Abstract
Mycobacterium tuberculosis (Mtb) has evolved multiple strategies to counter the human immune system. The effectors of Mtb play important roles in the interactions with the host. However, because of the lack of highly efficient strategies, there are only a handful of known Mtb effectors, thus hampering our understanding of Mtb pathogenesis. In this study, we probed Mtb proteome microarray with biotinylated whole-cell lysates of human macrophages, identifying 26 Mtb membrane proteins and secreted proteins that bind to macrophage proteins. Combining GST pull-down with mass spectroscopy then enabled the specific identification of all binders. We refer to this proteome microarray-based strategy as SOPHIE (Systematic unlOcking of Pathogen and Host Interacting Effectors). Detailed investigation of a novel effector identified here, the iron storage protein BfrB (Rv3841), revealed that BfrB inhibits NF-κB-dependent transcription through binding and reducing the nuclear abundance of the ribosomal protein S3 (RPS3), which is a functional subunit of NF- κB. The importance of this interaction was evidenced by the promotion of survival in macrophages of the mycobacteria, Mycobacterium smegmatis, by overexpression of BfrB. Thus, beyond demonstrating the power of SOPHIE in the discovery of novel effectors of human pathogens, we expect that the set of Mtb effectors identified in this work will greatly facilitate the understanding of the pathogenesis of Mtb, possibly leading to additional potential molecular targets in the battle against tuberculosis.
Mycobacterium tuberculosis (Mtb)1 is the causative agent of tuberculosis (TB). There were an estimated 1.4 million TB deaths in 2015, and an additional 400, 000 deaths resulting from the co-infection of TB and HIV (1). Owing especially to the emergence of highly drug-resistant strains of Mtb, including multidrug-resistant (MDR) and extensively drug- resistant (XDR) strains, as well as co-infection with HIV, TB is still a serious infectious disease world-wide (2, 3). Thus there is great urgency to identify novel proteins that can be targeted by drugs for effective treatment of TB, and especially MDR-TB and XDR-TB.
Mtb is a highly successful human pathogen, surviving and multiplying within macrophages of infected people (4, 5). Host innate immune responses play crucial roles in the early protection against Mtb infection (6). For one, the signaling pathway of the transcription factor NF-κB regulates innate immunity by controlling the expression of various immune-regulatory molecules and thus modulates the intracellular survival of pathogens (7, 8). On the pathogen side, Mtb membrane proteins and secreted proteins are important effectors that may play important roles in host-pathogen interactions (9). Previous work has indeed shown that Mtb has evolved several highly effective strategies including the secretion of specific effectors into the host to subvert the signaling pathways of the innate immune system, thereby allowing Mtb to avoid elimination and survive within macrophages for long periods of time (10, 11). The presently best-characterized survival mechanisms of Mtb are the inhibition of phagosomal acidification and maturation, and the inhibition of apoptosis of infected macrophages (12, 13).
However, there are presently only a limited number of well- characterized Mtb effectors that have been described, and only a few whose corresponding host-interacting proteins have been identified (14–23) (supplemental Table S1). These interactions were mainly discovered by yeast two-hybrid (Y2H) (21, 24) and affinity purification followed by mass spectrometry analysis (AP/MS) (22, 25). For example, Mehra et al. applied a high- throughput Y2H platform to identify Mtb secreted virulence factors and Mtb-host interacting proteins (21). However, one of the disadvantages of Y2H is that it detects only about 20% of all interactions because of its high false-negative rate (21, 26, 27). AP/MS can detect proteins in complexes but is unable to distinguish between direct from indirect interactions when several proteins are purified together (24, 26). Thus, because of the intrinsic limitations of Y2H and AP/MS, these methods are not well-suited for the global screening of protein-protein interactions (PPI) between Mtb and the host.
Systematic identification of Mtb and macrophage interactions holds the promise for revealing the complex mechanism underlying pathogenesis of Mtb (28, 29). Because of the lack of suitable proteome-scale techniques, we do not yet have a global picture of the Mtb-host interactions at the protein level.
Proteome microarrays, usually composed of thousands of proteins from one species that are affinity purified and functionally active, are high- throughput platforms for the global profiling of thousands of molecular interactions in a single experiment (30). They provide a versatile platform for investigating many aspects at the systems-level, such as discovering serum biomarkers for various diseases (31) and globally investigating interactions with proteins (32), DNA (33), RNA (34), lipids (35), and small molecules (36, 37).
Recently, we constructed a functional Mtb proteome microarray (with >95% coverage of the Mtb proteome) (38). The power of this Mtb proteome microarray has already been demonstrated in investigations of PPI (39) and small molecule/drug-protein binding (40).
Herein, we took advantage of this Mtb proteome microarray and established a strategy for the global identification of the Mtb proteins involved in Mtb-macrophage interactions. With this, we identified a total of 26 Mtb membrane proteins and secreted proteins that may play important roles in Mtb-host interactions. We focused on BfrB (Rv3841), a ferritin-like protein that was known to play a key role in the maintenance of Mtb iron homeostasis (41). Interestingly, BfrB was recently identified as a secreted protein that is enriched from infected macrophages and is also present in the exosomes of patients, suggesting that it may play additional roles in the interactions with the host (42–44). However, the mechanism by which BfrB functions in the modulation of the host immune response is poorly known. Therefore, we sought to identify crucial host target proteins to which BfrB binds. We found that BfrB subverts NF-κB function by binding and reducing the nuclear abundance of its subunit RPS3 (45) to inhibit host immune defense. Thus, our findings reveal a previously unknown mechanism by which BfrB, as a newly identified effector, subverts innate immunity.
EXPERIMENTAL PROCEDURES
Global Profiling of Mtb Proteins Interact with Proteins from Macrophage
THP-1 and U937 cell were differentiated into macrophage-like cells using 100 ng/ml PMA (Sigma-Aldrich, Saint Louis, MO) for 24 h. Then the medium was replaced and cells cultured in RPMI 1640 medium supplemented with 10% FBS for 24 h. The cells were washed twice, and the cell lysates were prepared using cell lysis buffer as described (46). The whole cell lysates were biotinylated using EZ-Link Sulfo-NHS-LC-Biotin protein biotinylation kit (Thermo Scientific, Bremen, Germany) according to the manual. Mtb proteome microarrays were blocked for 1 h at room temperature with shaking in blocking buffer as described (46). Microarrays were incubated in the biotinylated whole cell lysates for 12 h at 4 °C with shaking, then washed three times in 1× PBS (pH 7.4) with 0.1% Tween 20 (PBST). Arrays were probed with Cy3-streptavidin and incubated in a humidified chamber for 1 h at room temperature. After washing three times in PBST and once ddH2O, microarrays were dried in a SlideWasher (CapitalBio, Beijing, China) and then scanned with a GenePix 4200A microarray scanner (Molecular Devices, CA). Data were analyzed with GenePix Pro 6.1 (Molecular Devices, CA).
GST Pull-down Assay
To identity the host targets of Mtb effectors, GST-tagged BfrB were purified as previously described (38). PMA-differentiated THP-1 cells lysed in Nonidet P-40 lysis buffer (Beyotime, Shanghai, China) supplemented with 0.5 mm PMSF (Sigma-Aldrich). The GST pull- down was performed as described (11). The pull-down proteins were eluted by 2× SDS sampling buffer and denatured at 95 °C for 10 min and analyzed by standard silver staining.
Proteins Identification and LC-MS/MS Analysis
To determine the proteins interacted with BfrB, the human proteins enriched by BfrB were subjected for electrophoresis and silver staining, the major bands in the gel were excised, destained and reduced by incubation in a solution of 10 mm DTT at 60 °C for 20 min, followed by alkylating in a solution of 25 mm IAM at room temperature for 15 min. The pieces were digested in-gel with trypsin (Promega, Madison, WI) overnight at 37 °C. The tryptic peptide digests of the proteins were analyzed using a LC system (Nano Pump, Ultimate 3000, Dionex, Thermofisher) coupled with an ESI- Q-TOF mass spectrometer (maXis Impact, Bruker Daltonik, Bremen Area, Germany).
Tandem mass spectra were extracted, charge state deconvoluted and deisotoped by Compass Data Analysis version 4.1 (Bruker Daltonics). The peak list was directly generated from raw data using centroid algorithm with peak width set as 0.1 m/z and intensity above 100. No peak smooth or filter process was applied. After the charge states were calculated, the de-isotoped. Peak list was exported as mgf file for further mascot search. Mascot (version 2.4, Matrix Science, Boston, MA) was set up to search the SwissProt 2013_12 Homo sapiens database (20,274 entries). The following parameters were considered for the searches: peptide mass tolerance was set to 20 ppm, fragment mass tolerance was set to 0.05 Da, and a maximum of two missed cleavage of trypsin was chosen. Carbamidomethyl (C) was set as fixed modification, and oxidation (M) was set as variable modifications. The threshold score was greater than or equal to 30, expectation value was 0.01 and the number of peptides was more than 2 for accepting individual spectra.
Monitoring the Kinetics of BfrB and RPS3 Interaction by BLI
The binding kinetics of BfrB and RPS3 were measured using ForteBio Octet system (Pall, Menlo Park, USA). Affinity purified BfrB was biotinylated using EZ-Link Sulfo-NHS-LC-Biotin protein biotinylation kit as described above. Biotinylated BfrB was tethered on the tip surface of a streptavidin-coated sensor. The binding partner, RPS3, in S.D. buffer (1× PBS [pH 7.4] with 0.02% Tween-20 and 0.1% BSA) was then exposed to tethered biotinylated BfrB, and binding was measured by coincident change in the interference pattern.
Cell Transfection, Immunoblot Analysis, and Immunoprecipitation
Transfection was performed using Attractene Transfection Reagent (Qiagen, Hilden, Germany) according to the manufacturer's instruction. HEK293T cells were collected 24 h after transfection and lysed in Nonidet P-40 lysis buffer (Beyotime, Shanghai, China) supplemented with 0.5 mm PMSF (Sigma-Aldrich). The immunoprecipitation assay was performed as (11). The immunoblot analysis was followed as previously described (32). The results were then recorded by the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
RPS3 Nuclear Translocation
Cytosolic and nuclear protein extracts were prepared from HEK293T cells transfected with FLAG-BfrB/FLAG plasmids as previously described (48). All the following steps were carried out at 4 °C. TNF -α stimulation was used to promote RPS3 translocation into the nucleus with 30 ng/ml for 1 h.
RT-PCR
Total RNA was isolated with the RNAprep pure Cell/Bacteria Kit (TIANGEN, Beijing, China) and was reverse-transcribed into cDNA using the Quantscript RT Kit (TIANGEN, Beijing, China) with oligo (dT) primers. RT-PCR reactions were performed in triplicate with primers by using the SYBR Green PCR Master Mix (Roche, Basel, Switzerland) in the ABI StepOne Plus system (Applied Biosystems, Foster City, CA). The relative transcription level was calculated by using the 2−ΔΔCt method and were normalized to the expression of the control gene Gapdh.
Molecular Docking
Molecular docking were performed to investigate the binding mode between the Mtb BfrB to the human RPS3 using the ZDOCK server. The three-dimensional (3D) structures of the BfrB (PDB ID: 3QD8) and the RPS3 (PDB ID: 1WH9) were downloaded from Protein Data Bank. The top ranked pose as judged by the docking score was subjected to visual analysis using PyMoL 1.7.6 software.
Luciferase Reporter Assay
HEK293T cells were seeded in 6-well plates at 1 × 10 6 cells per well in the growth medium and cultured overnight. To measure NF-κB activation, HEK293T cells were co-transfected with 600 ng of pGL4.32 [luc2P/NF-κB-RE/Hygro] (Promega, Madison, WI) and 30 ng of pGL4.74 (hRluc/TK) (Promega) in the presence of 600 ng of FLAG-BfrB/FLAG plasmids. 24 h later, cells were treated with 10 ng/ml of TNF-α (PeproTech, Rocky Hill, NJ) for 6 h. Cell lysates were analyzed by using the Dual-Luciferase Kit (Promega). The fold induction was calculated as described (45).
CFU Counting
THP-1 cells were seeded at 3.5 × 10 6 cells/well in a 6-well plate and differentiated as described above. After 12 h, the cells were washed once and cultured in fresh RPMI 1640 medium for an additional 8 h before infection. The Msm infection and CFU counting assay performed as (11).
Experimental Design and Statistical Rationale
The proteome microarray data were extracted from the microarray images by GenePix Pro 6.1 (Molecular Devices, CA) and then processed using an R package developed in our lab (http://www.proteinmicroarray.cn/). Briefly, the SNR of each protein was averaged for the two duplicated spots on each microarray and was defined as the ratio of the median of foreground signal to the median of background signal. The SNRs for microarrays incubated with natural or denatured protein were set as SNR (+) and SNR (-), respectively. Another index, “Ratio” was defined as SNR (+)/SNR (-). To call the candidates, the cutoff was set as ratio ≥ 1.5 and p value ≤2 × 10−5.
RESULTS
Screening of Mtb Proteins that Interact with Macrophage Proteins Using the Mtb Proteome Microarray
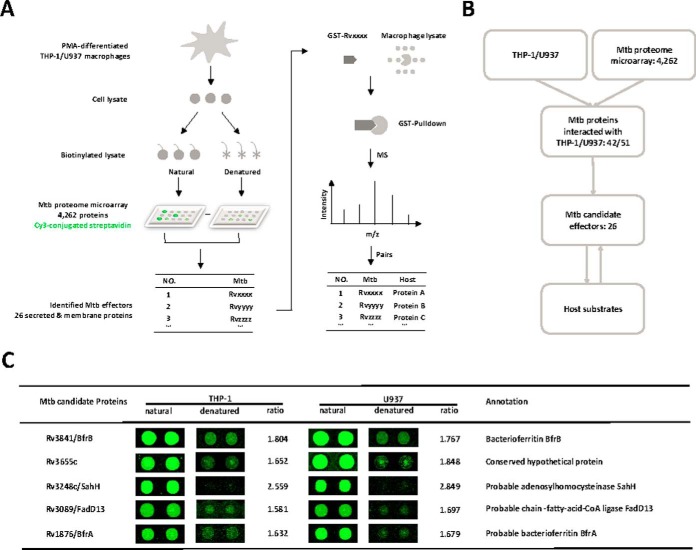
To globally identify the Mtb proteins that bind proteins from human macrophages, we developed the strategy, SOPHIE (Systematic unlOcking Pathogen and Host Interacting Effectors) (Fig. 1A). Briefly, the total cell lysate of human macrophages was prepared. After biotinylation, the lysate was incubated on the functional Mtb proteome microarray (38). The Mtb proteins that bind to proteins from the macrophage were then identified as potential effectors. These potential effectors were tagged with GST and used as bait to enrich for their binding partners from macrophage lysates through GST pull-down. Finally, the identities of the enriched proteins were determined by MS. As shown in the schematic diagram in Fig. 1A, two human monocytic cell lines, THP-1 and U937, were treated with Phorbol 12-myristate 13-acetate (PMA) to induce the differentiation into macrophages (Fig. 1B). Cell lysates were biotinylated and aliquoted. One aliquot of the cell lysates was denatured at 95 °C and included as a negative control. To identify potential candidates, the cut-off was set as the signal-to-noise ratio (SNR) > 1.5. In this way, we identified 51 and 42 proteins from the PMA-treated THP-1 and U937 cells, respectively, with 34 proteins in common between the two cell lines. This indicates a high reliability of the microarray experiments (Fig. 1B, supplemental Fig. S1 and supplemental Table S2).
Fig. 1.
The schematic of SOPHIE and the identification of Mtb effectors. A, The schematic diagram of SOPHIE strategy. Briefly, the biotinylated cell lysates of macrophage were probed on the Mtb proteome microarray. Mtb proteins that bind macrophage proteins were identified. Using the identified Mtb proteins as bait, the interacting macrophage proteins were determined. B, The workflow of this study. C, Representatives of the newly identified Mtb effectors. The cutoff for calling positive candidates was set as signal-to-noise (SNR) ratio > 1.5. Experiments were performed in two replicates.
Compared with other fractions of Mtb, the membrane proteins and secreted proteins have higher potential to interact with macrophage proteins (49, 50). Interestingly, of the 34 shared proteins (supplemental Fig. S1), 21 are membrane or secreted proteins. In addition, there are 2 (Rv0462, Rv3339c) and 3 (Rv3240c, Rv0203, and Rv2164c) unique membrane or secreted proteins that interact with proteins from the THP-1 or U937 cells, respectively. Thus, 26 Mtb membrane proteins or secreted proteins that are potential Mtb effectors were discovered with SOPHIE (Fig. 1C and supplemental Table S3).
Identification of the BfrB Binding Protein in Human Macrophages
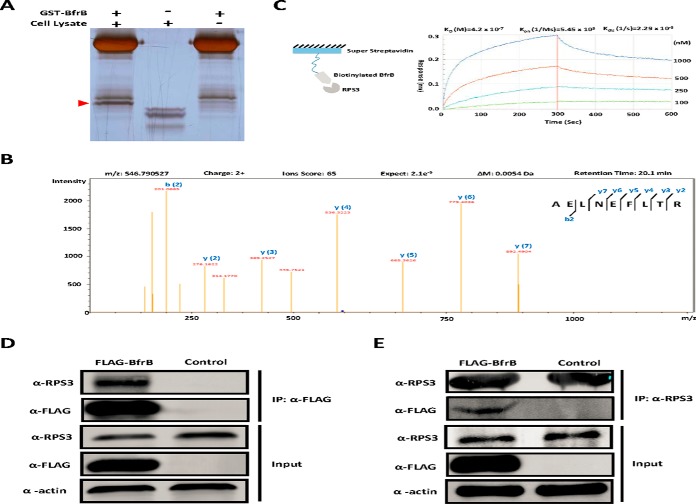
One of the effectors identified here, BfrB, was previously found to be required for Mtb survival in macrophages (41, 51) and is enriched from infected macrophages (43), suggesting that BfrB may play an important role in the interaction with the host. To identify the macrophage binding partners of BfrB, we purified GST-tagged BfrB and incubated it with PMA- treated THP-1 lysate before performing GST pull-down and silver staining (Fig. 2A). Subsequent MS identified a major pull-down protein of BfrB as RPS3 (Fig. 2B, supplemental Table S4), an essential subunit of NF-κB important for NF-κB binding to specific regulatory sites and for the inducible expression of a variety of genes (45).
Fig. 2.
Validation of the interaction between BfrB and RPS3. A, GST Pull-down of BfrB. GST tagged BfrB was incubated with PMA- differentiated THP-1 cell lysates followed by GST Pull-down, whereas GST only was included as the negative control. The unique band was then trypsin digested and subjected to MS/MS analysis. B, RPS3 was identified as the major binder of BfrB by LC-MS/MS analysis. C, The binding kinetics of BfrB to RPS3 measured by BLI assay. Biotinylated BfrB was immobilized on a streptavidin-coated biosensor and exposed to RPS3 in buffer. The binding was measured by coincident changes in the interference pattern. Results presented are representative of three experiments. D, Co-immunoprecipitation of RPS3 with FLAG-BfrB. Samples were immunoprecipitated with α-FLAG antibody to capture BfrB and immunoblotted with α-RPS3 to detect RPS3. Experiments were carried out in triplicate. E, Co-immunoprecipitation of FLAG-BfrB with RPS3. Samples were immunoprecipitated with α-RPS3 antibody to capture RPS3 and immunoblotted with α-FLAG antibody to detect BfrB. Experiments were carried out in triplicate.
To further validate the interaction of BfrB and RPS3, we examined the strength of their interaction using Bio-Layer Interferometry (BLI), finding that there is indeed a strong interaction between BfrB and RPS3 with an equilibrium dissociation constant (KD) value of 4.2 × 10−7 M (Fig. 2C).
To verify that RPS3 interacts with BfrB in vivo, a FLAG-tagged BfrB was overexpressed in HEK293T using an empty vector with only a FLAG epitope as control, followed by immune-precipitation with an anti-FLAG antibody. In this way, we found that endogenously expressed RPS3 could be enriched (Fig. 2D). Likewise, FLAG-tagged BfrB could also be enriched by immunoprecipitation with an anti-RPS3 antibody (Fig. 2E). Thus, consistent with the in vitro data, these results strongly suggest that BfrB interacts with RPS3 in vivo.
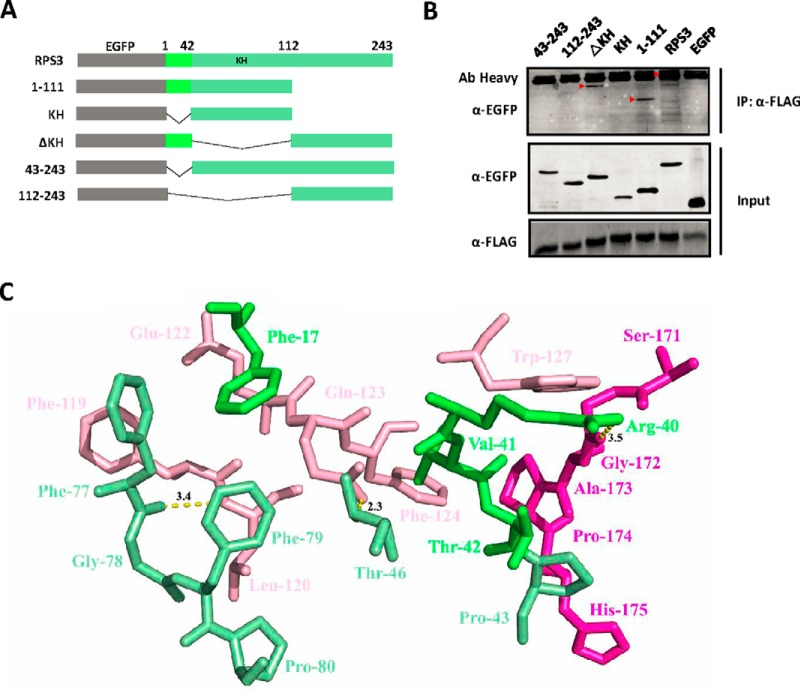
To map the binding domain of RPS3 on BfrB, we carried out a structure-function study with deletions of RPS3 that include its known functional domains (Fig. 3A). Co-immunoprecipitation revealed that the N-terminal region of RPS3 (amino acids 1–111), particularly residues 1–42, is required for RPS3 binding to BfrB (Fig. 3B).
Fig. 3.
BfrB binds to the N terminus of RPS3. A, Diagram of RPS3 and EGFP-tagged truncation mutants. KH, K homology. B, HEK293T cells transfected with constructs expressing FLAG-tagged BfrB and full-length or truncated EGFP-tagged RPS3 proteins, lysed after 48 h, and immunoblotted for EGFP-tagged RPS3 proteins after immunoprecipitation with FLAG antibody for BfrB in lysates. Experiments were carried out in triplicate. C, Detailed view of the interaction between the RPS3 (pale green) and BfrB (pink); Amino acids 164–181 of the BfrB were showed in rose red, and amino acids 1–42 of the RPS3 were displayed in green.
The crystal structures for both RPS3 (PDB ID: 1WH9) and BfrB (PDB ID: 3QD8) have been determined. To provide additional details of the binding mechanism, guided by the aforementioned binding assays (Fig. 3A and 3B) and the structure of RPS3 and BfrB, we identified a binding model through structure docking using ZDOCK server (http://zdock.umassmed.edu/). The interaction between RPS3 (pale green, residues 1–42 are colored in green) and BfrB (pink, residues 164–181 are colored in rose red) is shown in supplemental Fig. S2C. Detailed analysis showed that one hydrophobic interaction is observed among residues Phe-17, Phe-77, Gly-78, Phe-79, and Pro-80 of RPS3 and Phe-119 and Leu-120 of BfrB, whereas another hydrophobic interaction is observed between Val-41 of RPS3 and the Phe-124, Trp-127, Gly-172, Ala-173 and Pro-174 of BfrB, forming stable hydrophobic bindings (Fig. 3C). In addition, Phe-17 of RPS3 formed an anion-π interaction with Glu-122 of BfrB. We also identified three hydrogen bond interactions between Phe-77 of RPS3 and Leu-120 of the BfrB (bond length: 3.4 Å), Thr-46 of RPS3 and Gln-123 of BfrB (bond length: 2.3 Å), and Arg-40 of the RPS3 and Gly-172 of BfrB (bond length: 3.5 Å), which establish the main binding interaction between RPS3 and BfrB (Fig. 3C).
Mtb BfrB has an extended C termini (residues 164–181) that is unlike most other bacterial ferritins (52). Co-immunoprecipitation revealed that the C-terminal region of BfrB (residues 164–181) is important for the binding of BfrB to RPS3 (supplemental Fig. S2A and S2B) that is consistent with the aforementioned results.
BfrB Reduces the Nuclear Abundance of RPS3 and Alters Host NF-κB Activity
The RPS3 N-terminal (residues 1–42) contains a nuclear localization signal (NLS), which mediates translocation to the nucleus in response to TNF-α stimulation and DNA-damage (45, 53). We hypothesized that BfrB may mask the NLS of RPS3 by binding to the RPS3 N-terminal (residues 1–42).
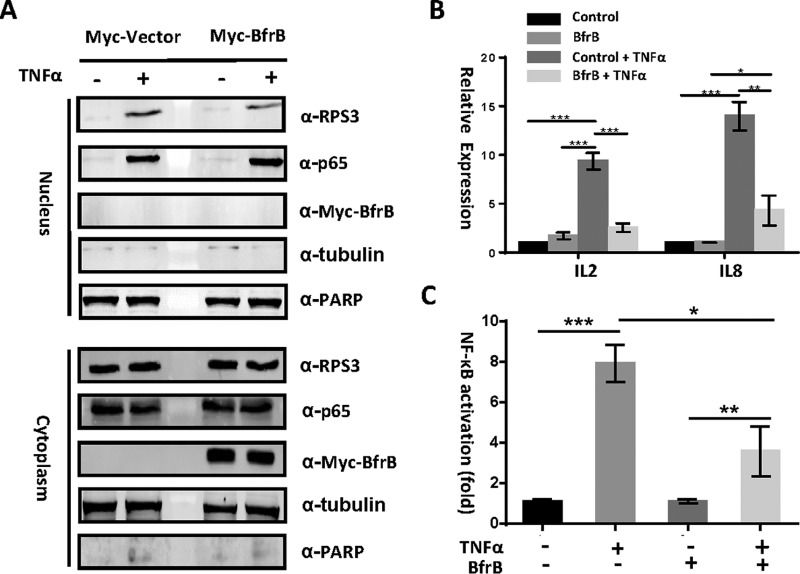
To determine if BfrB binding to RPS3 inhibits translocation of RPS3 into the nucleus, we detected the abundance of nuclear and cytoplasmic RPS3. HEK293T cells were transfected with bfrB, with or without stimulation with 100 ng/ml TNF-α for 1 h. The cells were fractionated to separate the nuclear- from the cytoplasmic-components. The lack of PARP and tubulin within the cytoplasmic and nuclear fractions, respectively, demonstrated the absence of cross-contamination among the fractions (Fig. 4A). We found that BfrB reduces the nuclear translocation of RPS3, suggesting that BfrB may also inhibit the host immune response by alteration of the transcription of NF-κB dependent genes.
Fig. 4.
BfrB reduces the nuclear abundance of RPS3 and alters NF- κB activity (A) Immunoblot analysis of cytoplasmic and nuclear fractions of HEK293T cells transfected with bfrB, or a FLAG control, in the presence or absence of TNF-α (100 ng/ml) stimulation for 1 h. A, Blots were probed with α-RPS3, α-p65, α-FLAG, α-tubulin, and α-PARP monoclonal antibodies. B, Luciferase assay of NF-κB activity. HEK293T cells were co- transfected with a firefly luciferase construct with a renilla luciferase plasmid, and bfrB, or a FLAG control driven by a consensus κB site. The cells were cultured for 24 h, in the presence or absence of TNF-α (100 ng/ml) stimulation for 1 h. Experiments were carried out in triplicate. C, Quantitative PCR analysis of IL2 mRNA and IL8 mRNA in HEK293T transfected with or without bfrB for 24 h. 0.01<*p < 0.05, 0.001<**p < 0.01, ***p < 0.001 (two-tailed unpaired t test). Data are representative of one experiment with three independent biological replicates.
To test this, we performed RT-PCR to assess the regulation of several RPS3-dependent genes (IL2, IL8), which are important to the innate response to infection (45). We indeed found that BfrB efficiently inhibits RPS3-dependent gene expression following the stimulation of TNF-α (Fig. 4B).
We also investigated whether Mtb BfrB modulates the NF-κB signaling pathway using a dual-luciferase reporter assay. Ectopic expression of Mtb bfrB in HEK293T cells efficiently blocked TNF-α induced NF-κB luciferase reporter activation (Fig. 4C). Further, BfrB affected neither the TNF-α (100 ng/ml, 1 h) induced phosphorylation of IκBα nor altered total IκBα concentrations (supplemental Fig. S2). Thus, these data suggest that BfrB plays a role in the reduction of RPS3 nuclear abundance, without significant impact on other NF-κB subunits.
BfrB Inhibits Cytokine Production of Macrophages and Promotes the Survival of Mycobacteria in Macrophages
We next investigated whether BfrB modulates the expression of inflammatory cytokines in macrophages by analyzing the expression of mRNA encoding TNF-α and IL-1β in macrophages infected with bfrB- overexpressing Mycobacterium smegmatis (Msm). We found that overexpression of bfrB in Msm significantly inhibits the expression of TNF and IL-1β mRNA and increased bacterial survival in THP-1 (Fig. 5). The expression of cytokines was markedly decreased in cells infected with bfrB-overexpressing Msm after 8 h, whereas the wild-type Msm and bfrB- overexpressing Msm strains had similar bacterial colony-forming units (CFU) until 12 h after infection. Thus, Mtb BfrB can promote survival of mycobacteria in macrophages by inhibiting cytokine production in host cells.
Fig. 5.
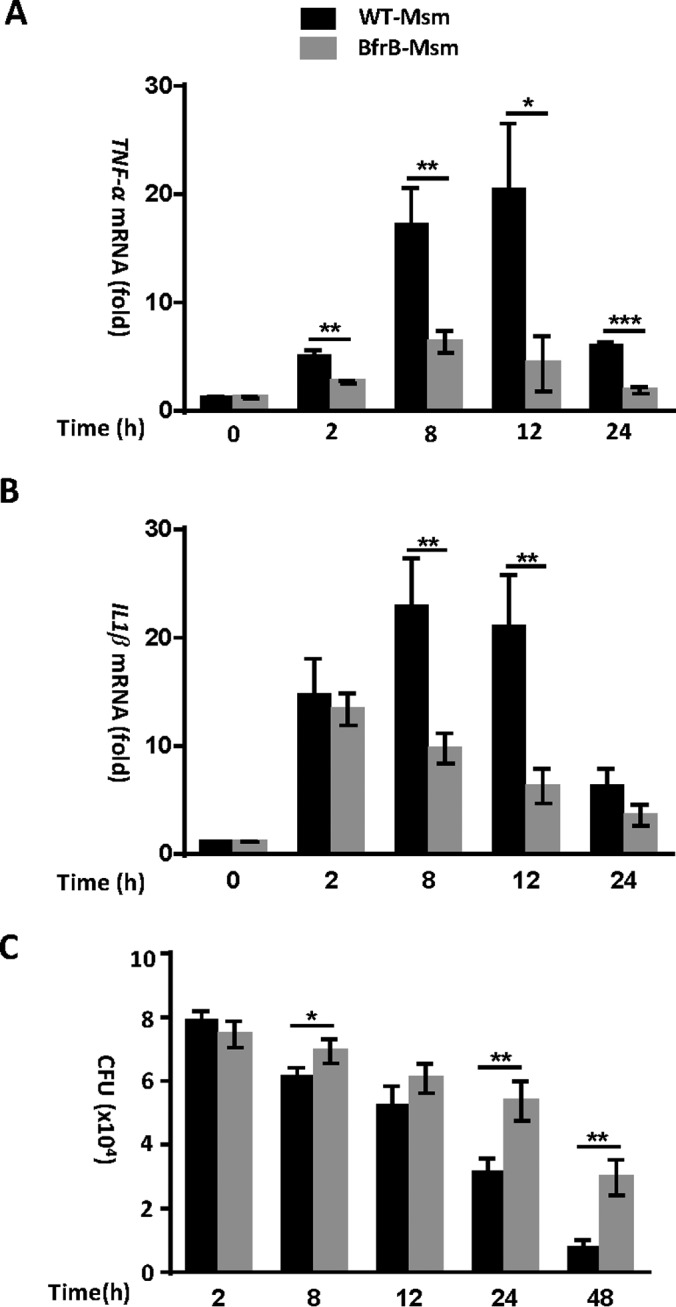
Overexpression of Mtb BfrB in Msm inhibits the expression of TNF-α and IL-1β and promotes the survival of mycobacteria in macrophages. A, B, Quantitative PCR analysis of TNF-α mRNA (A) and IL1β mRNA (B) in PMA-differentiated THP-1 macrophages infected for 0–24 h with wild-type Msm or Msm with bfrB-overexpressing. Experiments were carried out in triplicate. C, Survival of Msm in PMA-differentiated THP-1 macrophages infected for 0–48 h with wild-type Msm or Msm with bfrB-overexpressing 0.01<*p < 0.05, 0.001<**p < 0.01, ***p < 0.001 (two-tailed unpaired t test). Data are representative of one experiment with three independent biological replicates.
DISCUSSION
In this study, we developed a strategy that we call SOPHIE to identify potential effectors of Mtb. We identified 26 Mtb membrane and secreted proteins that interact with macrophage proteins. We found that BfrB, identified here for the first time as an Mtb effector, subverts the host innate immune system by binding the NF-κB subunit RPS3 and promotes the survival of mycobacteria in macrophages by inhibiting cytokine production in host cells.
One of the most important goals of present TB studies is to identify Mtb effectors that interact with macrophage proteins (54). The strategy of SOPHIE based on the Mtb proteome microarray has several advantages over more traditional approaches. First, the strategy can globally and unbiasedly profile the bindings of 4262 Mtb proteins with the macrophage cell lysate in a single experiment. Second, all the Mtb proteins on the microarray are expressed and affinity purified in a high-throughput manner following the same procedure. Thus, the local concentrations of proteins are relatively high that could greatly facilitate the identification of Mtb effectors. Third, as demonstrated in this study, the proteome-wide Mtb- macrophage PPI identification consumes only a limited number of microarrays in less than 18 h. In addition, the macrophage cell lysate was incubated on the Mtb proteome microarray, and so, the host proteins were maintained in a physiological condition that ensures more reliable results.
Of those potential effectors identified here, LpdC (Rv0462) (20, 55), SahH (Rv3248c) (22), Rv3655c (23), SecA1 (Rv3240c) (56), FadD13 (Rv3089) (57), BfrA (Rv1876) (41, 51) and BfrB (Rv3841) (41, 51, 58) are known to play critical roles for Mtb survival in the host. For Mtb proteins, LpdC, SahH and Rv3655c, their host interacting proteins had previously been identified. LpdC prevents phagosome-lysosome maturation by retaining the host factor, coronin 1 (20). SahH is one of the human IL-8-binding effectors and contributes to Mtb entry into human neutrophils (22). Rv3655c interacts with the human AL017 protein and suppresses apoptosis by blocking the extrinsic pathway (23).
Mtb BfrB is a ferritin that exists as a macromolecular assembly composed 24 identical subunits (52). BfrB is required for Mtb to overcome iron limitation and oxidative stress, and Mtb lacking BfrB were found to be unable to persist in mice (41). In addition, we note that iron is limited in the host cells (51). A recent report showed that pupylation of ferritin triggers the partial or complete disassembly of the 24-meric ferritin, which is required for adaption of Corynebacterium glutamicum to iron limitation (59). The BfrB of Msm has been shown to be pupylated at Lys-10 that is located on the outer surface of its crystal structure (60). Previous work has suggested that bacterial proteins smaller than 70 KDa can cross the host phagosomal membrane (61). Although the exact mechanism by which BfrB secreted across the Mtb membrane to macrophage remains unclear (43, 44), the disassembly of Mtb BfrB triggered by pupylation may be important in this regard.
Our results show that BfrB is a novel effector that inhibits the immune system by binding the NF-κB functional subunit, RPS3. The NF-κB signaling pathway is one of the most attractive targets exploited by pathogens to modulate host cell events (48, 62). For example, Mtb PtpA can block the host adaptor TAB3 mediated NF-κB signaling by competitively binding to the ubiquitin-interacting domain of TAB3 (11). The Escherichia coli strain O157:H7 effector NleH1 specifically binds and inhibits the phosphorylation of RPS3 at Ser-209, thus retarding its nuclear translocation and subsequent NF-κB function (48, 63). In this work, we found that BfrB interacts with the NLS region of RPS3 and reduces the nuclear abundance of RPS3 to reduce host transcriptional output. Mtb BfrB participation in the host immune response may provide a new insight into why BfrB is essential for Mtb during chronic infection of mice (41, 51). In addition, we also showed that Mtb BfrB promotes survival of mycobacteria in macrophages by inhibiting cytokine production in host cells.
We note that the secreted proteins of Mtb have not yet been comprehensively determined. A variety of studies have identified a series of secreted proteins through different strategies, e.g. detecting Mtb proteins in culture filtrate (CF) (42, 64–66), prediction by bioinformatics (67), or individual in-depth functional studies (11, 23). However, proteins may be in the CF because of bacterial lysis rather than secretion, and bioinformatics predictions may not be accurate. Thus, other proteins that were identified on the microarray but not included in the 26 protein-list may also possess important interactions with host proteins.
Finally, we believe that the 26 Mtb membrane or secreted proteins identified in this study could serve as a valuable resource for further basic research. In addition, some of these proteins may potentially be promising targets for Mtb drug development, because inhibitors targeting secreted or membrane proteins do not need to penetrate the unique and extremely impermeable mycobacterial cell wall (68–70). For example, some of the targets of the first- and second-line TB drugs, such as isoniazid and ethambutol (71, 72), are the proteins necessary for the biosynthesis and assembly of the mycobacterial cell wall and outer membrane (73). In our list, some proteins may be potential targets of anti-TB drug development. For example, SecA1, as the component of the canonical mycobacteria Sec secretion system, is essential and responsible for the bulk of proteins export (74). FadD13 is involved in maintaining the appropriate cell wall architecture of Mtb on exposure to the acidic pH encountered within macrophages (75). Mtb BfrB plays an important role in the interaction with the NF-κB subunit RPS3, suggesting a potential TB treatment via targeting of BfrB-RPS3 interfaces. Because these potential drug targets are different from the known ones, once effective drugs for these targets are successfully developed, they should have novel mode of action (MOA), thus, providing new “bullets” for combating with MDR and XDR TB.
In summary, we have performed a comprehensive survey of the effectors of Mtb using our novel strategy, SOPHIE. It is expected that these effectors will be a valuable resource for basic Mtb study and drug development. In addition, we fully anticipate that SOPHIE will become a highly effective strategy to identify effectors in other human pathogens, thus providing a versatile method in drug development strategies more generally.
DATA AVAILABILITY
The mass spectrometry data have been deposited to the Proteome-Xchange Consortium via the PRIDE (47) partner repository with the data set identifier PXD006927, 10.6019/PXD006927 and PXD006929, 10.6019/PXD006929.
Supplementary Material
Acknowledgments
We thank Daniel M. Czajkowsky for critical reading and editing. We also thank the Instrumental Analysis Center of Shanghai Jiao Tong University for the LC-MS/MS and data analysis.
Footnotes
Author Contributions: SCT conceived and designed the project; XH, HWJ and HC performed most of the experiments and XH and MKY carried out most of the computational analyses; SJG, WY, HNZ, YL, ZWX and FLW contributed reagents or provided laboratory assistance; JLH performed the mass spectrometry analysis; XH and SCT interpreted results and wrote the manuscript; JYD, LJB and XEZ provided key reagents and commented on the manuscript.
* This study was supported in part by The National Key Research and Development Program of China Grant 2016YFA0500600, National Natural Science Foundation of China Grants 31670831 and 31370813. This study was also supported by the National Natural Science Foundation of China 31471232, Science and Technology Planning Projects of Guangdong Province 2014B030301040 and the Chinese Academy of Sciences grant KJZD-EW-TZ-L04 to X.-e.Z.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- Mtb
- Mycobacterium tuberculosis
- MOA
- Mode of action
- SOPHIE
- Systematic unlOcking Pathogen and Host Interacting Effectors
- Msm
- Mycobacterium smegmatis
- TB
- tuberculosis
- RPS3
- ribosomal protein S3
- BLI
- Bio-Layer Interferometry
- PPI
- protein- protein interactions
- NF-κB
- Nuclear factor-kappa B.
REFERENCES
- 1. World Health Organization (WHO). (2016) Global tuberculosis report 2016. WHO Press [Google Scholar]
- 2. Gandhi N. R., Shah N. S., Andrews J. R., Vella V., Moll A. P., Scott M., Weissman D., Marra C., Lalloo U. G., Friedland G. H., Tugela Ferry C., and Research C. (2010) HIV coinfection in multidrug- and extensively drug-resistant tuberculosis results in high early mortality. Am. J. Respir. Crit. Care Med. 181, 80–86 [DOI] [PubMed] [Google Scholar]
- 3. Goldberg D. E., Siliciano R. F., and Jacobs W. R. Jr. (2012) Outwitting evolution: fighting drug- resistant TB, malaria, and HIV. Cell 148, 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jean P., and John G. (2002) Hijacking the host: survival of pathogenic mycobacteria inside macrophages. Trends Microbiol. 10, 142–146 [DOI] [PubMed] [Google Scholar]
- 5. Cambier C. J., Falkow S., and Ramakrishnan L. (2014) Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 159, 1497–1509 [DOI] [PubMed] [Google Scholar]
- 6. MacMicking J. D. (2014) Cell-autonomous effector mechanisms against Mycobacterium tuberculosis. Cold Spring Harb. Perspect. Med. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Behar S. M., Martin C. J., Booty M. G., Nishimura T., Zhao X., Gan H. X., Divangahi M., and Remold H. G. (2011) Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 4, 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu G., Wang J., Gao G. F., and Liu C. H. (2014) Insights into battles between Mycobacterium tuberculosis and macrophages. Protein Cell 5, 728–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poirier V., and Av-Gay Y. (2012) Mycobacterium tuberculosis modulators of the macrophage's cellular events. Microbes Infect. 14, 1211–1219 [DOI] [PubMed] [Google Scholar]
- 10. Kim K. H., An D. R., Song J., Yoon J. Y., Kim H. S., Yoon H. J., Im H. N., Kim J., Kim D. J., Lee S. J., Kim K. H., Lee H. M., Kim H. J., Jo E. K., Lee J. Y., and Suh S. W. (2012) Mycobacterium tuberculosis Eis protein initiates suppression of host immune responses by acetylation of DUSP16/MKP-7. Proc. Natl. Acad. Sci. U.S.A. 109, 7729–7734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J., Li B. X., Ge P. P., Li J., Wang Q., Gao G. F., Qiu X. B., and Liu C. H. (2015) Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat. Immunol. 16, 237–245 [DOI] [PubMed] [Google Scholar]
- 12. Liu M., Li W., Xiang X., and Xie J. (2015) Mycobacterium tuberculosis effectors interfering host apoptosis signaling. Apoptosis 20, 883–891 [DOI] [PubMed] [Google Scholar]
- 13. Wong D., Bach H., Sun J., Hmama Z., and Av-Gay Y. (2011) Mycobacterium tuberculosis protein tyrosine phosphatase (PtpA) excludes host vacuolar-H+-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. U.S.A. 108, 19371–19376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang J., Teng J. L., Zhao D., Ge P., Li B., Woo P. C., and Liu C. H. (2016) The ubiquitin ligase TRIM27 functions as a host restriction factor antagonized by Mycobacterium tuberculosis PtpA during mycobacterial infection. Sci. Rep. 6, 34827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bach H., Papavinasasundaram K. G., Wong D., Hmama Z., and Av-Gay Y. (2008) Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe 3, 316–322 [DOI] [PubMed] [Google Scholar]
- 16. Sun J., Wang X., Lau A., Liao T. Y., Bucci C., and Hmama Z. (2010) Mycobacterial nucleoside diphosphate kinase blocks phagosome maturation in murine RAW 264.7 macrophages. PLoS ONE 5, e8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vergne I., Chua J., Lee H. H., Lucas M., Belisle J., and Deretic V. (2005) Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 4033–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sreejit G., Ahmed A., Parveen N., Jha V., Valluri V. L., Ghosh S., and Mukhopadhyay S. (2014) The ESAT-6 protein of Mycobacterium tuberculosis interacts with beta-2-microglobulin (beta2M) affecting antigen presentation function of macrophage. PLoS Pathog. 10, e1004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mueller-Ortiz S. L., Wanger A. R., and Norris S. J. (2001) Mycobacterial protein HbhA binds human complement component C3. Infect. Immun. 69, 7501–7511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deghmane A.-E., Soualhine H., Bach H., Sendide K., Itoh S., Tam A., Noubir S., Talal A., Lo R., Toyoshima S., Av-Gay Y., and Hmama Z. (2007) Lipoamide dehydrogenase mediates retention of coronin-1 on BCG vacuoles, leading to arrest in phagosome maturation. J. Cell Sci. 120, 3489–3489 [DOI] [PubMed] [Google Scholar]
- 21. Mehra A., Zahra A., Thompson V., Sirisaengtaksin N., Wells A., Porto M., Koster S., Penberthy K., Kubota Y., Dricot A., Rogan D., Vidal M., Hill D. E., Bean A. J., and Philips J. A. (2013) Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking. PLoS Pathog. 9, e1003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dziadek B., Brzostek A., Grzybowski M., Fol M., Krupa A., Kryczka J., Plocinski P., Kurdowska A., and Dziadek J. (2016) Mycobacterium tuberculosis AtsG (Rv0296c), GlmU (Rv1018c) and SahH (Rv3248c) proteins function as the human IL-8-binding effectors and contribute to pathogen entry into human neutrophils. PLoS ONE 11, e0148030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Danelishvili L., Yamazaki Y., Selker J., and Bermudez L. E. (2010) Secreted Mycobacterium tuberculosis Rv3654c and Rv3655c proteins participate in the suppression of macrophage apoptosis. PLoS ONE 5, e10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rajagopala S. V., Sikorski P., Kumar A., Mosca R., Vlasblom J., Arnold R., Franca-Koh J., Pakala S. B., Phanse S., Ceol A., Hauser R., Siszler G., Wuchty S., Emili A., Babu M., Aloy P., Pieper R., and Uetz P. (2014) The binary protein-protein interaction landscape of Escherichia coli. Nat. Biotech. 32, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jäger S., Cimermancic P., Gulbahce N., Johnson J R, McGovern K E, Clarke S C, Shales M., Mercenne G., Pache L., Li K., Hernandez H., Jang G. M., Roth S. L., Akiva E., Marlett J., Stephens M., D'Orso I., Fernandes J., Fahey M., Mahon C., O'Donoghue A. J., Todorovic A., Morris J. H., Maltby D. A., Alber T., Cagney G., Bushman F. D., Young J. A., Chanda S. K., Sundquist W. I., Kortemme T., Hernandez R. D., Craik C. S., Burlingame A., Sali A., Frankel A. D., and Krogan N. J. (2011) Global landscape of HIV- human protein complexes. Nature 481, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mendez-Rios J., and Uetz P. (2010) Global approaches to study protein-protein interactions among viruses and hosts. Future Microbiol. 5, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu H. Y., Braun P., Yıldırım M. A., Lemmens I., Venkatesan K., Sahalie J., Hirozane-Kishikawa T., Gebreab F., Li N., Simonis N., Hao T., Rual J. F., Dricot A., Vazquez A., Ryan Murray R., Christophe S., Tardivo L., Tam S., Svrzikapa N., Fan C., de Smet A., Motyl. A., Hudson M., Park J., Xin X., Cusick M., Moore T., Boone C., Snyder M., Roth F., Barabási A., Tavernier J., Hill D., and Vida M. (2008) High- quality binary protein interaction map of the yeast interactome network. Science 322, 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boshoff H. I., and Lun D. S. (2010) Systems biology approaches to understanding mycobacterial survival mechanisms. Drug Discov. Today Dis. Mech. 7, e75–e82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barczak A. K., Avraham R., Singh S., Luo S. S., Zhang W. R., Bray M.-A., Hinman A. E., Thompson M., Nietupski R. M., Golas A., Montgomery P., Fitzgerald M., Smith R. S., White D. W., Tischler A. D., Carpenter A. E., and Hung D. T. (2017) Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog. 13, e1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., and Snyder M. (2001) Global Analysis of Protein Activities Using Proteome Chips. Science 293, 2101–2105 [DOI] [PubMed] [Google Scholar]
- 31. Gnjatic S., Ritter E., Buchler M. W., Giese N. A., Brors B., Frei C., Murray A., Halama N., Zornig I., Chen Y. T., Andrews C., Ritter G., Old L. J., Odunsi K., and Jager D. (2010) Seromic profiling of ovarian and pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A. 107, 5088–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deng R. P., He X., Guo S. J., Liu W. F., Tao Y., and Tao S. C. (2014) Global identification of O- GlcNAc transferase (OGT) interactors by a human proteome microarray and the construction of an OGT interactome. Proteomics 14, 1020–1030 [DOI] [PubMed] [Google Scholar]
- 33. Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., Wan J., Tao S. C., Qian J., Zhao Y., Boeke J. D., Berger S. L., and Zhu H. (2009) Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136, 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu J., Gopinath K., Murali A., Yi G., Hayward S. D., Zhu H., and Kao C. (2007) RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. U.S.A. 104, 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu K. Y., Tao S. C., Tzu-Ching Y., Yu-Hsuan H., Chia-Hsien L., Chen-Ching L., Hsueh-Fen J., Hsuan Cheng - H, Chin-Yu Y., Ming-Shuo C., Yu-Yi L., Jin-Ying L., Heng Z., and Chien-Sheng C. (2012) Profiling lipid–protein interactions using nonquenched fluorescent liposomal nanovesicles and proteome microarrays. Mol. Cell. Proteomics 11, 1177–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang H. N., Yang L., Jian-ya, Czajkowsky L. D. M., Wang J. F., Zhang X. W., Zhou Y. M., Ge FYang M-k Xiong Q, Guo S. J., Lea H. Y., Wu S. F., Yan W., Liu B., Zhu H., Chen Z., and Tao S. C. (2015) Systematic identification of arsenic-binding proteins reveals that hexokinase-2 is inhibited by arsenic. Proc. Natl. Acad. Sci. U.S.A. 112, 15084–15089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang J., Zhu H., Haggarty S. J., Spring D. R., Hwang H., Jin F., Snyder M., and Schreiber S. L. (2004) Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. U.S.A. 101, 16594–16599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng J. Y., Bi L. J., Zhou L., Guo S. J., Fleming J., Jiang H. W., Zhou Y., Gu J., Zhong Q., Wang Z. X., Liu Z., Deng R. P., Gao J., Chen T., Li W., Wang J. F., Wang X., Li H., Ge F., Zhu G., Zhang H. N., Gu J., Wu F. L., Zhang Z., Wang D., Hang H., Li Y., Cheng L., He X., Tao S. C., and Zhang X. E. (2014) Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Rep. 9, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 39. Wu Fan-Lin Yin Liu He-Wei Jiang Yi-Zhao Luan Hai-Nan Zhang Xiang He Zhao-Wei Xu Jing-Li Hou Ji L.-Y., Zhi Xie Daniel Czajkowsky M., Wei Yan Jiao-Yu Deng Li-Jun Bi Xian-En Zhang and Tao S.-C. (2017) The Ser/Thr protein kinase protein-protein interaction map of M.tuberculosis. Mol. Cell. Proteomics 16, 1491–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang H.-N., Zhao-wei, Jiang X. H. W., Fan-Lin He WXYin Guo L. S. J., Yang Bi L. L., Deng J., Xian-en Z, Tao S. C. (2017) Cyclic di-GMP regulates Mycobacterium tuberculosis resistance to ethionamide. Sci Rep. 7, 5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pandey R., and Rodriguez G. M. (2012) A ferritin mutant of Mycobacterium tuberculosis is highly susceptible to killing by antibiotics and is unable to establish a chronic infection in mice. Infect. Immun. 80, 3650–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mattow J., Schaible U. E., Schmidt F., Hagens K., Siejak F., Brestrich G., Haeselbarth G., Muller E. C., Jungblut P. R., and Kaufmann S. H. (2003) Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis 24, 3405–3420 [DOI] [PubMed] [Google Scholar]
- 43. Chande A. G., Siddiqui Z., Midha M. K., Sirohi V., Ravichandran S., and Rao K. V. S. (2015) Selective enrichment of mycobacterial proteins from infected host macrophages. Sci Rep. 5, 13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kruh-Garcia N. A., Wolfe L. M., Chaisson L. H., Worodria W. O., Nahid P., Schorey J. S., Davis J. L., and Dobos K. M. (2014) Detection of Mycobacterium tuberculosis peptides in the exosomes of patients with active and latent M.tuberculosis infection using MRM-MS. PLoS ONE 9:e103811, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., and Lenardo M. J. (2007) Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 46. Schroder C., Alhamdani M. S., Fellenberg K., Bauer A., Jacob A., and Hoheisel J. D. (2011) Robust protein profiling with complex antibody microarrays in a dual-colour mode. Protein Microarrays 785, 203–221 [DOI] [PubMed] [Google Scholar]
- 47. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao X., Wan F., Mateo K., Callegari E., Wang D., Deng W., Puente J., Li F., Chaussee M. S., Finlay B. B., Lenardo M. J., and Hardwidge P. R. (2009) Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog. 5, e1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li W., Fan X., Long Q., Xie L., and Xie J. (2015) Mycobacterium tuberculosis effectors involved in host-pathogen interaction revealed by a multiple scales integrative pipeline. Infect. Genet. Evol. 32, 1–11 [DOI] [PubMed] [Google Scholar]
- 50. Queiroz A., and Riley L. W. (2017) Bacterial immunostat: Mycobacterium tuberculosis lipids and their role in the host immune response. Revista da Sociedade Brasileira de Medicina Tropical 50, 9–18 [DOI] [PubMed] [Google Scholar]
- 51. Reddy P. V., Puri R. V., Khera A., and Tyagi A. K. (2012) Iron storage proteins are essential for the survival and pathogenesis of Mycobacterium tuberculosis in THP-1 macrophages and the guinea pig model of infection. J. Bacteriol. 194, 567–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khare G., Gupta V., Nangpal P., Gupta R. K., Sauter N. K., and Tyagi A. K. (2011) Ferritin structure from Mycobacterium tuberculosis: comparative study with homologues identifies extended C-terminus involved in ferroxidase activity. PLoS ONE 6, e18570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yadavilli S., Hegde V., and Deutsch W. A. (2007) Translocation of human ribosomal protein S3 to sites of DNA damage is dependant on ERK-mediated phosphorylation following genotoxic stress. DNA Repair 6, 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Espitia C., Rodríguez E., Ramón-Luing L., Echeverría-Valencia G., and Vallecillo J. A. (2012) Host- pathogen interactions in tuberculosis. Understanding tuberculosis-analyzing the origin of Mycobacterium tuberculosis pathogenicity, Pere-Joan Cardona, ed. (InTech), pp 43–76 [Google Scholar]
- 55. Venugopal A., Bryk R., Shi S., Rhee K., Rath P., Schnappinger D., Ehrt S., and Nathan C. (2011) Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe 9, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guo X. V., Monteleone M., Klotzsche M., Kamionka A., Hillen W., Braunstein M., Ehrt S., and Schnappinger D. (2007) Silencing essential protein secretion in Mycobacterium smegmatis by using tetracycline repressors. J. Bacteriol. 189, 4614–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cheruvu M., Plikaytis B. B., and Shinnick T. M. (2007) The acid-induced operon Rv3083-Rv3089 is required for growth of Mycobacterium tuberculosis in macrophages. Tuberculosis 87, 12–20 [DOI] [PubMed] [Google Scholar]
- 58. Subbian S., Pandey R., Soteropoulos P., and Rodriguez G. M. (2015) Vaccination with an Attenuated Ferritin Mutant Protects Mice against Virulent Mycobacterium tuberculosis. J. Immunol. Res. 385402, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Küberl A., Polen T., and Bott M. (2016) The pupylation machinery is involved in iron homeostasis by targeting the iron storage protein ferritin. Proc. Natl. Acad. Sci. U.S.A. 113, 4806–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Watrous J., Burns K., Liu W. T., Patel A., Hook V., Bafna V., CEBarry 3rd, Bark S., and Dorrestein P. C. (2010) Expansion of the mycobacterial “PUPylome”. Mol. Biosyst. 6, 376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Teitelbaum R., Cammer M., Maitland M. L., Freitag N. E., Condeeli J., and Bloom B. R. (1999) Mycobacterial infection of macrophages results in membrane-permeable phagosomes. Proc. Natl. Acad. Sci. U.S.A. 96, 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Asrat S., Davis K. M., and Isberg R. R. (2015) Modulation of the host innate immune and inflammatory response by translocated bacterial proteins. Cell Microbiol. 17, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wan F., Weaver A., Gao X., Bern M., Hardwidge P. R., and Lenardo M. J. (2011) IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Malen H., Berven F. S., Fladmark K. E., and Wiker H. G. (2007) Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7, 1702–1718 [DOI] [PubMed] [Google Scholar]
- 65. de Souza G. A., Leversen N. A., Malen H., and Wiker H. G. (2011) Bacterial proteins with cleaved or uncleaved signal peptides of the general secretory pathway. J. Proteomics 75, 502–510 [DOI] [PubMed] [Google Scholar]
- 66. Okkels L. M., Muller E. C., Schmid M., Rosenkrands I., Kaufmann S. H., Andersen P., and Jungblut P. R. (2004) CFP10 discriminates between nonacetylated and acetylated ESAT-6 of Mycobacterium tuberculosis by differential interaction. Proteomics 4, 2954–2960 [DOI] [PubMed] [Google Scholar]
- 67. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 68. Koul A., Arnoult E., Lounis N., Guillemont J., and Andries K. (2011) The challenge of new drug discovery for tuberculosis. Nature 469, 483–490 [DOI] [PubMed] [Google Scholar]
- 69. Wong D., Chao J. D., and Av-Gay Y. (2013) Mycobacterium tuberculosis-secreted phosphatases: from pathogenesis to targets for TB drug development. Trends Microbiol. 21, 100–109 [DOI] [PubMed] [Google Scholar]
- 70. Favrot L., and Ronning D. R. (2012) Targeting the mycobacterial envelope for tuberculosis drug development. Expert Rev. Anti. Infect. Ther. 10, 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Belanger A. E., Besra G. S., Ford M. E., Mikusová K., Belisle J. K., Brennan P. J., and Inamine J. M. (1996) The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. U.S.A. 93, 11919–11924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Heath R. J., White S. W., and Rock C. O. (2001) Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 40, 467–497 [DOI] [PubMed] [Google Scholar]
- 73. Aggarwal A., Parai M. K., Shetty N., Wallis D., Woolhiser L., Hastings C., Dutta N. K., Galaviz S., Dhakal R. C., Shrestha R., Wakabayashi S., Walpole C., Matthews D., Floyd D., Scullion P., Riley J., Epemolu O., Norval S., Snavely T., Robertson G. T., Rubin E. J., Ioerger T. R., Sirgel F. A., van der Merwe R., van Helden P. D., Keller P., Böttger E. C., Karakousis P. C., Lenaerts A. J., and Sacchettini J. C. (2017) Development of a novel lead that targets M. tuberculosis Polyketide Synthase 13. Cell 170, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feltcher M. E., Sullivan J. T., and Braunstein M. (2010) Protein export systems of Mycobacterium tuberculosis: novel targets for drug development? Future Microbiol. 5, 1581–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jatana N., Jangid S., Khare G., Tyagi A. K., and Latha N. (2011) Molecular modeling studies of Fatty acyl-CoA synthetase (FadD13) from Mycobacterium tuberculosis-a potential target for the development of antitubercular drugs. J. Mol. Model 17, 301–313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data have been deposited to the Proteome-Xchange Consortium via the PRIDE (47) partner repository with the data set identifier PXD006927, 10.6019/PXD006927 and PXD006929, 10.6019/PXD006929.