Abstract
The ubiquitous mold Aspergillus fumigatus threatens immunosuppressed patients as inducer of lethal invasive aspergillosis. A. fumigatus conidia are airborne and reach the alveoli, where they encounter alveolar epithelial cells (AEC). Previous studies reported the importance of the surfactant-producing AEC II during A. fumigatus infection via in vitro experiments using cell lines. We established a negative isolation protocol yielding untouched primary murine AEC II with a purity >90%, allowing ex vivo analyses of the cells, which encountered the mold in vivo. By label-free proteome analysis of AEC II isolated from mice 24h after A. fumigatus or mock infection we quantified 2256 proteins and found 154 proteins to be significantly differentially abundant between both groups (ANOVA p value ≤ 0.01, ratio of means ≥1.5 or ≤0.67, quantified with ≥2 peptides). Most of these proteins were higher abundant in the infected condition and reflected a comprehensive activation of AEC II on interaction with A. fumigatus. This was especially represented by proteins related to oxidative phosphorylation, hence energy production. However, the most strongly induced protein was the l-amino acid oxidase (LAAO) Interleukin 4 induced 1 (IL4I1) with a 42.9 fold higher abundance (ANOVA p value 2.91−10). IL4I1 has previously been found in B cells, macrophages, dendritic cells and rare neurons. Increased IL4I1 abundance in AEC II was confirmed by qPCR, Western blot and immunohistology. Furthermore, A. fumigatus infected lungs showed high levels of IL4I1 metabolic products. Importantly, higher IL4I1 abundance was also confirmed in lung tissue from human aspergilloma. Because LAAO are key enzymes for bactericidal product generation, AEC II might actively participate in pathogen defense. We provide insights into proteome changes of primary AEC II thereby opening new avenues to analyze the molecular changes of this central lung cell on infectious threats. Data are available via ProteomeXchange with identifier PXD005834.
Aspergillus fumigatus is an ubiquitous saprophytic fungus (1) that grows on decaying organic matter. Due to their small size and high surface charge, conidia, the spores of A. fumigatus, are airborne and hundreds are inhaled by humans daily together with the breathing air (2). Although healthy individuals quickly destroy inhaled conidia via immune-related mechanisms individuals suffering from immune suppression, e.g. because of chemotherapy or after solid organ transplantation, cannot effectively clear the pathogen and a life-threatening infection termed invasive aspergillosis (IA)1 establishes. Despite the existence of antifungal drugs IA is still a major cause of infection-related deaths on clinical wards treating immune-compromised patients (3, 4) which is because of the poor diagnosis and often in-effective therapies (5).
The lung-resident immune system of healthy people effectively controls the growth of inhaled conidia. Under healthy conditions the lung respiratory surface is populated with alveolar macrophages (AM) that phagocytose conidia and serve as sentinel cells for the recruitment of neutrophil granulocytes (Nφ) from the peripheral blood (6, 7). Nφ are extremely effective in phagocytosing conidia (8, 9), which is the reason why especially lack or dysfunction of Nφ is associated with a highly increased risk for IA development (10–12). Although these phagocytotic members of the lung-associated immune system are all well studied, it is obvious, that alveolar epithelial cells (AEC), that make up the entire more than 100 m2 large respiratory surface of the lung (12) have a much higher likelihood of physically engaging freshly inhaled conidia before any other cell of the immune system (13). However, the effects of this interaction are not known.
AEC are the most abundant cells of the lung epithelium. There are two types: AEC I and II. AEC I are very flat cells that form the respiratory epithelium (14). AEC II are cuboid cells located in corners of alveoli which are essential to produce surfactant, a protein-rich liquid covering the inner alveolar surface (15). In addition, AEC II have already been shown to have immunological functions (16, 17). The fact that AEC are pioneer cells to interact with inhaled conidia has led to many studies investigating their interaction with the fungus (summarized in (14)). Due to the lack of methods for isolation and culture there are no reports investigating murine AEC I. But also, most studies investigating the effect of A. fumigatus interaction with AEC II employed in vitro experiments using cell lines (primarily the A549 human alveolar carcinoma line) or primary human, rat and murine cells cultured in vitro for several days. This has uncontrollable effects on the physiology of the cultured cells and therefore requires careful interpretation of the data and verification of the results in more physiological settings (see (14) and references therein). Furthermore, only very few studies were undertaken to obtain a comprehensive characterization of the molecular response of AEC II to the fungal insult (18).
Because direct isolation of AEC II from infected human patients is not possible, the closest available alternative is the isolation of primary AEC II from a suitable animal model. Murine models of A. fumigatus infection are well established and closely recapitulate the human condition (10). However, available approaches for isolating untouched primary AEC II from murine lung require flow-cytometry-based cell sorting, which by itself massively influences cellular physiology (16, 17).
Quantitative proteome analysis has been used intensively to study models of infection and host-pathogen interaction. Thereby unbiased analysis of activated immune cells or pathogen-challenged cells revealed previously unknown players involved in immune defense and pathogenic mechanisms (19, 20). In this study, we established a novel protocol for the negative immunomagnetic isolation of untouched AEC II from murine lung. We then used quantitative label-free proteome analysis to investigate proteome changes in these cells induced on infection with A. fumigatus conidia. Hence, we provide the first comprehensive characterization of proteome changes in primary AEC II during A. fumigatus infection.
MATERIALS AND METHODS
Ethics Statement
All animal experiments were in accordance with German guidelines and were approved by the relevant local authorities in Essen.
Animals
All animals were OlaHsd C57BL/6 female and between 8 and 12 weeks of age, purchased from ENVIGO (Horst, The Netherlands) or were bred at the animal facility of the University Essen, Germany.
Aspergillus fumigatus Strains
Wild-type A. fumigatus conidia ATCC46645 were used throughout the project (21). A transgenic tdTomato expressing A. fumigatus strain was used for microscopy purposes. It was generated and kindly provided by Prof. Dr. Sven Krappmann. This strain expresses tdTomato fluorescent protein in the cytoplasm under control of the GAPDH promoter (22).
EXPERIMENTAL PROCEDURES
Intratracheal Infection
C57BL/6 mice were anesthetized (20 mg/ml Ketamin, Inresa Arzneimittel GmbH, Freiburg im Breisgau, Germany, 2 mg/ml Xylazin 2%, Ceva, Düsseldorf, Germany, in NaCl 0.9%, B. Braun, Melsungen, Germany) and hanged by their front incisors vertically. The tongue was pulled aside by a laryngoscope and a 22 G Intravenous (IV) catheter was placed into the trachea. Subsequently, 50 μl A. fumigatus conidia with the concentration of 1 × 108/ml were instilled into the lung.
Negative Immunomagnetic Isolation of Primary Murine AEC II
The lung tissue was digested in 2 ml (100 caseinolytic units) dispase (BD Biosciences, Heidelberg, Germany, Cat # 354235) for 45 min at room temperature and the bronchi were cut out by a pair of very fine surgery scissors (Fine Science Tools, Heidelberg, Germany, Cat #14568–09). The digestion process, single cell suspension preparation and immunomagnetic cell separation was performed as previously described by Gereke et al. (23) with modifications in the antibody mixture design. 5 μl (2 μg) of each of the following biotin-coupled antibodies (all purchased from Biolegend, San Diego, CA) were mixed in 460 μl PBS: α-CD11b (Clone: M1/70), α-CD11c (Clone: N418), α-CD19 (Clone: 6D5), α-F4/80 (Clone: BM8), α-CD31 (Clone: 390), α-CD45 (Clone: 30-F11), α-Sca-1 (Clone: D7), and α-T1α (Clone: 8.1.1) (Table I). Next, the cells were resuspended in 300 μl MACS buffer (PBS supplemented with 1% V/V FCS and 2 mm EDTA) and 200 μl antibiotin microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany, Cat # 130–090-485). Finally, the suspension was transferred onto an LS magnetic column (Miltenyi Biotec, Cat # 130–042-401) and the column was washed 3 times with 3 ml MACS buffer, each. Untouched AEC II were collected in a 15 ml PE tube and centrifuged at 4 °C, 250 G for 10 min (Fig. 1). The pellets were kept at −80 °C.
Table I. Components of the antibody cocktail. AEC II were purified from murine lungs via immunomagnetic negative isolation by applying the antibody cocktail listed below.
| Target | Clone | μg antibody per 107 cells |
|---|---|---|
| Integrin alpha-M (CD11b) | M1/70 | 2 |
| Integrin alpha-X (CD11c) | N418 | 2 |
| B-lymphocyte antigen CD19 (CD19) | 6D5 | 2 |
| Platelet endothelial cell adhesion molecule (CD31) | 390 | 2 |
| Receptor-type tyrosine-protein phosphatase C (CD45) | 30-F11 | 2 |
| Adhesion G protein-coupled receptor E1 (F4/80) | BM8 | 2 |
| Podoplanin (T1α) | 8.1.1 | 2 |
| Lymphocyte antigen 6A-2/6E-1 (α-Sca-1) | D7 | 2 |
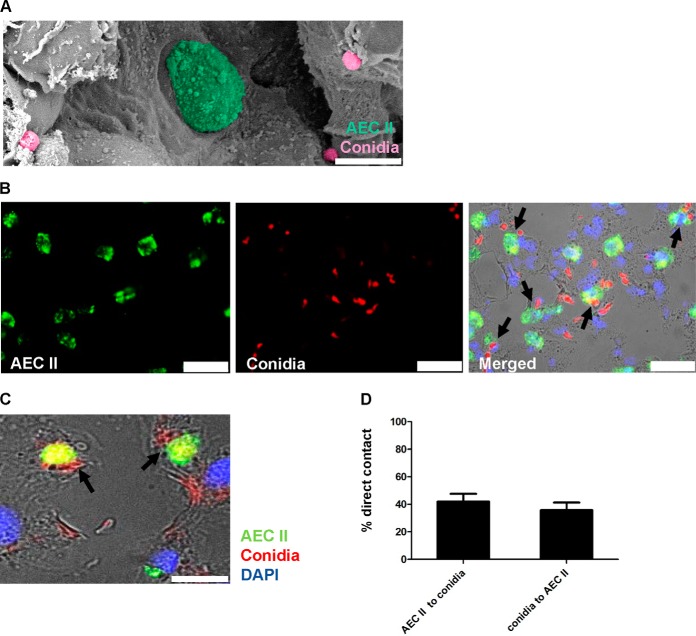
Fig. 1.
AEC II interact with A. fumigatus conidia. A, Scanning Electron Microscopy (S.E.M.) image of an alveolus in a murine lung 24 h post A. fumigatus infection. AEC II (green) and A. fumigatus conidia (pink) are in juxtaposition. Scale bar 10 μm. B, Immunofluorescent staining of a murine lung 24 h post A. fumigatus infection. AEC II (anti-Pro surfactant protein C (Alexa Fluor 488) green), transgenic conidia (tdTomato (red)) and nuclei (DAPI, blue). Direct interactions of AEC II and A. fumigatus conidia can be seen (black arrows). Scale bars 25 μm. C, Higher magnification of a direct contact between conidia and AEC II. Scale bar 10 μm. D, 42% of all AEC II were in direct contact to conidia and 35.8% of all conidia were in direct contact to AEC II. Data were from 5 independent fields of view analyzed from one infected lung.
Proteomics Experimental Design and Statistical Rationale
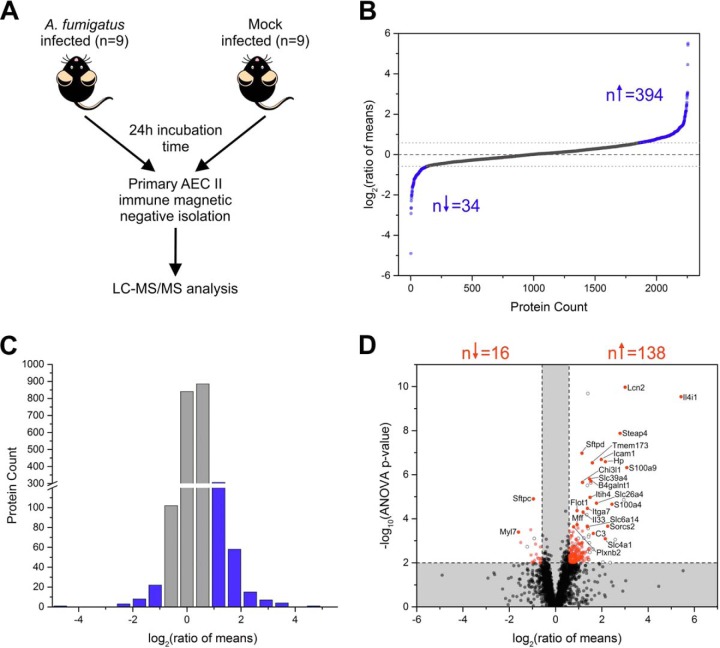
Isolated primary AEC II from A. fumigatus infected and mock (sterile tap water) infected C57BL/6 mice were collected. In preliminary experiments using 2-D DIGE we had established that the maximum change in protein composition between AEC II from uninfected versus infected lungs occurred at 24 h after infection (data not shown). Thus, we chose this time point for an in depth analysis of the AEC II proteome using mass spectrometry. For each sample 9 biological replicates were analyzed by label-free LC-MS/MS in alternating order (Fig. 3A). Statistical analysis was performed using ANOVA implemented in the Progenesis LC-MS software (ver. 4.1, Nonlinear Dynamics Ltd., Newcastle on Tyne, U.K.). We performed preexperiments with 5 replicates available for each experimental condition. Only 37% of proteins with an ANOVA p value below 0.05 had a power of at least 0.8 = 1-β. For the main study, we aimed at a percentage of at least 90% of the respective protein subset to have a power of at least 80%. According to the calculations, 8 samples were required to fulfill these criteria. Hence, we analyzed 9 replicates per group to account for the possible necessity to exclude a measurement for technical reasons.
Fig. 3.
Results of label-free quantitative proteome analysis of isolated primary AEC II. A, Schematic representation of the applied experimental set up. Primary AEC II were isolated from A. fumigatus infected and mock infected animals (9 replicates per group) and analyzed by label-free LC-MS/MS. B, Log2 transformed ratios of mean protein abundances of all quantified proteins (n = 2256). The dashed line indicates a log2 (ratio of means) of 0 (corresponding to a fold change of 1). The dotted lines represent the applied ratio of means cut offs of 1.5 and 0.67, respectively. 394 proteins showed a higher abundance in the A. fumigatus infected groups whereas only 34 were higher abundant in mock infected AEC II. C, Histogram showing the distribution of log2 transformed ratios of means for all quantified proteins. Blue bars represent proteins passing the applied ratio of means thresholds of 1.5 and 0.67. The histogram shows a shift toward positive ratios, corresponding to higher protein abundance in the A. fumigatus infected group. D, Volcano plot illustrating the results of the differential study. Ratios of means are displayed log2 transformed, ANOVA p values are presented -log10 transformed. Dashed lines represent the applied significance thresholds of an ANOVA p value ≤ 0.01 and a ratio of means ≥1.5 or ≤0.67 and proteins passing this threshold are highlighted in red (n = 154). Proteins displayed as full gray circle did not pass the significance threshold and proteins displayed as empty circle were quantified with only one peptide. The gene names are given for the top 25 proteins according to the Euclidian distance.
Tryptic Digestion
According to previous optimization experiments 4.6 × 105 AEC II were purified from each murine lung and lysed in 30 μl 50 mm ammonium bicarbonate with 0.1% RapiGest SF Surfactant (Cat# 186001861, Waters, Eschborn, Germany). The cells were briefly vortexed and sonicated 2 times for 1 min on ice in an ultrasonic bath. The protein concentration was determined using the Bradford assay (Bio-Rad, Hercules, CA). Four μg proteins in 20 μl were incubated at 95 °C for 5 min and the disulfide bonds were reduced with 5 mm DTT at 60 °C for 30 min. Thereafter, the samples were alkylated with 15 mm iodoacetamide in the dark at room temperature and 50 ng trypsin (SERVA Electrophoresis, Heidelberg, Germany) were added for enzymatic protein digestion. The samples were incubated at 37 °C for 16 h followed by 30 min incubation with 0.5% TFA at 37 °C for acidification. Precipitated RapiGest was removed by centrifugation before the samples were vacuum dried and subsequently dissolved in 0.1% TFA. The peptide concentration was determined by quantitative amino acid analysis as described previously (24).
LC-MS/MS Analysis
The RPLC-MS/MS analysis was performed using an Ultimate 3000 RSLCnano system online coupled to an Orbitrap Q-Exactive mass spectrometer (both Thermo Scientific, Bremen, Germany) as described earlier (19). Briefly, 350 ng peptides were injected in a volume of 15 μl and preconcentrated with 0.1% TFA on a trap column for 7 min (Acclaim PepMap 100, 300 μm × 5 mm, C18, 5 μm, 100 Å; flow rate 30 μl/min). Subsequently, the peptides were transferred to the analytical column (Acclaim PepMap RSLC, 75 μm × 50 cm, nano Viper, C18, 2 μm, 100 Å) and separated by a gradient from 5% to 40% solvent B over 98 min (solvent A: 0.1% FA, solvent B: 0.1% FA, 84% acetonitrile; flow rate 400 nL/min; column oven temperature 60 °C). The mass spectrometer was operated in data-dependent mode and full scan mass spectra were acquired at a resolution of 70,000 at 200 m/z within a mass range of 350–1400 m/z. The spray voltage was 1600 V (+), and the capillary temperature was 250 °C. The ten most abundant precursors (charge state + 2, + 3, + 4) were selected for MS/MS analysis, fragmented by higher-energy collisional dissociation (HCD) at a normalized collision energy of 27% and a fixed first mass at 130 m/z and measured in the orbitrap mass analyzer (m/z values initiating MS/MS were set on a dynamic exclusion list for 30 s, 35,000 resolution at 200 m/z).
Peptide Identification
Peptide identification was performed using Proteome Discoverer Software (ver. 1.4.1.14, Thermo Fisher Scientific, Rockford, IL). The mass spectra were searched against UniProtKB/Swiss-Prot database restricted to mus musculus (Uniprot/Swissprot-Release 2013_10; 541,561 entries, 16641 after taxonomy restriction) using the Mascot search engine (Matrix Sciences Ltd., London, UK, version 2.3). The mass tolerance was set to 5 ppm for precursor ions and 20 mmu for fragment ions. One tryptic missed cleavage was considered as well as chemical modifications of methionine (oxidation, dynamic) and cysteine (carbamidomethyl, static). The target decoy PSM validator implemented in proteome discoverer was used to estimate the peptide confidence and peptides that passed a false discovery rate < 1% (q-value < 0.01) were considered for analysis.
Protein Quantification
Ion intensity-based label-free quantification was performed using Progenesis LC-MS software. To account for retention time shifts the LC-MS runs were aligned to one run automatically chosen by the software. A master list of features considering retention time and m/z was generated in which only peptide ions with minimum three isotopic peaks and charges states +2, +3, and +4 were considered. The peptide identifications (peptides spectrum matches) from Proteome Discoverer were imported into the software and matched to the respective features. The protein abundances were calculated considering the normalized ion intensities of all nonconflicting peptides of a protein. An ANOVA p value ≤ 0.01, a ratio of means (A. fumigatus infected/Mock infected) ≥ 1.5 or ≤0.67 and a minimum number of two quantified peptides per protein were set as significance threshold. Proteins passing the threshold were significantly differentially abundant among the experimental groups and considered for further analysis. The significantly differentially abundant proteins were ranked according to the Euclidian distance which accounts for both the p value and the fold change:
| (Eq. 1) |
Confidence intervals were calculated using the normalized protein abundances derived from the Progenesis QI software. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD005834 and 10.6019/PXD005834.
Functional Annotation and Analysis
Proteins that passed the applied significance threshold were analyzed using PANTHER version 11 (25) and STRING v10.0 (26). In PANTHER the statistical overrepresentation test was performed while in STRING enrichment analysis was carried out. Annotations were based on gene ontology terms and KEGG pathways. For STRING network analysis the minimum required interaction score was set to 0.90 (highest confidence) and text mining was disabled as a source of interaction.
RT-PCR
mRNA was isolated from primary AEC II following the protocol from the Qiagen RNeasy kit (Qiagen, Venlo, The Netherlands, Cat # 74104). QuantiTect reverse transcription kit (Qiagen, Cat # 205310) was used for generation of cDNA from the isolated mRNA according to the manufacturer's protocol. RT-PCR was performed using gene specific forward and reverse primers for IL4I1 (Qiagen, Cat # QT00169505) expression relative to GAPDH (Qiagen, Cat # QT01658692) as the housekeeping gene and SYBR Green PCR master mix kit (Qiagen, Cat # 204074).
Immunoblot Analysis
Isolated primary AEC II were resuspended in 50 μl lysis buffer (PBS supplemented with 300 mm NaCl, 1% V/V Triton X-100 and 2 mm EDTA) per 1 × 106 cells and kept on ice for 30 min. After 15 min centrifugation at 4 °C and 14,000 rpm (centrifuge rotor FA-45–48-11, Eppendorf, Hamburg, Germany), protein concentration of the samples was measured by a BCA kit (Thermo Scientific, Cat # 23225). 20 μg of each sample was loaded per lane of a 10% SDS gel electrophoresis. The gel was blotted onto a PVDF membrane (Cat # 88518, Thermo Scientific) and the protein bands were detected by 1 μg/ml of a self-developed α-IL4I1 antibody (Rabbit monoclonal antibody developed by Epitomics, Inc. (Burlingame, CA). It is directed against a 17 amino acid epitope in the central part of the protein. It was tested and selected initially against the HEK cell line expressing IL4I1 or the nonexpressing cell line as described previously (27) and subsequently verified on human lymph node sections.) followed by an ECL kit application according to the manufacturer's protocol (SuperSignal WestDura Extended Duration Substrate, Thermo Scientific, Cat # 34075). The visualization of the bands was performed by an Imagequant LAS-4000 gel documentation system (GE Healthcare, Chicago, IL) with cooled CCD camera (Fujifilm, Tokyo, Japan).
Immunohistochemistry
C57Bl/6 mice were infected with 5 × 106 transgenic tdTomato conidia intratracheally as described (28) and the mice were sacrificed by CO2 asphyxiation 24 h post A. fumigatus infection. The lungs were dissected and fixed in 4% PFA overnight at 4 °C. Tissue dehydration was performed in the spin tissue processor (STP 120, Thermo Scientific) and the paraffin wax embedding process was performed using the tissue embedder (EG1150, Leica, Wetzlar, Germany). Finally, 5 μm sections were cut (microtome, HM 340 E, Thermo Scientific) and placed on Histobond slides (76 × 26 × 1 mm) (Marienfeld superior, Lauda-Königshofen, Germany). After the deparaffinization, antigen retrieval was performed (PBS supplemented with 10 mm Tris and 1 mm EDTA) at 98 °C for 30 min. The sections were blocked in 3% BSA and stained with 1 μg/ml α-IL4I1 antibody followed by biotinylated secondary antibody (1:200, Cat # BA-1000, Vector laboratories, Burlingame, CA) and an AF488 Streptavidin conjugate (1:500, Cat # S11223, Thermo Scientific). For AEC II staining 2 μg/ml α-proSP-C antibody (Cat# AB3786, Merck Millipore, MA) and goat α-rabbit-Alexa Fluor 488 (1:500, Cat # A11008, Invitrogen, Carlsbad, CA) were used. Samples were analyzed using a Leica DMI6000 microscope with a DFC365 FX monochrome digital camera and with a 100 x oil objective (HCX PL APO 100x/1.40–0.70, Leica). The tdTomato-fluorescence was detected with a 605/26 filter, AF488-fluorescence with a 525/18 filter and DAPI with 455/25 filter.
Analysis of the Branched-chain α-Ketoacids
Fifty milligrams of homogenized lung tissue was extracted with 300 μl cold 1 m perchloric acid. Insoluble material was removed by centrifugation for 10 min at 25,000 × g. For derivatization with OPD (o-phenylendiamine) reagent, 150 μl of the resulting supernatant was mixed with an equal volume of 25 mm OPD solution and derivatized by incubation at 50 °C for 30 min. After centrifugation for 10 min, the derivatized ketoacids were separated by reversed phase chromatography on an Acquity HSS T3 column (100 mm × 2.1 mm, 1.7 μm, Waters) connected to an Acquity H-class UPLC system. Prior separation, the column was heated to 40 °C and equilibrated with 5 column volumes of solvent A (0.1% formic acid in 10% acetonitrile) at a flow rate of 0.55 ml/min. Separation of ketoacid derivatives was achieved by increasing the concentration of solvent B (acetonitrile) in solvent A as follows: 2 min 2% B, 5 min 18% B, 5.2 min 22% B, 9 min 40% B, 9.1min 80% B and hold for 2 min, and return to 2% B in 2 min.
Scanning Electron Microscopy
Twenty-four hours post A. fumigatus infection the entire lung was dissected and fixed in 2.5% PFA overnight at 4 °C. The left lobe was segmented into four pieces and dehydration process with 30%, 50%, 70%, 80%, 96%, and 2 × 100% ethanol was performed, each step 15 min. Next, critical point drying (CPD7501, Polaron/Quorum Technologies Ltf., Lewes, UK) was carried out. The lung pieces were fixed on a sample holder (12.7 mm) and coated with a 7-nm thick layer of Platinum/Palladium with a Sputter Coater (Cressington Scientific Instruments Ltd., Hill, UK). The samples were visualized by the scanning electron microscope Hitachi S4000 (Hitachi High Technologies, Chiyoda, Tokyo Prefecture, Japan).
Confocal Microscopy
Five micron paraffin embedded infected lung slices were analyzed by a Leica TCS SP8 confocal microscope after IL4I1-specific staining. The system was installed with an Argon Ion (65 mW), HeNe (10 mW) and DPSS (20mW) lasers and a 25× (N2.1HCX IRAPO) objective and PMTs or hybrid nondescanned detectors (HyD). For visualization of IL4I1 the Argon laser and the PMT2 (497 nm–539 nm) detector were set. For visualization of tdTomato conidia the DPSS laser and a HyD (572 nm–654 nm) were used.
Human Aspergilloma Lung Tissue Immunohistochemistry
Ten cases of pulmonary aspergilloma in humans were detected by a fulltext search from the archive of the institute of pathology. All patients had given their informal consent to surgical resection and the study was approved by the local ethical board under the number 14–5845-BO. Tissue was fixed by buffered formaldehyde solution (4.5%) and cut according to the guidelines of surgical pathology. Slides were reviewed and one paraffin block of each case was selected showing aspergillus mycelium and surrounding tissue reaction. Further, 5 cases with bronchiectasis and other 5 cases with organizing pneumonia were selected as controls. Testing of blocks for scientific purposes was allowed by a decision of the ethics committee. For immunohistochemistry slides were prepared as usual. Staining was performed by an automatic unit (DAKO AutostainerPlus, Dako Colorado Inc., Fort Collins, CO) using a commercial antibody diluent and detection system (DAB Polymer, Zytomed Systems Berlin, Germany). The self-developed α-IL4I1 antibody was used in a dilution 1:500.
RESULTS
A. fumigatus conidia Interact with AEC II
It has been shown that AEC II play significant roles in association with A. fumigatus infection, albeit mostly using in vitro studies with cell lines (29–33). To get a more accurate insight, we studied the response of AEC II in lungs of mice after intratracheal infection with fungal spores. Fig. 1 A is a scanning electron microscopy (S.E.M.) image of AEC II 24 h post A. fumigatus infection showing conidia as well as AEC II (34). Immunohistochemistry of infected lungs confirmed a direct interaction between AEC II and conidia in vivo (Fig. 1B, 1C). To determine how many conidia interact with AEC II, the percentage of the AEC II in direct contact to conidia as well as the percentage of the conidia in direct contact to AEC II was quantified, which indicated that this event occurs frequently in the infected lungs (Fig. 1D).
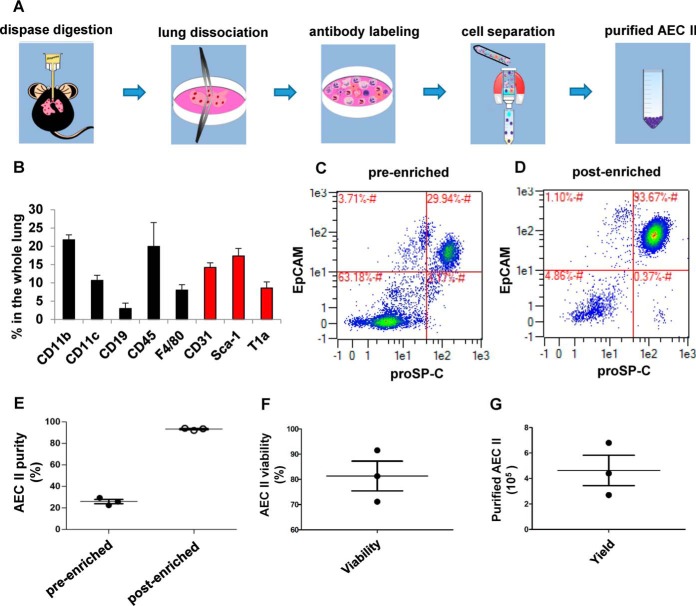
Primary AEC II Immunomagnetic Negative Isolation
Most existing studies on the molecular response of AEC II toward A. fumigatus infection were performed with the A549 cell line (35) whereas primary AEC II have not been studied so far. To reach this goal, we established an isolation protocol of AEC II from murine lung using immuno-magnetic negative isolation rather than the more intense single cell sorting approach (23). To allow tagging of all existing unwanted cell types in the whole lung with magnetically labeled antibodies, we used single cell suspensions of lungs after dispase digestion for cellular identification (Fig. 2A). We found that a mixture initially described by Gereke et al. (23) for cell sorting of AEC II (black bars in Fig. 2B) was insufficient for high quality negative isolation. Only after adding antibodies to α-Sca-1, α-T1α and α-CD31 to this mixture (red bars in Fig. 2B) we were able to label all the unwanted cells (Table I). The antibodies in this mixture were then prepared as biotin conjugates and used for immunomagnetic negative isolation together with anti-biotin magnetic microbeads. The data indicated that the population of AEC II (EpCAM+, proSP-C+) constitutes 29.9% ± S.E. of the whole lung in the presort state (Fig. 2C) and could be reproducibly enriched to >90% (Fig. 2D, 2E). The average AEC II viability after isolation was 81% (Fig. 2F) and the number of the AEC II yielded from this isolation protocol was 4.9 × 105 per murine lung on average (Fig. 2G).
Fig. 2.
Immunomagnetic negative isolation of primary AEC II. A, The schematic image shows the major steps of primary murine AEC II isolation. The lungs were digested by dispase and after dissociation all unwanted cell types in the single cell suspention were labeled by the designed biotinylated antibody mixture. The untouched AEC II were purified through immunomagnetic separation. B, Whole lung single cell suspension antibody staining and flow cytometry reveals the main cell types present in a murine lung after dispase digestion (black bars from an antibody mixture described in (23), red bars from antibodies found additionally in this study). Data are mean ± S.E. of three independent analyses. C, AEC II (EpCAM+ProSP-C+ cells) in a whole lung single cell suspension measured by flow cytometry. D, AEC II were isolated from murine lungs by negative immunomagnetic isolation using the antibody mixture defined in (B). Analysis of the resulting cells by flow cytometry as described in (C). E, The purity of AEC II before and after negative immunomagnetic isolation is highly reproducible. Each dot represents one indepentent analyzed mouse lung. F, Reproducibly high AEC II viability after the isolation process from the lungs in (E). G, Reproducible AEC II yield after the isolation process from the lungs in (E).
Label-free Proteome Analysis
AEC II were purified from A. fumigatus and mock infected animals 24 h post infection and they were further analyzed by LC-MS (Fig. 3A). In total, 20,750 peptides were identified that correspond to 2,703 protein groups (3,005 proteins) of which 2019 were identified with 2 unique peptides (supplemental Table S1). 2256 proteins were quantified using the Progenesis QI software (supplemental Table S2). 1668 proteins were quantified with minimum 2 peptides (as counted by the Progenesis software i.e. peptides measured with modifications or various charge states are counted multiple times). The median ratio of all quantified proteins was 1.05 with a maximum of 45.63 and a minimum of −29.85 (Fig. 3B). 394 proteins with a higher abundance in the A. fumigatus infected group showed a ratio greater than 1.5. Only 34 proteins that were higher abundant in the mock infected group passed the threshold of 1.5 thus illustrating a striking shift toward higher protein abundance in the A. fumigatus infected group (Fig. 3B and 3C). 154 proteins passed the applied significance filter criteria and were considered for further analyses. 138 of these proteins were higher abundant in the group of A. fumigatus infected AEC II whereas only 16 proteins showed a higher abundance in the mock infected group (Fig. 3D). The significantly differentially abundant proteins were ranked according to the Euclidian distance (supplemental Table S3). The 25 proteins with the highest Euclidian distance are summarized in Table II and highlighted in Fig. 3D. The protein showing the most remarkably increased abundance in A. fumigatus infected AEC II was an l-amino acid oxidase, also known as Interleukin 4 Induced-1 (O09046, IL4I1, ANOVA p value 2.91E-10, ratio of means 42.94, quantified with 6 unique peptides, supplemental Table S5). Therefore, based on the proteome analysis and the conceptual relevance of IL4I1 as an active player during different immune responses (36, 37) we selected IL4I1 for independent verification experiments.
Table II. Top25 significantly differentially abundant proteins. Isolated primary AEC II from A. fumigatus-infected and mock-infected mice were analyzed using label-free quantitative proteomics. The protein list shows the Top25 proteins according to the Euclidian distance.
| UniProt Accession | Gene | Protein | ANOVA p value | Fold Changea | Mean +/− 95% CI Mock infected groupb | Mean +/− 95% CI A. fumigatus infected group2 | deuclc |
|---|---|---|---|---|---|---|---|
| O09046 | IL4I1 | l-amino-acid oxidase | 2.91E-10 | 42.94 | 8624.18 [+/− 5931.43] | 370356.39 [+/− 118323.65] | 10.97 |
| P11672 | Lcn2 | Neutrophil gelatinase-associated lipocalin | 1.08E-10 | 8.06 | 1031450.00 [+/− 285770.00] | 8311040.00 [+/− 1824460.00] | 10.41 |
| Q923B6 | Steap4 | Metalloreductase STEAP4 | 1.34E-08 | 6.92 | 66085.60 [+/− 21384.02] | 457432.17 [+/− 125652.65] | 8.35 |
| P50404 | Sftpd | Pulmonary surfactant-associated protein D | 1.07E-07 | 2.21 | 1975420.00 [+/− 330780.00] | 4360470.00 [+/− 545120.00] | 7.07 |
| P31725 | S100a9 | Protein S100-A9 | 4.78E-07 | 8.46 | 37955.95 [+/− 20097.61] | 321132.66 [+/− 98290.51] | 7.03 |
| P13597 | Icam1 | Intercellular adhesion molecule 1 | 2.06E-07 | 3.94 | 777314.87 [+/− 260005.13] | 3063350.00 [+/− 574190.00] | 6.97 |
| Q61646 | Hp | Haptoglobin | 2.58E-07 | 4.46 | 286190.30 [+/− 121723.63] | 1277260.00 [+/− 365220.00] | 6.93 |
| Q3TBT3 | Tmem173 | Stimulator of interferon genes protein | 2.92E-07 | 3.01 | 57301.22 [+/− 9673.70] | 172515.91 [+/− 35899.68] | 6.73 |
| Q78IQ7 | Slc39a4 | Zinc transporter ZIP4 | 1.58E-06 | 2.78 | 384125.49 [+/− 82143.61] | 1067720.00 [+/− 232640.00] | 5.99 |
| Q09200 | B4galnt1 | Beta-1,4 N-acetylgalactosaminyltransferase 1 | 1.98E-06 | 2.89 | 125044.33 [+/− 23328.29] | 361341.13 [+/− 96077.11] | 5.90 |
| Q61362 | Chi3l1 | Chitinase-3-like protein 1 | 2.25E-06 | 2.24 | 4970190.00 [+/− 844150.00] | 11157000.00 [+/− 2003300.00] | 5.77 |
| P27005 | S100a8 | Protein S100-A8 | 2.21E-05 | 5.43 | 242576.79 [+/− 92733.75] | 1316290.00 [+/− 823660.00] | 5.26 |
| A6X935 | Itih4 | Inter alpha-trypsin inhibitor, heavy chain 4 | 1.08E-05 | 2.82 | 131852.24 [+/− 25175.45] | 371453.29 [+/− 131722.70] | 5.19 |
| Q9R155 | Slc26a4 | Pendrin | 1.98E-05 | 3.41 | 38027.57 [+/− 20490.86] | 129805.62 [+/− 26243.77] | 5.03 |
| P21841 | Sftpc | Pulmonary surfactant-associated protein C | 1.26E-05 | 0.52 | 46854400.00 [+/− 8163400.00] | 24177000.00 [+/− 3882500.00] | 4.99 |
| Q61738 | Itga7 | Integrin alpha-7 | 3.40E-05 | 2.60 | 335197.85 [+/− 122747.89] | 872546.66 [+/− 109406.50] | 4.68 |
| O08917 | Flot1 | Flotillin-1 | 4.35E-05 | 1.90 | 161653.03 [+/− 40381.33] | 307831.45 [+/− 42515.77] | 4.46 |
| Q8BVZ5 | Il33 | Interleukin-33 | 5.08E-05 | 2.29 | 1361550.00 [+/− 205950.00] | 3112180.00 [+/− 1051480.00] | 4.46 |
| Q9EPR5 | Sorcs2 | VPS10 domain-containing receptor SorCS2 | 0.0002 | 4.77 | 15232.56 [+/− 11822.13] | 72633.77 [+/− 24979.04] | 4.30 |
| Q9JMA9 | Slc6a14 | Sodium- and chloride-dependent neutral and basic amino acid transporter B(0+) | 0.0002 | 2.60 | 44186.70 [+/− 23923.40] | 114773.00 [+/− 18011.89] | 3.88 |
| Q6PCP5 | Mff | Mitochondrial fission factor | 0.0002 | 1.90 | 91102.58 [+/− 24768.45] | 173528.95 [+/− 32773.39] | 3.84 |
| P04919 | Slc4a1 | Band 3 anion transport protein | 0.0008 | 4.45 | 197822.54 [+/− 286988.43] | 879608.93 [+/− 684861.07] | 3.77 |
| Q9QVP4 | Myl7 | Myosin regulatory light chain 2, atrial isoform | 0.0004 | 0.33 | 821689.24 [+/− 442970.76] | 270501.63 [+/− 90726.22] | 3.75 |
| B2RXS4 | Plxnb2 | Plexin-B2 | 0.0002 | 1.72 | 566770.22 [+/− 142301.89] | 972886.08 [+/− 104653.92] | 3.71 |
| P01027 | C3 | Complement C3 | 0.0005 | 3.10 | 1381810.00 [+/− 321080.00] | 4289030.00 [+/− 2433310.00] | 3.71 |
aAll displayed proteins were quantified with minimum two unique peptides. Ratios of means were calculated A. fumigatus infected/Mock infected.
bThe 95% confidence intervals were calculated using the normalized protein abundances. Displayed are the mean protein abundance and the difference to the upper and lower bound, respectively.
cThe protein list was ranked according to the Euclidian distance
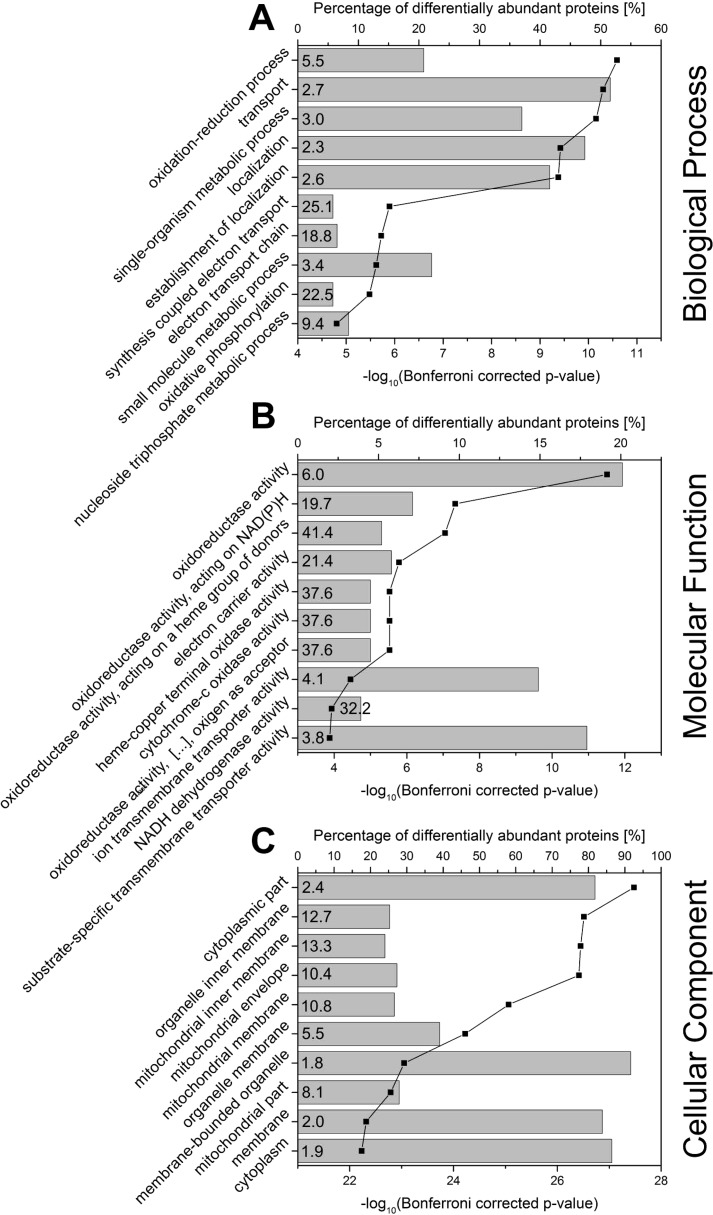
Functional Characterization of Differentially Abundant Proteins
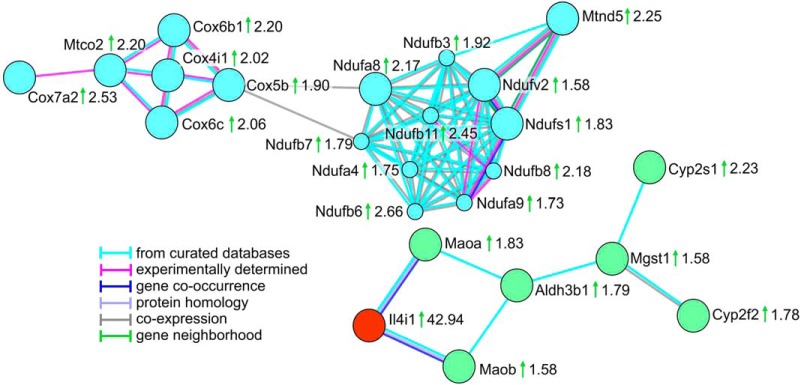
Using PANTHER, we performed the statistical overrepresentation test based on the GO annotations for biological process, molecular function and cellular component (Fig. 4, supplemental Table S4). For both, the biological process and the molecular function, the most significantly enriched GO terms were related to oxidation and reduction processes. Inspection of the proteins in the enriched categories revealed an obvious relation to oxidative phosphorylation. Several enriched GO terms like electron transport chain (biological process), electron carrier activity and NADH dehydrogenase activity (molecular function) also contained proteins related to this process. Oxidative phosphorylation was also represented by the enrichment analysis for the cellular component, because membrane proteins, especially of the mitochondrial membrane were strongly enriched. The enrichment analysis carried out with STRING showed similar results (supplemental Fig. S1). Analysis based on KEGG pathways demonstrated the enrichment of oxidative phosphorylation (pathway ID 00190, 17 proteins in the category, false discovery rate 2.66E-15). The STRING network analysis resulted in two main clusters in which all proteins also were related to oxidation-reduction processes. One of them contained proteins involved in oxidative phosphorylation and the electron transport chain (Fig. 5, blue circles, supplemental Fig. S2, supplemental Table S4). The second cluster (green circles) contained IL4I1 (red) with its direct interactors (Maoa, Q64133; Maob, Q8BW75) connected for their common function in amino acid metabolism. The more distant interactions in the cluster represented common functions in either amino acid metabolism or xenobiotic metabolism. However, none of the interactors in the cluster showed a similarly strongly increased abundance as IL4I1, thereby suggesting a specific role for IL4I1.
Fig. 4.
Results of Statistical Overrepresentation Test Based on Gene Ontology Annotations. A statistical overrepresentation test based on gene ontologies was performed for significantly differentially abundant proteins (n = 154) using PANTHER. Given are the top 10 overrepresented ontologies for (A) the biological process, (B) the molecular function, and (C) the cellular component. Bars represent the percentage of significantly differentially abundant proteins (n = 154) annotated with the enriched GO term. Data points and lines represent the corresponding Bonferroni-corrected p value. The enrichment factor (fold enrichment) calculated by PANTHER is displayed inside of the bars.
Fig. 5.
Main Protein Clusters Resulting from String Network Analysis. STRING network analysis was performed for significantly differentially abundant proteins (n = 154) considering only highest confidence interactions (interaction score 0.9, text mining as a source of interaction was disabled). The analysis resulted in two main protein clusters, one of them representing proteins related to oxidative phosphorylation (blue circles). The second cluster (green circles) contained proteins related to oxidation-reduction processes, one of them the most strikingly upregulated protein IL4I1 (highlighted in red). Gene names are given next to the protein symbols, ratios of means are indicated next to the gene names, green arrows indicate higher protein abundance in the A. fumigatus infected group. Edges represent protein-protein interactions with the color code given next to the network.
Increased IL4I1 Abundance is Confirmed on the Protein and Transcript Level
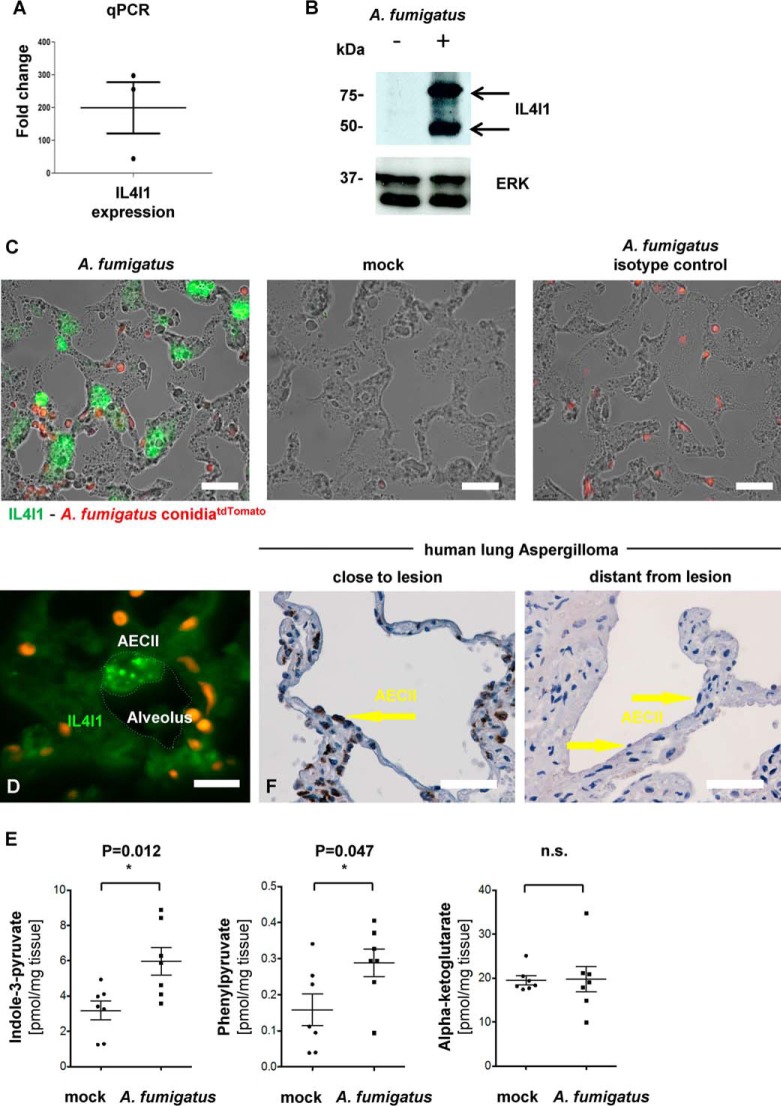
mRNA isolated from purified AEC II of both experimental groups was analyzed by RT-PCR for the level of IL4I1 transcript to confirm the results of mass spectrometry analysis. A 199 fold upregulation of IL4I1 in AEC II isolated from A. fumigatus infected mice compared with AEC II from mock treated controls was observed (Fig. 6A).
Fig. 6.
Independent confirmation of IL4I1 upregulation in AEC II during A. fumigatus infection. A, qPCR of IL4I1 mRNA in isolated AEC II from infected compared noninfected mice. The analysis of the data was performed by ΔΔ CT calculation against the mock infected sample and the housekeeping gene GAPDH. Data are mean ± S.E. from three independent experiments. B, Western blot analysis of IL4I1 abundance in AEC II isolated from control or A. fumigatus infected lungs. ERK was used as a loading control. C, Immunofluorescence of lungs infected (left) or not (middle) with tdTomato-transgenic A. fumigatus were stained with green fluorescent anti-IL4I1 antibody. A strong IL4I1 signal (green) and tdTomato conidia (red, black arrows) can be seen in the infected samples, only. An infected sample stained with isotype control only shows the red signal of the transgenic fungus (right). D, Confocal image of an A. fumigatus infected lung section stained with a green fluorescent anti-IL4I1 antibody shows a signal (green) in vesicles in AEC II. Conidia can be seen in tdTomato (red). E, HPLC analysis of whole lung lysates for the presence of the indicated metabolites of IL4I1 enzyme activity. Data are mean ± S.E. from 5 replicates per condition. F, IL4I1 immunohistochemistry of human lung tissue suffering from aspergilloma. Please note IL4I1 positive AEC II in an area close to the lesion (yellow arrow). AEC II in regions more distant from mycetoma are negative. Scale bars: 10 μm (C), 5 μm (D), 50 μm (F).
Following the confirmation of IL4I1 upregulation on the mRNA level, we extracted the protein from purified AEC II of both experimental groups and studied the presence of IL4I1 protein by Western blotting using an IL4I1 specific antibody and could indeed confirm the presence of the protein in the A. fumigatus infected sample (Fig. 6B). Thereby, the double band most likely demonstrated the existence of cleavage products from the original transcript or partial glycosylation. It is currently unknown, whether these protein variants have different functions.
Subsequently, the presence of IL4I1 was visualized in lung slices from animals infected or not with a transgenic strain of A. fumigatus expressing the red fluorescent protein tdTomato in its conidia and hyphae (38). For this analysis, we performed fluorescence microscopy employing the same anti-IL4I1 antibody that was used for Western blotting. A signal was indeed, detected in the corners of the alveoli where AEC II are located (34) (Fig. 6C). Moreover, the IL4I1 signal was often co-localized with the presence and in some cases even germination of the tdTomato-expressing A. fumigatus conidia (Fig. 6C, black arrows). Importantly, no IL4I1 signal was detected in mock-infected lung slices or isotype controls from infected samples (Fig. 6C).
When the intracellular localization of IL4I1 was inspected by 3-D confocal microscopy we found the protein to be localized in vesicles (Fig. 6D, supplemental Movie S1), which is reminiscent of the staining pattern of IL4I1 found in fibroblasts (39) or in lymph node macrophages (36).
Having shown the high abundance of IL4I1 mRNA and protein in AEC II in vivo after A. fumigatus infection we next wished to demonstrate the presence of IL4I1 enzymatic activity in the infected lungs. This was performed by measuring the main products of IL4I1 mediated l-amino acid metabolism. Previously it has been reported that IL4I1 prefers aromatic amino acids such as l-phenylalanine and l-tryptophan (27, 39). Indeed, the key metabolites of IL4I1 namely Indole-3-pyruvate (I3P) and Phenylpyruvate (PhePyr), the respective breakdown products of l-tryptophan and l-phenylalanine, were more abundant in the whole lung of A. fumigatus infected mice relative to the mock infected controls. As a control Alpha Ketoglutarate, an IL4I1 non-related keto acid, was not increased under the same circumstances (Fig. 6E). These data are further confirming an induction of IL4I1 activity in the lung within 24 h after A. fumigatus infection.
Collectively these data showed proteome changes in AEC II exposed to an infection with A. fumigatus with IL4I1 showing the most prominent increase in abundance both, in terms of relative abundance as well as activity.
Finally, to verify the relevance of our findings for human disease conditions we investigated the presence of IL4I1 by immunohistology of lung tissue from human aspergilloma. Interestingly, the AEC II near aspergilloma lesions were clearly positive for IL4I1. In contrast, tissue from the same lung but more distant from mycetoma was negative for IL4I1 signal (Fig. 6 F). This finding was highly reproducible in all ten cases (Table III). Cases with bronchiectasis and pneumonia revealed similar changes at the sites of inflammation. In both diseases AEC II showed positive antigen reactions. The severity correlated with the intensity of inflammation. AEC II in normal lung tissue without inflammation was completely negative (supplemental Fig. S3), as already seen in aspergilloma.
Table III. Informaton of the aspergilloma patients. Ten cases of pulmonary human aspergilloma from the archive of the institute of pathology at Essen University Hospital.
| Number | Resection | Age | Sex | Aspergilloma | Associated lung tissue |
|---|---|---|---|---|---|
| 1 | inferior lobe | 63 | female | positive | missing |
| 2 | upper lobe | 65 | female | positive | positive |
| 3 | wedge | 42 | male | positive | positive |
| 4 | upper lobe | 68 | male | positive | positive |
| 5 | segment | 52 | female | positive | positive |
| 6 | explant | 53 | female | positive | positive |
| 7 | upper lobe | 40 | male | positive | positive |
| 8 | wedge | 58 | female | positive | positive |
| 9 | wedge | 48 | male | positive | positive |
| 10 | wedge | 59 | male | positive | positive |
DISCUSSION
A. fumigatus infection is a life-threatening condition for immunosuppressed patients around the globe, because it can develop into invasive aspergillosis, which eventually can lead to death. Immunocompetent individuals, on the other hand, fight this infection very efficiently and easily (40). During an A. fumigatus infection different phagocytes play a decisive role in clearing up the fungus. From an immunological perspective, these cells can be divided into two groups: professional and nonprofessional phagocytes (35). Classical innate immune cells such as monocytes/macrophages, neutrophil granulocytes and dendritic cells are placed in the first group of phagocytes whereas alveolar epithelial cells (AEC) are regarded to belong to the second group (41). Previously, it has been reported that isolated rat primary AEC II could internalize conidia 24 h after culture (29). Simultaneously, another study demonstrated conidial uptake by the A549 cell line (33). Knowing the importance of the AEC II as one of the earliest cell types that conidia physically engage after entering the alveolus (14), proteome analysis of these cells and the proteome changes on A. fumigatus infection are of great interest and might gain new insights into the relevance of AEC II in lung immunity.
To be closer to the physiological condition, performing the molecular studies on purified primary cells is preferred. The Isolation of the murine primary AEC II dates to 1987 (42). Since then different studies have focused on designing protocols for purification of this cell type using different techniques (23, 43–45). Although the enrichment has been improved over time in yield and purity, all these protocols including the latest, strongly stress the isolated cells (23). In this study we concentrated on establishing an immunomagnetic protocol for negative isolation of primary AEC II with the least distress and best purity for the cells. Therefore, the antibody mixture for AEC II isolation with a flow cytometric approach suggested by Gereke et al. (23) was considered as the basis. This antibody mixture could only exclude immune cell types. Hence many different nonimmune cell types such as endothelial cells, epithelial cells and resident cells in bronchi, intralobular bronchioles and the bronchoalveolar duct junction (BADJ) (Clara cells, Basal cells, ciliated cells, neuroendocrine cells, Goblet cells, and epithelial progenitor cells (46)) were not excluded. Moreover, antibodies able to bind to specific antigens on the surfaces of these cells do not exist. To overcome this complication, bronchi were cut out by a pair of very fine surgery scissors after the lung digestion step and before lung dissociation with forceps. With this morphological approach many of the contaminating cell types efficiently got excluded. Additionally, some of the bigger cells such as goblet and ciliated cells (∼50 μm) could be eliminated during cell straining process. Subsequently, the biotinylated antibody mixture for negative isolation with routinely >90% pure AEC II was generated (Table I). This protocol turned out to be ideal for unbiased LC-MS/MS analysis of AEC II.
Quantitative proteome analysis of the isolated primary AEC II from A. fumigatus infected and mock infected mice and further functional annotation and enrichment analysis of the proteome data revealed a strikingly increased abundance of the proteins related to oxidation and reduction and especially oxidative phosphorylation, which points to extensive energy production. This suggests an activation of AEC II in response to the contact with A. fumigatus conidia, which is also reflected by the predominant increase of protein abundance in the A. fumigatus infected group (Fig. 3B, 3C, 3D). Inspection of the proteins showing the highest Euclidian distance revealed that Lipocalin 2 (Lcn 2, P11672) with a p value of 1.08 × 10−10 was 8 times higher abundant in AEC II during A. fumigatus infection compared with the mock infected control. This protein inhibits bacterial growth in AEC II during Mycobacterial infection (47) but no pro-inflammatory role against A. fumigatus infection has been reported for Lcn2 yet. Therefore, it is a promising candidate for further investigation. Metalloreductase Steap4 (Q923B6) showed a 6.9-fold higher abundance on A. fumigatus infection and a p value of 1.34 × 10−8. It is known to be an iron regulator during inflammatory responses (48). Both, Lipocalin and Steap4are able to limit the availability of free iron, which in turn would massively impair growth of A. fumigatus (49). Thus, AEC II might use iron sequestration to control fungal growth. Additionally, we found higher abundance of ICAM-1 (P13597) with a p value of 2.06 × 10−7 and a max ratio of means of 3.94. This integrin ligand is essential for leukocyte/endothelial transmigration (50). The increased abundance of ICAM-1 might thus enable more effective leukocyte extravasation from underlying blood vessels (Table II). It is noteworthy that several protein species of ICAM-1 have been reported in the literature (51). These cannot be distinguished by the applied proteomics approach and therefore require further investigations. It is noteworthy that protein speciation may affect the functional relevance of the described proteins and is generally neglected by the performed bottom-up approach. Further investigations are needed to elucidate the impact of protein speciation in AEC II during A. fumigatus infection.
The protein with the most remarkable elevation of abundance in the A. fumigatus infected group was IL4I1 which belongs to the family of LAAO (39). The general function of this family of enzymes is catalysis of LAAO deamination and the amino acid which IL4I1 catalyzes most is known to be Phenylalanine (27). IL4I1 was initially reported to be abundant in B-cells through STAT6 activation (52). In addition to B-cells, IL4I1 was found highly abundant in antigen presenting cells, such as dendritic cells and macrophages (36, 37). Here, we show for the first time that IL4I1 is present in primary murine AEC II during A. fumigatus infection. IL4I1 transcript expression and protein abundance were illustrated on the mRNA and protein level, respectively, in the purified AEC II 24 h post infection. Furthermore, this data was confirmed by demonstrating the presence of IL4I1 enzymatic activity in the infected lungs by measuring the main products of IL4I1 mediated l-amino acid metabolism using a novel HPLC approach in a pilot study (data not shown). Because AEC II were also reported to function as an antigen presenting cell type makes this data compatible with the previous findings (53). IL4I1 abundance in monocytic phagocytes has been shown to have a direct bactericidal activity dependent on H2O2 production amplified by basification because of the concomitant accumulation of ammonia (54). Thus, increased IL4I1 abundance in AEC II might have a host protective role participating to A. fumigatus infection elimination in immunocompetent individuals. Further investigations will be necessary to clarify this concept and to elucidate the impact of the two reported IL4I1 variants in A. fumigatus infection (55).
Regarding the quantification of the metabolic products of IL4I1 in the infected lungs, it was not possible to test the activity of IL4I1 from the isolated AEC II in these measurements because of the nature of the analysis. Instead we had to measure entire lung lysates. It might, therefore, be possible that IL4I1 from other cells than AEC II contributed to the observed generation of the metabolic products. However, our immunohistological analysis implies that AEC II were the cells with the highest levels of IL4I1 in murine lung slices. Also, the measurement of IL4I1 mRNA by qPCR provided the highest values when measured from purified AEC II and significantly lower levels when measured from entire lung lysates (data not shown), thus further supporting the concept that AEC II are the main source of this protein in the A. fumigatus infected murine lung. It is interesting to note, that a lung infection is not per se increasing IL4I1 abundance in AEC II, because we could not detect very high levels of these data in transcriptomic profiles of AEC II after influenza infection (16).
Importantly, an increase in IL4I1 abundance appears to be an evolutionarily conserved mechanism of anti-Aspergillus responses in AEC II. However, the human studies also suggest that the increase of IL4I1 in AEC II is more generally induced on microbial infection and might also be a side effect of generalized lung inflammation. Future work should, therefore, clarify the functional significance of this molecule for host defense.
Collectively this study suggests that the response of AEC II on the proteome level is highly responsive to an infectious threat and warrants further investigation into the molecular response of this key cell type of the lung other inflammatory conditions. The work presented here lays the technological basis for such endeavors.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD005834 and 10.6019/PXD005834.
Supplementary Material
Acknowledgments
We thank Kristin Rosowski and Birgit Korte for their excellent technical assistance. The IMCES (Imaging Center Essen) is acknowledged for expert technical support in imaging experiments. The authors thank the PRIDE Team for their assistance during the data upload.
Footnotes
Author contributions: P.S., T.B., M.S., A.K., G.P., T.H., D.T., C.O., A.A.B., and B.S. performed research; P.S., T.B., M.S., J.W., A.H., A.A.B., B.S., M.H., and M.G. analyzed data; P.S., T.B., B.S., M.H., and M.G. wrote the paper; T.B., V.M.-F., F.C., O.K., J.W., and A.H. contributed new reagents/analytic tools; B.S., M.H., and M.G. designed research.
* This work was supported by the International Leibniz Research School (Jena, Germany) and by PURE (Protein research Unit Ruhr within Europe) funded by the Ministry of Science, North Rhine-Westphalia, Germany.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- IA
- Invasive Aspergillosis
- AEC
- alveolar epithelial cell
- A. fumigatus
- Aspergillus fumigatus
- AM
- alveolar macrophage
- CA assay
- bicinchoninic acid assay
- BSA
- bovine serum albumin
- CCD camera
- charge coupled device camera
- ΔΔ Ct
- ΔΔ cycle threshold
- EpCAM
- epithelial cell adhesion molecule
- ECL
- enhanced chemiluminescence
- FDR
- False Discovery Rate
- HCD
- higher-energy collisional dissociation
- HPLC
- high performance liquid chromatography
- I3P
- indole-3-pyruvate
- IL4I1
- Interleukin 4 induced-1
- LAAO
- L-amino acid oxidase
- LC-MS
- Liquid Chromatography Mass Spectrometry
- ACS
- Magnetic Activated Cell Sorting
- mRNA
- messenger Ribonucleic Acid
- Nφ
- Neutrophil granulocyte
- PD
- O-phenylenediamine
- PFA
- Paraformaldehyde
- PhePyr
- Phenylpyruvate
- proSP-C
- proSurfactant Protein-C
- PVDF membrane
- Polyvinylidene difluoride membrane
- TdTomato
- Tandem Tomato fluorescent protein
- TFA
- Trifluoroacetic Acid.
REFERENCES
- 1. Latge J. P. (1999) Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12, 310–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aimanianda V., Bayry J., Bozza S., Kniemeyer O., Perruccio K., Elluru S. R., Clavaud C., Paris S., Brakhage A. A., Kaveri S. V., Romani L., and Latge J. P. (2009) Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460, 1117–1121 [DOI] [PubMed] [Google Scholar]
- 3. Rolle A. M., Hasenberg M., Thornton C. R., Solouk-Saran D., Männ L., Weski J., Maurer A., Fischer E., Spycher P. R., Schibli R., Boschetti F., Stegemann-Koniszewski S., Bruder D., Severin G. W., Autenrieth S. E., Krappmann S., Davies G., Pichler B. J., Gunzer M., and Wiehr S. (2016) ImmunoPET/MR imaging allows specific detection of Aspergillus fumigatus lung infection in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, E1026–E1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., and White T. C. (2012) Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113. [DOI] [PubMed] [Google Scholar]
- 5. Osherov N., and Kontoyiannis D. P. (2016) The anti-Aspergillus drug pipeline: Is the glass half full or empty? Med Mycol. 55, 118–124 [DOI] [PubMed] [Google Scholar]
- 6. Männ L., Kochupurakkal N., Martin C., Verjans E., Klingberg A., Sody S., Kraus A., Dalimot J., Bergmuller E., Jung S., Voortman S., Winterhager E., Brandau S., Garbi N., Kurrer M., Eriksson U., Gunzer M., and Hasenberg M. (2016) CD11c.DTR mice develop a fatal fulminant myocarditis after local or systemic treatment with diphtheria toxin. Eur. J. Immunol. 46, 2028–2042 [DOI] [PubMed] [Google Scholar]
- 7. Mircescu M. M., Lipuma L., Van R. N., Pamer E. G., and Hohl T. M. (2009) Essential role for neutrophils but not alveolar macrophages at early time points following Aspergillus fumigatus infection. J Infect. Dis. 200, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruns S., Kniemeyer O., Hasenberg M., Aimanianda V., Nietzsche S., Thywiβen A., Jeron A., Latge J. P., Brakhage A. A., and Gunzer M. (2010) Production of Extracellular Traps against Aspergillus fumigatus in vitro and in Infected Lung Tissue is Dependent on Invading Neutrophils and Influenced by Hydrophobin RodA. PLoS Pathog. 6, e1000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Behnsen J., Narang P., Hasenberg M., Gunzer F., Bilitewski U., Klippel N., Rohde M., Brock M., Brakhage A. A., and Gunzer M. (2007) Environmental Dimensionality Controls the Interaction of Phagocytes with the Pathogenic Fungi Aspergillus fumigatus and Candida albicans. PLoS Pathog. 3, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hasenberg A., Hasenberg M., Männ L., Neumann F., Borkenstein L., Stecher M., Kraus A., Engel D. R., Klingberg A., Seddigh P., Abdullah Z., Klebow S., Engelmann S., Reinhold A., Brandau S., Seeling M., Waisman A., Schraven B., Göthert J. R., Nimmerjahn F., and Gunzer M. (2015) Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat. Methods 12, 445–452 [DOI] [PubMed] [Google Scholar]
- 11. Gunzer M. (2014) Traps and hyper inflammation - new ways that neutrophils promote or hinder survival. Br. J. Haematol 164, 188–199 [DOI] [PubMed] [Google Scholar]
- 12. Hasenberg M., Stegemann-Koniszewski S., and Gunzer M. (2013) Cellular immune reactions in the lung. Immunol. Rev. 251, 189–214 [DOI] [PubMed] [Google Scholar]
- 13. Pollmacher J., and Figge M. T. (2014) Agent-based model of human alveoli predicts chemotactic signaling by epithelial cells during early Aspergillus fumigatus infection. PLoS ONE 9:e111630, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Croft C. A., Culibrk L., Moore M. M., and Tebbutt S. J. (2016) Interactions of Aspergillus fumigatus Conidia with Airway Epithelial Cells: A Critical Review. Front. Microbiol. 7, 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chroneos Z. C., Sever-Chroneos Z., and Shepherd V. L. (2010) Pulmonary surfactant: an immunological perspective. Cell Physiol. Biochem. 25, 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stegemann-Koniszewski S., Jeron A., Gereke M., Geffers R., Kroger A., Gunzer M., and Bruder D. (2016) Alveolar Type II Epithelial Cells Contribute to the Anti-Influenza A Virus Response in the Lung by Integrating Pathogen- and Microenvironment-Derived Signals. MBio 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gereke M., Jung S., Buer J., and Bruder D. (2009) Alveolar type II epithelial cells present antigen to CD4(+) T cells and induce Foxp3(+) regulatory T cells. Am. J. Respir. Crit. Care Med. 179, 344–355 [DOI] [PubMed] [Google Scholar]
- 18. Chen F., Zhang C., Jia X., Wang S., Wang J., Chen Y., Zhao J., Tian S., Han X., and Han L. (2015) Transcriptome Profiles of Human Lung Epithelial Cells A549 Interacting with Aspergillus fumigatus by RNA-Seq. PLoS ONE 10, e0135720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Witzke K. E., Rosowski K., Muller C., Ahrens M., Eisenacher M., Megger D. A., Knobloch J., Koch A., Bracht T., and Sitek B. (2017) Quantitative Secretome Analysis of Activated Jurkat Cells Using Click Chemistry-Based Enrichment of Secreted Glycoproteins. J. Proteome Res. 16, 137–146 [DOI] [PubMed] [Google Scholar]
- 20. Rattay S., Trilling M., Megger D. A., Sitek B., Meyer H. E., Hengel H., and Le-Trilling V. T. (2015) The Canonical Immediate Early 3 Gene Product pIE611 of Mouse Cytomegalovirus Is Dispensable for Viral Replication but Mediates Transcriptional and Posttranscriptional Regulation of Viral Gene Products. J. Virol. 89, 8590–8598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hearn V. M., and Mackenzie D. W. R. (1980) Mycelial Antigens from 2 Strains of Aspergillus-Fumigatus - an Analysis by Two-Dimensional Immunoelectrophoresis. Mykosen 23, 549–562 [PubMed] [Google Scholar]
- 22. Krappmann S., Bayram Ö., Braus G. H. (2005) Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryotic Cell 4, 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gereke M., Autengruber A., Grobe L., Jeron A., Bruder D., and Stegemann-Koniszewski S. (2012) Flow cytometric isolation of primary murine type II alveolar epithelial cells for functional and molecular studies. J. Vis. Exp. 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Megger D. A., Bracht T., Kohl M., Ahrens M., Naboulsi W., Weber F., Hoffmann A. C., Stephan C., Kuhlmann K., Eisenacher M., Schlaak J. F., Baba H. A., Meyer H. E., and Sitek B. (2013) Proteomic differences between hepatocellular carcinoma and nontumorous liver tissue investigated by a combined gel-based and label-free quantitative proteomics study. Mol. Cell. Proteomics 12, 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mi H. Y., Huang X. S., Muruganujan A., Tang H. M., Mills C., Kang D., and Thomas P. D. (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45, D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K. P., Kuhn M., Bork P., Jensen L. J., and von Mering C. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boulland M. L., Marquet J., Molinier-Frenkel V., Moller P., Guiter C., Lasoudris F., Copie-Bergman C., Baia M., Gaulard P., Leroy K., and Castellano F. (2007) Human IL4I1 is a secreted L-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood 110, 220–227 [DOI] [PubMed] [Google Scholar]
- 28. Rolle A. M., Hasenberg M., Thornton C. R., Solouk-Saran D., Mann L., Weski J., Maurer A., Fischer E., Spycher P. R., Schibli R., Boschetti F., Stegemann-Koniszewski S., Bruder D., Severin G. W., Autenrieth S. E., Krappmann S., Davies G., Pichler B. J., Gunzer M., and Wiehr S. (2016) ImmunoPET/MR imaging allows specific detection of Aspergillus fumigatus lung infection in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, E1026–E1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paris S., Boisvieux-Ulrich E., Crestani B., Houcine O., Taramelli D., Lombardi L., and Latgé J. P. (1997) Internalization of Aspergillus fumigatus conidia by epithelial and endothelial cells. Infection Immunity 65, 1510–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wasylnka J. A., and Moore M. M. (2002) Uptake of Aspergillus fumigatus conidia by phagocytic and nonphagocytic cells in vitro: Quantitation using strains expressing green fluorescent protein. Infection Immunity 70, 3156–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wasylnka J. A., and Moore M. M. (2003) Aspergillus fumigatus conidia survive and germinate in acidic organelles of A549 epithelial cells. J. Cell Sci. 116, 1579–1587 [DOI] [PubMed] [Google Scholar]
- 32. Han X., Yu R., Zhen D., Tao S., Schmidt M., and Han L. (2011) beta-1,3-Glucan-induced host phospholipase D activation is involved in Aspergillus fumigatus internalization into type II human pneumocyte A549 cells. PloS one 6, e21468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeHart D. J., Agwu D. E., Julian N. C., and Washburn R. G. (1997) Binding and germination of Aspergillus fumigatus conidia on cultured A549 pneumocytes. J. Infect Dis. 175, 146–150 [DOI] [PubMed] [Google Scholar]
- 34. Fehrenbach H. (2001) Alveolar epithelial type II cell: defender of the alveolus revisited. Respir. Res. 2, 33–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Osherov N. (2012) Interaction of Aspergillus fumigatus with host cells. Mycoses 55, 8–8 [Google Scholar]
- 36. Marquet J., Lasoudris F., Cousin C., Puiffe M. L., Martin-Garcia N., Baud V., Chereau F., Farcet J. P., Molinier-Frenkel V., and Castellano F. (2010) Dichotomy between factors inducing the immunosuppressive enzyme IL-4-induced gene 1 (IL4I1) in B lymphocytes and mononuclear phagocytes. Eur. J. Immunol. 40, 2557–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chu C. C., Chavan S. S., Naidu M. D., Mason J. M., Porti D., Barcia M., and Teichberg S. (2004) Interleukin-four induced gene-1 (I14i1 or Fig 1) expressed primarily in antigen presenting cells. Faseb. J. 18, A37–A37 [Google Scholar]
- 38. Hasenberg A., Hasenberg M., Mann L., Neumann F., Borkenstein L., Stecher M., Kraus A., Engel D. R., Klingberg A., Seddigh P., Abdullah Z., Klebow S., Engelmann S., Reinhold A., Brandau S., Seeling M., Waisman A., Schraven B., Gothert J. R., Nimmerjahn F., and Gunzer M. (2015) Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat. Methods 12, 445-+ [DOI] [PubMed] [Google Scholar]
- 39. Mason J. M., Naidu M. D., Barcia M., Porti D., Chavan S. S., and Chu C. C. (2004) IL-4-induced gene-1 is a leukocyte L-amino acid oxidase with an unusual acidic pH preference and lysosomal localization. J. Immunol. 173, 4561–4567 [DOI] [PubMed] [Google Scholar]
- 40. Mansour M. K., Tam J. M., and Vyas J. M. (2012) The cell biology of the innate immune response to Aspergillus fumigatus. Ann. NY Acad. Sci. 1273, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rabinovitch M. (1995) Professional and non-professional phagocytes: an introduction. Trends Cell Biol. 5, 85–87 [DOI] [PubMed] [Google Scholar]
- 42. Massey T. E., Geddes B. A., and Forkert P. G. (1987) Isolation of nonciliated bronchiolar epithelial (Clara) cells and alveolar type II cells from mouse lungs. Can. J. Physiol. Pharmacol. 65, 2368–2372 [DOI] [PubMed] [Google Scholar]
- 43. Corti M., Brody A. R., and Harrison J. H. (1996) Isolation and primary culture of murine alveolar type II cells. Am. J. Respiratory Cell Mol. Biol. 14, 309–315 [DOI] [PubMed] [Google Scholar]
- 44. Harrison J. H. Jr, Porretta C. P., and Leming K. (1995) Purification of murine pulmonary type II cells for flow cytometric cell cycle analysis. Exp. Lung Res. 21, 407–421 [DOI] [PubMed] [Google Scholar]
- 45. Kumar R. K., Li W., and O'Grady R. (1995) Maintenance of differentiated phenotype by mouse type 2 pneumocytes in serum-free primary culture. Exp. Lung Res. 21, 79–94 [DOI] [PubMed] [Google Scholar]
- 46. Rock J. R., and Hogan B. L. (2011) Epithelial progenitor cells in lung development, maintenance, repair, and disease. Ann. Rev. Cell Develop. Biol. 27, 493–512 [DOI] [PubMed] [Google Scholar]
- 47. Saiga H., Nishimura J., Kuwata H., Okuyama M., Matsumoto S., Sato S., Matsumoto M., Akira S., Yoshikai Y., Honda K., Yamamoto M., and Takeda K. (2008) Lipocalin 2-dependent inhibition of mycobacterial growth in alveolar epithelium. J. Immunol. 181, 8521–8527 [DOI] [PubMed] [Google Scholar]
- 48. Ohgami R. S., Campagna D. R., McDonald A., and Fleming M. D. (2006) The Steap proteins are metalloreductases. Blood 108, 1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schrettl M., Bignell E., Kragl C., Joechl C., Rogers T., Arst H. N. Jr, Haynes K., and Haas H. (2004) Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J. Exp. Med. 200, 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sans E., Delachanal E., and Duperray A. (2001) Analysis of the roles of ICAM-1 in neutrophil transmigration using a reconstituted mammalian cell expression model: Implication of ICAM-1 cytoplasmic domain and Rho-dependent signaling pathway. J. Immunol. 166, 544–551 [DOI] [PubMed] [Google Scholar]
- 51. Ramos T. N., Bullard D. C., and Barnum S. R. (2014) ICAM-1: isoforms and phenotypes. J. Immunol. 192, 4469–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chu C. C., and Paul W. E. (1997) Fig 1, an interleukin 4-induced mouse B cell gene isolated by cDNA representational difference analysis. P Natl Acad Sci U.S.A. 94, 2507–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gereke M., Jung S., Buer J., and Bruder D. (2009) Alveolar type II epithelial cells present antigen to CD4(+) T cells and induce Foxp3(+) regulatory T cells. Am. J. Respir. Crit Care Med. 179, 344–355 [DOI] [PubMed] [Google Scholar]
- 54. Puiffe M. L., Lachaise I., Molinier-Frenkel V., and Castellano F. (2013) Antibacterial properties of the mammalian L-amino acid oxidase IL4I1. PLoS ONE 8, e54589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Molinier-Frenkel V., Mestivier D., and Castellano F. (2016) Alterations of the immunosuppressive IL4I1 enzyme activity induced by naturally occurring SNP/mutations. Genes Immun. 17, 148–152 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD005834 and 10.6019/PXD005834.