence or absence of 20 μM KU55933 (ATMi). The proteins were separated by SDS-PAGE and visualized by phosphorimaging. (B) A two-step in vitro ATM kinase assay was performed in the presence of Ku/DNA-PKcs with DNA-PK inhibitor (DNA-PKi) added in the first or second incubation as indicated. DNA-PKcs-mediated phosphorylation events (Incubation 1, Inc. 1) and the phosphorylation of p53 substrate by ATM (Incubation 2, Inc. 2) were separated in two different incubations (Inc.). (C) A two-step ATM kinase assay was performed with DNA-PKi added in the first or second incubation as indicated. (D) A two-step ATM kinase assay as in (B) was performed with Ku or DNA-PKcs alone. (E) Schematic diagram of the ATM Y2755A analog-sensitive ATM mutant that accepts the bulky ATP analog N6-Fu-ATP. (F) An in vitro kinase assay with analog-sensitive ATM mutant (Y2755A) in the presence of [γ-32P]-labeled bulky ATP analog γ-N6-Fu-ATP was performed. The protein products were separated by SDS-PAGE and visualized by phosphorimaging.
DNA-PKcs Inhibits the Activation of ATM by MRN/DNA in Vitro
To determine the effect of DNA-PKcs mediated phosphorylation of ATM and MRN has any effect on ATM kinase activity, we performed in vitro kinase assays using GST-p53 as a substrate (Lee and Paull, 2005). Because p53 can be phosphorylated by both ATM and DNA-PKcs in vitro, we used a two-step procedure in which the phosphorylation of ATM and MRN by DNA-PKcs and the phosphorylation of p53 substrate by ATM were separated into two incubations. During the first incubation, ATM was incubated with Ku/DNA-PKcs, MRN and DNA in the presence or absence of DNA-PKi. In reactions where DNA-PKi was added, DNA-PKcs-mediated phosphorylation of the p53 substrate was blocked (Figure 2B, lanes 5 and 6). During the second incubation, p53 substrate was added to all reactions and DNA-PKi was added to those reactions where it was absent in the first incubation, ensuring that p53 substrate could only be phosphorylated by ATM. Notably, pre-incubation of ATM with active DNA-PK complex in the first step impairs ATM activation, as shown by the decreased phosphorylation level of p53 substrate compared with the reaction in which DNA-PKcs kinase activity is blocked by DNA-PKi during the first incubation (lanes 2 and 3). This effect is most likely to be mediated by active DNA-PK complex, as pre-incubation of ATM with DNA-PKi itself, Ku or DNA-PKcs alone does not affect ATM activity (Figure 2C, D).
The inhibitory effect of DNA-PKcs on ATM activity was further confirmed by using the ATP analog-sensitive ATM mutant Y2755A, which has a cavity within the ATP-binding pocket and can utilize bulky ATP analog N6-(furfuryl)-ATP for phosphorylation of p53 substrate (Figure 2E)(Kodama et al., 2010; Lee et al., 2013). Since the ATP analog is labeled with 32P, ATM-mediated phosphorylation of p53 can be observed directly by phosphorimaging while p53 phosphorylation by DNA-PKcs will not be 32P-labeled. ATM(Y2755A) strongly phosphorylates p53 using the radioactive bulky ATP analog as a phosphate donor (Figure 2F, lane 1), while DNA-PKcs activity with this analog is minimal (lane 5). Consistent with results obtained from two-step kinase assays (Figure 2B), active DNA-PK complex reduces ATM(Y2755A)-mediated phosphorylation of p53 substrate in a dose-dependent manner (Figure 2F, lane 2 and 3). Taken together, these results demonstrate that DNA-PKcs inhibits ATM kinase activity in vitro.
DNA-PKcs Inhibits ATM Kinase Activity through Phosphorylation of ATM
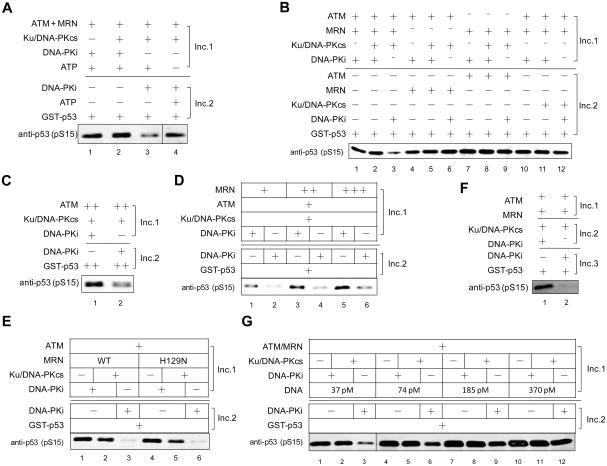
We further examined if the inhibitory effect of DNA-PKcs on ATM is mediated by phosphorylation. A two-step kinase assay was performed with ATP either being added to the first incubation or to the second incubation. Elimination of ATP during the first incubation overcomes the inhibitory effect of DNA-PKcs on ATM (Figure 3A), suggesting that this effect is indeed related to DNA-PKcs-mediated phosphorylation events occurring in the first incubation.
Figure 3. DNA-PKcs Inhibits ATM Kinase Activity by Phosphorylating ATM in Vitro.
(A) A two-step ATM kinase assay was performed as in Fig. 2B, with ATP added at the 1st incubation (reactions 1, 2 and 3) or the 2ndincubation (reaction 4). (B) A two-step ATM kinase assay was performed as in Fig. 2B, with various components added as indicated. (C) The basal activity of ATM was examined with high levels of ATM protein (2 nM) and GST-p53 substrate (250 nM) in the absence of MRN. (D) Two-step in vitro ATM kinase assays as in Figure 2B, with a titration of purified recombinant MRN (5 nM, 10 nM and 20 nM) in the assay. (E) Two-step in vitro ATM kinase assays as in Fig. 2B, using 20 nM purified recombinant wild-type MRN complex (WT) or nuclease-deficient MRN mutant (H129N) in the assay. (F) ATM was pre-activated by MRN and DNA (Inc. 1) and then subjected to the two-step kinase assay. (G) A two-step ATM kinase assay was performed as in (A) but with a titration of linearized DNA (moles of DNA molecules, as indicated).
Since both MRN and ATM can be phosphorylated by DNA-PKcs (Figure 2A), we asked which phosphorylation event actually leads to the DNA-PKcs inhibition of ATM activity. To answer this question, DNA-PKcs, MRN or ATM was eliminated from the first incubation. As expected, DNA-PKcs does not show any effect on ATM activity when it is only added to the second incubation in the presence of DNA-PK inhibitor (Figure 3B, lane 12). No decrease in ATM activation is observed when ATM protein is absent from the first incubation (Figure 3B, lane 9), suggesting that the inhibitory effect of DNA-PKcs on ATM is not caused by DNA-PKcs-mediated phosphorylation of MRN. Interestingly, inhibition of ATM activity is also not observed when MRN is absent from the first incubation (lane 6), likely due to the fact that ATM is poorly phosphorylated by DNA-PKcs in the absence of MRN (Figure 2A, lane 7).
To determine conclusively whether the presence of MRN is essential for the inhibitory effects of DNA-PKcs on ATM, we made use of the fact that high levels of ATM kinase by itself yield some basal activity in vitro when the substrate concentration is also high (data not shown). A kinase assay was performed with high concentrations of ATM and GST-p53 substrate in the absence of MRN. The result clearly shows that DNA-PKcs is still able to inhibit ATM activity without MRN (Figure 3C), suggesting that the inhibitory effect of DNA-PKcs on ATM is caused by DNA-PKcs phosphorylation of ATM, although MRN strongly promotes this effect in a dose-dependent manner (Figure 3D). The nuclease activity of MRN is not required for DNA-PKcs inhibition of ATM, because a nuclease-deficient mutant M(H129N)RN shows a similar effect as WT MRN (Figure 3E).
In resting cells, ATM exists as an inactive non-covalent dimer which is recruited to DNA ends by the MRN complex and converted to active monomers (Bakkenist and Kastan, 2003; Lee and Paull, 2005). We examined whether DNA-PKcs can affect the activity of pre-activated ATM monomers using a three-step procedure. ATM is pre-incubated with MRN and DNA, where ATM undergoes monomerization and activation, before proceeding to the two-step kinase assay. Incubation of the pre-activated ATM with DNA-PKcs still leads to impaired ATM activation (Figure 3F), suggesting that DNA-PKcs can inactivate the kinase activity of ATM monomers through phosphorylation.
We also performed a DNA titration experiment and found that the decrease in ATM kinase activity is less pronounced with increased levels of linearized DNA in the reaction (Figure 3G), likely because ATM and DNA-PKcs are binding to different DNA molecules and are less likely to contact each other in the presence of more DNA ends.
The role of ATM autophosphorylation in ATM function has remained controversial (Paull, 2015). We hypothesized that autophosphorylation of ATM might repress DNA-PKcs-mediated phosphorylation and hence overcome DNA-PKcs inhibition of ATM, as it is not a rare case that one phosphorylation event inhibits further phosphorylation of the same protein in the cellular signaling network. An autophosphorylation-mimetic ATM mutant (S367D/S1893D/S1981D/S2996D) was purified and tested in the in vitro kinase assay in comparison with WT ATM protein. The results clearly show that DNA-PKcs can still phosphorylate this mutant and inhibit its kinase activity (Figure S2A), suggesting that ATM autophosphorylation at the four identified sites does not affect DNA-PKcs phosphorylation or inhibition of ATM. In addition, this ATM mutant (S367D/S1893D/S1981D/S2996D) can still undergo autophosphorylation in vitro (Figure S2B), suggesting the existence of other uncharacterized autophosphorylation sites in ATM.
Identification of target site(s) responsible for the inhibitory effect of DNA-PKcs on ATM kinase activity
To identify target ATM site(s) responsible for DNA-PKcs inhibition of ATM, purified ATM protein was incubated with MRN and DNA in the presence or absence of DNA-PKcs and subjected to mass spectrometry analysis (Table S1). Based on these results and the known preference of DNA-PKcs for phosphorylation of S/TQ sites, we made a series of phospho-blocking (S/T→A) and phospho-mimetic (S→D, T→E) ATM mutants and tested their kinase activities in vitro (summarized in Table S2).
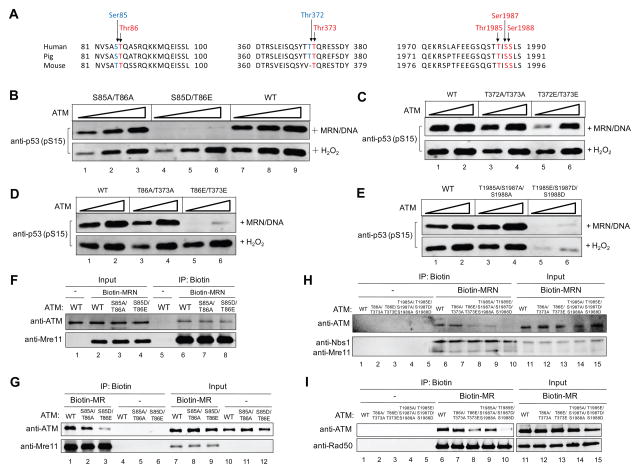
The screening led to the identification of three clusters of sites of potential interest: S85/T86, T372/T373 and S1985/T1987/T1988, all of which are conserved in mammalian species (Figure 4A). Phospho-mimetic mutations at these sites all result in decreased ATM kinase activity in the presence of MRN/DNA, while phospho-blocking mutations (alanine substitutions) at the same sites do not affect ATM activation (Figure S3A–E, Table S2). Interestingly, combinations of adjacent phospho-mimetic mutations render ATM more severely impaired in MRN/DNA-dependent activation (Table S2, Figure 4B–E). Both the S85D/T86E mutant and the S1985D/T1987E/T1988E mutant are kinase-deficient (Figure 4B and E). In contrast, the activation of the T372E/T373E mutant is less severely impaired (Figure 4C). Considering the similar amino acid sequence pattern of the S85/T86 cluster and the T372/T373 cluster (ST/Q and TT/Q, respectively), we also introduced S85/T372 and T86/T373 combined mutations into ATM. As expected, the MRN/DNA-dependent activity of both S85D/T372E and T86E/T373E mutants are much lower than WT ATM or corresponding phospho-blocking mutants (Figure S3F and Figure 4D).
Figure 4. Characterization of ATM Phosphorylation Site Mutants Identifies Candidates With Reduced Affinity for MRN.
(A) Alignment of ATM sequences from mammalian species. The highlighted residues which are mutated in this study are conserved. (B) In vitro kinase assays were performed with recombinant wild-type ATM (WT), phospho-mimetic ATM mutant S85D/T86E, or phospho-blocking ATM mutant S85A/T86A, in the presence of MRN/DNA or H2O2. (C) In vitro kinase assays were performed with wild-type ATM (WT), T372E/T373E ATM, or T372A/T373A ATM, in the presence of MRN/DNA or H2O2. (D) In vitro kinase assays were performed with wild-type ATM (WT), T86E/T373E ATM, or T86A/T373A ATM, in the presence of MRN/DNA or H2O2. (E) In vitro kinase assays were performed with wild-type ATM (WT), T1985E/S1987D/S1988D ATM, or T1985A/S1987A/S1988A ATM, in the presence of MRN/DNA or H2O2. (F) In vitro assay for ATM-MRN association with biotinylated MRN (Biotin-MRN), wild-type ATM (WT), ATM S85AT86A and ATM S85DT86E mutant proteins. Streptavidin beads were used to isolate Biotin-MRN and associated ATM protein. Proteins on the beads were eluted and analyzed by western blotting. (G) A similar experiment was performed as in (F), using Biotin-MR instead of Biotin-MRN. (H) Similar experiment was performed as in (F) for other ATM mutants: T86A/T373A, T86E/T373E, T1985A/S1987A/S1988A and T1985E/S1987D/S1988D. (I) A similar experiment was performed as in (H), using Biotin-MR instead of Biotin-MRN.
Another important pathway of ATM activation is dependent on oxidative stress (Guo et al., 2010). We also examined H2O2-induced ATM activation for the mutants mentioned above. Interestingly, the S85D/T86E, T372E/T373E and T86E/373E ATM mutants are all activated normally by H2O2, whereas the T1985E/S1987D/S1988D mutant is deficient in the presence of H2O2 in vitro (Figure 4B–E).
The regulation of ATM activation by phosphorylation at these sites was further examined in human HEK-293T cells in which the endogenous ATM was partially depleted by shRNA while shRNA-resistant WT or mutant ATM alleles were transiently over-expressed. The cells were treated with bleomycin to induce DSBs or H2O2 to induce oxidative stress, followed by detection of the phosphorylation levels of ATM targets (KAP1, Chk2 and p53). Cells expressing the phospho-mimetic mutants (S85D/T86E, T86E/T373E, or T1985E/S1987D/S1988D) clearly show decreased phosphorylation of ATM substrates as well as S1981 ATM autophosphorylation upon bleomycin treatment, while the activity of T372E/T373E is only slightly decreased (Figure S4). These data suggest that ATM activation is impaired by phosphorylation of the identified sites upon DNA damage in cells. With H2O2 treatment, S85D/T86E, T372E/T373E and T86E/T373E mutants are all similar to WT ATM (Figure S5). In contrast to the in vitro result (Figure 4E), the T1985E/S1987D/S1988D ATM mutant could be normally activated by H2O2 in cells (Figure S5B), possibly because of the action of other ATM stimulators such as ATMIN and NKX3.1 (Bowen et al., 2013; Kanu and Behrens, 2007).
DNA-PKcs Phosphorylation of ATM Inhibits MRN Binding
Since the ATM S85D/T86E mutant fails to be activated by MRN, we speculated that the ATM-MRN interaction, which is essential for ATM activation (Lee and Paull, 2004), might be impaired by the phospho-mimetic mutations. To test this idea, we performed a protein-protein interaction assay using purified biotinylated MRN complex (Biotin-MRN). Surprisingly, the ATMS85D/T86E mutant can interact normally with MRN (Figure 4F). ATM has been shown to interact with both MR and Nbs1 in vitro (Lee and Paull, 2004). Therefore, we performed protein interaction assays with biotinylated MR (Biotin-MR) and found that the interaction between ATM S85D/T86E mutant and MR is significantly impaired (Figure 4G), suggesting that the ATM-MR interaction and the ATM-Nbs1 interaction are both essential for ATM activation. We also examined the interaction between biotin-MRN and other ATM mutants and found that the T86A/T373A mutant binds to MRN normally while the kinase-deficient T86E/T373E mutant shows decreased affinity to MRN (Figure 4H). Lastly, the ATM T1985E/S1987D/S1988D mutant exhibits essentially no binding to MRN (Figure 4H, lane 10). Similar results were observed in a binding assay using Biotin-MR as bait (Figure 4I), suggesting that phosphorylation of ATM at these sites may cause a conformational change of the protein and hence lead to decreased affinity to MR in the MRN complex.
Kinase-deficient Phospho-mimetic ATM Mutants Fail to Support Cell Survival upon DNA Damage in Human Cells
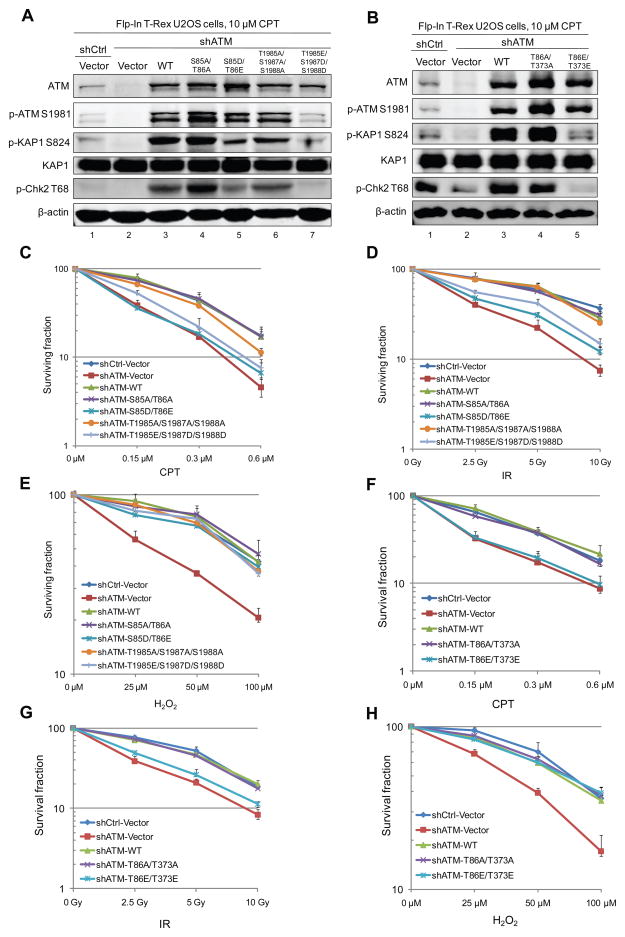
To examine the effects of ATM phosphorylation in human cells, we utilized U2OS cells containing the mutant or wild-type ATM alleles under doxycycline control (Flp-In™ T-Rex™ system, Invitrogen). Endogenous ATM was depleted using shRNA, and shRNA-resistant wild-type ATM or mutant ATM was induced to express from a single genomic locus upon doxycycline treatment (Figure 5A, B). In agreement with observations in vitro and in 293T cells (Figure 4 and Figure S4), the activation of ATM S85D/T86E, T86E/T373E and T1985E/S1987D/S1988D by camptothecin (CPT)-induced DNA damage is deficient in comparison to WT ATM, as shown by the reduced phosphorylation of ATM targets (KAP1 and Chk2) and by ATM autophosphorylation (Figure 5A, B).
Figure 5. Kinase-deficient Phospho-mimetic ATM Mutants Fail to Support Cell Survival upon DNA Damage in Human Cells.
(A) Expression of WT ATM and ATM mutants, including S85A/T86A, S85D/T86E, T1985A/S1987A/ S1988A and T1985E/S1987D/S1988D, were induced by doxycycline in U2OS Flp-In T-Rex cells in which endogenous ATM was depleted by shRNA. Cells were treated with 10 μM camptothecin (CPT) for 1 h and the phosphorylation KAP1 and Chk2 was examined by western blotting. (B) Expression of WT ATM and ATM mutants were induced by doxycycline in U2OS Flp-In T-Rex cells depleted for endogenous ATM by shRNA. Cells were treated with 10 μM camptothecin (CPT) for 1 h and the phosphorylation of KAP1 and Chk2 was examined by western blotting. (C) Cells grown as in (A) were subjected to survival analysis (clonogenic assay) after 1 h treatment with various concentrations of CPT. (D) Cells grown as in (A) were subjected to survival analysis after various doses of ionizing radiation (IR) treatment. (E) Cells grown as in (A) were subjected to survival analysis after various doses of H2O2 treatment. (F) Cells grown as in (B) were subjected to survival analysis after 1 h treatment with various concentrations of CPT. (G) Cells grown as in (B) were subjected to survival analysis after various doses of ionizing radiation (IR) treatment. (H) Cells grown as in (B) were subjected to survival analysis after various doses of H2O2 treatment.
Consistent with the critical regulatory roles of ATM in DNA damage repair and oxidative stress responses, we found that depletion of ATM leads to decreased cell survival when cells are treated with CPT, IR or H2O2 (Figure 5C–E). With CPT or IR treatment, the phospho-mimetic S85D/T86E and T1985E/S1987D/S1988D mutants both fail to restore cell survival as wild-type ATM does, while the corresponding phospho-blocking ATM mutants (S85A/T86A and T1985A/S1987A/ S1988A) can efficiently support cell survival upon DNA damage (Figure 5C, D). In contrast, the S85D/T86E and T1985E/S1987D/S1988D alleles both rescue cell survival upon H2O2 treatment (Figure 5E), suggesting that ATM phosphorylation at these sites does not affect ATM function in oxidative stress responses. Similar effects on cell survival are also observed for the combined ATM mutants T86A/T373A and T86E/T373E (Figure 5F–H).
To determine the effect of DNA-PKcs phosphorylation on endogenous ATM activity, we introduced T1991E/S1993D/S1994D mutations, which correspond to the T1985E/S1987D/S1988D mutations in human ATM, into the Atm gene in mouse cells. The mutations were targeted to the WT ATM allele in ROSA26+/Cre-ERT2ATMC/+ murine embryonic stem (ES) cells along with a Neo resistant cassette flanking by FRT sequences (Figure S6A–C). After expression of FLP recombinase, we obtained several independent clones of the Rosa26+/Cre-ERT2AtmC/(T1991E/S1993D/S1994D) ES cells. The Rosa26+/Cre-ERT2allele expresses a Cre recombinase fused with estrogen receptor ligand binding domain, which mediates effective 4-OHT induced nuclear translocation of the Cre recombinase. 4-OHT treatment generated several Atm−/(T1991E/S1993D/S1994D) clones. The ATM T1985E/S1987D/S1988D mutant protein was expressed at a similar level as the WT protein in both clones (Figure S6E). However, IR-induced phosphorylation of Kap1 was lower in Atm−/( T1991E/S1993D/S1994D) cells than Atm−/+ controls (Figure S6E), consistent with observations in human cells.
Phosphorylation of ATM T86 occurs in human cells
To better understand the function of ATM phosphorylation at the defined sites, we developed a polyclonal phospho-specific antibody against an ATM peptide containing pS85pT86. As side products, ATM pS85 and ATM pT86 antibodies were also obtained and affinity purified using pS85 and pT86 peptides, respectively. The pS85pT86, pS85 and pT86 antibodies had reasonable specificity, as they could recognize exogenous WT ATM, but not S85A/T86A or S85D/T86E ATM mutants in 293T cells (Figure S7A). Since the ATM pT86 antibody seemed to work best, we focused on this antibody for further study. We then treated 293T cells with bleomycin and harvested cells at various time points after bleomycin treatment. Interestingly, we saw an obvious increase of ATM pT86 signal at 2 h and 4 h of recovery, which was largely blocked by the pT86 peptide but not by the pS85 peptide (Figure S7B), suggesting that the antibody was indeed against the ATM pT86 site. However, the phosphorylation signal was only partially removed by phosphatase treatment (Figure S7C), thus the antibody can also recognize non-phophorylated ATM. Despite this, background issue, it is still clear that ATM T86 phosphorylation increased 2–4 h after DNA damage and then decreased to the background level (Figure S7D, E). We also observed that the increase in phospho-ATM T86 correlated with a decrease in KAP1 phosphorylation (Figure S7B, E). Overall, results with the T86 antibody indicate that the phosphorylation of ATM does occur in human cells and that the phosphorylation peaks at 2 to 4 h after DNA damage.
The ATM phospho-blocking mutants T86A/T373A and T1985A/S1987A/S1988A are resistant to DNA-PKcs inhibition
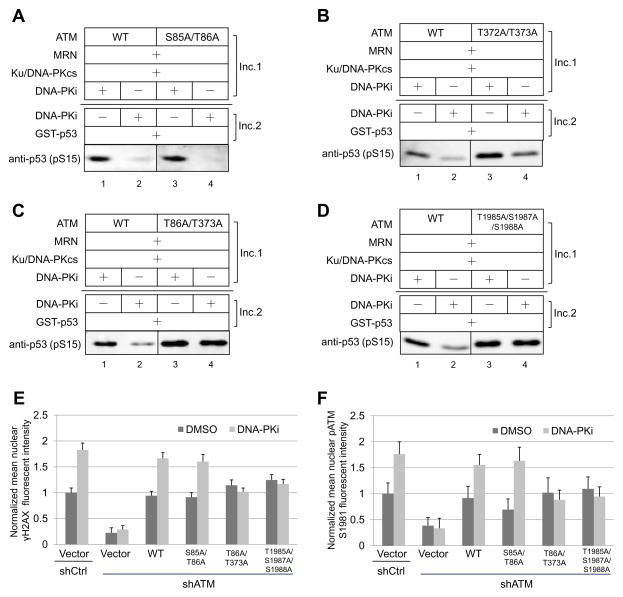
Based on our in vitro results, the potential sites that are responsible for the inhibitory effect of DNA-PKcs on ATM kinase activity should meet two criteria: 1) phospho-mimetic mutations at these sites should decrease kinase activity, and 2) phospho-blocking mutations at these sites should render ATM resistant to DNA-PKcs inhibition. Therefore, we further tested the phospho-blocking ATM mutants in the two-step kinase assay described above. Neither the S85A/T86A mutant nor the T372A/T373A mutant shows complete resistance to DNA-PKcs inhibition (Figure 6A, B). However, the combined T86A/T373A mutant is clearly resistant to DNA-PKcs inhibition, while the S85A/T372A mutant is not (Figure 6C, Table S2). In addition, the triple ATM mutant T1985A/S1987A/S1988A is also completely resistant to DNA-PKcs inhibition in vitro (Figure 6D).
Figure 6. Identification of Sites Responsible for the Inhibitory Effect of DNA-PKcs on ATM Kinase Activity.
(A) A two-step in vitro ATM kinase assay was performed as in Fig. 2B, using purified recombinant wild-type ATM (WT) or S85A/T86A ATM. (B) A two-step in vitro ATM kinase assay was performed using wild-type ATM or T372A/T373A ATM. (C) A two-step in vitro ATM kinase assay was performed using purified recombinant wild-type ATM orT86A/T373A ATM. (D) A two-step in vitro ATM kinase assay was performed using wild-type ATM orT1985A/S1987A/S1988A ATM. (E, F) Expression of WT ATM and ATM mutants as indicated were induced by doxycycline treatment in Flp-In T-Rex U2OS cells expressing ER-AsiSI (Zhou et al., 2014). Cells were treated with 600 nM 4-OHT for 4 h, fixed and stained with phospho-ATM S1981 and γH2AX antibodies. The intensities of nuclear γH2AX foci signal (E) and phospho-ATM S1981 foci signal (F) per cell were quantitated from at least 200 cells for each cell line with the value of control cells (Vector + shCtrl + DMSO) normalized to 1. Error bars: S.E.M.
We further tested whether the T86A/T373A andT1985A/S1987A/S1988A ATM mutants are resistant to DNA-PKcs inhibition in human cells. We used γH2AX and phospho-ATM S1981 foci as readouts of ATM activation upon DNA damage in the presence or absence of DNA-PK inhibitor. Unlike cells expressing WT ATM which show elevated γH2AX signal and phospho-ATM S1981 signal when DNA-PKcs kinase activity is inhibited, cells expressing the T86A/T373A or T1985A/S1987A/S1988A ATM mutants fail to respond to inhibition of DNA-PKcs activity (Figure 6E, F), confirming that these sites are indeed responsible for DNA-PKcs inhibition of ATM kinase activity.
DSB end resection and cell cycle checkpoint activation upon DNA damage are impaired by the phospho-mimetic mutations of ATM in human cells
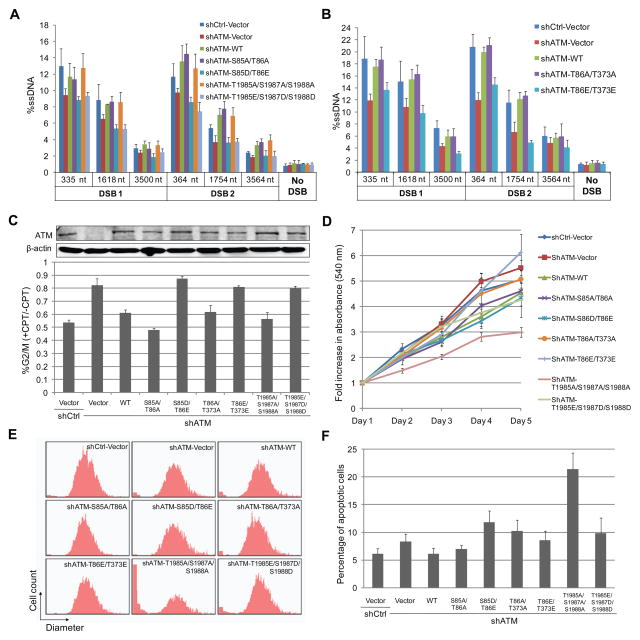
ATM activity is required for efficient use of the HR pathway, primarily because of its effects on DSB resection. ATM phosphorylates DNA end resection factors and modulates their activities, including CtIP and hSSB1 (Makharashvili et al., 2014; Richard et al., 2008). The effect of the phospho-blocking/mimetic ATM mutations on DSB end resection was examined using a quantitative PCR-based method described previously (Zhou et al., 2014). Consistent with the effect of ATM inhibition on resection (Zhou and Paull, 2013), depletion of ATM leads to decreased resection, which can be rescued by expression of WT ATM (Figure 7A, B). The phospho-blocking ATM mutants (S85A/T86A, T86A/T373A and T1985A/S1987A/S1988A) rescued resection similar to WT ATM, but the phospho-mimetic ATM mutants (S85D/T86E, T86E/T373E and T1985E/S1987D/S1988D) did not (Figure 7A, B), indicating that phosphorylation of ATM at these sites leads to reduced end resection and may in this way inhibit HR.
Figure 7. The Effects of ATM Phosphorylation Site Mutants on Cellular Responses to DNA damage and normal growth.
(A, B) The effects of ATM mutations on DSB end resection were examined by measuring ssDNA intermediates of resection at various locations adjacent to two DSBs generated by AsiSI in U2OS Flp-In T-Rex cells. (C) Cell lines were synchronized by aphidicolin and analyzed for intra-S-phase checkpoint activation after 1 μM CPT treatment for 1 h. Error bars indicates standard deviation from three independent experiments. (D) Growth of U2OS Flp-In T-Rex cell lines expressing ATM mutants were analyzed with the MTT cell proliferation assay. Error bars indicate standard deviation from three independent experiments. (E)U2OS cell lines were trypsinized and the size distribution of cells was determined using Scepter™ 2.0 Handheld Automated Cell Counter. (F) U2OS Flp-In T-Rex cell lines expressing ATM mutants as indicated were tested for apoptosis using a fluorescent Annexin V conjugate. Error bars indicate standard deviation from three independent experiments.
ATM activation is also essential for cell cycle checkpoint activation upon DNA damage. We examined the effects of the ATM mutants on checkpoint activation by exposing the cells to CPT during S phase and measuring the percentage of cells that bypass the intra-S phase checkpoint and traverse into G2/M phase. As expected, depletion of ATM leads to deficient intra-S-phase checkpoint activation, as a higher percentage of cells proceed to G2/M phase in the presence of DNA damage compared to WT cells (Figure 7C). Checkpoint activation is rescued by the expression of wild-type ATM, or the phospho-blocking ATM mutants (S85A/T86A, T86A/T373A and T1985A/S1987A/S1988A). However, cells expressing the kinase-deficient phospho-mimetic ATM mutants (S85D/T86E, T86E/T373E and T1985E/S1987D/S1988D) fail to activate the intra-S-phase checkpoint (Figure 7C), likely due to the deficiency of these ATM mutants in activation upon DNA damage.
The ATM T1985A/S1987A/S1988A mutations lead to reduced growth and elevated apoptosis in human cells
We further investigated whether the expression of these ATM mutants affects normal growth of cells (Figure 7D). Interestingly, while most ATM mutants do not significantly affect cell growth, cells expressing the ATM T1985A/S1987A/S1988A mutant show a reduced rate of cell proliferation (Figure 7D). We also observed a clear increase in the percentage of extremely small cells that is normally consistent with cell death, a change that is only observed with the T1985A/S1987A/S1988A mutant and not the phospho-mimeticT1985E/S1987D/S1988D allele or the other mutant alleles, and is dependent on doxycycline induction (Figure 7E and data not shown). To determine if this shift is due to programmed cell death responses, we measured apoptosis and found that expression of the T1985A/S1987A/S1988A allele in cells with no other perturbation exhibit high levels of apoptosis (Figure 7F). These results suggest that an inability to restrain ATM activity through DNA-PKcs results in a significantly higher frequency of cell death in every cell cycle.
DISCUSSION
ATM is a central player in the DNA damage response, but the mechanisms regulating ATM activity are still being elucidated. Numerous studies have shown the involvement of phosphorylation in ATM activation (Paull, 2015). Although the functional importance of ATM autophosphorylation remains controversial, phosphorylation of ATM by other kinases, including CDK5, EGFR and Aurora B, has been implicated in the regulation of ATM activation upon DNA damage or mitosis (Lee et al., 2015; Tian et al., 2009; Yang et al., 2011). However, no phosphorylation events that negatively regulate ATM kinase activity have been reported so far.
DNA-PKcs has been shown to be a phosphorylation target of ATM, and ATM-mediated phosphorylation of DNA-PKcs at Thr2609 is critical for DNA-PKcs function in DNA repair (Chen et al., 2007). Our study shows that DNA-PKcs also phosphorylates ATM and inhibits ATM kinase activity. It should be noted that the inhibitory effect of DNA-PKcs on ATM kinase activity is stronger with a limited amount of DNA ends than with excess DNA ends, presumably because the presence of excess DNA substrate is likely to reduce the co-localization of DNA-PKcs and ATM on DNA ends. A widely accepted model suggests that Ku and MRN, which recruit DNA-PKcs and ATM respectively, compete with each other for DSB end binding (Paull, 2010). However, our study indicates that DNA-PKcs and ATM are simultaneously recruited and activated at the same DSB ends. Surprisingly, pre-incubation of ATM with chemically inhibited DNA-PKcs results in normal ATM activation, suggesting that the presence of Ku/DNA-PKcs protein per se does not affect the recruitment of MRN/ATM to DNA ends and that the inhibition of ATM kinase activity by active DNA-PKcs is not simply an issue of DNA end competition between Ku/DNA-PKcs and MRN/ATM. This conclusion is supported by a recent study showing that MRN and Ku do not affect the recruitment of each other to DSBs (Hartlerode et al., 2015). ChIP-chip profiles also show that activated ATM and DNA-PKcs are similarly enriched at AsiSI-induced DSBs in U2OS cells (Caron et al., 2015). Therefore, it is possible that Ku/DNA-PKcs and MRN/ATM could form a large complex at DSB ends in which DNA-PKcs and ATM can contact and phosphorylate each other.
The mechanisms for choice between NHEJ and HR in S/G2 phase in mammalian cells are not fully understood. Previous studies suggest that the NHEJ factor DNA-PKcs and HR factor ATM may coordinate with one another to regulate the choice of DSB repair pathways. ATM has been implicated in the regulation of NHEJ through phosphorylation of DNA-PKcs and Artemis (Chen et al., 2007; Riballo et al., 2004). DNA-PKcs is involved in inhibitition of DSB end resection while ATM overcomes this inhibition by phosphorylating DNA-PKcs and promoting DNA-PKcs dissociation from DNA ends (Zhou and Paull, 2013). The current study demonstrates that DNA-PKcs can also directly phosphorylate ATM at multiple sites and hence inhibit ATM kinase activity, which provides an important regulatory mechanism for pathway choice as inhibition of ATM kinase activity impairs DSB end resection and HR (Zhou and Paull, 2013).
DNA-PKcs Inhibition of ATM Activity through Phosphorylation
Seven sites of interest are identified in ATM in the current study, including S85, T86, T372, T373, T1985, S1987 and S1988. Phospho-mimetic mutations at these sites all reduce ATM catalytic activity by varying degrees, and hence lead to impaired DNA damage responses in cells. Notably, the S85D/T86E mutant, which is kinase-deficient in the presence of MRN/DNA, binds normally to the intact MRN complex but shows decreased affinity to MR, suggesting that ATM must interact with both Nbs1 and MR to achieve activation. In contrast, the T86E/T373E and T1985E/S1987D/S1988D mutants show decreased binding to both MRN and MR. This can also explain why the phospho-mimetic mutants are deficient in the MRN/DNA pathway in vitro. Previous mass spectrometry analysis has identified phosphorylation of ATM at S85, T86, T373 and S1985 upon DNA damage in human cells (Kettenbach et al., 2011; Lee et al., 2015; Matsuoka et al., 2007; Sharma et al., 2014), suggesting that ATM is indeed phosphorylated at these sites in cells.
Our study reveals a complex mechanism for regulation of ATM activation by DNA-PKcs. The single T86A and T373A mutants are not resistant to DNA-PKcs inhibition while the T86A/T373A double mutant is, suggesting the necessity of phosphorylation at both sites for DNA-PKcs regulation of ATM activity. In addition, T86A/T373A and T1985A/S1987A/S1988A both show resistance to DNA-PKcs inhibition, suggesting that phosphorylation of these two clusters can regulate ATM activity independently from each other. The S85/T86 and T372/T373 sites in ATM are both located in N-terminal HEAT repeats, and they share a similar amino acid pattern: STQ and TTQ. Several other phosphorylation sites that are involved in ATM activation, including T370, S794 and S1403, are also located in the HEAT repeats, suggesting the importance of this region for regulation of ATM activity. We have also tested another two clusters of sites with a similar pattern: S474/S475(Q) and S2591/S2592(Q) (Table S2). Phospho-mimetic mutations at S475 and S2592 have no effect on ATM activation, although phosphorylation of both sites has been identified (this study and (Lee et al., 2015)). The cluster of T1985/S1987/S1988 sites is localized in the FAT domain and sits very close to the ATM autophosphorylation site S1981, but mutations at these three sites do not affect ATM S1981 autophosphorylation in vitro (data not shown). In cells expressing the phospho-mimetic T1985E/S1987D/S1988D allele there is reduced S1981 phosphorylation in the presence of DNA damage. But it remains to be investigated whether the phospho-mimetic mutations directly inhibit S1981 phosphorylation and hence repress ATM activation. The fact that mutations in all of the autophosphorylation sites fail to affect DNA-PKcs inhibition of ATM activity suggests that these autophosphorylation sites act independently from the sites identified in this work.
Compared with cells expressing WT ATM, cells expressing the phospho-mimetic ATM mutants S85D/T86E, T1985E/S1987D/S1988D and T86E/T373E show decreased ATM activation and DNA resection at DSBs, impaired checkpoint activation, and increased sensitivity to DNA damage, suggesting that the phosphorylation of ATM at these sites plays an important role in regulation of DNA damage response. The observation that alanine mutations at T1985/S1987/S1988 and T86/T373 block the inhibitory effect of DNA-PKcs on ATM kinase activity in vitro and in human cells strongly suggests that DNA-PKcs regulates all of these cellular responses to DNA damage indirectly through its control of ATM.
Previous studies have shown that several phosphatases associate with ATM: PP2A and Wip1 prior to DNA damage and PP5 after DNA damage (Ali et al., 2004; Goodarzi et al., 2004; Shreeram et al., 2006). It was proposed that, in resting cells, ATM kinase activity is inhibited by serine/threonine phosphorylation of certain sites, while PP5 phosphatase binds to ATM and removes these inhibitory phosphates from ATM upon DNA damage, leading to ATM activation (Ali et al., 2004). There is also evidence showing that some ATM phosphorylation sites are dephosphorylated in response to DNA damage (Kozlov et al., 2003). Therefore, it is possible that some of the sites identified in this study are constitutively phosphorylated in undamaged cells, but dephosphorylated by PP5 or other phosphatases in order to activate ATM in response to DNA damage. By developing phospho-specific antibodies, we showed that ATM phosphorylation at T86 peaks at 2–4 h after DNA damage, which correlates well with the decrease of ATM activity, suggesting that T86 phosphorylation may serve as a critical mechanism to inactivate ATM after DSB repair.
A requirement for DNA-PKcs regulation of ATM during normal cell growth
Unlike the T1985E/S1987D/S1988D allele of ATM, expression of the T1985A/S1987A/S1988A allele produces normal ATM activation in vitro and WT levels of activity in response to DSBs and oxidative stress in human cells. These observations show that the regulation of ATM by DNA-PKcs is not essential for the activation of ATM through either pathway. However, we also found that the phospho-blocking allele induced cell death through apoptosis at a significantly higher rate than with the phospho-mimetic allele or the WT allele. The simplest interpretation of this result is that spontaneous activation of ATM occurs under conditions of normal growth, and that the inhibitory effect of DNA-PKcs is necessary to restrain this activation. In addition to being activated by DSBs, both ATM and DNA-PKcs have been shown to be activated in mitosis (Lee et al., 2011; Yang et al., 2011). Therefore, the regulatory effect of DNA-PKcs on ATM activity might be critical for the DNA damage response as well as for normal cell growth. From this point of view, it is perhaps significant that no patients with deletions or complete loss of DNA-PKcs have ever been identified, although patients with missense mutations and varying levels of DNA-PKcs protein have been identified with immunodeficiency and in some cases profound neurological impairment (Mathieu et al., 2015; van der Burg et al., 2009; Woodbine et al., 2013). It is possible that the dysregulation of ATM, which would occur in the absence of DNA-PKcs, is responsible for the severe phenotype in humans.
In summary, our study shows that DNA-PKcs, a key NHEJ factor, negatively regulates ATM activity upon DNA damage through phosphorylation of ATM. This function of DNA-PKcs is not essential for the basic mechanisms of ATM activation but plays important roles in regulating ATM under physiological conditions. Considering the critical role of ATM in HR, this study also provides an important mechanism for DNA repair pathway choice upon DNA damage as well as for ATM inactivation after DNA repair.
EXPERIMENTAL PROCEDURES
See also Supplemental Experimental Procedures for details about mutagenesis, cell culture, protein purification, protein-protein interaction assay, reagents, resection, western blotting, clonogenic assays, and immunofluorescence
In Vitro ATM Kinase Assay
To examine the activation of purified recombinant ATM protein by MRN and DNA, ATM protein(0.05 nM to 0.2 nM) was incubated at 37°C for 1 h in reaction buffer containing 25 mM MOPS, 1 mM DTT, 5 mM MgCl2, 1 mM ATP, 70 mM NaCl, 50 μg/ml BSA and 1 ng 4.4 kb linearized DNA substrate (37 pM) in the presence of 20 nM MRN and 100 nM GST-p53 substrate. To examine the activation of ATM by H2O2, ATM protein was incubated with 817 μM H2O2 at 37°C for 1 h in reaction buffer except without additional DTT. For the two-step kinase assay, the first incubation was performed at 37 °C for 45 min, followed by a 1 hr second incubation at 37°C. 3.5 nM DNA-PKcs, 7 nM Ku and 200 μM DNA-PK inhibitor NU7026 (DNA-PKi) were used to examine the effect of Ku/DNA-PKcs on ATM activity in these reactions. The reaction mixture was mixed with 5X SDS loading buffer and boiled for 5 min. The reaction products were detected by Western blotting using phospho-p53 (S15) antibody (see Supp. Exp. Procedures) and imaged using the Licor Odyssey system.
Intra-S-phase Checkpoint Assay
U2OS cells (~50% confluency) were treated with 2 μg/ml aphidicolin overnight in order to be synchronized in G1/early S phase. DNA damage was induced with 1 μM CPT for 1 h or with mock treatment. After treatment, cells were incubated with 400 ng/ml nocodazole for 17 h, followed by fixation with 75% cold ethanol for > 24 h. Cells were washed with PBS twice and stained with 40 μg/ml propidium iodide solution at 4 °C overnight. The intra-S-phase checkpoint efficiency was determined by calculating the ratio between the percentage of cells in G2/M phase with CPT treatment and the percentage of cells in G2/M phase without CPT treatment.
Supplementary Material
Table 1 related to Figures 4 to 7.
Summary of all ATM mutants regarding kinase activity and resistance to DNA-PKcs
| ATM mutants$ | Kinase activity (MRN/DNA)* | Activity repressed by DNA-PKcs?# | |
|---|---|---|---|
| 1 |
|
= | Yes |
| 2 | S85A/T372A/S1885A | = | Yes |
| 3 | S85D/T372E/S1885D | ↓↓↓ | |
| 4 | S85A | = | Yes |
| 5 | S85D | ↓↓ | |
| 6 | T372A | = | Yes |
| 7 | T372E | ↓ | |
| 8 | S85A/T372A | = | Yes |
| 9 | S85D/T372A | ↓↓↓ | |
| 10 | S1885A | = | Yes |
| 11 | S1885D | = | |
| 12 | T86A | = | Yes |
| 13 | T86E | ↓↓ | |
| 14 | T373A | = | Yes |
| 15 | T373E | ↓ | |
| 16 | T86A/T373A | = | No |
| 17 |
|
↓↓↓ | |
| 18 | S85A/T86A | = | Yes |
| 19 |
|
↓↓↓↓ | |
| 20 |
|
↓↓ | Yes |
| 21 | S85D/T86A | ↓↓ | Yes |
| 22 | T372A/T373A | = | Yes |
| 23 |
|
↓↓ | |
| 24 | S474A/S475A | = | Yes |
| 25 | S475A/S2592A | = | Yes |
| 26 |
|
= | |
| 27 | T86A/T373A/S475A/S2592A | = | No |
| 28 |
|
↓↓↓ | |
| 29 | S440A | = | Yes |
| 30 | S440D | = | |
| 31 | S200D | = | |
| 32 | T297E | = | |
| 33 | S1878D | = | |
| 34 | S85A/T86A/S440A | = | Yes |
| 35 |
|
↓↓↓ | |
| 36 | T1985E | ↓ | |
| 37 | S1987D | ↓ | |
| 38 | S1988D | ↓ | |
| 39 | T1985A/S1987A/S1988A | = | No |
| 40 | T1985E/S1987D/S1988D | ↓↓↓ |
The kinase activity of each ATM mutants in the presence of MRN/DNA was tested in comparison to the activity of wild-type (WT) ATM. “=” activity similar to WT; “↓” decreased activity compared with WT.
The effect of DNA-PKcs on the kinase activity of ATM mutants is tested in vitro using recombinant proteins and summarized here as “Yes” for repression and “No” for no detectable inhibitory effect.
Phospho-mimetic mutations at SQ/TQ sites are highlighted in red.
Acknowledgments
Work in the Paull laboratory was supported by the Cancer Research and Prevention Institute of Texas (RP110465-P4). Work in the Zha laboratory was supported by the National Institutes of Health (NCI R01CA184187). We are grateful to Blerta Xhelmace, Eric Hendrickson, Gaelle Legube and Susan Lees-Miller for reagents.
Footnotes
Author Contributions
Y.Z. designed and performed experiments, analyzed results, and wrote the manuscript. J.-H.L. designed and performed experiments. W.J. and J.L.C carried out experiments using mouse ES cells and analyzed results. S.Z designed experiments and contributed to the editing of the manuscript. T.T.P. analyzed results and wrote the manuscript.
References
- Ali A, Zhang J, Bao S, Liu I, Otterness D, Dean NM, Abraham RT, Wang XF. Requirement of protein phosphatase 5 in DNA-damage-induced ATM activation. Genes Dev. 2004;18:249–254. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bowen C, Ju JH, Lee JH, Paull TT, Gelmann EP. Functional activation of ATM by the prostate cancer suppressor NKX3.1. Cell Rep. 2013;4:516–529. doi: 10.1016/j.celrep.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron P, Choudjaye J, Clouaire T, Bugler B, Daburon V, Aguirrebengoa M, Mangeat T, Iacovoni JS, Alvarez-Quilon A, Cortes-Ledesma F, et al. Non-redundant Functions of ATM and DNA-PKcs in Response to DNA Double-Strand Breaks. Cell Rep. 2015;13:1598–1609. doi: 10.1016/j.celrep.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Pellegrini M, Lee JH, Paull TT, Feigenbaum L, Nussenzweig A. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J Cell Biol. 2008;183:777–783. doi: 10.1083/jcb.200805154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzel A, Grybowski A, Strasen J, Cristiano E, Loewer A. Hyperactivation of ATM upon DNA-PKcs inhibition modulates p53 dynamics and cell fate in response to DNA damage. Mol Biol Cell. 2016;27:2360–2367. doi: 10.1091/mbc.E16-01-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Jonnalagadda JC, Douglas P, Young D, Ye R, Moorhead GB, Lees-Miller SP, Khanna KK. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- Hartlerode AJ, Morgan MJ, Wu Y, Buis J, Ferguson DO. Recruitment and activation of the ATM kinase in the absence of DNA-damage sensors. Nat Struct Mol Biol. 2015;22:736–743. doi: 10.1038/nsmb.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jette N, Lees-Miller SP. The DNA-dependent protein kinase: A multifunctional protein kinase with roles in DNA double strand break repair and mitosis. Prog Biophys Mol Biol. 2015;117:194–205. doi: 10.1016/j.pbiomolbio.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanu N, Behrens A. ATMIN defines an NBS1-independent pathway of ATM signalling. EMBO J. 2007;26:2933–2941. doi: 10.1038/sj.emboj.7601733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama M, Otsubo C, Hirota T, Yokota J, Enari M, Taya Y. Requirement of ATM for rapid p53 phosphorylation at Ser46 without Ser/Thr-Gln sequences. Mol Cell Biol. 2010;30:1620–1633. doi: 10.1128/MCB.00810-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov S, Gueven N, Keating K, Ramsay J, Lavin MF. ATP activates ataxia-telangiectasia mutated (ATM) in vitro. Importance of autophosphorylation. J Biol Chem. 2003;278:9309–9317. doi: 10.1074/jbc.m300003200. [DOI] [PubMed] [Google Scholar]
- Kozlov SV, Graham ME, Jakob B, Tobias F, Kijas AW, Tanuji M, Chen P, Robinson PJ, Taucher-Scholz G, Suzuki K, et al. Autophosphorylation and ATM activation: additional sites add to the complexity. J Biol Chem. 2011;286:9107–9119. doi: 10.1074/jbc.M110.204065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov SV, Graham ME, Peng C, Chen P, Robinson PJ, Lavin MF. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006;25:3504–3514. doi: 10.1038/sj.emboj.7601231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Lan L, Peng G, Chang WC, Hsu MC, Wang YN, Cheng CC, Wei L, Nakajima S, Chang SS, et al. Tyrosine 370 phosphorylation of ATM positively regulates DNA damage response. Cell Res. 2015;25:225–236. doi: 10.1038/cr.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Mand MR, Deshpande RA, Kinoshita E, Yang SH, Wyman C, Paull TT. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J Biol Chem. 2013;288:12840–12851. doi: 10.1074/jbc.M113.460378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Lin YF, Chou HY, Yajima H, Fattah KR, Lee SC, Chen BP. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem. 2011;286:12796–12802. doi: 10.1074/jbc.M110.212969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, et al. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell. 2014;54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu AL, Verronese E, Rice GI, Fouyssac F, Bertrand Y, Picard C, Chansel M, Walter JE, Notarangelo LD, Butte MJ, et al. PRKDC mutations associated with immunodeficiency, granuloma, and autoimmune regulator-dependent autoimmunity. J Allergy Clin Immunol. 2015;135:1578–1588. e1575. doi: 10.1016/j.jaci.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair (Amst) 2010;9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem. 2015;84:711–738. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature. 2006;443:222–225. doi: 10.1038/nature05112. [DOI] [PubMed] [Google Scholar]
- Peng Y, Woods RG, Beamish H, Ye R, Lees-Miller SP, Lavin MF, Bedford JS. Deficiency in the catalytic subunit of DNA-dependent protein kinase causes down-regulation of ATM. Cancer Res. 2005;65:1670–1677. doi: 10.1158/0008-5472.CAN-04-3451. [DOI] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Richard DJ, Bolderson E, Cubeddu L, Wadsworth RI, Savage K, Sharma GG, Nicolette ML, Tsvetanov S, McIlwraith MJ, Pandita RK, et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature. 2008;453:677–681. doi: 10.1038/nature06883. [DOI] [PubMed] [Google Scholar]
- Sharma K, D’Souza RC, Tyanova S, Schaab C, Wisniewski JR, Cox J, Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell. 2006;23:757–764. doi: 10.1016/j.molcel.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Tian B, Yang Q, Mao Z. Phosphorylation of ATM by Cdk5 mediates DNA damage signalling and regulates neuronal death. Nat Cell Biol. 2009;11:211–218. doi: 10.1038/ncb1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Burg M, Ijspeert H, Verkaik NS, Turul T, Wiegant WW, Morotomi-Yano K, Mari PO, Tezcan I, Chen DJ, Zdzienicka MZ, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest. 2009;119:91–98. doi: 10.1172/JCI37141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viniegra JG, Martinez N, Modirassari P, Hernandez Losa J, Parada Cobo C, Sanchez-Arevalo Lobo VJ, Aceves Luquero CI, Alvarez-Vallina L, Ramon y Cajal S, Rojas JM, et al. Full activation of PKB/Akt in response to insulin or ionizing radiation is mediated through ATM. J Biol Chem. 2005;280:4029–4036. doi: 10.1074/jbc.M410344200. [DOI] [PubMed] [Google Scholar]
- Woodbine L, Neal JA, Sasi NK, Shimada M, Deem K, Coleman H, Dobyns WB, Ogi T, Meek K, Davies EG, et al. PRKDC mutations in a SCID patient with profound neurological abnormalities. J Clin Invest. 2013;123:2969–2980. doi: 10.1172/JCI67349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Tang X, Guo X, Niikura Y, Kitagawa K, Cui K, Wong ST, Fu L, Xu B. Aurora-B mediated ATM serine 1403 phosphorylation is required for mitotic ATM activation and the spindle checkpoint. Mol Cell. 2011;44:597–608. doi: 10.1016/j.molcel.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Paull TT. DNA-dependent protein kinase regulates DNA end resection in concert with Mre11-Rad50-Nbs1 (MRN) and ataxia telangiectasia-mutated (ATM) J Biol Chem. 2013;288:37112–37125. doi: 10.1074/jbc.M113.514398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.