Abstract
GOLPH3 is the first example of a Golgi resident oncogene protein. It was independently identified in multiple screens; first in proteomic-based screens as a resident protein of the Golgi apparatus, and second as an oncogene product in a screen for genes amplified in cancer. A third screen uncovered the association of GOLPH3 with the Golgi resident phospholipid, phosphatidyl inositol 4 phosphate (PI4P) to maintain the characteristic ribbon structure of the Golgi apparatus favoring vesicular transport of secretory proteins.
The organization of the Golgi apparatus is unique to each cell type of every organ in the body. In many cell types it consists of stacks of cisternae connected by membranous tubules and defined as the Golgi ribbon (1). Although somehow linked to its central role in coordinating the process of protein secretion, a mechanistic rationale for the basic structure of the Golgi apparatus has eluded cell biologists (2). Proteomics has provided an unprecedented window into the protein makeup of the Golgi apparatus that is slowly resolving the puzzle of its structure and functions (3–5).
Early efforts that characterized abundant proteins of isolated Golgi fractions by mass spectrometry (6) led to the discovery of a previously unreported protein named GPP341 for Golgi Peripheral Protein of 34 kDa. Renamed GOLPH3 (GOLgi PHosphoprotein3), studies of its function have provided mechanistic insight into Golgi apparatus structure and cell homeostasis that may be informative for strategies to unravel the functions of the thousands of proteins discovered by -omics technologies (7).
Isolation of the Golgi: A Key Step to Characterize GOLPH3 (Formerly GPP34)
GOLPH3 was first characterized in a proteomics screen of highly enriched Golgi fractions (6). Detailed electron microscope morphometry of liver parenchymal cells in situ, had revealed that the Golgi accounted for only 1% of the volume of the hepatocyte (8). Therefore, isolation of the Golgi apparatus from rat liver homogenates (Fig. 1) was necessary to assure the characterization of Golgi resident proteins given the low dynamic range and low sensitivity of mass spectrometry at that time. To enrich further for Golgi membrane proteins, phase partitioning of the isolated Golgi fraction with the detergent Triton X-114 was performed to select for membrane proteins (9), then followed by protein separation through SDS-PAGE (6).
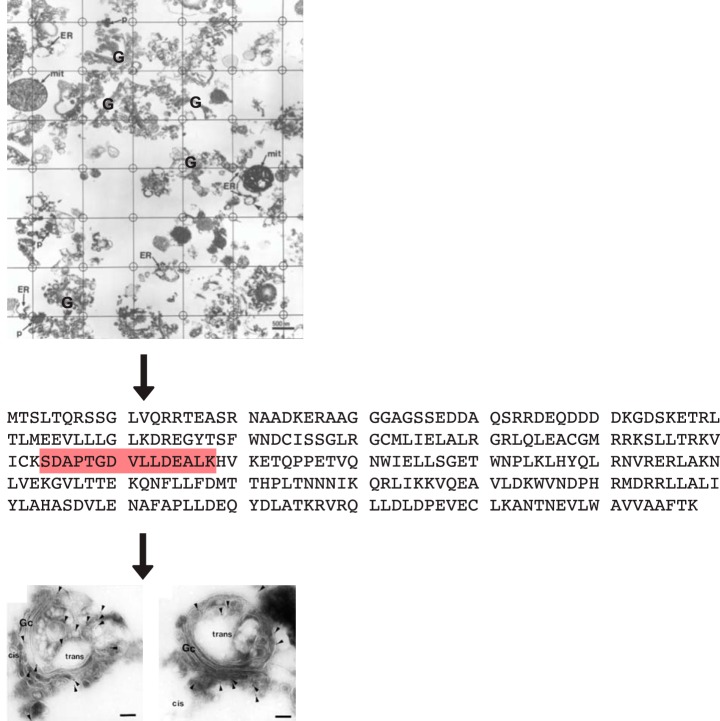
Fig. 1.
Characterization and localization of GOLPH3. A, Morphometric grid used for quantification of Golgi apparatus in isolated Golgi fractions of the liver as performed by Bell et al. (6). Most profiles from n = 3 Golgi fractions (84%) are of stacked Golgi cisternae (e.g. G) with endoplasmic reticulum (ER), mitochondrial (mit), and peroxisomal (p) contaminants indicated. B, Primary sequence of human GOLPH3 with the tryptic peptide characterized by tandem mass spectrometry. C, EM of cryosections of isolated Golgi labeled with gold decorated antibodies to monospecific anti-GOLPH3. Gold particles (indicated by arrowheads) are at the periphery of stacked Golgi cisternae. The cis and trans side of the stacked Golgi cisternae are indicated. Modified from (6).
Eighty-one proteins were characterized by mass spectrometry (MALDI and nanoelectrospray MS/MS) complemented by N-terminal sequencing (Edman degradation); of these 24 were contaminants. As expected, most of the 57 genuine Golgi proteins revealed primary sequences with transmembrane domains or known lipid anchors. However, one protein that was named GPP34 (GOLPH3) had not been characterized previously. By both immunofluorescence and analytical subcellular fractionation, GOLPH3 was localized to the Golgi apparatus and cytosol. By cryo-immuno electron microscopy, gold-labeled antibodies revealed GOLPH3 localization at the periphery of isolated stacked Golgi cisternae (Fig. 1) (6).
The complete protein sequence of GOLPH3 was inferred from database searches (6). Initially, a perfect match of the sequence of one tryptic fragment was found in the human EST database corresponding to a previously unidentified protein that was identified as GOLPH3 (Fig. 1). The complete sequence of GOLPH3 corresponded to 2 human gene products, 2 mouse gene products and one gene product in D. melanogaster fruit flies, the worm C. elegans, and the budding yeast S. cerevisiae. There were no motifs or transmembrane domains to explain the localization of GOLPH3 to the Golgi or its partitioning into the detergent phase of Triton X-114.
Wu et al. coincidentally discovered a protein they named GMX33 (10) as a Golgi matrix protein (hence GMX) of identical sequence to GOLPH3. GMX33 was also found to partition into Triton-X-114 and further characterized as a phosphoprotein. This was deduced from multiple spots for GMX33 on 2D gels that resolved into one after alkaline phosphatase treatment (10).
One of the 2 gene products for GOLPH3 in mice and humans corresponded to a related sequence but different gene product (6, 10) named GOLPH3L. In human cells, GOLPH3L reveals 68% identity and 78% similarity in primary sequence to GOLPH3. Furthermore expression of GOLPH3L is restricted to a subset of secretory cell types with a function deduced to be antagonistic to GOLPH3 in these cell types (e.g. (11)). This review will focus on GOLPH3.
Yeast Genetics Provides Insight: A Role in Mannosylation and Golgi Localization of Mannosyl Transferases
Both Bell et al. (6) and Wu et al. (10) found a similarity between the primary sequence of mammalian GOLPH3 and the predicted yeast protein Vps74p. Budding yeast are a powerful model system to solve the mechanisms of proteins linked to the secretory pathway (12). Through yeast genetics, the first functions of GOLPH3 were deduced.
In 2008, Schmitz et al. (13) concluded that the VPS74 gene was required for correct mannosylation of N-linked and O-linked mannoproteins in the yeast Golgi. Mannosylation takes place in the yeast Golgi by mannosyltransferases that span the membrane with their N termini facing the cytosol and their C termini facing the lumen of the Golgi cisterna. Vps74p was observed to associate with the cytosolically exposed N-terminal amino acids of the mannosyl transferases. This direct interaction between Vps74p and the mannosyl transferase cytosolic domains was postulated to guide the mannosyl transferases to their correct yeast Golgi cisterna. In this way, the sequential, ordered, terminal glycosylation of newly synthesized secretory glycoproteins (14) would occur at the right place and right time in the cell during glycoprotein maturation. When the VPS74 gene was deleted a subset of Golgi resident mannosyl transferases was mislocalized leading to improper protein glycosylation.
Tu et al. (15, 16) then deduced that Vps74p monitored the localization of Golgi mannosyl transferases by regulating their retrograde trafficking through COPI coated vesicles. COPI coated vesicles were generated at the periphery of a Golgi cisterna where they captured the mannosyl tranferases. The COPI coated vesicle would then bud off and travel in a retrograde direction to the “earlier” more cis Golgi cisterna to deliver the mannosyl transferase (Golgi of budding yeast are unstacked but individual Golgi cisternae maintain a temporal relationship of “early” and “late” cisternae based on the transport of newly synthesized secretory cargo). Golgi cisternae with enclosed newly synthesized secretory mannoproteins would move in an anterograde direction to form “later” Golgi cisternae in this process of Golgi maturation (17–19).
Importantly, expression of the human orthologue, GOLPH3, of the yeast protein Vps74p, complemented the aberrant phenotypes of yeast deleted for the VPS74 gene. This was a rare occurrence of a human protein complementing a deleted yeast gene.
A Proteomics Screen and X-ray Crystallography Reveals GOLPH3 as a PI4P Binding Protein
Dippold et al. (20) tested the in vitro translation products of 4,000 different cDNAs from D. melanogaster for their ability to bind to a panel of phospholipids including multiple phosphatidyl inositols (PI). Drosophila GOLPH3 bound specifically to PI4P (phosphatidyl inositol 4 phosphate). It was also confirmed that the association of Vps74p and GOLPH3 with PI4P was specific (see (21)). PI4P is a resident phospholipid embedded in the cytosolic leaflet of the Golgi membrane phospholipid bilayer with GOLPH3 localization and function a consequence of Golgi localized PI4P levels (22).
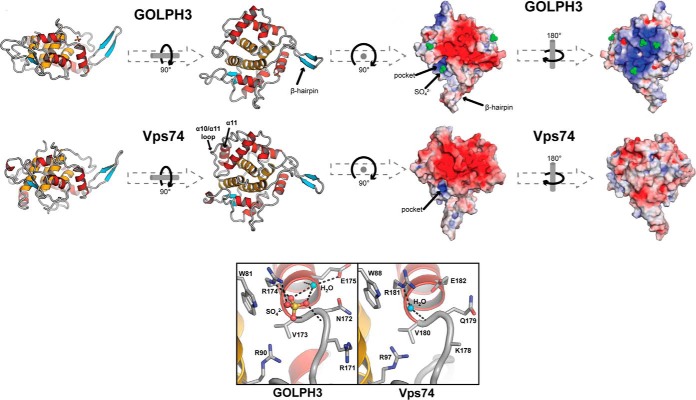
A breakthrough revealed a common Golgi recognition mechanism for yeast Vps74p and GOLPH3 (13, 21). Using X-ray crystallography, a common structure was observed (Fig. 2). A conserved sulfate-binding pocket was deduced as the site of association with the Golgi phospholipid, PI4P (phosphatidyl inositol 4- phosphate) (13, 20, 21). Later studies revealed a further association of Vps74p with the PI4P lipid phosphatase Sac1p to regulate Golgi PI4P levels (22). It is this interaction of Vps74p with Sac1p that was deduced to underlie the correct sorting of mannosyl transferases to Golgi cisternae.
Fig. 2.
Homology of GOLPH3 structure with Vps74p showing the lipid-binding pocket of each (in lower boxes). Taken from (21) with permission from the Journal of Cell Biology. (©2009 Woods et al. Journal of Cell Biology. 187:967–75. doi: 10.1083/jcb.200909063.)
Large-scale Screens Identify GOLPH3 as an Oncogene Product
A new paradigm emerged when Scott et al. (23) used an unbiased method to screen for gene amplification in solid tumors. Using genomic microarrays, 83 different human melanoma specimens were screened, as well as nonsmall cell lung cancer and colon adenocarcinomas. It was found that amplification occurred within chromosome 5p13 that harbored the GOLPH3 gene. It was further demonstrated that GOLPH3 corresponded to an oncogene, for growth of cells on soft agar, proliferation of nonsmall cell lung cancer cells, and larger melanoma tumors in nude mice. Gene amplified GOLPH3 has been repeatedly observed in several human cancers (24, 25).
GOLPH3 Regulation of the Golgi Ribbon: DNA Protein Kinase Signals DNA Damage to the Golgi Apparatus Through GOLPH3 Phosphorylation
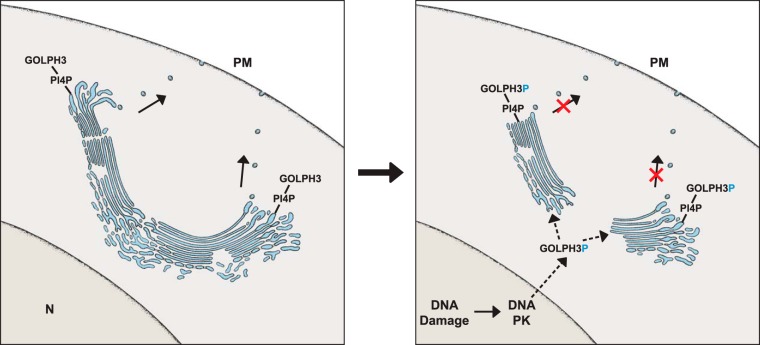
Knockdown of GOLPH3 altered the structure of the Golgi ribbon, a name used to define the single continuous nature of the Golgi apparatus (27), which in mammals is attained by tubules that interconnect multiple adjacent stacked Golgi cisternae (Fig. 3, left side). GOLPH3 knockdown caused scission of the interconnecting tubules of the Golgi ribbon leading to the separation of the individual stacks of Golgi cisternae. Vesicular transport emanating from these disconnected stacks of Golgi cisternae was reduced (20).
Fig. 3.
Left side, Illustration of a Golgi ribbon with PI4P and GOLPH3 during normal secretion. The Golgi is formed of adjacent stacks of cisternae whose edges are connected by tubules. Right side, DNA damage induced activation of DNA PK, the proposed phosphorylation of GOLPH3, the scission of the Golgi ribbon at the level of the interconnecting tubules resulting in a reduction of secretion.
A further breakthrough occurred in 2014 when Farber-Katz et al. (26) documented two sites of phosphorylation on human GOLPH3 by the DNA damage protein kinase, DNA PK. GOLPH3 phosphorylation by DNA PK was observed to result in the same phenotype as GOLPH3 knockdown (illustrated in Fig. 3, right side (26)).
DNA PK localizes to cell nuclei for DNA double strand repair after DNA damage (27). It is likely the cytosolic pool of GOLPH3 that accesses cell nuclei to become phosphorylated. The phosphoGOLPH3 would then be transported from the cell nuclei to the cytosol then the Golgi ribbon (Fig. 3 right side). Increased levels of phosphoGOLPH3 coincides remarkably with the protection of cells suffering DNA damage from cell death (26). In this way, GOLPH3 senses DNA damage through DNA PK, to coordinate Golgi structure with vesicular transport from the Golgi to the plasma membrane, that somehow protects from cell death caused by DNA damage that frequently occurs in cancer (26).
A further link between GOLPH3 and cancer (Halberg et al. (27)) is through PITPNC1. The PITPNC1 gene is amplified in several metastatic breast, melanoma and colon cancers. Like GOLPH3, PITPNC1 (PhosphoInositide Transfer ProteiN, Cytoplasmic 1) associates with PI4P. However, when expressed during gene amplification, the higher levels of PITPNC1 also result in increased RAB1 recruitment to the Golgi ribbon. Increased RAB1 levels increase PI4P in the Golgi, and an increased recruitment of GOLPH3 to the Golgi from the cytosol.
However, this affects the morphology of the Golgi ribbon in a different way than GOLPH3 knockdown. The ribbon is extended and there is no severance of the interconnecting tubules. Because the Golgi ribbon is not severed but elongates in size, increased vesicular transport of secretory cargo from the Golgi stacked cisternae is observed (27).
Five proteins in metastatic cells were discovered whose secretion was increased in this way, all of which were shown to enhance metastasis and angiogenesis (27). The 5 proteins FAM3C (Family with sequence similarity 3), MMP1 (Matrix Metallopeptidase 1), HTRA1 (High Temperature Requirement A1 serine protease), PDGFA (Platelet derived growth factor subunit A), and ADAM10 (A Disintegrin And Metalloprotease 10) were required for the movement of cancer cells through the extracellular matrix for metastasis, and for endothelial cells to vascularize the tumors (summarized in Table I).
Table I. Inhibition of metastasis (Matrigel), endothelial recruitment (Endothelial) and lung colonization (Lung) after knockdown of FAM3C(ILEI), MMP1, HTRA1, PDGFA, or ADAM10 in PITPNC1 overexpressing cells (summarized from ref 28).
| Name | Matrigel | Endothelial | Lung |
|---|---|---|---|
| FAM3C (ILEI) | + | + | + |
| MMP1 | + | + | + |
| HTRA1 | + | + | + |
| PDGFA | + | + | + |
| ADAM10 | + | + | + |
The work of Halberg et al. (27) reveals an opposite phenotype to that of Farber-Katz et al. (26) (the latter is shown in Fig. 3). These two opposite effects on the Golgi ribbon, by amplification of the PITPNC1 gene (Golgi ribbon elongation and increased secretion) in some cancer cells and amplification of the GOLPH3 gene (disconnected Golgi stacks and decreased secretion) in others, are each linked to cancer. The former may enhance metastasis with the latter protecting from DNA damage and cell death (28).
Other reasons for the oncogenic properties of gene amplified GOLPH3 have been proposed (29), including the trafficking of overexpressed GOLPH3 to endosomes and signaling through mTOR (23). It has also been proposed that GOLPH3 gene amplification in cancer leads to aberrant sorting of Golgi glycosyl transferases (29). The speculation is that this scenario would affect the extent of glycosylation of newly synthesized protein cargo transiting through the Golgi before vesicular transport to the cell surface. Somehow, the altered modification of secretory proteins under this condition would be linked to cancer.
Differential Expression of GOLPH3 in Cell Differentiation
GOLPH3 expression during cell differentiation (spermatogenesis) (4) is especially prominent when early spermatids form the acrosome (step 1–7), the major secretory product of male germ cells. It is during acrosome formation that the Golgi ribbon is at its most elaborate. A burst of expression is also observed at the last step (step 19) of spermatid development, before sperm release from the testis. There is no Golgi ribbon in step 19 spermatids; only individual unstacked Golgi cisternae are observed (4) (similar to Golgi apparatus structure in budding yeast (30)). Although the high expression of GOLPH3 during acrosome formation is expected (Fig. 3, left side), the function of GOLPH3 in step 19 spermatids is as yet unknown.
While studying spermatocytes in Drosophila melanogaster, Sechi et al. (31) uncovered a new function for GOLPH3. They showed GOLPH3 localization to the Golgi and the cleavage furrow of dividing meiotic spermatocytes. It was proposed that PI4P linked to GOLPH3 and myosin II created a tensile action at the cleavage furrow for actomyosin mediated cleavage, furrow constriction, and cytokinesis at telophase. These studies have been extended (32) to show in spermatocytes the requirement of the GTPase RAB1B for GOLPH3 recruitment to Golgi apparatus (see as well (27)) and the cleavage furrow during cytokinesis (32). Speculatively, this new function of GOLPH3 may be related to the requirement of spermatids to cleave the intercellular bridges that interconnect the step 19 spermatids. Abscission of these bridges is necessary for sperm to be released as individual spermatozoa (4).
Another function for GOLPH3 has been documented in regulating the reorganization of Golgi apparatus to the leading edge of migrating cells by affecting membrane trafficking and secretion (33).
Clearly the discoveries (Table II) regarding this protein continue to expand its versatility. The link between the Golgi structure and function has focused on the role of the Golgi ribbon in mammalian cells. The tensile forces acting on the Golgi to create the Golgi ribbon are now under scrutiny (34). Exactly how GOLPH3 affects tensile forces acting on the Golgi ribbon may resolve several of the questions posed by cell biologists still puzzled by the Golgi apparatus structure (2).
Table II. Functions of GOLPH3. Selected highlights of functions deduced for GOLPH3.
| Function | Reference |
|---|---|
| Required for correct Golgi localization of mannosyl transferases in budding yeast | (13, 15, 16, 22) |
| Associates with Golgi PI4P in budding yeast and mammalian cells | (20, 21) |
| Regulates structure of Golgi ribbon in mammalian cells - knockdown and overexpression of GOLPH3 | (20, 26, 27) |
| Gene amplified and shown to be an oncogene in several human cancers | (23–25) |
| Phosphorylated by DNA Protein kinase leading to breakdown of Golgi ribbon and protection from cell death consequent to DNA damage | (26) |
CONCLUSION
Through rigorous purification of Golgi liver fractions by two different groups, GOLPH3 was discovered through proteomics. Once established as a Golgi resident protein, multiple discoveries from independent unbiased screens dovetailed to complete our understanding of its significance in cell function and relevance to cancer.
Yeast genetic screens led to the elucidation of GOLPH3 involvement in localization of the Golgi mannosyl transferases by regulating their retrograde trafficking through COPI vesicles. Unbiased proteomics screens uncovered the link between GOLPH3 and the Golgi resident phospholipid PI4P. An independent large-scale screen for genes amplified in cancer uncovered GOLPH3 as a resident Golgi oncogene. The impetus to uncover mechanisms for GOLPH3 led to further links with DNA damage, cell death, and in mammalian cells, alterations in the structure of the Golgi ribbon.
Fortunately, the lack of any membrane localization domain or motif, or lack of any other domain to predict location and function, did not lead to the rejection of GOLPH3 as a potentially important protein. Indeed, the unpredicted pathway of the GOLPH3 discovery trail indicates that a good deal of time and effort may be needed following a proteomics discovery before its significance may be uncovered.
Acknowledgments
We thank Abe Fuks (McGill University), Peter McPherson (McGill University), Richard Rachubinski (University of Alberta) and Michel Desjardins (University of Montreal) for their helpful comments. We are grateful to Dr. Kathryn Ferguson (Yale), and Dr. Christopher Burd (Yale), as well as the Journal of Cell Biology for permission to use the published structures of GOLPH3 and Vps74p (©2009 Woods et al. Journal of Cell Biology. 187:967–75. doi: 10.1083/jcb.200909063.)
Footnotes
Author contributions: J.J.B., C.E.A., D.Y.T., and L.H. wrote the paper.
* This review is supported by Canadian Institutes of Health Research grant MOP 5605 (to J.J.M.B.) and is dedicated to the memory of Drs. Yves Clermont, a pioneer of Golgi apparatus morphology and the Golgi ribbon.
1 The abbreviations used are:
- GPP34
- golgi peripheral protein of 34kDa
- ADAM10
- A disintegrin and metalloprotease 10
- COPI
- coatomer protein complex I
- DNA-PK
- DNA protein kinase
- EST
- expressed sequence tags
- FAM3C
- family with sequence similarity 3
- GMX33
- golgi matrix protein of 33kDa
- GOLPH
- golgi phosphoprotein
- HTRA1
- high temperature requirement A1 serine protease
- MALDI
- matrix-assisted laser desorption/ionization
- MMP1
- matrix metallopeptidase 1
- mTOR
- mechanistic target of rapamycin
- PDGFA
- platelet derived growth factor subunit A
- PI4P
- phosphatidyl inositol 4- phosphate
- PITPNC1
- phosphoinositide transfer protein, cytoplasmic 1
- RAB
- Ras in the brain GTPase
- SAC1
- suppressor of the temperature-conditional act1–1 allele
- SDS PAGE
- sodium dodecyl sulphate polyacrylamide gel electrophoresis
- VPS
- vacuolar protein sorting.
REFERENCES
- 1. Rambourg A., Clermont Y., and Hermo L. (1979) Three-dimensional architecture of the golgi apparatus in Sertoli cells of the rat. Am. J. Anat. 154, 455–476 [DOI] [PubMed] [Google Scholar]
- 2. Emr S., Glick B. S., Linstedt A. D., Lippincott-Schwartz J., Luini A., Malhotra V., Marsh B. J., Nakano A., Pfeffer S. R., Rabouille C., Rothman J. E., Warren G., and Wieland F. T. (2009) Journeys through the Golgi–taking stock in a new era. J. Cell Biol. 187, 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilchrist A., Au C. E., Hiding J., Bell A. W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S. J., Hallett M., Paiement J., Kearney R. E., Nilsson T., and Bergeron J. J. (2006) Quantitative proteomics analysis of the secretory pathway. Cell 127, 1265–1281 [DOI] [PubMed] [Google Scholar]
- 4. Au C. E., Hermo L., Byrne E., Smirle J., Fazel A., Simon P. H., Kearney R. E., Cameron P. H., Smith C. E., Vali H., Fernandez-Rodriguez J., Ma K., Nilsson T., and Bergeron J. J. (2015) Expression, sorting, and segregation of Golgi proteins during germ cell differentiation in the testis. Mol. Biol. Cell 26, 4015–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Au C. E., Hermo L., Byrne E., Smirle J., Fazel A., Kearney R. E., Smith C. E., Vali H., Fernandez-Rodriguez J., Simon P. H., Mandato C., Nilsson T., and Bergeron J. J. (2015) Compartmentalization of membrane trafficking, glucose transport, glycolysis, actin, tubulin and the proteasome in the cytoplasmic droplet/Hermes body of epididymal sperm. Open Biol. 5, 150080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bell A. W., Ward M. A., Blackstock W. P., Freeman H. N., Choudhary J. S., Lewis A. P., Chotai D., Fazel A., Gushue J. N., Paiement J., Palcy S., Chevet E., Lafreniere-Roula M., Solari R., Thomas D. Y., Rowley A., and Bergeron J. J. (2001) Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 276, 5152–5165 [DOI] [PubMed] [Google Scholar]
- 7. Gaudet P., Michel P. A., Zahn-Zabal M., Britan A., Cusin I., Domagalski M., Duek P. D., Gateau A., Gleizes A., Hinard V., de Laval V. R., Lin J., Nikitin F., Schaeffer M., Teixeira D., Lane L., and Bairoch A. (2016) The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 45, D177–D182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blouin A., Bolender R. P., and Weibel E. R. (1977) Distribution of organelles and membranes between hepatocytes and nonhepatocytes in the rat liver parenchyma. A stereological study. J. Cell Biol. 72, 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bordier C. (1981) Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256, 1604–1607 [PubMed] [Google Scholar]
- 10. Wu C. C., Taylor R. S., Lane D. R., Ladinsky M. S., Weisz J. A., and Howell K. E. (2000) GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic 1, 963–975 [PubMed] [Google Scholar]
- 11. Ng M. M., Dippold H. C., Buschman M. D., Noakes C. J., and Field S. J. (2013) GOLPH3L antagonizes GOLPH3 to determine Golgi morphology. Mol. Biol. Cell 24, 796–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schekman R., and Sudhof T. (2014) An interview with Randy Schekman and Thomas Sudhof. Trends Cell Biol. 24, 6–8 [DOI] [PubMed] [Google Scholar]
- 13. Schmitz K. R., Liu J., Li S., Setty T. G., Wood C. S., Burd C. G., and Ferguson K. M. (2008) Golgi localization of glycosyltransferases requires a Vps74p oligomer. Developmental Cell 14, 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dean N. (1999) Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426, 309–322 [DOI] [PubMed] [Google Scholar]
- 15. Tu L., Tai W. C., Chen L., and Banfield D. K. (2008) Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science 321, 404–407 [DOI] [PubMed] [Google Scholar]
- 16. Tu L., Chen L., and Banfield D. K. (2012) A conserved N-terminal arginine-motif in GOLPH3-family proteins mediates binding to coatomer. Traffic 13, 1496–1507 [DOI] [PubMed] [Google Scholar]
- 17. Ishii M., Suda Y., Kurokawa K., and Nakano A. (2016) COPI is essential for Golgi cisternal maturation and dynamics. J. Cell Sci. 129, 3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Papanikou E., Day K. J., Austin J., and Glick B. S. (2015) COPI selectively drives maturation of the early Golgi. eLife 4, e13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Papanikou E., and Glick B. S. (2009) The yeast Golgi apparatus: insights and mysteries. FEBS Lett. 583, 3746–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dippold H. C., Ng M. M., Farber-Katz S. E., Lee S. K., Kerr M. L., Peterman M. C., Sim R., Wiharto P. A., Galbraith K. A., Madhavarapu S., Fuchs G. J., Meerloo T., Farquhar M. G., Zhou H., and Field S. J. (2009) GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell 139, 337–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wood C. S., Schmitz K. R., Bessman N. J., Setty T. G., Ferguson K. M., and Burd C. G. (2009) PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J. Cell Biol. 187, 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wood C. S., Hung C. S., Huoh Y. S., Mousley C. J., Stefan C. J., Bankaitis V., Ferguson K. M., and Burd C. G. (2012) Local control of phosphatidylinositol 4-phosphate signaling in the Golgi apparatus by Vps74 and Sac1 phosphoinositide phosphatase. Mol. Biol. Cell 23, 2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scott K. L., Kabbarah O., Liang M. C., Ivanova E., Anagnostou V., Wu J., Dhakal S., Wu M., Chen S., Feinberg T., Huang J., Saci A., Widlund H. R., Fisher D. E., Xiao Y., Rimm D. L., Protopopov A., Wong K. K., and Chin L. (2009) GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 459, 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buschman M. D., Rahajeng J., and Field S. J. (2015) GOLPH3 links the Golgi, DNA damage, and cancer. Cancer Res. 75, 624–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scott K. L., and Chin L. (2010) Signaling from the Golgi: mechanisms and models for Golgi phosphoprotein 3-mediated oncogenesis. Clin. Cancer Res. 16, 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farber-Katz S. E., Dippold H. C., Buschman M. D., Peterman M. C., Xing M., Noakes C. J., Tat J., Ng M. M., Rahajeng J., Cowan D. M., Fuchs G. J., Zhou H., and Field S. J. (2014) DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell 156, 413–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Halberg N., Sengelaub C. A., Navrazhina K., Molina H., Uryu K., and Tavazoie S. F. (2016) PITPNC1 recruits RAB1B to the golgi network to drive malignant secretion. Cancer Cell 29, 339–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Makowski S. L., Tran T. T. T., and Field S. J. (2017) Emerging themes of regulation at the Golgi. Current Opinion Cell Biol. 45, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rizzo R., Parashuraman S., D'Angelo G., and Luini A. (2016) GOLPH3 and oncogenesis: What is the molecular link? Tissue Cell 49, 170–174 [DOI] [PubMed] [Google Scholar]
- 30. Losev E., Reinke C. A., Jellen J., Strongin D. E., Bevis B. J., and Glick B. S. (2006) Golgi maturation visualized in living yeast. Nature 441, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 31. Sechi S., Colotti G., Belloni G., Mattei V., Frappaolo A., Raffa G. D., Fuller M. T., and Giansanti M. G. (2014) GOLPH3 is essential for contractile ring formation and Rab11 localization to the cleavage site during cytokinesis in Drosophila melanogaster. PLoS genetics 10, e1004305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sechi S., Frappaolo A., Fraschini R., Capalbo L., Gottardo M., Belloni G., Glover D. M., Wainman A., and Giansanti M. G. (2017) Rab1 interacts with GOLPH3 and controls Golgi structure and contractile ring constriction during cytokinesis in Drosophila melanogaster. Open Biol. 7, 160257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xing M., Peterman M. C., Davis R. L., Oegema K., Shiau A. K., and Field S. J. (2016) GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol. Biol. Cell 27, 3828–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bruun K., Beach J. R., Heissler S. M., Remmert K., Sellers J. R., and Hammer J. A. (2017) Re-evaluating the roles of myosin 18Aalpha and F-actin in determining Golgi morphology. Cytoskeleton 74, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]