Abstract
Viral DNA genomes replicating in cells encounter a myriad of host factors that facilitate or hinder viral replication. Viral proteins expressed early during infection modulate host factors interacting with viral genomes, recruiting proteins to promote viral replication, and limiting access to antiviral repressors. Although some host factors manipulated by viruses have been identified, we have limited knowledge of pathways exploited during infection and how these differ between viruses. To identify cellular processes manipulated during viral replication, we defined proteomes associated with viral genomes during infection with adenovirus, herpes simplex virus and vaccinia virus. We compared enrichment of host factors between virus proteomes and confirmed association with viral genomes and replication compartments. Using adenovirus as an illustrative example, we uncovered host factors deactivated by early viral proteins, and identified a subgroup of nucleolar proteins that aid virus replication. Our data sets provide valuable resources of virus-host interactions that affect proteins on viral genomes.
Viruses create conditions conducive to their own replication by shaping the cellular environment. Viral proteins achieve this by manipulating multiple cellular processes to promote progeny generation and to evade host defenses. Despite maximizing their coding capacity, viruses must rely on cellular proteins to fulfill functions required during infection. Active recruitment of host factors to viral genomes functions to promote viral gene expression, DNA replication and virion assembly, whereas preventing access to the viral genome by cellular sensors and repressors can help to counteract antiviral defenses. Consequently, the outcome of infection is governed by dynamic protein interactions taking place on viral genomes. Identifying host factors that associate with viral genomes during virus replication may provide insights into the virus-host interactions that are critical in regulating the infectious process.
Viral DNA genomes are diverse in size, structure and site of replication. Most eukaryotic DNA viruses, including adenovirus type 5 (Ad5)1 and herpes simplex virus type 1 (HSV-1), replicate their genomes in the cell nucleus where they can use host enzymes to express viral genes and replicate viral DNA. In contrast poxviruses, such as vaccinia virus (VACV), replicate in the cytoplasm of infected cells. The VACV genome is a linear double stranded (ds) DNA molecule ∼191 Kb, that encodes several proteins involved in viral replication and assembly (1). For instance, the VACV genome encodes at least nine proteins involved in viral DNA synthesis (2, 3), making VACV independent from host replication machinery and thus the cell nucleus. In contrast, the 36 Kb Ad5 linear dsDNA genome encodes only three proteins required for viral DNA replication (4), and HSV-1 has a 150 Kb dsDNA genome encoding seven proteins that participate in viral DNA synthesis (5). These differences in coding capacity and site of replication correlate with the degree of dependence on host machinery, the availability of host factors to be exploited, and the antiviral responses to be counteracted by each virus.
DNA viruses form compartmentalized structures within the host cell referred to as viral replication factories or compartments (VRCs) where they carry out viral processes fundamental to their life cycle (6–9). VRCs accumulate viral and cellular proteins that interact directly or indirectly with the viral genome to support viral DNA replication, late gene transcription and formation of packaged viral progeny (6). Some host factors recruited to VRCs are known to contribute to these viral processes. Cellular proteins have been identified by affinity purification with viral DNA and replication proteins, or by specific candidate-retrieval approaches (10–17). However, the proteomic landscape of cellular proteins associated with viral DNA replicating in VRCs to facilitate the viral life cycle remains largely unknown. In addition to factors recruited to VRCs, there are cellular proteins actively excluded from these structures. There is a temporal cascade of viral gene expression during the infectious lifecycle. The first proteins to be expressed are from immediate early and early viral genes, and they function to promote viral gene expression and mediate viral DNA replication. Viral proteins expressed early during the infectious process also frequently alter the cellular environment to promote viral replication by inactivating host factors and preventing their association with viral DNA in VRCs where they could limit gene transcription or replication of viral genomes. Late viral genes produce proteins that are structural or involved in packaging the viral genome into virion particles, as well as proteins that can counter host immune responses.
Defining proteomes associated with viral genomes replicating in VRCs provides an opportunity to uncover previously unknown cellular factors and pathways manipulated by DNA viruses to advance their life cycle. Recently, several proteomics approaches have been developed to identify proteins on replicating DNA (18–20). These approaches rely on labeling replicating DNA with deoxy-thymidine analogs including 5-ethynyl-2′-deoxyuridine (EdU). The EdU-labeled DNA is then biotinylated via click chemistry and purified along with bound proteins using streptavidin beads (21, 22). Replicating viral DNA can also be labeled using ethynyl-modified nucleosides (23). A recent study employed an elegant approach with a mutant HSV-1 virus to adapt the cellular proteomics technique to identify viral and cellular proteins interacting with genomes of HSV-1 replicated during infection (24). In our current study, we expand this approach to profile the proteomes associated with replicated DNA recovered from cells infected with multiple wild-type viruses. We employed the technique of Isolation of Proteins on Nascent DNA coupled with Mass Spectrometry (iPOND-MS) to define proteomes associated with newly synthesized DNA (18, 19, 21, 22, 25). By comparing proteomes associated with replicated DNA from cells uninfected or infected with Ad5, HSV-1 and VACV, we provide a comprehensive description of host factors and cellular processes potentially exploited by each virus during infection. Comparative gene ontology analysis revealed global similarities and differences in the manipulation of cellular processes between viruses and differentiates nuclear-replicating (Ad5 and HSV-1) from cytoplasmic-replicating viruses (VACV). To illustrate the power of our profiling strategy to identify host factors exploited or inactivated, we further analyzed relative protein abundance within the Ad5 proteome. We then combined relative abundance with the fold change between Ad5 and Host proteomes to reveal potential targets of early viral proteins. This analysis showed that i) TFII-I, a cellular transcription regulator, is inactivated by early viral proteins, ii) DNA repair proteins such as SLX4 can associate with viral replication centers and promote viral DNA replication, and iii) several nucleolar proteins, including TCOFI, are recruited to VRCs to promote viral replication. Overall, our unbiased proteomics approach generated comprehensive databases that represent new resources to probe virus-host interactions and a framework for understanding how viruses manipulate host factors to redirect cellular processes and promote viral replication.
EXPERIMENTAL PROCEDURES
Additional methodology is described in the supplemental Experimental Procedures.
Cell Culture
Experiments were performed using U2OS, A549, HFF and HeLa cells cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (v/v) Fetal Bovine Serum (Seradigm, Radnor, PA), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco, Waltham, MA). Human small airway epithelial cells (SAECs) were obtained from the American Tissue Culture Collection (ATCC, Manassas, VA) and grown according to the provider's instructions. U2OS cells that express E1B55K were described previously (26). Immortalized RA3331 FA-P cells (SLX4-deficient fibroblasts) and matched cells expressing wild-type SLX4 were cultured in DMEM supplemented with 15% FBS, 100 U/ml penicillin, 100 U/ml streptomycin, and non-essential amino acids (Thermo Fisher Scientific, Waltham, MA), as previously described (27).
Viruses and Infections
Infections were performed according to standard procedures using the following viruses: wild-type Ad5, HSV-1 strain 17syn+ and VACV strain WR. E4 mutant Ad viruses were described previously (28–30).
iPOND-MS
We adapted the iPOND protocol previously described (22) to include infection conditions. Per condition, eight 15 cm cell culture dishes containing U2OS at ∼90 confluence (∼1.8 × 107 cells) were mock-infected or infected with Ad5 (multiplicity of infection (MOI) 40), HSV-1 (MOI 3), or VACV (MOI 3–7). Cells were incubated with infection media (serum free DMEM plus antibiotics) for 1 h (HSV-1) or 2 h (Ad5 and VACV) at 37 °C. Following adsorption, cells were topped-off with fresh complete culture media (DMEM plus 10% FBS and antibiotics) incubated at 37 °C for 24 h (Ad5), 8 h (HSV-1) or 6 h (VACV) before pulsing with 10 μm EdU (Invitrogen, Waltham, MA) for 15 min at 37 °C. Cells were fixed with 1% paraformaldehyde in PBS for 20 min at room temperature, crosslinking was quenched with 125 mm glycine and cells were scraped and harvested. To facilitate subsequent iPOND steps, cells from four plates of the same condition were combined and pelleted together generating two cell pellets per condition. Samples were then processed for iPOND as described previously (22, 25), with the following adaptations: After click chemistry reaction, cell pellets were resuspended in 0.5 ml of Lysis buffer (20 mm HEPES pH 7.9, 400 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Triton X-100) supplemented with 1 mm DTT and protease inhibitors (Complete protease inhibitor mixture tablets (Roche) and 1 mm phenylmethylsulfonyl fluoride) and sonicated with a Bioruptor (Diagenode, Denville, NJ) for 20 min in 30 s on/off cycles at a high intensity. Capture of DNA-protein complexes was carried out by incubating lysates with streptavidin magnetic beads (Dynabeads M-280, Invitrogen) for 16–18 h at 4 °C. Beads were washed once in Lysis buffer, once in 1 m NaCl, four times in Wash buffer (20 mm HEPES pH7.4, 110 mm KOAc, 2 mm MgCl2, 0.1% Tween-20, 0.1% Triton-X-100,150 mm NaCl) and once in PBS. To combine samples from the same condition, proteins from one of the samples were eluted in 60 μl of 1X LDS sample buffer (Invitrogen) containing 10% DTT by incubating at 95 °C for 10 min. Then, the same 60 μl of sample buffer used to elute proteins from the first sample were used for the elution of sample two of the same condition. Finally, eluted proteins were boiled at 95 °C for 45 min to reverse crosslinks.
iPOND isolates were separated on ∼0.8 cm on a 10% Bis-Tris Novex mini-gel (Invitrogen) using the MOPS buffer system. The gel was stained with Coomassie and excised into four equal 2 × 7 mm segments. Gel segments were destained with 50% methanol/1.25% acetic acid, reduced with 5 mm DTT (Thermo Fisher Scientific), and alkylated with 40 mm iodoacetamide (Sigma-Aldrich, St. Louis, MO). Gel pieces were then washed with 20 mm ammonium bicarbonate (Sigma) and dehydrated with acetonitrile (Thermo Fisher Scientific). Trypsin (Promega, Madison, WI) (5 ng/μl in 20 mm ammonium bicarbonate) was added to the gel pieces and proteolysis was allowed to proceed overnight at 37 °C. Peptides were extracted with 0.3% triflouroacetic acid (J.T.Baker, Phillipsburg, NJ), followed by 50% acetonitrile. Extracts were combined and the volume was reduced by vacuum centrifugation. Tryptic digests were analyzed by LC-MS/MS on a hybrid LTQ Orbitrap Elite mass spectrometer (Thermo) coupled with a nanoLC Ultra (Eksigent Technologies, Dublin, CA). Peptides were separated by reverse phase (RP)-HPLC on a nanocapillary column, 75 μm id x 15 cm Reprosil-pur 3 μM, 120 A (Dr. Maisch, HPLC GmbH, Germany) in a Nanoflex chip system (Eksigent). Mobile phase A consisted of 1% methanol (Fisher)/0.1% formic acid (Thermo Fisher Scientific) and mobile phase B of 1% methanol/0.1% formic acid/80% acetonitrile. Peptides were eluted into the mass spectrometer at 300 nl/min with each RP-LC run comprising a 90-min gradient from 10 to 25% B in 65 min, 25–40% B in 25 min. The mass spectrometer was set to repetitively scan m/z from 300 to 1800 (r = 240,000 for LTQ-Orbitrap Elite) followed by data-dependent MS/MS scans on the twenty most abundant ions, with a minimum signal of 1500, dynamic exclusion with a repeat count of 1, repeat duration of 30 s, exclusion size of 500 and duration of 60 s, isolation width of 2.0, normalized collision energy of 33, and waveform injection and dynamic exclusion enabled. FTMS full scan AGC target value was 1 × 106, whereas MSn AGC was 1 × 104, respectively. FTMS full scan maximum fill time was 500 ms, whereas ion trap MSn fill time was 50 ms; microscans were set at one. FT preview mode; charge state screening, and monoisotopic precursor selection were all enabled with rejection of unassigned and 1+ charge states. The MS raw files associated with this manuscript have been deposited to the public database ProteomeXchange (Project number: PXD007741).
MS Data Processing and Database Searching
MS raw files were analyzed by MaxQuant software (31) version 1.5.2.8. MS/MS spectra were searched by the Andromeda search engine (32) against the Human UniProt FASTA database (9606; 136,251 entries) (version July 2014). Viral proteins were identified using Adenovirus C serotype 5 (28285; 31 entries), Human Herpesvirus strain 17 (10299; 73 entries), and Vaccinia virus (10245; 230 entries) UniProt FASTA database (version January 2015). Additionally, the database included 247 common contaminants, discarded during data analysis. The search included variable modifications of methionine oxidation and N-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Trypsin was specified as the digestive enzyme. Minimal peptide length was set to seven amino acids and a maximum of two missed cleavages was allowed. The false discovery rate (FDR) was set to 0.01 for peptide-spectrum matches (PSMs) and protein identifications. Protein grouping was enabled. Peptide identification was performed with an allowed precursor mass deviation up to 4.5 ppm after time-dependent mass calibration and an allowed fragment mass deviation of 20 ppm. Protein identification required at least one unique or razor peptide per protein group. Label-free quantification in MaxQuant was performed using the intensity-based absolute quantification (iBAQ) algorithm (33). The human proteome was searched using the match-between-runs functionality with the retention time alignment window set to 20 min and the match time window to 1 min. Match-between-runs was not used when searching the viral proteomes. Protein tables were filtered to eliminate the identifications from the reverse database, only identified by site and common contaminants. Protein tables are given as supplemental Tables S1, S2, and S3.
Virus Genome Accumulation in SLX4-deficient Cells
RA3331 FA-P cells complemented with wild-type SLX4 or with empty vector were infected with Ad5 (MOI 20) in 12-well plates and harvested by Trypsin at the indicated time points. Total DNA was isolated using the PureLink Genomic DNA kit (Invitrogen). Quantitative PCR was performed using primers specific for viral DBP (5′ gccattgcgcccaagaagaa and 5′ ctgtccacgattacctctggtgat) or cellular tubulin (5′ ccagatgccaagtgacaagac and 5′ gagtgagtgacaagagaagcc). Values for DBP were normalized internally to tubulin and to the 4 h time point to control for any variation in virus input. Quantitative PCR was performed using the standard protocol for Sybr Green (Thermo) and analyzed using the ViiA 7 Real-Time PCR System (Thermo). The experiment was completed in triplicate, and statistical analysis was performed using Prism v7 (GraphPad Software, Inc., La Jolla, CA).
Virus Genome Accumulation in TCOF1-depleted Cells
A549 cells in 12-well plates were transfected with siRNA against TCOF1 (s13921) or negative control (Scrmbl). 72 h post transfection, cells were infected with Ad5 (MOI 25) and harvested at the indicated time points. Total DNA isolation and quantitative PCR were performed as described above. The experiment was completed in triplicate, and statistical analysis was performed using Prism v7 (GraphPad).
Virus Yield in SLX4-deficient Cells
RA3331 FA-P cells complemented with wild-type SLX4 or with empty vector were infected with HSV-1 (MOI 0.1), and supernatants were collected at the indicated time points. Viral yield was measured by plaque assay on monolayers of U2OS cells. Statistical analysis was performed using Prism v7 (GraphPad).
Virus Yield in TCOF1-depleted Cells
A549 cells in 12-well plates were transfected with siRNAs and infected with Ad5 as above. Infected cells were harvested at the indicated time points and lysed by three cycles of freeze-thawing. Virus yield was determined by plaque assay as follows: cell lysates were diluted serially in DMEM supplemented with 2% FBS and antibiotics to infect HEK293 cells in 12-well plates. After virus adsorption for 2 h, cells were overlaid with complete culture media containing 0.45% SeaPlaque agarose (Lonza, Walkersville, MD). Plaques were stained with crystal violet 7 days postinfection. Statistical analysis was performed using Prism v7 (GraphPad).
Experimental Design and Statistical Rationale
For most iPOND-MS experiments, four conditions were included per experiment: (1) infection plus biotin, (2) infection minus biotin, (3) mock plus biotin, and (4) mock minus biotin. Minus biotin samples were used to identify background/contaminant proteins within our system. Three independent biological replicate experiments were performed per condition, except for “mock plus biotin” samples with fifteen replicates and “VACV infection minus biotin” samples with two replicates. Resulting iBAQ values from replicates were combined for our statistical analysis. The sample size was chosen to provide enough statistical power to apply parametric tests (either homoscedastic or heteroscedastic one-tailed t test, depending on the statistical value of the F-test; heteroscedastic if F-test p value<0.05). The t test was considered as valuable statistical test because binary comparisons were performed and the number of replicates was limited to some conditions. No samples were excluded as outliers (this applies to all proteomics analyses described in this manuscript). Proteins with t test p value smaller than 0.05 were considered as significantly altered between the two tested conditions. Data distribution was assumed to be normal but this was not formally tested. Findings from proteomics analysis were validated and further studied using immunoblotting and immunofluorescence among other molecular biology techniques.
RESULTS
Optimizing Conditions to Isolate Proteins on DNA Replicated During Virus Infection
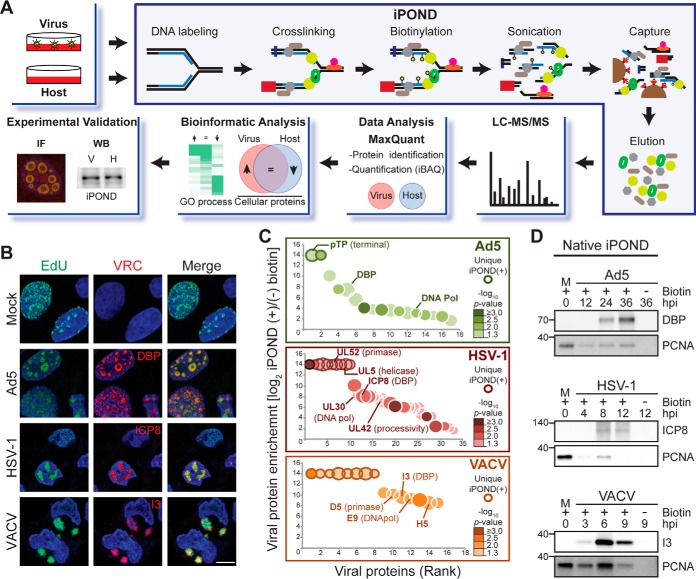
To profile proteins associated with viral DNA replicated during infection with Ad5, HSV-1 and VACV, we optimized the EdU labeling and proteomic techniques (Fig. 1A). The timing of EdU pulse during infection was examined using immunofluorescence (Fig. 1B and supplemental Fig. S1). We assessed accumulation of EdU signal in VRCs at multiple different hours post infection (hpi) during infection with Ad5, HSV-1 and VACV. When infected cells were treated with a 15 min EdU pulse at the peak of viral DNA synthesis (Ad5 at 24 hpi, HSV-1 at 8 hpi and VACV at 6 hpi), we observed a high number of cells with strong EdU signal (Fig. 1B and supplemental Fig. S1). Each of these viruses express an early viral protein that has single-stranded DNA binding activity and marks VRCs (DBP for Ad5, ICP8 for HSV-1, and I3 for VACV), and the EdU labeling in infected cells was predominantly colocalized with these viral replication proteins (Fig. 1B). EdU patterns observed within VRCs resembled those previously reported for in situ hybridization using genomic viral DNA probes (34–36) or 5-Bromo-2′deoxyuridine (BrdU) (37) to label VRCs of Ad5, HSV-1, and VACV. In contrast, EdU incorporation in uninfected cells appeared in granular patterns characteristic of cellular replication foci (38, 39). All three viruses included in this study are capable of shutting down host cellular replication during infection (9, 37, 40, 41), suggesting that at peak viral replication there is minimal host replication. When EdU pulses were given at earlier times during infection, the EdU signal was dispersed or localized at small VRCs. In contrast, pulses at later infection times resulted in EdU signal accumulated in patterns reminiscent of the late stages of infection (supplemental Fig. S1A–S1C). We tested different labeling periods (supplemental Fig. S1D), and chose 15 min as appropriate for labeling replicating viral DNA. Using our optimized conditions, we performed iPOND on uninfected cells and cells infected with Ad5, HSV-1, and VACV and sequenced the genomes recovered from each condition. Sequence alignment of total reads from each condition showed that most of the DNA recovered from virus infections was labeled viral genomes, and in each case less than 50% of the labeled DNA was human: ∼28.5% in Ad5, 45.6% in HSV-1, and 43.5% in VACV (supplemental Fig. S1E). Although the iPOND technique was originally developed to identify proteins on nascent cellular DNA (21, 22), the asynchronous nature of infection, simultaneous viral origin firing, and differences in genome size between viruses, all act to preclude analysis of proteins only at replication forks. Therefore, applying this technique to infected cells will identify proteins on replicated DNA throughout the virus genome.
Fig. 1.
Profiling proteomes on DNA replicated during virus infection. A, Schematic of workflow. B, EdU incorporation (green) into replicating genomes in uninfected U2OS cells (Mock) and cells infected with Ad5, HSV-1, or VACV. Single-strand DNA binding proteins for each virus mark viral replication compartments (VRCs). DAPI stains nuclei. Scale bar, 10 μm. C, Bubble plot showing the enrichment of viral proteins isolated on Virus replicated-DNA proteomes. The log2 fold change in protein intensity between iPOND(+) and iPOND(-) biotin samples (y axis) is plotted against the rank of the protein (x axis). Dot size indicates the average abundance of each protein in iPOND(+). Color gradient indicates significance of the observed fold change (-log10 p value; one-tailed, t test). Only proteins with a significant fold change (-log10 p value ≤1.3) were plotted. Viral proteins identified uniquely in iPOND(+) samples are highlighted by a bolded edge. Viral replicative proteins are labeled. (D) Immunoblotting of DBP (Ad5), ICP8 (HSV-1), and I3 (VACV) confirms enrichment of viral DNA-interacting proteins in native iPOND isolates. Cellular PCNA is a positive control in uninfected cells. Molecular weight markers (kDa) are marked on left. See also supplemental Fig. S1 and supplemental Table S1.
Viral Proteins Identified on Replicated DNA from Infected Cells
Using our optimized conditions, we isolated proteins on replicated DNA from uninfected (Host) or infected (Virus) cells. Protocols were performed with and without biotin and the resulting proteomes were profiled using liquid chromatography-tandem mass spectrometry (LC-MS/MS) and label-free analysis by MaxQuant software (31) as shown in Fig. 1A. To validate that our approach enabled capture and identification of proteins associated with replicated viral DNA, we first asked whether known viral DNA-interacting proteins were recovered from virus-infected samples. Within the proteomes isolated from infected cells we identified viral proteins: 25 for Ad5, 55 for HSV-1, and 81 for VACV (supplemental Fig. S2A and supplemental Table S1). Isolated viral proteins included DNA-interacting proteins, which were uniquely found (bolded edge) or significantly enriched (p value <0.05) on Virus replicated-DNA proteomes isolated in the presence of biotin compared with no biotin controls (Fig. 1C). Enriched DNA-interacting viral proteins included viral DNA replication factors, transcription factors, transcription regulators and proteins involved in viral genome packaging. Immunoblotting of proteins on replicated DNA isolated from infected cells demonstrated a gradual increase in levels of the early viral single-strand DNA binding proteins DBP, ICP8 and I3 over time of infection with Ad5, HSV-1 and VACV respectively (Fig. 1D). The cellular protein PCNA was used as a positive control for the isolation of proteins associated with replicating DNA in uninfected cells. We also observed enrichment of this core component of the cellular replication fork on replicated DNA isolated from infected cells (Fig. 1D). The levels of PCNA recovered from infected cells increased gradually over the course of infection, peaking at a time when DNA replication is at its maximum for each virus. This observation suggests the association of PCNA with replicating viral DNA. Together, these results demonstrate isolation of viral proteins associated with replicating genomes during virus infection, and provide feasibility to identify host proteins associated with replicated DNA during virus infections.
Host Proteins Identified on Replicated DNA During Virus Infections
To uncover host factors potentially associating with viral genomes during infection, we analyzed the cellular proteins identified in the replicated DNA proteomes recovered from Virus and Host samples. Proteins isolated exclusively in the no biotin controls were considered contaminants and excluded from our analysis (supplemental Fig. S2B and supplemental Table S2). Within the replicated DNA proteome isolated from infected cells we quantified a total of 1792 cellular proteins for Ad5, 1747 for HSV-1, 1618 for VACV and 2123 in Host (supplemental Fig. S2A and supplemental Table S3). Pearson correlation coefficients between biological replicates of these samples confirmed quality and reproducibility of our data (∼0.90 for Host and ∼0.86 for Viruses; supplemental Fig. S2C). The observed lower correlation between different virus biological samples may reflect the asynchronous nature of infection. Correlations were more diverse between different conditions. Ad5 showed highest similarity to Mock (PCCAd5 ∼0.84) compared with HSV-1 (PCCHSV-1 ∼0.79) and VACV (PCCVACV ∼0.71), which may highlight greater independence of HSV-1 and VACV viruses from host replication machinery. This was expected, as HSV-1 and VACV genomes encode more proteins than Ad5, thus they use fewer host enzymes to express viral genes and replicate viral DNA. On the other hand, the high correlation of Ad5 and HSV-1 virus (PCCAd5/HSV ∼0.82) as compared with VACV (PCCAd5/VACV ∼0.71; PCCHSV-1/VACV ∼0.58) may reflect differences in sites where these viruses replicate within the infected cell. Ad5 and HSV-1 replicate their genomes in the cell nucleus, whereas VACV replicates in the cytoplasm of infected cells. Surprisingly, Ad5 was more like VACV than HSV-1, suggesting perhaps more commonalities than previously thought in the way these two viruses manipulate host cell processes to promote their replication. Principal component analysis (PCA) showed distinct proteomes were isolated for each condition (supplemental Fig. S2D).
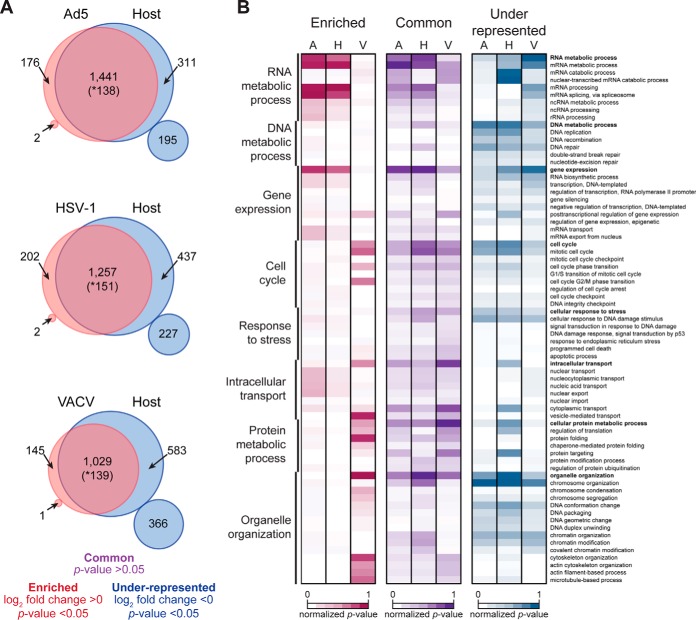
The profiles obtained by analyzing cellular proteins on replicated DNA purified from Ad5, HSV-1 and VACV infections suggest host factors and potential cellular processes are manipulated by these viruses to promote their efficient replication. To define differences between proteins on replicated DNA isolated from uninfected (Host) and infected (Virus) cells, we compared proteomes and classified cellular proteins into three enrichment categories: “common,” “enriched” on virus, or “under-represented” on virus (Fig. 2A and supplemental Table S3). Although our comparative analysis does not account for differences in DNA replication efficiencies between each virus and the cell, this classification highlights quantitative differences between the composition of Virus and Host proteomes, which may reflect manipulation of specific host pathways during virus infection. Host proteins in the “common” category are those with no significant difference (p value >0.05) in abundance between Virus and Host proteomes. These proteins (purple) were found at similar levels on the replicated DNA isolated from uninfected and infected cells. We suggest that many proteins in this category are specifically associated with the replicated viral DNA extracted from virus-infected samples. The “enriched” proteins are those found to be significantly (p value <0.05) more abundant (log2 fold change >0) within Virus proteomes (red). Because these proteins were present at higher levels on replicated DNA isolated from infected cells compared with uninfected cells, we suggest these proteins are associated with viral DNA and might represent cellular pathways used in viral processes. In contrast to the “enriched” proteins, host proteins assigned “under-represented” were those significantly less abundant (log2 fold change <0) within proteomes recovered from infected cells when compared with proteomes on replicated DNA from uninfected cells (blue). Proteins in this category may include host factors that are either used by the virus but found at lower levels on viral DNA compared with replicated cellular DNA, or cellular factors that are not harnessed during virus infection. Furthermore, proteins “under-represented” on replicated DNA during infections might also highlight cellular processes that are specifically inactivated by early viral proteins. Interestingly, the percent of cellular proteins found “under-represented” in Virus DNA proteomes ranged from ∼23.8% (506 proteins) for Ad5 (∼36Kb), to ∼31% (664 proteins) for HSV-1 (∼150Kb), and ∼44.7% (949 proteins) for VACV (∼190Kb). This trend in the number of “under-represented” host proteins for each virus, along with the number of viral proteins identified within each Virus DNA proteome, supports the notion that viruses with larger viral genomes are less dependent on host machinery.
Fig. 2.
Differential enrichment of cellular proteins on genomes replicated during infection. A, Venn diagrams summarize enrichment differences of cellular factors between Host and Virus replicated-DNA proteomes from Ad5, HSV-1 and VACV. Proteins significantly different (one-tailed, t test; p value <0.05) are considered “enriched” if log2 fold change >0 or “under-represented” if <0. Small circles show the number of proteins found only in iPOND samples of the indicated condition with one-tailed, t test; p value <0.05. These significantly different proteins in small circles are considered unique to each proteome. Proteins not significantly different (one-tailed, t test; p value >0.05) are considered “common” between proteomes. * Indicates number of proteins considered not significantly different, but found only in uninfected samples. These proteins are characterized by being uniquely identified in very few Host (+)Biotin samples (mainly in 1, 2, or 3 replicates) at highly variable levels and thus cannot be considered unique to the Host proteome. B, Annotation matrix of GO biological processes represented by host proteins found enriched (red), common (purple) or under-represented (blue) in each Virus proteome (A: Ad5; H: HSV-1; V: VACV). Color gradient heat map represents normalized p value (-log10) from MetaCore analysis (false discovery rate (FDR) <5%). See also supplemental Figs. S2 and supplemental Tables S2 and S3.
To gain insight into the biological processes potentially manipulated during viral infection and to illustrate global similarities and differences between viruses, we performed comparative gene ontology (GO) enrichment analysis of host proteins in each category for each Virus-Host comparison (Fig. 2B). We observed that ontologies represented by “enriched” proteins revealed distinctions in processes harnessed and differentiated viruses that are nuclear-replicating (Ad5 and HSV-1) from cytoplasmic-replicating (VACV). For example, host proteins “enriched” in Ad5 and HSV-1 proteomes include factors involved in mRNA processing and export, whereas proteins “enriched” in VACV represent processes such as vesicle-mediated transport and protein folding. We observed that most ontologies represented by “enriched” proteins point to cellular pathways potentially harnessed to contribute to viral processes taking place after viral DNA synthesis such as gene expression, mRNA synthesis, splicing and transport for Ad5 and HSV-1 and protein folding, processing and transport for VACV.
Comparative GO enrichment analysis of proteins classified as “under-represented” on replicated DNA proteomes from virus infected cells also showed distinctions between viruses, especially between HSV-1 and VACV. For example, proteins found “under-represented” in HSV-1 reflected processes such as cytoplasmic transport and protein targeting, whereas in VACV, “under-represented” proteins highlighted processes including mRNA processing and covalent chromatin modification. Our global comparison also shows biological processes “under-represented” in all three viruses including DNA replication, DNA repair, chromatin organization and cell cycle. These cellular processes commonly “under-represented” in all three Virus proteomes might highlight those that are fine-tuned during virus infection: components detrimental to virus replication may be deactivated whereas components that are beneficial are exploited. Altogether, this analysis provides an overview of cellular processes commonly or specifically manipulated by each virus based on protein abundance differences found between each Virus DNA-replicated proteome relative to that of the Host.
Association of Cellular DNA Replication Factors with Viral DNA
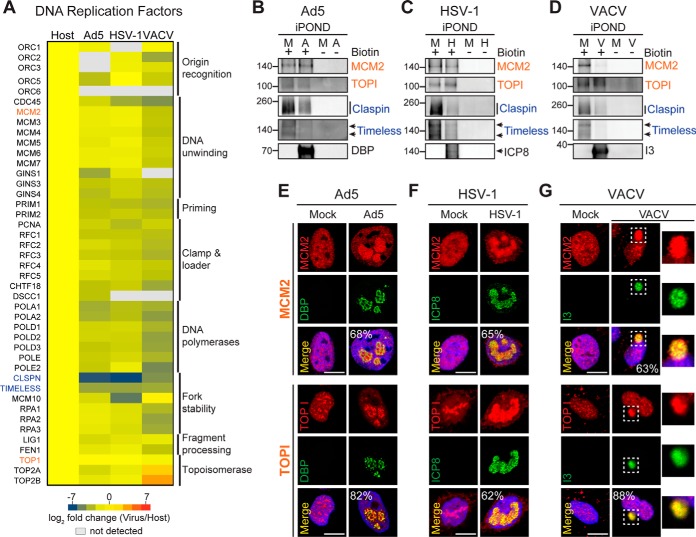
Our global GO enrichment analysis showed that cellular factors involved in DNA replication and repair were found in the “under-represented” category in the proteomes recovered from all three viruses when compared with Host. However, several of these factors were enriched in Virus replicated DNA proteomes when compared with no biotin controls, suggesting these proteins are associated with the DNA replicated in infected cells even though their abundance is lower on viral DNA than cellular DNA (supplemental Table S4). To gain a deeper understanding into how each virus could use cellular DNA replication and repair proteins for viral processes, we compared protein abundance of specific host DNA replication and repair factors between the replicated DNA proteomes isolated from each virus infection (Ad5, HSV-1, and VACV). We hypothesized that the activity of some of these host factors may be beneficial for viral DNA synthesis by one virus, whereas detrimental for another virus. A global overview of the enrichment differences of core cellular DNA replication factors in each Virus compared with the Host replicated DNA proteome highlights host factors potentially used by Ad5, HSV-1 and VACV (Fig. 3A). We observed that the abundances of most host DNA replication factors are only slightly lower on Virus proteomes compared with Host, as shown by the prevalence of yellow and light green shades in the heat map. Remarkably, all subunits of the mini-chromosome maintenance (MCM2–7) replicative helicase complex were isolated at similar levels within the Virus and Host DNA proteomes (Fig. 3A). Similarly, topoisomerases I, IIα, and IIβ were found at comparable levels in Virus and Host DNA proteomes, except that both type II topoisomerases were found highly enriched (log2 fold change ≥0) in the VACV proteome. This result is consistent with a previous report showing type II topoisomerases are recruited to VACV cytoplasmic VRCs to facilitate viral replication (15). In contrast, most subunits of the replicative DNA polymerases are under-represented on replicated DNA from infected cells, likely reflecting that each virus encodes its own viral DNA polymerase and therefore is not reliant on cellular DNA polymerases (2, 42, 43). Claspin, Timeless, and MCM10 showed the largest negative fold change in the DNA proteomes on Virus compared with Host, suggesting these proteins do not associate with replicating viral DNA perhaps because of their activity being detrimental to viral replication.
Fig. 3.
Host DNA replication factors are recruited to sites of viral DNA replication. A, Heat map highlighting differences in enrichment of cellular replisome components between each Virus replicated-DNA proteome compared with Host. B–D, Immunoblotting of proteins recovered on replicated DNA from cells infected with Ad5 (A), HSV-1 (H), VACV (V) or mock (M). MCM2, TOPI, Claspin and Timeless were examined to validate fold change differences (orange: log2 FC near zero, blue: log2 FC far under zero). Viral proteins DBP, ICP8, and I3 confirmed isolation of viral genomes. Molecular weight markers (kDa) are marked on left. E–G, Immunofluorescence showing localization of MCM2 and TOPI in uninfected U2OS cells (Mock) or cells infected with (E) Ad5, (F) HSV-1, and (G) VACV. Representative images showing MCM2 and TOPI colocalization with viral replication compartments in all three viruses were quantified. Percentages shown correspond to the number of infected cells with the observed pattern. DAPI stains nuclei. Scale bar, 10 μm. Insets show VACV cytoplasmic VRCs. See also supplemental Fig. S3 and supplemental Table S4.
We further investigated enrichment and localization of cellular replicative factors during virus infection by immunoblotting and immunofluorescence. Consistent with our proteomics analysis, by immunoblot we detected both MCM2 and topoisomerase I (TOPI) at nearly similar levels in replicated-DNA samples recovered from cells uninfected or infected with Ad5 (Fig. 3B) and HSV-1 (Fig. 3C), and at slightly lower levels in VACV (Fig. 3D). In contrast, Claspin and Timeless were detected at significantly reduced levels in DNA samples recovered from infected cells (Fig. 3B–3D). Despite differences in sensitivity between techniques, our immunoblotting results globally reflect our proteomics observations. Immunofluorescence analysis of infected cells confirmed MCM2, TOPI and PCNA localized to sites of viral replication (Fig. 3E–3G and supplemental Fig. S3A). We observed strong signals for accumulation of MCM2, TOPI and PCNA at VRCs marked by DBP for Ad5, ICP8 for HSV-1 and I3 for VACV (see quantification in Fig. 3E–3G). Although a less quantitative technique than iPOND, immunofluorescence analysis of selected host DNA replication factors reflected our proteomics data and confirmed that MCM2, TOPI, and PCNA accumulate at sites where viral DNA is being synthesized. The accumulation of PCNA on VRCs further supports the association of this host protein with replicating viral DNA previously suggested by immunoblotting of iPOND samples recovered from infected cells (Fig. 1D). Together, these results suggest that our comparative proteomic analysis highlights host factors associated with viral DNA during infection.
Selective Enrichment of Host DNA Damage/Repair Factors on Viral DNA
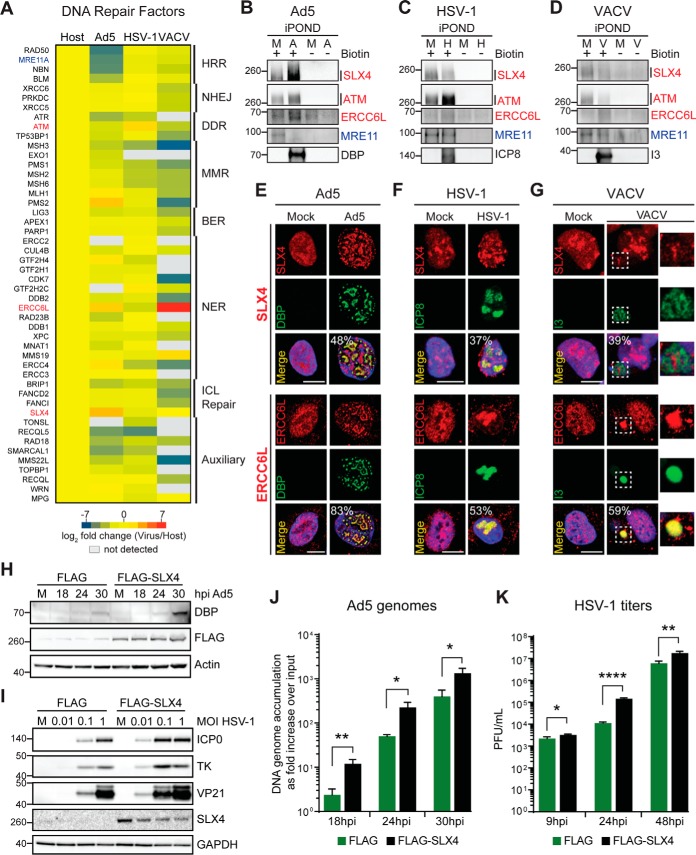
Viruses manipulate DNA damage signaling and repair pathways to promote their own replication (44–46). To provide a comprehensive overview of host DNA repair factors potentially exploited or deactivated by Ad5, HSV-1, and VACV, we also examined the presence and abundance of DNA repair factors within the DNA proteomes of each virus compared with Host (Fig. 4A and supplemental Table S4). Like DNA replication factors, host DNA repair factors were observed within DNA proteomes of all three viruses at levels similar to or higher than Host. These included proteins involved in Non-Homologous End Joining (NHEJ), Base Excision Repair (BER), Nucleotide Excision Repair (NER), and Interstrand Crosslink (ICL) Repair. This observation suggests Ad5, HSV-1 and VACV coopt these pathways to promote their replication. We also found host factors differentially enriched between the proteomes recovered from each virus infection. For example, proteins implicated in Homologous Recombination Repair (HRR), including Bloom's helicase (BLM), MRE11, RAD50, and NBS1 (NBN), were found at lower levels in Ad5 than in HSV-1 or VACV DNA proteomes. This result is consistent with previous reports that these four HRR factors are targeted for degradation and/or mislocalization away from VRCs by Ad5 early viral proteins (30, 47–49), but are harnessed by HSV-1 to promote viral replication (10, 11, 50). Interestingly, a role for cellular HRR factors in VACV infection has not been described. Our analysis showed most HRR factors were found in the VACV DNA proteome at levels comparable to Host, suggesting that these factors may be recruited similarly to HSV-1, potentially to aid viral DNA replication.
Fig. 4.
Selective association of DNA repair factors within viral DNA. A, Heat map comparing enrichment of host DNA repair factors within Virus replicated-DNA proteomes compared with Host. B–D, Immunoblotting of proteins captured from cells infected with Ad5 (A), HSV-1 (H), VACV (V) or mock (M). SLX4, ATM, ERCC6L and MRE11 are shown to validate association of differentially “enriched” (red) and “under-represented” (blue) host DNA repair factors with viral DNA. Viral proteins DBP, ICP8 and I3 confirmed viral genomes were recovered. Molecular weight markers (kDa) are marked on left in all immunoblots in this figure. E–G, Immunofluorescence showing localization of SLX4 and ERCC6L in Mock or cells infected with Ad5 (E), HSV-1 (F), and VACV (G). SLX4 localized clearly to VRCs in Ad5 infections only, whereas ERCC6L localized to VRCs in Ad5 and VACV infections. Representative images were quantified. Percentages shown correspond to the number of infected cells with the observed pattern. DAPI stains nuclei. Scale bar, 10 μm. Insets zoom into VACV cytoplasmic VRCs. H–K, Hypomorphic SLX4 cells and matched cells complemented with FLAG-SLX4 were infected with Ad5 or HSV-1 to examine the effect of SLX4 on viral processes. H, Immunoblots of lysates at the indicated time points demonstrating increased Ad5 DBP levels in the presence of SLX4 (FLAG). Actin serves as a loading control. I, Immunoblots demonstrating increased HSV-1 protein levels (ICP0, TK and VP21) in the presence of SLX4 at low MOI. SLX4 does not affect HSV-1 protein levels at high MOI. GAPDH serves as a loading control. J, The presence of SLX4 results in an increase in Ad5 genome accumulation as assessed by qPCR at indicated time points during infection. Accumulation of viral DNA is represented as fold increase over input viral DNA as determined at the 4 h time point, and error bars represent standard deviation for three biological replicates. *p < 0.05, **p < 0.01. K, HSV-1 viral yield was measured by plaque assay at indicated time points. Viral yield is indicated as plaque-forming units (PFU) per ml and is shown to increase in the presence of SLX4. The average of three biological replicates is shown with error bars displaying S.D. of triplicates. *p < 0.05, **p < 0.01, ****p < 0.0001. See also supplemental Fig. S3 and supplemental Table S4.
Enrichment and localization of selected DNA repair factors were examined further using immunoblotting and immunofluorescence to validate our proteomics analysis. Immunoblotting confirmed MRE11 association with DNA isolated from cells infected with HSV-1 and VACV, but not Ad5 (Fig. 4B–4D). Among host DNA repair factors, we observed high enrichment of SLX4, ATM and ERCC6L within proteomes of Ad5, HSV-1, and VACV, respectively. We detected high levels of SLX4 on Ad5 (Fig. 4B), ATM on HSV-1 (Fig. 4C), and ERCC6L on both Ad5 and VACV (Fig. 4D) by immunoblotting. Immunofluorescence analysis of infected cells confirmed localization of SLX4, ERCC6L, KU70 (XRCC6), and MRE11 to sites of viral DNA replication as predicted by our proteomics data (Fig. 4E–4G and supplemental Fig. S3B–S3C). A strong signal was detected for SLX4 at Ad5 VRCs marked by DBP, but SLX4 was more difficult to observe at HSV-1 and VACV VRCs (see quantification in Fig. 4E–4G). In contrast, overlapping signals for ERCC6L and KU70 with the viral proteins marking VRCs were easily observed for all three viruses. In addition, immunofluorescence was consistent with our proteomics data and revealed MRE11 localized to cytoplasmic replication sites during VACV infection (supplemental Fig. S3C), recruited to HSV-1 VRCs, and depleted in Ad5 infected cells. These data support the hypothesis that DNA viruses selectively harness host DNA repair factors.
We propose that host factors classified by our comparative proteomic analysis as “common” or “enriched” on replicated DNA from virus-infected cells may be exploited for viral DNA replication. Conversely, proteins found significantly “under-represented” within proteomes associated with replicated DNA from virus infections may not be required for viral DNA replication, or may be actively targeted by viral proteins to avoid their association with viral nascent DNA. For instance, our comparative analysis categorized SLX4 as “enriched” in Ad5 and “common” in HSV-1, suggesting both viruses coopt this host factor to enhance their replication. Following our rationale, SLX4 depletion should have a negative impact on Ad5 and HSV-1 replication. To validate our proposal, we examined the effect of SLX4 on the replication of Ad5 and HSV-1. We examined infection with Ad5 (Fig. 4H and 4J) or HSV-1 (Fig. 4I and 4K) in SLX4-deficient cells (27) complemented with empty vector (FLAG) or with SLX4 (FLAG-SLX4). We assessed viral protein production by immunoblotting (Fig. 4H and I), viral DNA replication by quantitative PCR (Fig. 4J), and viral progeny production by plaque assay (Fig. 4K). We observed augmented viral protein levels for both Ad5 and HSV-1 in the presence of SLX4 (Fig. 4H and 4I). Similarly, we observed a significant enhancement in viral genome accumulation for Ad5 (Fig. 4J) and viral titer for HSV-1 (Fig. 4K) in deficient cells complemented with SLX4. Together these data demonstrate the strength of our comparative proteomic approach in predicting host factors harnessed by DNA viruses during infection.
Host Factors Depleted from Viral Nascent DNA Proteomes
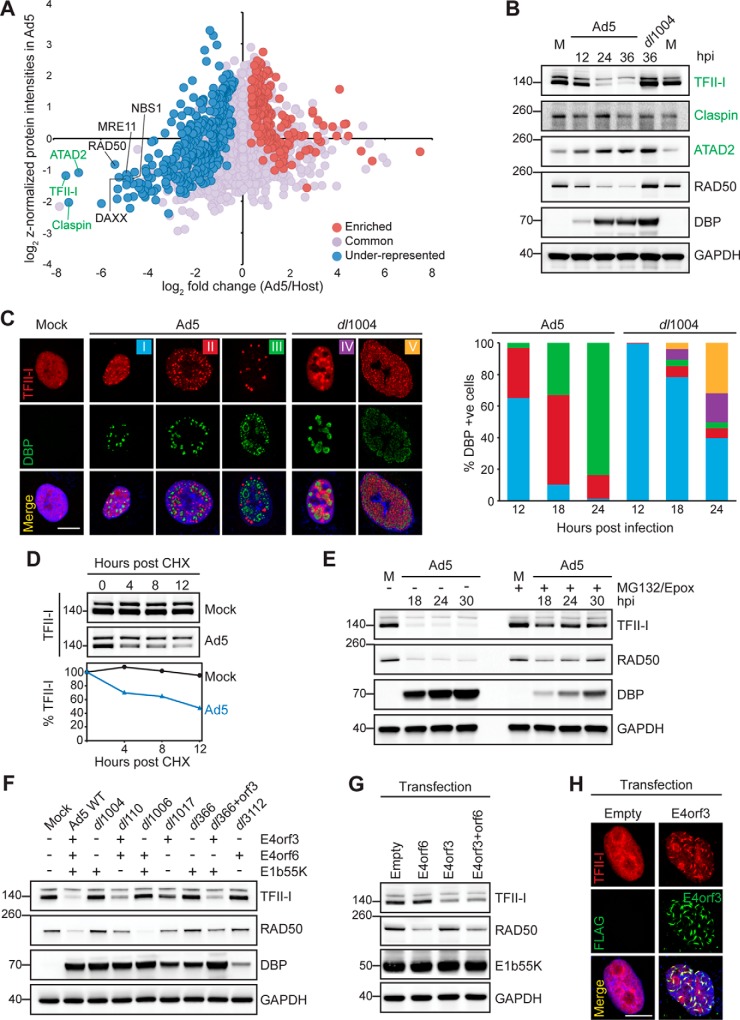
We hypothesized that proteins classified as “under-represented” within the replicated-DNA proteomes from virus-infected cells would include host factors potentially deactivated by early viral proteins during infection. To illustrate that our proteomics approach facilitates identification of previously unknown targets of early viral proteins, we further analyzed the abundance of cellular proteins within the Ad5 replicated-DNA proteome. The early viral proteins E1b55K, E4orf3 and E4orf6 of Ad5 play important roles in promoting viral DNA replication by counteracting intrinsic cellular responses to infection (51). E4orf3 protein acts by re-localizing key components of these responses to nuclear protein scaffolds known as E4orf3 tracks (52), whereas E1b55K and E4orf6 interact with specific cellular proteins to form E3 ubiquitin ligase complexes that direct the proteasome-mediated degradation of repressive host factors (reviewed in (53). Members of the MRN complex are known targets of Ad5 E4orf3, E1b55K, and E4orf6 (26, 30, 48, 49, 54). We observed the MRN complex components (MRE11 RAD50, and NBS1) under-represented in the Ad5 proteome compared with Host, and with an abundance lower than the mean of all host proteins within the Ad5 proteome (Fig. 5A). Interestingly, ATAD2, TFII-I, and Claspin, showed a similar pattern to MRN components within the Ad5 proteome, suggesting these proteins might also be targets of early viral proteins. As predicted by our proteomics analysis, immunofluorescence of Ad5-infected cells showed that ATAD2, TFII-I, and Claspin do not localize to VRCs marked by DBP (supplemental Fig. S4A). We therefore examined total protein levels during infection with wild-type Ad5 or a mutant virus lacking the E4 region (dl1004). This mutant virus is missing E4orf3 and E4orf6 and thus cannot promote degradation or re-localization of cellular factors targeted by these viral proteins. We observed protein levels of TFII-I, but not Claspin and ATAD2, decreased dramatically during infection with wild-type Ad5. TFII-I levels were rescued when infecting with the E4-deleted virus (Fig. 5B). TFII-I is a transcriptional regulator that functions as a repressor in multiple cellular processes (55). Association of TFII-I with the Ad5 genome could therefore have a negative impact on viral transcription, and may explain why TFII-I is targeted by early viral proteins. We then further examined TFII-I localization during infection with wild-type Ad5 and E4-deleted mutant. In uninfected cells TFII-I localized throughout the nucleoplasm but in Ad5 infected cells the TFII-I immunofluorescence signal decreased, and remaining protein was re-localized into large foci distinct from sites of virus replication (Fig. 5C and supplemental Fig. S4A). In cells infected with E4-deleted mutant the TFII-I signal did not decrease, and TFII-I was found at or around DBP staining VRCs. We quantitated TFII-I patterns over a time-course of infection with both wild-type Ad5 and E4-deleted mutant (Fig. 5C). Together these results suggest that Ad5 infection results in mislocalization and degradation of TFII-I protein because of expression of viral E4 gene products.
Fig. 5.
TFII-I is a novel target of Ad5 early viral proteins. A, Scatter plot comparing cellular protein enrichment of Host replicated-DNA proteome (x axis: log2 FC Ad5/Host) compared with protein abundance within the Ad5 replicated-DNA proteome (y axis: z-normalized protein intensities in Ad5). Host factors known as Ad5 early viral proteins targets (black) and potential new substrates (green) are highlighted. B, Immunoblotting for TFII-I, Claspin and ATAD2 in mock (M) or infected cells over time of infection with wild-type (Ad5) or E4-deleted mutant (dl1004). RAD50 is a control for E4-dependent degradation, GAPDH is a loading control and DBP controls for infection. C, Immunofluorescence shows representative distribution patterns (I-V) of TFII-I localization during infection by Ad5 and dl1004. Scale bar, 10 μm. The five patterns of TFII-I staining (I-V) were quantified over time of infection. D, Cycloheximide (CHX) treatment at 10 hpi shows increased turnover of TFII-I during Ad5 infection. E, TFII-I turnover is proteasome-dependent. F, Infections with mutant viruses at 24 hpi demonstrate requirement for E4orf3 to decrease TFII-I abundance. A (+) or (-) sign depicts whether the indicated viral protein is encoded by each mutant virus (G) E4orf3 expression by transfection reduces TFII-I levels but not RAD50. E4orf6 expression reduces RAD50 but not TFII-I. H, E4orf3 expression by transfection forms track-like structures and redistributes endogenous TFII-I. Scale bar, 10 μm. See also supplemental Fig. S4.
The kinetics observed for TFII-I resemble RAD50 degradation (Fig. 5B), which is proteasome-mediated and E4-dependent (26, 30, 54). We confirmed by immunoblotting of Ad5-infected cells treated with protein synthesis or proteasome inhibitors that TFII-I levels decreased because of enhanced protein turnover (Fig. 5D) mediated by the proteasome (Fig. 5E). Further, we showed that virus-induced decrease in TFII-I levels appeared most significant when E4orf3 was expressed in the presence of E1b55K (Fig. 5F) using a panel of mutant viruses that lack E1b55K, E4orf3 and E4orf6 genes individually or in different combinations (28, 56, 57). We then expressed E4orf3 and E4orf6 in the presence of E1b55K by transfection of HEK293 cells and demonstrated that E4orf3 expression led to degradation of TFII-I (Fig. 5G). Expression of E4orf3 in transfection experiments was also sufficient to rearrange TFII-I into nuclear track-like structures (Fig. 5H), as previously observed for PML and the MRN complex (30, 48, 52, 58). Similar results were observed in multiple cell types (supplemental Fig. S4B–S4D). These results indicated that gene products expressed from early regions of Ad5 induce redistribution and degradation of TFII-I, and result in exclusion from replicated DNA at viral replication centers. These findings also demonstrate that the enrichment scheme resulting from our comparative analysis of Virus and Host proteomic profiles enables identification of host targets of viral proteins.
Host Factors Involved in Ribosome Biogenesis Associate with viral Genomes
To provide insights into pathways harnessed by Ad5, we further examined the cellular proteins classified as “enriched” within the Ad5 replicated-DNA proteome. GO cellular compartment enrichment analysis revealed that 60% of these proteins were of nucleolar origin (Fig. 6A). Components of the RNA polymerase I (POL I) and the small-subunit (SSU) processome had the highest enrichment levels among “enriched” proteins. During ribosome biogenesis, POL I transcribes a long ribosomal RNA (rRNA) precursor (47S) from which three out of four mature rRNAs (18S, 5.8S, and 28S) are derived (59, 60). The SSU processome is a ribonucleoprotein complex involved in processing, assembly and maturation of the ribosomal small subunit RNA (18S). The SSU processome is composed of distinct subcomplexes, including the small nucleolar riboprotein (snoRNP) complexes U3, Box C/D and Box H/ACA; the UTP-A, UTP-B, UTP-C, Mpp10, and Bms1/Rcl1 complexes and several individual proteins (61). Analysis of the full proteome identified on Ad5 replicated-DNA revealed additional POL I and SSU processome protein components (Fig. 6B). We also identified the same group of proteins within the HSV-1 proteome (supplemental Fig. S5A), but few proteins in VACV (supplemental Fig. S5B). These results suggest that POL I and multiple protein components of the SSU processome are recruited by DNA viruses that replicate in the cell nucleus. These results are also consistent with our observation that RNA processing pathways are potentially exploited by nuclear-replicating viruses (Ad5 and HSV-1), but not for VACV (Fig. 2B).
Fig. 6.
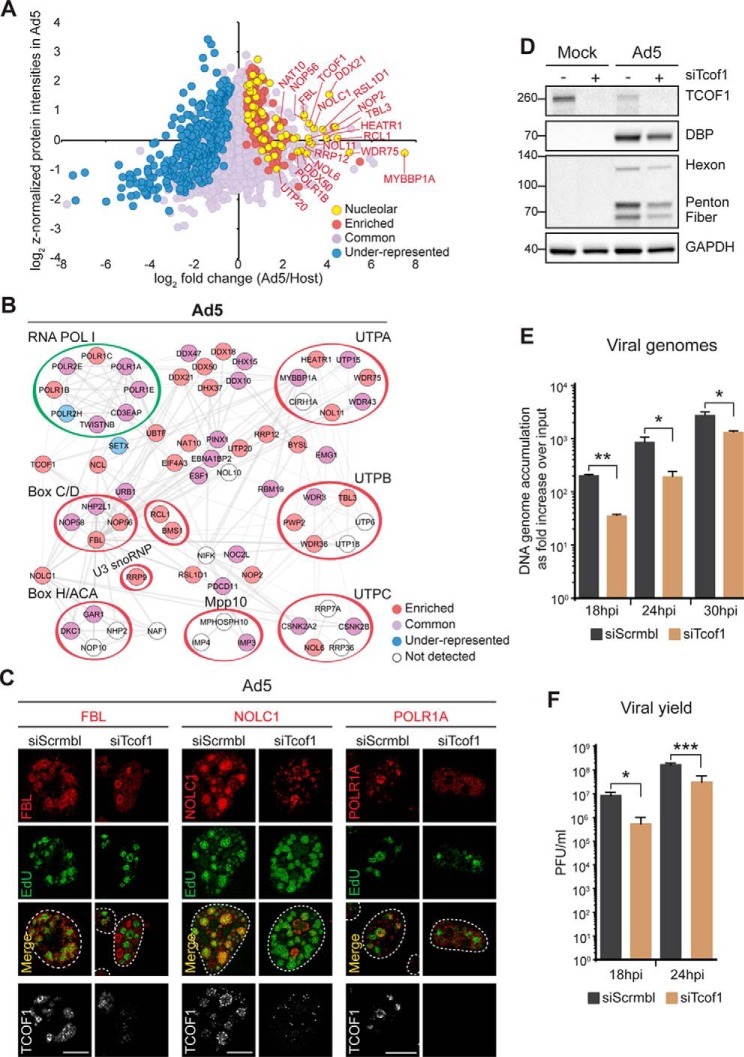
TCOF1-dependent recruitment of nucleolar proteins to viral DNA is required for efficient viral replication. A, Representative nucleolar proteins enriched in Ad5 replicated-DNA proteome highlighted in the same scatter plot shown in Fig. 5A. B, Protein network analysis of all nucleolar proteins identified in the Ad5 replicated-DNA proteome that are associated with the RNA POL I complex and the SSU processome. Components of characterized subcomplexes are circled. Protein color reflects enrichment status as in (A). C, Localization of nucleolar proteins on TCOF (gray) knockdown and infected with Ad5 at 24hpi, EdU marks VRCs. Scale bar, 10 μm. D, TCOF1 depletion reduced late protein production. E, TCOF1 depletion reduced viral DNA replication. Ad5 genome accumulation was assessed by qPCR at indicated time points during infection of cells with TCOF1 reduced by siRNA compared with control siScrbl. Accumulation of viral DNA is represented as fold increase over input viral DNA as determined at the 4 h time point, and error bars represent standard deviation for three biological replicates. F, TCOF1 depletion reduced progeny production. Ad5 viral yield was measured by plaque assay at indicated time points. Viral yield is indicated as plaque-forming units (PFU) per ml and is shown to decrease in the absence of TCOF1. The average of four biological replicates is shown with error bars displaying S.D. of quadruplicates. ***p < 0.001, **p < 0.01 and *p < 0.05. See also supplemental Fig. S5 and S6.
To validate our proteomics analysis, we examined subcellular localization of representative POL I and SSU processome proteins during Ad5 virus infection. We observed that POLR1A, NOP56, Fibrillarin (FBL), Dyskerin (DKC1), MYBBP1A, DDX21 and NAT10 localized to VRCs marked by DBP in three distinct patterns: (1) diffuse signal with puncta around the VRC edges (TCOF1, NOLC1, NOP56, FBL, and DKC1), (2) signal overlapping with DBP (POLR1A and MYBBP1a), and (3) diffuse nucleoplasmic signal partially overlapping with VRCs (DDX21 and NAT10) (Fig. 6C and supplemental Fig. S5C). These results demonstrate that nucleolar proteins partially accumulate at sites where viral DNA is replicated during Ad5 infection, suggesting an important role in the viral life cycle.
TCOF1 is Required for Protein Recruitment and Efficient Ad5 Replication
Among nucleolar proteins enriched in the proteome isolated by Ad5 replicated-DNA we identified TCOF1 (Fig. 6A). TCOF1 regulates ribosomal DNA (rDNA) transcription and preribosomal RNA processing through its interaction with POL I and protein components of the Box C/D snoRNPs (62–64). Werner et al. recently showed that TCOF1 and NOLC1 are monoubiquitinylated to form a scaffold that recruits POL I and the SSU processome subcomplexes, Box C/D and H/ACA snRNPs (65). Based on these data and our observation of TCOF1 enrichment in the Ad5 proteome, we hypothesized that the localization of POL I and SSU processome protein components to Ad5 VRCs is dependent on TCOF1. We observed that depletion of TCOF1 by siRNA in uninfected cells did not impact levels of NOLC1, FBL, POLR1A and DKC1 (supplemental Fig. S6A), but did alter their localization pattern from nucleolar to diffuse in the nucleoplasm (supplemental Fig. S6B). In Ad5 infected cells TCOF1 depletion impacted localization of NOLC1, FBL, and POLR1A to sites of Ad5 DNA replication (Fig. 6C). DKC1 localization was not affected by TCOF1 depletion (supplemental Fig. S6C). These results suggest that TCOF1 plays a role in recruiting NOLC1, POL I, and Box C/D snoRNPs to sites of Ad5 viral replication.
To determine the impact of TCOF1 depletion on Ad5 infection, we examined viral late protein production by immunoblotting (Fig. 6D), viral DNA replication by quantitative PCR (Fig. 6E), and viral progeny production by plaque assay (Fig. 6F). We observed a reduction in viral late protein levels in cells with TCOF1 knockdown (Fig. 6D). This appeared to be specific to TCOF1 because depletion of other nucleolar proteins, including NAT10, did not affect viral protein levels (supplemental Fig. S6D). We also observed that TCOF1 knockdown impacted viral genome accumulation (Fig. 6E) and viral yield (Fig. 6F). Together these results suggest that TCOF1 plays a positive role in efficient viral infection, likely through recruitment of POL I and SSU processome subcomplexes that promote viral protein production.
DISCUSSION
Virus replication is regulated by interactions of cellular proteins with viral proteins and the viral DNA genome. Defining the proteome associated with the genomes replicated during virus infection can reveal proteins participating in these interactions and highlight important cellular factors harnessed or inactivated by DNA viruses to advance progeny production. In this study, we applied a proteomics approach to determine proteins associated with DNA replicating in human cells in the absence (Host) or presence (Virus) of infection by Ad5, HSV-1 and VACV. The resulting proteomes provide unique protein profiles for each virus that facilitate interrogation of cellular factors impacting the virus life cycle. Comparative analysis of Virus and Host protein profiles, allowed us to generate enrichment signatures for host proteins within each Virus proteome. These enrichment signatures enabled us to (1) predict common and specific cellular factors and processes manipulated by the three DNA viruses; (2) identify TFII-I as a target of Ad5 early viral proteins; and (3) uncover a role for TCOF1 in recruiting nucleolar proteins to VRCs to aid Ad5 replication. The data presented here provide a comprehensive resource for exploration of the impact of host factor activities during viral replication. To promote and facilitate the use of our data in generating hypotheses regarding the potential impact of host factors on viral replication, a guide to navigate our data tables has been included in the supplementary Information.
Our approach to profiling proteins on viral DNA is amenable to studies that compare infection in different cell types, time points during infection, and between different viral species or mutants. The isolation of proteins on labeled DNA could also be combined with biochemical approaches to fractionate VRCs (66). In addition, our approach may be adapted to profile proteins associated with incoming viral DNA genomes (23) to identify cellular sensors and early cellular responses. Overall, our results illustrate the power of proteomics for in-depth profiling of proteins accumulated on viral DNA genomes in a reproducible quantitative and qualitative fashion. A technique has recently been developed to identify host factors associated with viral RNA and involved in amplification of viral RNA genomes (67). Further work using complementary approaches will continue to provide a broad view of virus-host interactions on viral nascent genomes.
The three viruses compared in this study are known to limit host replication in favor of their own replication. Consistent with this, sequencing of total DNA isolated by iPOND from infected cells demonstrated that labeled DNA was predominantly viral (supplemental Fig. S1E). Labeling host cellular DNA could be further minimized by synchronizing infections or using non-replicating cells. A recent study used a similar approach to identify viral and cellular proteins associated with viral HSV-1 genomes in resting fibroblast cells infected with a mutant virus lacking UL2 (dUTPase) and UL50 (uracil glycosylase) (24). In our study we specifically used proliferating cells to define the Host replicated-DNA proteome in the same cells used for virus infections and to enable our comparative analysis. Therefore, a caveat to our approach is that some proteins in the Virus proteomes were isolated on labeled cellular DNA resulting from uninfected cells within the population that were replicating normally. However, these proteins are expected to be notably less abundant in Virus proteomes compared with Host because a multiplicity of infection sufficient to infect at least 90% of cells was used for each virus. Despite using proliferating cells, our study confirmed the enrichment on HSV-1 genomes at 8 hpi of the 96 viral and cellular proteins previously reported by Dembowski and DeLuca (24). Furthermore, our approach revealed new virus-host interactions for Ad5, HSV-1 and VACV that were validated using immunoblotting and immunofluorescence. Future studies could incorporate immunopreciptation approaches to assess relative association with viral or cellular DNA. This could be combined with high resolution imaging to reveal spatial arrangements of host factors within subdomains of VRCs (34, 66, 68), where they may have specific temporal activities that aid viral infection.
Comparing protein profiles highlighted a high degree of overlap between Virus and Host proteomes. Interestingly, the overlap between VACV and Host proteomes was almost 50%. This finding is consistent with previous reports showing that despite its cytoplasmic location, VACV replication factories recruit host nuclear proteins, including transcription factors, RNA polymerases, histone deacetylases, DNA ligases and topoisomerases (14–16). Our results complement the list of nuclear factors shown to be required for productive infection and proper morphology of VACV virions in two recent RNAi screens (69, 70), underscoring the importance of host nuclear proteins in the VACV lifecycle. Despite the surprising similarity in “enriched” factors, our overview also portrays clear distinctions between the nuclear- and cytoplasmic-replicating DNA viruses. For example, mRNA processing is among the most highly enriched processes for the nuclear-replicating Ad5 and HSV-1 viruses, but was not enriched for VACV. The process of vesicle-mediated transport is highly enriched within the proteome on VACV DNA but not Ad5 and HSV-1, consistent with VACV DNA replication depending on the endoplasmic reticulum (ER) membrane wrapping around cytoplasmic VRCs (9). Another biological process highly represented in VACV, but not in the two nuclear viruses is the mitotic cell cycle process. Proteins enriched in VACV and assigned to this category include members of the condensing complex and both isoforms of topoisomerase type 2, which might be recruited to help condense the 191 kb genome of this virus. Interestingly, the rest of the host factors found within this category have been implicated in the formation and stabilization of kinetochore fibers. These proteins include Tubulin alpha and beta, which are major structural components of microtubules (71) and Clathrin and ch-TOG, which along with TACC3 form a complex that bridges microtubules to stabilize the kinetochore fibers during chromosome segregation (72). The elevated association of this set of proteins with replicating and replicated VACV genomes may hint at a mechanism by which VACV translocates its large genome within cytoplasmic VRCs during replication.
Among the processes commonly manipulated by all three viruses were cellular DNA replication and/or DNA repair. Of all host DNA replication factors identified, Claspin was the most depleted in the replicated-DNA proteomes of Ad5 and HSV-1 and only second to ORC2 in the proteome of VACV. Consistent with these data, we observed Claspin excluded almost completely from VRCs in cells infected with Ad5 by immunofluorescence analysis (supplemental Fig. S4). Claspin associates with Chk1 on replication stress and DNA damage to activate Chk1 kinase activity, which can lead to the slowing or stalling of DNA replication (73–75). Hence, the interaction of Claspin and Chk1 on replicating viral genomes and the resulting signaling cascade could potentially impact the efficiency of viral replication. Our results may reflect an active effort of DNA viruses to keep Claspin from associating with their genome to facilitate the replication process. Moreover, there appears to be differential usage of host DNA replication and repair machinery between the viruses tested. For example we found HRR factors recruited to HSV-1 replicated genomes, where they have been reported to promote virus infection (10, 11, 50). In contrast, some HRR components were excluded from Ad5 replicated genomes, consistent with reports that the MRN complex inhibits virus replication (48, 49, 76). Despite some HRR complexes being depleted during Ad5 infection, we found SLX4 recruited to Ad5 VRCs. We showed that SLX4 promotes Ad5 genome accumulation and protein production. Similarly, we found SLX4 associated with HSV-1 genomes and showed that SLX4 promotes viral protein production and progeny production during HSV-1 infection. SLX4 is a multifunctional protein that promotes the repair of several types of DNA lesions (77). SLX4 functions include coordinating structure-specific endonucleases (78–82) and SUMO E3 ligase activity (83). Although it remains to be determined what this multifunctional protein does during virus infection, our results provide evidence that Ad5 and HSV-1 coopt SLX4 to advance viral processes. VACV replication factories accumulated both topoisomerase I and ERCC6L, which have recently been shown to be involved in proper chromosome segregation (84), and it will be interesting to determine how they are used for viral replication.
One notable finding of our study was the enrichment of nucleolar proteins on Ad5 viral DNA. Although previous reports have suggested manipulation of some nucleolar components by Ad (85–88), our study provides a comprehensive list of nucleolar proteins that accumulate on replicated viral genomes. Our data clearly show enrichment for components of the SSU processome. Functionally, many SSU processome components participate in posttranscriptional modification of rRNA, such as pseudouridylation and 2′O-ribose methylation (89). It was recently demonstrated that box H/ACA and C/D snoRNPs associated with RNA POL I to change ribosome translational output during neural crest specification (65). This association was driven by the formation of a TCOF1-NOLC1 protein scaffold induced on mono-ubiquitination of both proteins. We demonstrated that core proteins of both types of snoRNP complexes, RNA POL I, TCOF1, and NOLC1, all accumulated at Ad5 replication centers. Except for DKC1, a component of box H/ACA snoRNPs, the localization was dependent on TCOF1. Furthermore, we observed that TCOF1 depletion reduced levels of Ad5 late proteins and affected viral DNA accumulation and viral yield. SSU processome components may be exploited by Ad5 to modify viral mRNA and promote translation and production of viral late proteins. SSU processome protein components were also “enriched” on the nascent genomes of HSV-1, but not VACV suggesting a global role in the life cycle of DNA viruses that replicate in the nucleus of infected cells.
Our analysis identified TFII-I as a target of early Ad5 proteins. TFII-I functions as both an activator and repressor of gene expression, and has been implicated in the DNA damage response as well as interferon responses to virus infections (55). Two recent proteomic analyses of proteins associated with nascent cellular DNA also identified TFII-I among enriched factors, and depletion suggested a potential role in DNA replication (19, 20). In addition, TFII-I was identified as a substrate for post-translational SUMOylation induced by the Ad5 E4orf3 protein (90), and as a negative regulator of the viral L4 promoter (91, 92). TFII-I was recently shown to target the CCCTC-binding factor CTCF to gene promoters (93), and because CTCF may represent a common repressor of DNA viruses (94), TFII-I may be targeted as a means of preventing repression. The emerging model from our work and these recent reports is that E4orf3 is required to induce proteasome-mediated degradation of TFII-I. This promotes exclusion of TFII-I from Ad5 DNA to relieve repression of viral promoters during infection. Comparing replicated-DNA proteomes recovered from infections with wild-type and mutant viruses lacking viral proteins that antagonize intrinsic defenses could make this approach even more powerful for probing how host factors associating with viral genomes can restrict infection.
In conclusion, our study outlines a robust, unbiased proteomics approach to identify and classify host factors associated with replicated DNA during virus infection. Our pan-viral comparison of DNA viruses with distinct coding capacity and replication characteristics provides a framework for probing virus-host interactions taking place on replicating viral genomes. This work highlights the common and distinct strategies employed by different viruses to redirect cellular machinery toward viral DNA replication. We anticipate that our proteomic approach will be easily adapted to other replicating DNA viruses. Our findings regarding Ad5 infection demonstrate the use of our proteomic data sets, which will likely yield additional valuable insights into virus-host interactions. Further investigation of the mechanistic basis for how host factors are targeted or recruited will provide critical insight into virus replication and host defense strategies that are inactivated. The proteome profiles generated provide a rich resource to identify cellular factors that functionally associate with viral nucleic acids and uncover a wealth of new virus-host interactions and insights into key host components regulating virus DNA replication.
DATA AVAILABILITY
The MS raw files associated with this manuscript have been deposited to the public database ProteomeXchange. This data can be found under project number: PXD007741 at the following website: http:www.ebi.ac.uk/pride/archive/projects/PXD007741.
Supplementary Material
Acknowledgments
We thank members of the Weitzman Lab for insightful discussions and input. We thank Ariella Sasson, Pichai Raman and Perry Evans for help with bioinformatics. We thank Christopher McKennan for help with MS analysis. We thank Annie Chen, Frances Taschuk, Shubham Chattopadhyay, Cipriano Zuluaga, Micah Weitzman, Mansee Patel and Morgann Reed for technical assistance. We thank the Penn CDB Microscopy Core for imaging assistance. We are grateful to Arnold Levine, Thomas Dobner, James Wilson, David Knipe and David Evans for generously sharing antibodies. We thank Agata Smogorzewska for SLX4 cells. We thank Chris Boutell, Eric Brown, Paul Lieberman, Adam Resnick, and Jonathan Weitzman for critical reading of the manuscript.
Footnotes
Author contributions: M.D.W. conceived the project and directed the study. E.D.R. and M.D.W designed experiments; E.D.R., N.J.P., L.N.A., D.C.A., E.T.K., D.B., and S.K. performed the experiments; K.K. and S.H.S. performed MS analysis; K.K. and E.D.R. performed bioinformatics analysis; S.N.I. assisted with VACV infections. E.T.K., N.J.P., S.N.I., S.H.S., and B.A.G. advised on experimental design and interpretation; E.D.R. and M.D.W. interpreted the data and wrote the manuscript and all authors were involved in editing the manuscript.
* This work was supported by grants to M.D.W. from the National Institutes of Health (NS082240, CA097093 and AI115104), a Catalyst Grant from the Institute for Immunology of the University of Pennsylvania, and the Foerderer Award from the Children's Hospital of Philadelphia. B.A.G acknowledges funding from the National Institutes of Health (AI118891 and GM110174). E.D.R. was supported by an Academic Diversity Postdoctoral Fellowship from the Office of the Vice Provost for Research at the University of Pennsylvania and by an NIH Research Supplement to Support Diversity (NS082240). N.J.P. was supported in part by T32 NS007180. D.C.A. was supported in part by T32 CA115299 and F32 GM112414. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Ad5
- adenovirus C type 5
- E1
- adenovirus early coding region 1
- E4
- adenovirus early coding region 4
- HSV-1
- herpes simplex virus type-1
- VACV
- vaccinia virus
- VRCs
- viral replication compartments
- MOI
- multiplicity of infection
- hpi
- hours post infection
- EdU
- 5-ethynyl-2′-deoxyuridine
- BrdU
- 5-bromo-2′-deoxyuridine
- DAPI
- 4′,6-diamidino-2-phenylindole, dihydrochloride
- CHX
- cycloheximide
- iPOND
- isolation of proteins on nascent DNA
- NHEJ
- nonhomologous end joining
- BER
- base excision repair
- NER
- nucleotide excision repair
- ICL
- interstrand crosslink repair
- HRR
- homologous recombination repair
- VRCs
- viral replication centers
- GO
- gene ontology
- KOAc
- potassium acetate
- DTT
- dithiothreitol
- iBAQ
- intensity-based absolute quantification.
REFERENCES
- 1. Goebel S. J., Johnson G. P., Perkus M. E., Davis S. W., Winslow J. P., and Paoletti E. (1990) The complete DNA sequence of vaccinia virus. Virology 179, 247–266, 517–563 [DOI] [PubMed] [Google Scholar]
- 2. Moss B. (2013) Poxvirus DNA replication. Cold Spring Harb. Perspect. Biol. 5, a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boyle K. A., Greseth M. D., and Traktman P. (2015) Genetic confirmation that the H5 protein is required for vaccinia virus DNA replication. J. Virol. 89, 6312–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Challberg M. D., and Kelly T. J. (1989) Animal virus DNA replication. Annu. Rev. Biochem. 58, 671–717 [DOI] [PubMed] [Google Scholar]
- 5. Challberg M. D. (1986) A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl. Acad. Sci. U.S.A. 83, 9094–9098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmid M., Speiseder T., Dobner T., and Gonzalez R. A. (2014) DNA virus replication compartments. J. Virol. 88, 1404–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Puvion-Dutilleul F., and Puvion E. (1990) Replicating single-stranded adenovirus type 5 DNA molecules accumulate within well-delimited intranuclear areas of lytically infected HeLa cells. Eur. J. Cell Biol. 52, 379–388 [PubMed] [Google Scholar]
- 8. Quinlan M. P., Chen L. B., and Knipe D. M. (1984) The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36, 857–868 [DOI] [PubMed] [Google Scholar]
- 9. Tolonen N., Doglio L., Schleich S., and Krijnse Locker J. (2001) Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 12, 2031–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor T. J., and Knipe D. M. (2004) Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78, 5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilkinson D. E., and Weller S. K. (2004) Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78, 4783–4796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ebert S. N., Subramanian D., Shtrom S. S., Chung I. K., Parris D. S., and Muller M. T. (1994) Association between the p170 form of human topoisomerase II and progeny viral DNA in cells infected with herpes simplex virus type 1. J. Virol. 68, 1010–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katsafanas G. C., and Moss B. (2007) Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2, 221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oh J., and Broyles S. S. (2005) Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J. Virol. 79, 12852–12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin Y.-C. J., Li J., Irwin C. R., Jenkins H., DeLange L., and Evans D. H. (2008) Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J. Virol. 82, 5922–5932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paran N., De Silva F. S., Senkevich T. G., and Moss B. (2009) Cellular DNA ligase I is recruited to cytoplasmic vaccinia virus factories and masks the role of the vaccinia ligase in viral DNA replication. Cell Host Microbe 6, 563–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Jong R. N., and van der Vliet P. C. (1999) Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene 236, 1–12 [DOI] [PubMed] [Google Scholar]
- 18. Sirbu B. M., McDonald W. H., Dungrawala H., Badu-Nkansah A., Kavanaugh G. M., Chen Y., Tabb D. L., and Cortez D. (2013) Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 288, 31458–31467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopez-Contreras A. J., Ruppen I., Nieto-Soler M., Murga M., Rodriguez-Acebes S., Remeseiro S., Rodrigo-Perez S., Rojas A. M., Mendez J., Muñoz J., and Fernandez-Capetillo O. (2013) A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 3, 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alabert C., Bukowski-Wills J.-C., Lee S.-B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., and Groth A. (2014) Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sirbu B. M., Couch F. B., Feigerle J. T., Bhaskara S., Hiebert S. W., and Cortez D. (2011) Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25, 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sirbu B. M., Couch F. B., and Cortez D. (2012) Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7, 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang I.-H., Suomalainen M., Andriasyan V., Kilcher S., Mercer J., Neef A., Luedtke N. W., and Greber U. F. (2013) Tracking viral genomes in host cells at single-molecule resolution. Cell Host Microbe 14, 468–480 [DOI] [PubMed] [Google Scholar]
- 24. Dembowski J. A., and DeLuca N. A. (2015) Selective recruitment of nuclear factors to productively replicating herpes simplex virus genomes. PLoS Pathog. 11, e1004939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dungrawala H., Rose K. L., Bhat K. P., Mohni K. N., Glick G. G., Couch F. B., and Cortez D. (2015) The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carson C. T., Schwartz R. A., Stracker T. H., Lilley C. E., Lee D. V., and Weitzman M. D. (2003) The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22, 6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim Y., Lach F. P., Desetty R., Hanenberg H., Auerbach A. D., and Smogorzewska A. (2011) Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 43, 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bridge E., and Ketner G. (1989) Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63, 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bridge E., and Ketner G. (1990) Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174, 345–353 [DOI] [PubMed] [Google Scholar]
- 30. Stracker T. H., Carson C. T., and Weitzman M. D. (2002) Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418, 348–352 [DOI] [PubMed] [Google Scholar]
- 31. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 32. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 33. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 34. Pombo A., Ferreira J., Bridge E., and Carmo-Fonseca M. (1994) Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13, 5075–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hancock M. H., Corcoran J. A., and Smiley J. R. (2006) Herpes simplex virus regulatory proteins VP16 and ICP0 counteract an innate intranuclear barrier to viral gene expression. Virology 352, 237–252 [DOI] [PubMed] [Google Scholar]
- 36. Lin Y.-C. J., and Evans D. H. (2010) Vaccinia virus particles mix inefficiently, and in a way that would restrict viral recombination, in coinfected cells. J. Virol. 84, 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Bruyn Kops A., and Knipe D. M. (1988) Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55, 857–868 [DOI] [PubMed] [Google Scholar]
- 38. Nakamura H., Morita T., and Sato C. (1986) Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res. 165, 291–297 [DOI] [PubMed] [Google Scholar]
- 39. Leonhardt H., Rahn H. P., Weinzierl P., Sporbert A., Cremer T., Zink D., and Cardoso M. C. (2000) Dynamics of DNA replication factories in living cells. J. Cell Biol. 149, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Halbert D. N., Cutt J. R., and Shenk T. (1985) Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56, 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Joklik W. K., and Becker Y. (1964) The replication and coating of vaccinia DNA. J. Mol. Biol. 10, 452–474 [DOI] [PubMed] [Google Scholar]
- 42. Hoeben R. C., and Uil T. G. (2013) Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 5, a013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weller S. K., and Coen D. M. (2012) Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 4:a013011, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Weitzman M. D., Lilley C. E., and Chaurushiya M. S. (2010) Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64, 61–81 [DOI] [PubMed] [Google Scholar]
- 45. Luftig M. A. (2014) Viruses and the DNA damage response: activation and antagonism. Annu. Rev. Virol. 1, 605–625 [DOI] [PubMed] [Google Scholar]
- 46. Turnell A. S., and Grand R. J. (2012) DNA viruses and the cellular DNA-damage response. J. Gen. Virol. 93, 2076–2097 [DOI] [PubMed] [Google Scholar]
- 47. Orazio N. I., Naeger C. M., Karlseder J., and Weitzman M. D. (2011) The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J. Virol. 85, 1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Evans J. D., and Hearing P. (2005) Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79, 6207–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mathew S. S., and Bridge E. (2007) The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology 365, 346–355 [DOI] [PubMed] [Google Scholar]
- 50. Lilley C. E., Carson C. T., Muotri A. R., Gage F. H., and Weitzman M. D. (2005) DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U.S.A. 102, 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weitzman M. D., and Ornelles D. A. (2005) Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24, 7686–7696 [DOI] [PubMed] [Google Scholar]
- 52. Doucas V., Ishov A. M., Romo A., Juguilon H., Weitzman M. D., Evans R. M., and Maul G. G. (1996) Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10, 196–207 [DOI] [PubMed] [Google Scholar]
- 53. Schreiner S., Wimmer P., and Dobner T. (2012) Adenovirus degradation of cellular proteins. Future Microbiol. 7, 211–225 [DOI] [PubMed] [Google Scholar]
- 54. Schwartz R. A., Lakdawala S. S., Eshleman H. D., Russell M. R., Carson C. T., and Weitzman M. D. (2008) Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 82, 9043–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Roy A. L. (2012) Biochemistry and biology of the inducible multifunctional transcription factor TFII-I: 10years later. Gene 492, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shepard R. N., and Ornelles D. A. (2003) E4orf3 is necessary for enhanced S-phase replication of cell cycle-restricted subgroup C adenoviruses. J. Virol. 77, 8593–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang M. M., and Hearing P. (1989) Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63, 2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Carson C. T., Orazio N. I., Lee D. V., Suh J., Bekker-Jensen S., Araujo F. D., Lakdawala S. S., Lilley C. E., Bartek J., Lukas J., and Weitzman M. D. (2009) Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 28, 652–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sloan K. E., Bohnsack M. T., Schneider C., and Watkins N. J. (2014) The roles of SSU processome components and surveillance factors in the initial processing of human ribosomal RNA. RNA 20, 540–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lafontaine D. L. J. (2015) Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22, 11–19 [DOI] [PubMed] [Google Scholar]
- 61. Phipps K. R., Charette J. M., and Baserga S. J. (2011) The small subunit processome in ribosome biogenesis—progress and prospects. RNA 2, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hayano T., Yanagida M., Yamauchi Y., Shinkawa T., Isobe T., and Takahashi N. (2003) Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J. Biol. Chem. 278, 34309–34319 [DOI] [PubMed] [Google Scholar]
- 63. Lin C.-I., and Yeh N.-H. (2009) Treacle recruits RNA polymerase I complex to the nucleolus that is independent of UBF. Biochem. Biophys. Res. Commun. 386, 396–401 [DOI] [PubMed] [Google Scholar]
- 64. Ciccia A., Huang J.-W., Izhar L., Sowa M. E., Harper J. W., and Elledge S. J. (2014) Treacher Collins syndrome TCOF1 protein cooperates with NBS1 in the DNA damage response. Proc. Natl. Acad. Sci. U.S.A. 111, 18631–18636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Werner A., Iwasaki S., McGourty C. A., Medina-Ruiz S., Teerikorpi N., Fedrigo I., Ingolia N. T., and Rape M. (2015) Cell-fate determination by ubiquitin-dependent regulation of translation. Nature 525, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hidalgo P., Anzures L., Hernández-Mendoza A., Guerrero A., Wood C. D., Valdés M., Dobner T., and Gonzalez R. A. (2016) Morphological, biochemical, and functional study of viral replication compartments isolated from adenovirus-infected cells. J. Virol. 90, 3411–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lenarcic E. M., Landry D. M., Greco T. M., Cristea I. M., and Thompson S. R. (2013) Thiouracil cross-linking mass spectrometry: a cell-based method to identify host factors involved in viral amplification. J. Virol. 87, 8697–8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sugimoto A., Kanda T., Yamashita Y., Murata T., Saito S., Kawashima D., Isomura H., Nishiyama Y., and Tsurumi T. (2011) Spatiotemporally different DNA repair systems participate in Epstein-Barr virus genome maturation. J. Virol. 85, 6127–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mercer J., Snijder B., Sacher R., Burkard C., Bleck C. K. E., Stahlberg H., Pelkmans L., and Helenius A. (2012) RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep. 2, 1036–1047 [DOI] [PubMed] [Google Scholar]
- 70. Sivan G., Martin S. E., Myers T. G., Buehler E., Szymczyk K. H., Ormanoglu P., and Moss B. (2013) Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 110, 3519–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cassimeris L., and Skibbens R. V. (2003) Regulated assembly of the mitotic spindle: a perspective from two ends. Curr. Issues Mol. Biol. 5, 99–112 [PubMed] [Google Scholar]
- 72. Booth D. G., Hood F. E., Prior I. A., and Royle S. J. (2011) A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 30, 906–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chini C. C. S., and Chen J. (2003) Human claspin is required for replication checkpoint control. J. Biol. Chem. 278, 30057–30062 [DOI] [PubMed] [Google Scholar]
- 74. Lin S.-Y., Li K., Stewart G. S., and Elledge S. J. (2004) Human Claspin works with BRCA1 to both positively and negatively regulate cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 101, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang Y., and Hunter T. (2014) Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 134, 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lakdawala S. S., Schwartz R. A., Ferenchak K., Carson C. T., McSharry B. P., Wilkinson G. W., and Weitzman M. D. (2008) Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J. Virol. 82, 8362–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kottemann M. C., and Smogorzewska A. (2013) Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493, 356–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Andersen S. L., Bergstralh D. T., Kohl K. P., LaRocque J. R., Moore C. B., and Sekelsky J. (2009) Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell 35, 128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fekairi S., Scaglione S., Chahwan C., Taylor E. R., Tissier A., Coulon S., Dong M.-Q., Ruse C., Yates J. R., Russell P., Fuchs R. P., McGowan C. H., and Gaillard P.-H. L. (2009) Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Muñoz I. M., Hain K., Déclais A.-C., Gardiner M., Toh G. W., Sanchez-Pulido L., Heuckmann J. M., Toth R., Macartney T., Eppink B., Kanaar R., Ponting C. P., Lilley D. M. J., and Rouse J. (2009) Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell 35, 116–127 [DOI] [PubMed] [Google Scholar]
- 81. Saito T. T., Youds J. L., Boulton S. J., and Colaiácovo M. P. (2009) Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 5, e1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Svendsen J. M., Smogorzewska A., Sowa M. E., O'Connell B. C., Gygi S. P., Elledge S. J., and Harper J. W. (2009) Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Guervilly J.-H., Takedachi A., Naim V., Scaglione S., Chawhan C., Lovera Y., Despras E., Kuraoka I., Kannouche P., Rosselli F., and Gaillard P.-H. L. (2015) The SLX4 complex is a SUMO E3 ligase that impacts on replication stress outcome and genome stability. Mol. Cell 57, 123–137 [DOI] [PubMed] [Google Scholar]
- 84. Nielsen C. F., Huttner D., Bizard A. H., Hirano S., Li T.-N., Palmai-Pallag T., Bjerregaard V. A., Liu Y., Nigg E. A., Wang L. H.-C., and Hickson I. D. (2015) PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat. Commun. 6, 8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Okuwaki M., Matsumoto K., Tsujimoto M., and Nagata K. (2001) Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Letters 506, 272–276 [DOI] [PubMed] [Google Scholar]
- 86. Lawrence F. J. (2006) Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis. J. Cell Sci. 119, 2621–2631 [DOI] [PubMed] [Google Scholar]
- 87. Hindley C. E., Davidson A. D., and Matthews D. A. (2007) Relationship between adenovirus DNA replication proteins and nucleolar proteins B23.1 and B23.2. J. Gen. Virol. 88, 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lam Y. W., Evans V. C., Heesom K. J., Lamond A. I., and Matthews D. A. (2010) Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol. Cell. Proteomics 9, 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maden B. E. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 39, 241–303 [DOI] [PubMed] [Google Scholar]
- 90. Sohn S.-Y., Bridges R. G., and Hearing P. (2015) Proteomic analysis of ubiquitin-like posttranslational modifications induced by the adenovirus E4-ORF3 protein. J. Virol. 89, 1744–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wright J., Atwan Z., Morris S. J., and Leppard K. N. (2015) The human adenovirus type 5 L4 promoter is negatively regulated by TFII-I and L4–33K. J. Virol. 89, 7053–7063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bridges R. G., Sohn S.-Y., Wright J., Leppard K. N., and Hearing P. (2016) The adenovirus E4-ORF3 protein stimulates SUMOylation of general transcription factor TFII-I to direct proteasomal degradation. mBio 7, e02184-15-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Peña-Hernández R., Marques M., Hilmi K., Zhao T., Saad A., Alaoui-Jamali M. A., del Rincon S. V., Ashworth T., Roy A. L., Emerson B. M., and Witcher M. (2015) Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc. Natl. Acad. Sci. U.S.A. 112, E677–E86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Pentland I., and Parish J. L. (2015) Targeting CTCF to control virus gene expression: a common theme amongst diverse DNA viruses. Viruses 7, 3574–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS raw files associated with this manuscript have been deposited to the public database ProteomeXchange. This data can be found under project number: PXD007741 at the following website: http:www.ebi.ac.uk/pride/archive/projects/PXD007741.