Cas9-expressing mosquito strains pave the way toward gene drives
SEM image of genome-edited adult female mosquito. Left compound eye is split up, and there are three maxillary palps instead of the normal two. Image courtesy of Michelle Bui and Alexander Knyshov (University of California, Riverside, CA).
Previous efforts to use genome editing to thwart the ability of mosquito vectors to spread pathogens have been hampered by low mutation rates, poor survival of edited mosquitoes, and inefficient transmission of disrupted genes to offspring. Ming Li et al. (pp. E10540–E10549) developed transgenic strains of Aedes aegypti mosquitoes—major carriers of dengue, chikungunya, yellow fever, and zika viruses—that stably express a bacterial Cas9 enzyme in the germline, enabling highly efficient genome editing in the mosquito species. CRISPR-based gene disruption in the transgenic strains resulted in mosquitoes with an extra eye, an extra maxillary palp, a nonfunctional proboscis, malformed wings, and chromatic defects in the eyes and cuticle—traits that affect vision, flight, or blood feeding. Compared with coinjection of guide RNAs and recombinant Cas9 into wild-type mosquito embryos, genome editing using the transgenic strains resulted in increased mosquito survival rates, mutagenesis efficiency, and heritable mutation rates. Further, mutagenesis rates and efficiency of germline transmission of disrupted genes were boosted when multiple guide RNAs were coinjected into one of the transgenic strains. Double and triple mutants could be easily generated in a single step by coinjection of multiplexed RNAs targeting different genes into the transgenic strains. According to the authors, the study lays the groundwork for the development of efficient gene drives to control the mosquito population. — P.N.
Enhancing neutrophil and macrophage antitumor activity
Immunotherapy has shown immense potential for treating cancers, but current immunotherapies fail to produce complete and durable responses in many cancer patients. Signal regulatory protein alpha (SIRPα) is a promising immunotherapy target that serves as a myeloid cell-specific immune checkpoint that engages the antiphagocytic signal CD47. Nan Guo Ring et al. (pp. E10578–E10585) developed the monoclonal antibody KWAR23, which binds SIRPα and disrupts its binding to CD47. The authors found that KWAR23 was inert when administered by itself, but when combined with tumor-opsonizing monoclonal antibodies, it greatly enhanced the killing of several human tumor-derived cell lines by inducing macrophage-dependent phagocytosis. KWAR23 enhanced the antitumor activity of neutrophils and macrophages both in vitro and in vivo. When the KWAR23 antibody was used to treat a mouse model with human SIRPα that had been knocked in, neutrophils and macrophages infiltrated a human Burkitt’s lymphoma xenograft and significantly reduced tumor growth. The authors created a bispecific anti-CD70/SIRPα antibody and found that it showed enhanced antitumor activity, compared with individually delivered antibodies in certain types of cancers. Using an antibody to block SIRPα led to increased antitumor activity by myeloid cell subsets that frequently infiltrate tumors, and KWAR23 might be a promising candidate for combination therapy, according to the authors. — S.R.
Molecular signatures across subtypes of Alzheimer’s disease patients
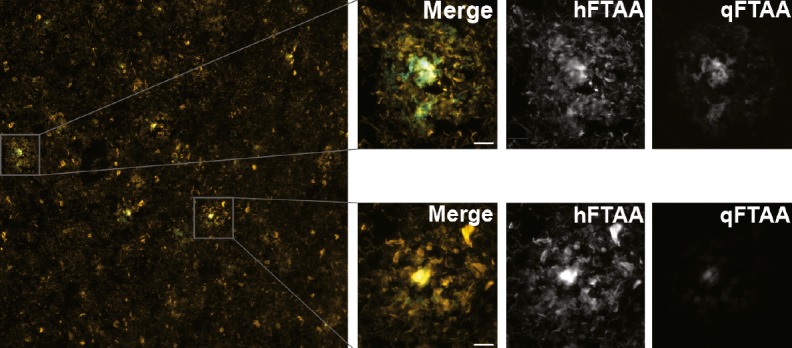
Amyloid plaques in brain tissue labeled by quadro-formyl thiophene acetic acid/hepta-formyl thiophene acetic acid fluorescent dyes.
One major hallmark of Alzheimer’s disease (AD) is the accumulation of the amyloid-β peptide into plaque deposits in the brain. Despite the common molecular hallmark of the disease, clinical symptoms and pathologic changes in brain tissues vary significantly across patients. Jay Rasmussen et al. (pp. 13018–13023) tested the hypothesis that the largely unexplained heterogeneity observed across patients may be influenced by diverse molecular structures of amyloid plaques in the brain. The authors analyzed postmortem tissue samples obtained from 40 clinically and pathologically diagnosed cases of AD. The authors labeled amyloid plaques in three brain regions—temporal, occipital, and frontal cortex—using a class of dyes called luminescent conjugated oligothiophenes, which bind to amyloid and emit variable fluorescence emission spectra that reflect the 3D structure of the protein aggregates. The spectral signatures of the amyloid plaque cores varied significantly across different patient subtypes, distinguishing carriers of certain types of mutations (e.g., APP V717I and PSEN1 A431E) from other groups. Within an individual brain, minor populations of amyloid-β aggregates had different molecular structures, even though the mean spectral properties of the amyloid plaques were similar across all three brain regions. According to the authors, the findings support the idea that the heterogeneous molecular structures of amyloid plaques in the brain might contribute to the diverse clinical and pathological disease manifestations observed across patients. — J.W.
Human population, climate, and food production
Radiocarbon dating is increasingly used to reconstruct past dynamics of human population. In Britain and Ireland, archaeological sites have yielded a high density of radiocarbon samples from botanical and faunal material. Andrew Bevan et al. (pp. E10524–E10531) used more than 30,000 archaeological radiocarbon dates from databases and published reports to study human population dynamics in various regions of Britain and Ireland during the middle and later Holocene. The authors report at least three instances of human population decline that coincided with periodic changes in climate and subsequent societal responses in food use. For instance, after an increase in human population during the Early Neolithic around 4000–3850 BCE, a population decline occurred from around 3500–3000 BCE. The population decline was consistent with reduced solar activity and changing salt input to the Greenland ice sheet; the decline was also likely associated with cold winters and wet summers. Moreover, food use during the Middle–Late Neolithic, around 3500–2500 BCE, moved toward hardy cereals, gathered resources, and pastoralism, with wheat giving way to increased barley cultivation, and later, an emphasis on gathered hazelnuts and cattle herding. According to the authors, the findings offer insight into the long-term dynamics of human population, food production, and climate change. — C.S.
California bird nesting and climate change

California Scrub-Jay nestlings. Photograph by Joseph S. Dixon; © 2007 Museum of Vertebrate Zoology.
Species respond to climate change by shifting their geographical ranges poleward or upward to remain in their preferred temperature zones, or by shifting the timing of life history events to mirror changes in resource availability. Jacob Socolar et al. (pp. 12976–12981) examined the relationship between temperature and breeding dates for 150 bird species in California’s Coast Range and 160 bird species in California’s Sierra Nevada. Pairing bird data collected from 1911–1940 with 2003–2010 resurveys performed at the same sites, the authors found that breeding dates for California birds have advanced by approximately 5–12 days. The shift in nesting dates reduced average temperatures during nesting by more than 1 °C—approximately the same magnitude that average regional temperatures have warmed over the same period. Using national nest monitoring data, the authors found that in the early summer across North America, warm temperatures are associated with high nest success in the cold parts of the birds’ ranges and low nest success in the warm parts of the birds’ ranges. By nesting earlier, California birds are nesting at temperatures similar to those at which they nested a century ago, thus reducing the need for range shifts, according to the authors. — L.C.



