Significance
USA300 is a hypervirulent, community-acquired, multidrug-resistant Staphylococcus aureus clone that started to spread in the United States around 17 years ago. Many studies detected it also in South America, Europe, and the Asia-Pacific region. In this study, we show that USA300 is also circulating in sub-Saharan Africa. Locating the temporal and spatial origin of clonal lineages is important with respect to epidemiology and molecular evolution of pathogens. We show that USA300 evolved from a less virulent and less resistant ancestor circulating in Central Europe around 160 years ago. Constant surveillance of pathogen transmission routes is vital to prevent and control potential outbreaks. Whole genome sequencing proved to be a useful tool for epidemiological surveillance.
Keywords: USA300, molecular evolution, CA-MRSA, comparative genomics, Africa
Abstract
USA300 is a pandemic clonal lineage of hypervirulent, community-acquired, methicillin-resistant Staphylococcus aureus (CA-MRSA) with specific molecular characteristics. Despite its high clinical relevance, the evolutionary origin of USA300 remained unclear. We used comparative genomics of 224 temporal and spatial diverse S. aureus isolates of multilocus sequence type (ST) 8 to reconstruct the molecular evolution and global dissemination of ST8, including USA300. Analyses of core SNP diversity and accessory genome variations showed that the ancestor of all ST8 S. aureus most likely emerged in Central Europe in the mid-19th century. From here, ST8 was exported to North America in the early 20th century and progressively acquired the USA300 characteristics Panton–Valentine leukocidin (PVL), SCCmec IVa, the arginine catabolic mobile element (ACME), and a specific mutation in capsular polysaccharide gene cap5E. Although the PVL-encoding phage ϕSa2USA was introduced into the ST8 background only once, various SCCmec types were introduced to ST8 at different times and places. Starting from North America, USA300 spread globally, including Africa. African USA300 isolates have aberrant spa-types (t112, t121) and form a monophyletic group within the clade of North American USA300. Large parts of ST8 methicillin-susceptible S. aureus (MSSA) isolated in Africa represent a symplesiomorphic group of ST8 (i.e., a group representing the characteristics of the ancestor), which are rarely found in other world regions. Isolates previously discussed as USA300 ancestors, including USA500 and a “historic” CA-MRSA from Western Australia, were shown to be only distantly related to recent USA300 clones.
The bacterium Staphylococcus aureus is an opportunistic pathogen that has the ability to colonize the skin and mucous membranes of humans and different animal species. It may cause mild to severe diseases, ranging from superficial wound infections or food poisoning to bacteremia and other systemic infections (1, 2). Multidrug resistance dramatically complicates infection treatment and can cause fatal outcomes. Especially, methicillin-resistant S. aureus (MRSA) is of major concern for public health. MRSA used to be limited to the hospital environment but has increasingly been identified in community-acquired (CA) infections within the past two decades. CA-MRSA isolates usually belong to particular clonal lineages and possess comparatively small staphylococcal cassette chromosome mec (SCCmec) elements of types IV or V (MRSA-IV/V), which confer methicillin resistance and the phage-encoded toxin Panton–Valentine leukocidin (PVL), which seems to play a role in enhanced skin and soft tissue infections (2). There are major variations in the spatial and temporal population structures and global distribution of CA-MRSA, with different clones dominating in different world regions (3–7). However, one clone is found worldwide in regionally varying frequencies: the multilocus sequencing (MLST) sequence type (ST) 8-MRSA-IV “USA300.”
USA300 is a hypervirulent clone that was first identified in the United States around the turn of the millennium (8). Initially, it was mainly isolated from certain risk groups, including athletes, children, prisoners, and military recruits (9–11), but soon spread epidemically in the general population and eventually became the dominant CA-MRSA circulating in North America (8, 12). In 2003, it was termed “USA300” based on its characteristic pulsed-field gel electrophoresis (PFGE) pattern (13). Moreover, the “classical” USA300 clone is characterized by specific genetic features: association with ST8 and spa-type t008, lack of a functional capsular polysaccharide due to point mutations in genes cap5D and cap5E (14), and possession of SCCmec IVa, PVL-encoding genes lukF-PV and lukS-PV, and the arginine catabolic mobile element (ACME) (8, 15). This clone is sometimes referred to as “USA300-0114” (12).
The epidemiology of ST8/USA300 differs remarkably among world regions. In Europe, ST8 is a common CA-MRSA, but most of them are non-USA300 (3). Repeated introductions of USA300 to the European population have been shown (16–18); however, the clone does not seem to spread in the general population. ST8 is very rare in Asia (4), with sporadic findings of USA300 reported from Japan (19), South Korea (20), and Pakistan (21). The clone CA-MRSA/J gained some attention in Japan over the past years; however, this clone was suggested to have evolved from a Japanese HA-MRSA rather than the classic USA300 (22). ST8 is also no major clone in the Asia-Pacific region, including Australia and New Zealand (5), although there are early reports of ST8 CA-MRSA from remote settlements in Western Australia (23–25). Since 2006, USA300 has also been increasingly detected in South American countries (26). Despite the concordant PFGE patterns, USA300 isolates from South America were found to possess different molecular features, compared with USA300-0114, including an SCCmec IVc element and a mobile genetic element conferring copper and mercury resistance (COMER) instead of ACME (27). The North and South American variants are now considered as two distinct genetic lineages termed USA300-NAE for “North American Epidemic” and USA300-SAE for “South American Epidemic” (27) (or USA300-LV for “Latin American Variant”) (12). Both lineages shared a common ancestor ∼40 y ago (27). However, the geographic and evolutionary origin of this ancestor is still unclear.
One suggested scenario hypothesized the evolution of USA300 from the North American HA-MRSA lineage USA500 (ST8-MRSA-IV), with subsequent acquisition of PVL and ACME (28). However, more recent genome-based analyses rather suggest the evolution from PVL-positive CA–methicillin-susceptible S. aureus (CA-MSSA) (27). Consequently, detection of high rates of PVL-positive ST8-MSSA in Trinidad and Tobago gave reason to speculate about a Caribbean origin of the USA300 lineage (29). When high rates of PVL-positive ST152-MSSA were detected in Haiti, their origin was linked with sub-Saharan Africa (SSA) because many people from the Caribbean have African ancestry (30, 31) and PVL is known to be present in SSA in very high rates (6, 7, 32). Like ST152, ST8 is a widespread clone in SSA. Even sporadic findings of USA300-like PVL- and ACME-positive ST8-MRSA-IV were reported from Cameroon (n = 1) (33), Gabon (n = 6) (34–37), Ghana (n = 1) (38), and two Swiss patients with travel or immigration history to/from Cameroon and Sierra Leone (17). However, most of these African isolates (9/10) did not belong to the classic USA300 spa-type t008, but to the closely related spa-types t112 (n = 3) and t121 (n = 6). Consequently, it was not certain if the African ST8 isolates shared the same evolutionary history as the North American USA300, or whether they had evolved from a local “African” S. aureus population. SSA was shown before to be the origin of a pandemic CA-MRSA lineage (CC80) (39). Thus, we have reason to hypothesize an African origin of the pandemic USA300 lineage. We focused our research on countries of SSA instead of including the entire African continent. This was done because the North Africa/Maghreb region is culturally and genetically stronger connected to Southern Europe and the Middle East than to African countries south of the Sahara (40). The division of the African continent into “North Africa” and “sub-Saharan Africa” also complies with United Nations classification schemes (https://unstats.un.org/unsd/methodology/m49/, accessed April 5, 2017). In this study, we performed whole genome sequencing (WGS) analysis of 224 diverse global ST8 isolates, allowing the interpretation of the temporal and spatial evolution of USA300.
Results
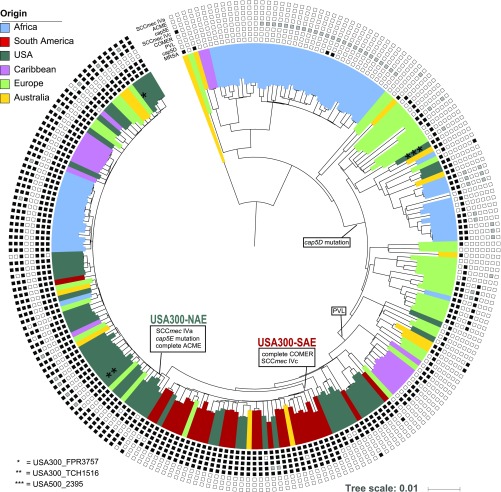
We performed whole genome sequencing (WGS) of 224 S. aureus isolates aimed to represent the genetic diversity of ST8 over space and time. These isolates differed in 12,403 (12,469 prepurging) core genome single nucleotide polymorphisms (SNPs). The concatenated SNPs were used to reconstruct the temporal and spatial evolution of S. aureus USA300 using maximum-likelihood (ML) (Fig. 1) and Bayesian statistics (Fig. 2). All major splits were supported by high bootstrap and posterior support values (Figs. S1C and S2).
Fig. 1.
Maximum-likelihood phylogeny of 224 ST8 S. aureus isolates based on 12,403 core genome single nucleotide polymorphisms. The colors represent the region of origin of each sample. Phylogenetic positions of the included NCBI RefSeq genomes (two USA300 and one USA500) are highlighted with asterisks. Information about the complete presence (black squares), partial presence (gray squares), or absence (white squares) of USA300-specific genetic features is given for each sample. Major genetic introduction events are indicated on the respective phylogenetic branches. (Scale bar: substitution per site.)
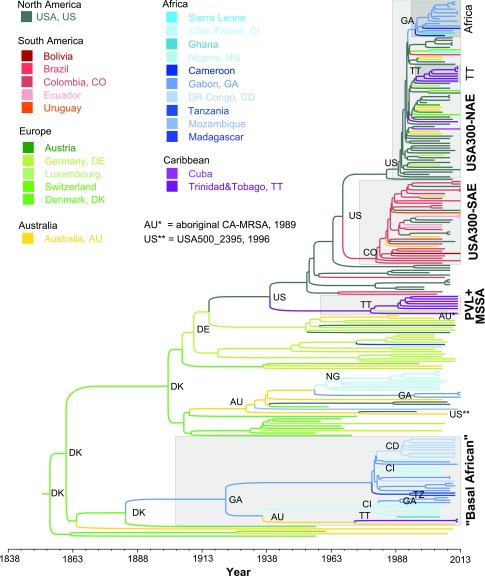
Fig. 2.
Bayesian maximum clade credibility tree calculated from 10,801 sampled trees. Branch colors represent the country of origin of each sample and their most recent common ancestor (MRCA), respectively. Major clade’s MRCA origins are verbalized using international country codes. Important groups like USA300 or the “Basal African” clade are highlighted with gray squares (annotation in bold). Time periods and age of clades can be inferred using the timeline at the bottom. The most recent isolate was collected in 2013; the MRCA of all isolates was dated to 1854 (HPD 95%, 1841 to 1865). MSSA, methicillin-susceptible S. aureus; PVL+, Panton–Valentine leukocidin positive; USA300-NAE, North American Epidemic USA300 clone; USA300-SAE, South American Epidemic USA300 clone. Detailed divergence timings and countries for each node are displayed in Fig. S1.
ST8 Emerged from Central Europe, USA300 from North America.
Both phylogenies confirm the separation of USA300 into two distinct lineages, USA300-NAE and USA300-SAE, which shared a common ancestor around 50 y ago [median 1967, highest probability density (HPD) 95%, 1963 to 1971]. All isolates with gene combinations characteristic for USA300-NAE and USA300-SAE, such as ACME, COMER, and cap5E mutations, were exclusively found in these two clades, confirming the exclusivity of these genetic combinations within the ST8 lineage (Fig. 1 and Dataset S1). However, partial elements of ACME and COMER were also found in isolates from other phylogenetic clades, albeit in different allelic variants. Nevertheless, we did not find evidence for successful spread of those clones possessing single genes of ACME or COMER. This indicates that, if these elements indeed confer an evolutionary advantage, as it was suggested in previous studies (15, 41), it is presumably the specific combination of genes or regulatory mechanisms that facilitate the success of USA300. According to our results, USA300-NAE acquired ACME, SCCmec IVa, and the cap5E mutation Asp75Tyr in the late 1980s (median 1989, HPD 95%, 1987 to 1991) (Fig. 2) while USA300-SAE acquired SCCmec IVc and the COMER element some years earlier (median 1980, HPD 95%, 1977 to 1983) (Fig. S1).
Our Results Suggest a Single Introduction of the PVL-Phage ϕSa2USA and Various Introductions of Different SCCmec Elements to the ST8 Background.
First, we determined the sequences of the PVL-encoding genes lukF-PV and lukS-PV, which were identical among all isolates (n = 120), except for one nonsynonymous SNP (Tyr208Cys) in lukF-PV in one Swiss isolate (Switzerland_2013_2). Reference mapping was performed for all isolates with available read data (66 of 120 isolates). Considering the results of both assembly methods, ϕSa2USA (5,189 bp, GenBank accession no. KX130855.1) and its 300 nucleotides 3′ and 5′ flanking regions were identified in all 120 samples. The exact reference sequence was identified in 95 samples, 19 samples possessed allelic variants with one to four point mutations, and five samples possessed allelic variants with frameshift mutations. In one isolate (Colombia_2006_4), 25 point mutations were identified within a stretch of 193 nucleotides. National Center for Biotechnology Information (NCBI) BLAST results suggested a recombination event, in which a part of the S. aureus peptidoglycan hydrolase gene was integrated into the ϕSa2USA amidase gene. The recombinant sequence and flanking regions, respectively, were identical to the peptidoglycan hydrolase and amidase genes of isolate CA12 (CP007672.1, Colombia_2007_2), which originates from the same region as Colombia_2006_4. As all flanking regions of the ϕSa2USA were identical in all isolates, our results suggest a single introduction of the PVL-phage ϕSa2USA to the ST8 background, which is in contrast to multiple introductions of different SCCmec elements at various times and places. Specifically, except for SCCmec IVa and IVc, we also identified six SCCmec IVb isolates from the United States (n = 5) and Austria (n = 1), two SCCmec IVd isolates from Australia (n = 1) and the United States (n = 1), eight SCCmec IVg isolates from the United States (n = 5) and Gabon (n = 3), two SCCmec IVi isolates from the United States, and one SCCmec V isolate from Australia (Dataset S1).
African USA300 and ST8 Isolates Are Unique.
All 10 African isolates previously described as USA300-like (ST8-MRSA, PVL-positive, ACME-positive, spa types t112 or t121) (17, 33–38) were found within the phylogenetic clade of USA300-NAE and could consequently be confirmed to be “true” USA300-NAE isolates, despite their non-t008 spa-types (Figs. 1 and 2). Noteworthy, nine of these isolates, collected in Gabon (n = 6), Cameroon (n = 2), and Ghana (n = 1), formed one distinct monophyletic “African USA300” clade, indicating a single introduction event to the African continent, followed by spread in the local population. In addition to these nine isolates previously described as USA300-like, six PVL- and ACME-positive isolates from Gabon also fell into this monophyletic African clade (Fig. 2). Our results indicate a transmission from the United States to Gabon in 1996 (HPD 95%, 1994 to 1997) (Figs. 2 and 3). All monophyletic African USA300 isolates belonged to spa-types t121 (n = 12) or t112 (n = 3). The only “nonmonophyletic” African USA300 isolate had spa-type t008 and was isolated in Switzerland from a person with travel or immigration history to/from Sierra Leone. This suggests that the epidemiological link to Sierra Leone was wrong and that this isolate should rather be considered as European.
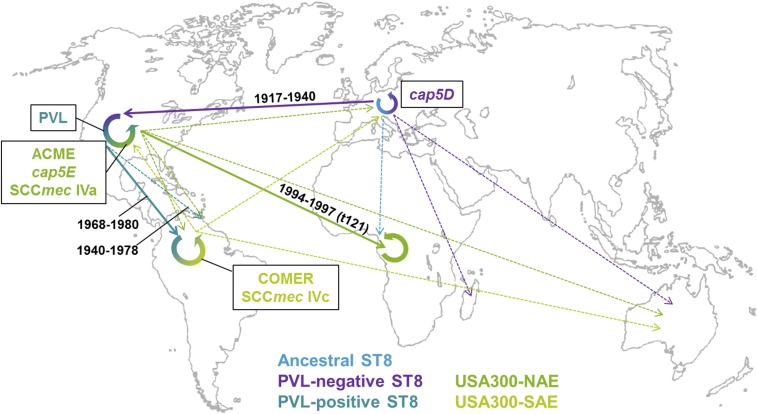
Fig. 3.
Phylogeographic evolution of S. aureus USA300. The figure shows a subset of the global transmission routes of different ST8 lineages, including USA300. Different lineages are highlighted in different colors; thick lines represent major transmission events during the evolution of USA300; dotted lines represent other transmissions. Transmission times are indicated on major routes. For a comprehensive view of all transmission routes, including single isolates, view Dataset S6 using Google Earth.
No African ST8 isolate was found in direct ancestry to the USA300 clades, consequently ruling out the hypothesized evolution of USA300 from SSA (Fig. 2). Likewise, USA500 and the aboriginal CA-MRSA from Western Australia were found in different phylogenetic clades and shared a common ancestor with USA300 that circulated in Central Europe ∼100 y ago. The isolates basal to both USA300 lineages comprised mainly PVL-positive (20/21) MRSA (n = 12) and MSSA (n = 9) from the United States (n = 11), Colombia (n = 2), Germany (n = 2), and Trinidad and Tobago (n = 6). The most recent common ancestor (MRCA) of all these isolates apparently circulated in the United States around 1940 (median 1939, HPD 95%, 1932 to 1946) (Fig. 2). However, North America does not appear to be the origin of the entire ST8 clone. Instead, our results suggest an emergence of ST8 in Europe during the mid-19th century (Denmark, 93.2%; median 1854, HPD 95%, 1841 to 1865).
Around 1900 (median 1900, HPD 95%, 1892 to 1907), the shortened cap5D allele (frameshift mutation in nucleotide position 994) (14) (Dataset S2) arose and spread in the European ST8 population. In the early 20th century, one cap5D-mutated European ST8 MSSA strain was exported to the United States, where it spread and diversified before eventually evolving into the epidemic USA300 clone by stepwise acquisition of PVL, ACME, and the cap5E mutation (Fig. 3). Later, this clone was reintroduced to Europe on multiple occasions (16, 17, 42).
Before the spread of the mutated cap5D in the European ST8 population, one isolate with WT cap5D was exported to Gabon around 1920 and founded a symplesiomorphic, mainly “African” ST8 sublineage (MRCA, Gabon, 1922, HPD 95%, 1912 to 1930) (Fig. 2). “Symplesiomorphic” means that this group does not possess any “derived” characteristics, like PVL or ACME, but represents the state of the common ancestor. The African clone spread and diversified in the Gabonese population and was transmitted to other SSA countries, including Côte d’Ivoire, the Democratic Republic of Congo, and Tanzania, as well as to Australia and Trinidad and Tobago (Fig. 2).
Identification of Markers for USA300.
We screened our collection for the unique presence or absence of genes and group-specific nonsynonymous SNPs for USA300-NAE, USA300–SAE, and the African USA300 clade (Fig. 2), respectively. Within our collection, no yet unknown USA300-specific unique genetic markers were found for the first two groups; for the African USA300 clade, however, we identified five nonsynonymous SNPs that were present in all 15 isolates of this clade in comparison with all other isolates investigated (Table 1).
Table 1.
Unique nonsynonymous SNPs within the 15 African USA300 clade isolates
| SNP position (locus tag)* | Gene product | SNP | Substitution |
| 328599 (SAUSA300_RS01475) | DUF5079 domain-containing protein | G → A | Glu 213 Lys |
| 992391 (SAUSA300_RS04860) | Hypothetical protein | A → G | Val 172 Ala |
| 2232409 (SAUSA300_RS11400) | Threonylcarbamoyl-AMP synthase | C → T | Ala 236 Thr |
| 2648462 (SAUSA300_RS13595) | MFS transporter | G → A | Pro 356 Leu |
| 2779237 (SAUSA300_RS14285) | Arginine-ornithine antiporter | T → G | Ile 30 Leu |
SNP localizations, locus tags, and gene products were given with reference to the genome of strain USA300_FPR3757 (GenBank accession no. NC_007793).
Discussion
The results and findings presented here shed light on the evolution of S. aureus ST8 and USA300. Our findings show that direct evolution of USA300 from USA500 is unlikely because their last common ancestor circulated in Europe around 100 y ago (Fig. 2). This corroborates previous findings (16, 27, 43). Similarly, direct evolution from ST8 CA-MRSA isolated from indigenous people in Western Australia, or PVL-positive MSSA from Trinidad and Tobago or Western Africa, was not supported by our data (Figs. 2 and 3 and Fig. S1). Instead, the most likely origin of USA300-NAE and USA300-SAE was calculated to be PVL-positive MSSA circulating within the United States. The respective PVL-negative ST8 ancestor appears to have been imported from Europe in the early 20th century during an era of increased emigration of people from Europe to the United States due to war, economic crisis, and political persecution (Fig. 3 and Fig. S1). This confirms the findings of Planet et al. (27), who stated that “members of the USA300 have been present in the Western hemisphere since the 1930’s.” Other pandemic S. aureus clones have likewise been shown to have originated in Europe: for example, ST22 EMRSA-15 (44).
Our analysis of representative ST8 isolates from different continents puts previous findings about the regional epidemiology of ST8 and USA300 into a broader context. ST8 isolates from Europe and Australia were found scattered across our phylogeny, with few clusters of closely related isolates, indicating either a diverse local population or multiple introductions with limited spread in the general population. This means that the ST8 S. aureus population in these continents consists of genetically diverse isolates that represent both the ancestral ST8 population as well as strains with apomorphies (i.e., derived characters) like PVL and ACME. This genetic diversity in European isolates was also observed in previous studies (16, 17).
In contrast to the European and Australian ST8, isolates from the Caribbean and from Africa were exclusively found in distinct “regional” monophyletic branches at distant positions in our phylogeny. This indicates lower genetic diversity of ST8 S. aureus in these regions, presumably caused by single introduction events from outside, followed by local diversification and regional spread of the respective clone. This might be due to significantly fewer travel and trade links of SSA countries with other continents, compared with connections among, for example, Europe and North America. However, ST8 S. aureus seems to spread successfully among SSA countries. Due to this “endemic” situation, we find locally evolved African ST8 clones that substantially differ from ST8 clones isolated in other regions: e.g., “wild-type” cap5D MSSA.
On the other hand, African USA300-like isolates were found to be very similar to North American USA300 isolates. Although the classical USA300 clone belongs to spa-type t008, some North American USA300 have been identified as spa-type t121 (45). Probably by chance, one isolate with this more unusual spa-type was imported to Africa and further evolved locally (founder effect). Consequently, most African USA300 isolates identified today either belong to spa-type t121 or to spa-type t112, which presumably evolved from t121 by duplication of the second spa repeat. In contrast to Europe, where it was repeatedly shown that imported USA300 isolates from overseas spread only very slowly and limitedly in the general population (16, 17), USA300 in Africa seems to have successfully spread in the local communities; at this point, we can only speculate whether this success is related to the unique nonsynonymous SNPs of the African USA300 isolates (Table 1). This is also indicated by the monophyletic subgroup of African USA300 isolates in our phylogeny (Fig. 2) and is emphasized by a suggested increase of the proportion of USA300 among MRSA carriers in the Gabonese population (34, 36), which is in contrast to an only marginal increase of the proportion of USA300 in Germany from 0.1% in 2004 to 0.6% in 2010/2011 (46). One USA300 isolate isolated in Switzerland (17) and epidemiologically related to Sierra Leone turned out to be more likely of European origin because it possesses a “non-African” spa-type and does not fall into the same monophyletic clade as the other nine African USA300 isolates. This is a good example of how WGS on global collections may help to facilitate epidemiological reconstructions and outbreak surveillance in the future. For example, detailed knowledge about regionally circulating sublineages enables an early detection of newly emerging clones. Moreover, WGS-based typing as a highly discriminatory method is capable of detecting clonal transmissions and facilitates the elucidation of potential transmission routes (47, 48).
Our results present both a temporal and spatial bias because concurrent representative isolates from all regions were not available, and the inclusion of additional isolates may shift the results regarding the origin and evolution of USA300 and ST8. However, vital timings in our analyses (Fig. S1) share a large resemblance to other studies focusing on the acquisitions of ACME, COMER, and PVL (15, 27). Similarly, the exclusive inclusion of an older isolate from Denmark could be reflected in the phylogeographic results that strongly indicated a Danish origin of ST8.
In conclusion, our phylogenomics analysis indicates that the origin of the epidemic USA300 clone lies in the mid-19th century ST8-MSSA population in Central Europe (Denmark). A cap5D mutated ST8-MSSA was exported to the United States in the early 20th century and progressively acquired PVL, ACME/COMER, SCCmec IVa/c, and cap5E Asp75Tyr within North and South America, and eventually spread to other continents. The existence of USA300-NAE in Africa has now been confirmed, despite unusual spa-types. It is important to conduct further studies about the factors contributing to the variable success of USA300 in different geographic regions to contain the health threat caused by this hypervirulent CA-MRSA clone.
Materials and Methods
Selection of Bacterial Isolates.
Our dataset includes a diverse subset of all S. aureus ST8 WGS sequences available in public databases until November 2015, without including all available USA300 genomes from the United States. It was completed by in-house Illumina shotgun-sequenced published isolates from diverse countries of sub-Saharan Africa, Western Australia, Trinidad and Tobago, and diverse European countries. Our isolates represent the previously reported diversity of ST8 (49). Even upon request, no ST8 isolates from Asian countries were available.
In total, 224 human ST8 S. aureus from the United States (n = 57), South America (n = 28), Australia (n = 14), Europe (n = 43), the Caribbean (n = 18), and Africa (n = 64) were analyzed by whole genome sequencing (WGS) (Table 2). The time span of the isolate collection ranges from 1957 to 2013. Twenty-two samples collected from patients in Switzerland were previously associated with overseas travel or immigration (17) and were treated according to their potential country of origin in all analyses.
Table 2.
Origin of the 224 included S. aureus isolates
| Global region (n isolates total) and country of origin | Year(s) of collection | No. of isolates | Reason to include | Genome sequenced for this study | Source |
| Africa (64) | |||||
| Cameroon | 2007 | 1 | African background ST8 | Yes | (33) |
| Cameroon | 2013 | 1 | Global USA300 | No | (17) |
| DR Congo | 2011 | 10 | African background ST8 | Yes | (65) |
| Gabon | 2005 | 1 | USA300-like | Yes | (34) |
| Gabon | 2005 | 1 | USA300-like | Yes | (37) |
| Gabon | 2008 to 2012 | 7 | African background ST8 | Yes | This study* |
| Gabon | 2009 | 4 | African background ST8 | Yes | (66) |
| Gabon | 2009 to 2012 | 4 | USA300-like | Yes | (36) |
| Gabon | 2009 to 2012 | 3 | African background ST8 | Yes | (36) |
| Gabon | 2012 | 2 | African background ST8 | No | (53) |
| Gabon | 2013 | 1 | USA300-like | Yes | (35) |
| Gabon | 2013 | 3 | African background ST8 | Yes | (35) |
| Ghana | 2011 | 1 | USA300-like | Yes | (38) |
| Ghana | 2005 | 1 | African background ST8 | Yes | This study† |
| Côte d’Ivoire | 2005 | 10 | African background ST8 | Yes | (65) |
| Madagascar | 2007 | 1 | African background ST8 | Yes | (33) |
| Mozambique | 2012 | 1 | African background ST8 | No | (53) |
| Nigeria | 2005 | 7 | African background ST8 | Yes | (67) |
| Nigeria | 2007 | 2 | Phylogeny of CC8 | Yes | (49) |
| Sierra Leone | 2013 | 1 | Global USA300 | No | (17) |
| Tanzania | 2011 | 2 | African background ST8 | No | (53) |
| Australia (14) | |||||
| Australia | 1989 | 1 | First ST8 CA-MRSA | Yes | (23) |
| Australia | 2004 to 2006 | 6 | Global ST8 diversity | Yes | (24) |
| Australia | 2003 to 2008 | 4 | Global ST8 diversity | Yes | (52) |
| Australia | 2008 to 2011 | 3 | Global ST8 diversity | Yes | This study‡ |
| Caribbean (18) | |||||
| Trinidad & Tobago | 2012 | 17 | Global ST8 diversity | Yes | (29) |
| Cuba | 2013 | 1 | Global USA300 | No | (17) |
| Europe (43) | |||||
| Austria | 2005 to 2006 | 3 | Phylogeny of CC8 | Yes | (49) |
| Denmark | 1957 to 1973 | 12 | Temporal collection | Yes | This study† |
| Germany | 2010 to 2011 | 6 | Global ST8 diversity | No | (53) |
| Germany | 2003 to 2008 | 18 | Phylogeny of CC8 | Yes | (49) |
| South America (28) | |||||
| Bolivia | 2013 | 1 | Global USA300 | No | (17) |
| Brazil | 2013 | 1 | Global USA300 | No | (17) |
| Colombia | 2006 to 2007 | 13 | USA300 NAE/SAE | No | (27) |
| Colombia | 2006 to 2007 | 2 | Global ST8 diversity | No | (27) |
| Colombia | 2013 | 6 | Global USA300 | No | (17) |
| Ecuador | 2006 | 2 | USA300 NAE/SAE | No | (27) |
| Ecuador | 2013 | 2 | Global USA300 | No | (17) |
| Uruguay | 2013 | 1 | Global USA300 | No | (17) |
| North America (United States) (57)§ | |||||
| FL, HI, IL, IN, MA, MS, MT, PA, SC, NY | 2001 to 2012 | 17 | USA300 NAE/SAE | No | (27) |
| NY | 2008 to 2011 | 17 | USA300 NAE/SAE | No | (50) |
| FL, MA, MS, TX, NY | 1999 to 2011 | 8 | Global ST8 diversity | No | (27) |
| NY | 2008 to 2010 | 7 | Global ST8 diversity | No | (50) |
| NY | 1996 | 1 | USA500 reference genome | No | (68) |
| CA | 2000 | 1 | USA300 reference genome | No | (69) |
| TX | 2003 | 1 | USA300 reference genome | No | (70) |
| Unknown | 2003, 2013 | 5 | Global USA300 | No | (17) |
Bacterial isolates obtained from the routine microbiology laboratory at the Albert Schweitzer Hospital in Lambaréné, Gabon.
Bacterial isolates were obtained from the S. aureus national surveillance unit at Statens Serum Institut, Copenhagen.
Bacterial isolates were obtained from the Australian Collaborating Centre for Enterococcus and Staphylococcus, Murdoch University, Murdoch, Australia.
FL, Florida; HI, Hawaii; IN, Indiana; MA, Massachusetts; MS, Mississippi; MT, Montana; PA, Pennsylvania; SC, South Carolina; NY, New York; TX, Texas.
Isolates from North and South America comprised a selection of published ST8 S. aureus genomes from two large phylogenetic studies by Planet et al. (n = 42) (27) and Uhlemann et al. (n = 24) (50). These studies represent the genetic diversity of USA300-NAE and -SAE and were complemented with some non-USA300 North American ST8. No extensive WGS study covering the diversity of ST8 was available from South America. Moreover, we were not able to include early isolates of aboriginal Canadians (51). Two NCBI reference genomes of USA300-NAE and one of USA500 were included (USA300_TCH1516, NC_010079.1; USA300_FPR3735, NC_007793.1; USA500_2395, NZ_CP007499.1).
Isolates from Australia comprised one early ST8 CA-MRSA collected in 1989 in a remote aboriginal community in Western Australia (23–25) and 13 diverse ST8 isolates, including PVL- and ACME-positives, isolated between 2003 and 2011 (24, 52).
European isolates mainly included representative isolates of the CC8 phylogeny (49) and were supplemented by six ST8 genomes from Germany (53) and three USA300 genomes from Switzerland (17). To increase the temporal span of the collection, 12 “historic” ST8 bacteremia isolates from Denmark isolated between 1957 and 1997 were added (all except one were isolated between 1957 and 1973).
The Caribbean samples comprised 17 diverse ST8 S. aureus isolates from Trinidad and Tobago, including PVL-positive ST8-MSSA and USA300 (29), and one USA300 isolate with epidemiological link to Cuba, which was isolated in Switzerland (17).
The African isolates included 10 MRSA isolates previously defined as USA300-like from Cameroon (n = 1) (33), Gabon (n = 6) (34–37), Ghana (n = 1) (38), and Sierra Leone (n = 1) (17) and 54 diverse ST8 MRSA and MSSA samples from different African countries from various small-scale studies (Table 2).
Ethical Statement.
This study was performed in accordance with guidelines approved by the Ethical Committee of the Medical Faculty of the University of Münster and of the Aerztekammer WestfalenLippe. Our institutional review board waived the need for written informed consent from the participants.
Whole Genome Sequencing and Genome Analysis.
Available WGS data from previous studies (n = 102) were retrieved in the form of assembled sequences (contigs) from NCBI GenBank or as reads from the NCBI Sequence Read Archive (SRA) or European Nucleotide Archive (ENA). They were included into our dataset provided they passed our internal WGS quality controls: i.e., confirmed affiliation to ST8 by in silico MLST and identification of ≥95% of 1,861 core genome MLST (cgMLST) targets (54). The remaining isolates (n = 122) were sequenced using Illumina technology and Nextera XT version 2 chemistry, either with 150-bp or 250-bp paired-end protocols on a NextSeq 500 or MiSeq sequencer (Illumina). All newly generated reads were deposited at ENA under study accession no. PRJEB14816. Quality trimming of fastq files (average base quality of 30, aiming for 100-fold coverage) and de novo assembly using Velvet 1.1.04 (55) were performed with SeqSphere+ (version 3.1; Ridom GmbH) as described recently (56). Only genomes that harbored ≥95% cgMLST targets passed the quality control; otherwise, sequencing was repeated until the criterion was met. Identification of USA300-characteristic genes and their allelic variants was performed using a BLAST search with ≥95% nucleotide identity and ≥99% length coverage against all nucleotide sequences listed in Dataset S2. SCCmec types were determined by the BLAST-based detection of specific SCCmec typing primer sequences (57, 58) and different combinations of cassette chromosome recombinases (ccr) gene complexes and the mec gene complex, according to the definitions of the International Working Group on the Staphylococcal Cassette Chromosome elements (IWG-SCC; www.sccmec.org) (Dataset S3).
PVL-Phage Identification.
All de novo assembled genomes positive for both PVL genes, i.e., lukF-PV and lukS-PV (n = 120), were screened for the presence of any of the seven phages known to carry the PVL genes (59, 60): (i) ϕ2958PVL (NC_011344.1, 47,342 bp), (ii) ϕPVL (NCBI GenBank accession no. AB009866.2, 41,401 bp), (iii) ϕPVL108 (NCBI GenBank accession no. NC_008689.1, 44,857 bp), (iv) ϕPVL-CN125 (NCBI GenBank accession no. FJ713816.1, 44,492 bp), (v) ϕSa2USA (NCBI GenBank accession no. KX130855.1, 5,189 bp), (vi) ϕSaMW2 (NCBI GenBank accession no. BA000033.2, 45,550 bp), and (vii) ϕSLT (NCBI GenBank accession no. NC_002661.2, 42,942 bp). Only ϕSa2USA was found in the de novo assembled isolates. Subsequently, to determine whether the phage was inserted at the same position in all isolates, we created a 5,787-bp reference sequence comprising the ϕ2SaUSA phage region plus additional 300 3′ and 5′ flanking nucleotides of USA300 RefSeq FPR3757 (NC_007793.1) and mapped all available reads of lukF/S-PV positive isolates (n = 66) against it. Reference mapping was conducted with CLC Genomics Workbench 8.5.1 (CLC Bio; Qiagen) using default parameters, with the exception of a length fraction value of “0.8” and a similarity fraction value of “0.95.” No reads (only contigs) were available for public isolates from refs. 17 and 27 (n = 54).
Single Nucleotide Polymorphism and Phylogenetic Analysis.
The WGS fastq data of 121 isolates were aligned against the chromosome of the S. aureus TCH1516 ST8 reference genome (GenBank accession no. CP000730) using the short-read alignment component of the Burrows–Wheeler Aligner (BWA) after removal of duplicated regions identified using NUCmer. Additionally, 103 fasta files containing contigs of the remaining isolates were aligned using MUMmer/Nucmer. Each alignment was analyzed for single nucleotide polymorphisms (SNPs) using NASP (https://github.com/TGenNorth/NASP). If either the minimum coverage of 10 was not met, or if the variant was present in <90% of the base calls, the respective SNPs were excluded. Indications of putative horizontal gene transfer events were defined as a unique SNPs cluster (≥3) in the NASP output and purged from the alignment. The concatenated sequence of SNPs can be found in Dataset S4. Based upon the purged SNP alignment, a phylogenetic tree was constructed using the maximum-likelihood algorithm implemented in PhyML (61) with Smart Model Selection (SMS) and the Bayesian Information Criterion (BIC) with 100 bootstrap replicates. For rooting, the chromosome of S. aureus isolates MW2 (ST1) (GenBank accession no. BA000033) was used as it represents a closely related outgroup to ST8 (62) (Fig. S2). The ML phylogeny, including each sample’s association with USA300-characteristic genes, was visualized using itol.embl.de/.
Ancestral Dating and Phylogeographic Analysis.
To explore the timing of diversification events, we used coalescence-based analyses to investigate evolutionary rates and timing of common ancestors using duplicate runs in BEAST v1.8.2 (63) at CIPRES Science Gateway (64,) (https://www.phylo.org). We compared the results of different models, including the general-time reversible (GTR) and the Hasegawa–Kishino–Yano (HKY) substitution models, under the assumption of both strict and relaxed molecular clock and different demographic models: i.e., Bayesian skyline, constant population, and exponentially growing population tree priors. For each analysis, 200 million steps were performed, and the chain was sampled every 20,000th step. The first 10% of each chain was discarded as burn-in. The best model was chosen based on the highest effective samples size (ESS) and Bayes factor (BF) values, resulting in the HKY, strict clock, constant population size model with uniform clock rate ([0,1], initial = 0.001). Data were analyzed with Tracer v1.6 and TreeAnnotator v1.8.2. The maximum clade credibility tree was visualized using FigTree v1.4.2.
A Bayesian analysis was also conducted to determine the most likely geographical origin of the most recent common ancestors (MRCAs) using an HKY, strict clock, constant population size model with uniform clock rate ([0,1], initial = 0.001) and a Markov chain Monte Carlo (MCMC) length of 120 million steps, with chain sampling every 10,000th step and burn-in at 10%. Taxa groups were defined for monophyletic groups of interest: e.g., USA300-NAE, USA300-SAE, or basal-African based on the ML data. Detailed parameters are provided in Dataset S5. Again, the analysis was run in BEAST v1.8.2 at CIPRES Science Gateway. The geographic origin of the entire dataset based on the .root BEAST output file was determined according to the instructions provided at bedford.io/projects/dynamics-practical/examine-the-phylogeographic-output.html (accessed April 2016). The maximum clade credibility (mcc) tree was calculated with TreeAnnotator v.1.8.2 and used for graphical representation of the data. A .kml file to visualize the phylogeographic spread using Google Earth was created from the mcc tree using the phylogeo.jar script developed by Trevor Bedford (https://github.com/trvrb/dynamics-practical/tree/master/scripts, accessed April 2016) (Dataset S6).
Identification of Genetic Markers.
We searched for the presence or absence of genes and unique nonsynonymous SNPs that might be associated with the evolutionary success of the two USA300 lineages (USA300-NAE and USA300–SAE) and the African USA300 clade. Therefore, we screened the genomes of all USA300-NAE isolates present in our dataset (n = 74) and the genomes of two closely related ST8 outgroup isolates (USA_2009_11 and USA_2009_12) for the presence of USA300-NAE core genome genes, including all single copy genes (genes with fragments that occur in multiple copies with an identity of ≥90% and >100-bp overlap were excluded) with a minimal length of 50 bp of NCBI RefSeq USA300 genome FPR3757 (NC_007793.1; 2,804 targets in total) (Dataset S7). Similarly, we screened the genomes of all USA300-SAE isolates of our dataset (n = 35) and the two outgroup strains for the presence of USA300-SAE–specific core genome genes derived from strain M121 (CP007670.1; 2,550 targets in total) (Dataset S7). Moreover, to identify the absence of genes in USA300, we screened both USA300-NAE and USA300–SAE isolates for the presence of core genome genes of one of the outgroup strains (strain C1768, GenBank accession no. LAMT01000000, syn. USA_2009_11; 2,678 targets in total) (Dataset S7). Candidate genes for USA300-specific yet unknown genetic markers were identified by either being present in all USA300-NAE or USA300–SAE isolates from our collection and absent in the outgroup, or being absent in all USA300-NAE or USA300–SAE isolates and present in both outgroup strains. The resulting candidate genes were subsequently queried against all other ST8 sequences of our dataset (n = 113). For the African USA300 clade, we performed the same procedures using isolate Gabon_2009_1 (this study, 2,643 targets in total) (Dataset S7) as representative for the African USA300 clade. The analyses were conducted with the aid of the SeqSphere+ software.
Supplementary Material
Acknowledgments
We thank Isabell Höfig, Ursula Keckevoet, Stefan Bletz, and Elvira Chapka for excellent technical assistance. Support of the Münster Graduate School of Evolution (MGSE) (L.S.) is gratefully acknowledged. This study was supported by the Deutsche Forschungsgemeinschaft (EI 247/8-1, Me3205/4-1). Funders had no role in study design or interpretation of results.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (accession no. PRJEB14816).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1702472114/-/DCSupplemental.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Otto M. Community-associated MRSA: What makes them special? Int J Med Microbiol. 2013;303:324–330. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolo J, et al. CONCORD Working Group High genetic diversity among community-associated Staphylococcus aureus in Europe: Results from a multicenter study. PLoS One. 2012;7:e34768. doi: 10.1371/journal.pone.0034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang Y-Y, Huang Y-C. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. 2013;13:698–708. doi: 10.1016/S1473-3099(13)70136-1. [DOI] [PubMed] [Google Scholar]
- 5.Williamson DA, Coombs GW, Nimmo GR. Staphylococcus aureus ‘Down Under’: Contemporary epidemiology of S. aureus in Australia, New Zealand, and the South West Pacific. Clin Microbiol Infect. 2014;20:597–604. doi: 10.1111/1469-0691.12702. [DOI] [PubMed] [Google Scholar]
- 6.Schaumburg F, Alabi AS, Peters G, Becker K. New epidemiology of Staphylococcus aureus infection in Africa. Clin Microbiol Infect. 2014;20:589–596. doi: 10.1111/1469-0691.12690. [DOI] [PubMed] [Google Scholar]
- 7.Abdulgader SM, Shittu AO, Nicol MP, Kaba M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: A systematic review. Front Microbiol. 2015;6:348. doi: 10.3389/fmicb.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: Origin and epidemiology. J Antimicrob Chemother. 2009;64:441–446. doi: 10.1093/jac/dkp241. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention (CDC) Methicillin-resistant Staphylococcus aureus infections among competitive sports participants–Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000-2003. MMWR Morb Mortal Wkly Rep. 2003;52:793–795. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC) Methicillin-resistant Staphylococcus aureus infections in correctional facilities—Georgia, California, and Texas, 2001-2003. MMWR Morb Mortal Wkly Rep. 2003;52:992–996. [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention (CDC) Methicillin-resistant Staphylococcus aureus skin or soft tissue infections in a state prison–Mississippi, 2000. MMWR Morb Mortal Wkly Rep. 2001;50:919–922. [PubMed] [Google Scholar]
- 12.Nimmo GR. USA300 abroad: Global spread of a virulent strain of community-associated methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2012;18:725–734. doi: 10.1111/j.1469-0691.2012.03822.x. [DOI] [PubMed] [Google Scholar]
- 13.McDougal LK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyle-Vavra S, et al. USA300 and USA500 clonal lineages of Staphylococcus aureus do not produce a capsular polysaccharide due to conserved mutations in the cap5 locus. MBio. 2015;6:e02585–e14. doi: 10.1128/mBio.02585-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Planet PJ, et al. Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine catabolic mobile element and speG-mediated adaptations for survival on skin. MBio. 2013;4:e00889–e13. doi: 10.1128/mBio.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser P, et al. Demography and intercontinental spread of the USA300 community-acquired methicillin-resistant Staphylococcus aureus lineage. MBio. 2016;7:e02183–e15. doi: 10.1128/mBio.02183-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Von Dach E, et al. Comparative genomics of community-associated methicillin-resistant Staphylococcus aureus shows the emergence of clone ST8-USA300 in Geneva, Switzerland. J Infect Dis. 2016;213:1370–1379. doi: 10.1093/infdis/jiv489. [DOI] [PubMed] [Google Scholar]
- 18.Aanensen DM, et al. European SRL Working Group Whole-genome sequencing for routine pathogen surveillance in public health: A population snapshot of invasive Staphylococcus aureus in Europe. MBio. 2016;7:e00444–e16. doi: 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura I, et al. Clinical aspects of infection with methicillin-resistant Staphylococcus aureus USA300 strain, generally regarded as community-acquired, in Japan. Jpn J Infect Dis. 2013;66:416–420. doi: 10.7883/yoken.66.416. [DOI] [PubMed] [Google Scholar]
- 20.Jung J, et al. Emergence of Panton-Valentine leucocidin-positive ST8-methicillin-resistant Staphylococcus aureus (USA300 clone) in Korea causing healthcare-associated and hospital-acquired bacteraemia. Eur J Clin Microbiol Infect Dis. 2016;35:1323–1329. doi: 10.1007/s10096-016-2668-y. [DOI] [PubMed] [Google Scholar]
- 21.Madzgalla S, et al. Molecular characterization of Staphylococcus aureus isolates causing skin and soft tissue infections in patients from Malakand, Pakistan. Eur J Clin Microbiol Infect Dis. 2016;35:1541–1547. doi: 10.1007/s10096-016-2695-8. [DOI] [PubMed] [Google Scholar]
- 22.Iwao Y, et al. The emerging ST8 methicillin-resistant Staphylococcus aureus clone in the community in Japan: Associated infections, genetic diversity, and comparative genomics. J Infect Chemother. 2012;18:228–240. doi: 10.1007/s10156-012-0379-6. [DOI] [PubMed] [Google Scholar]
- 23.Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- 24.Monecke S, Ehricht R, Slickers P, Tan H-L, Coombs G. The molecular epidemiology and evolution of the Panton-Valentine leukocidin-positive, methicillin-resistant Staphylococcus aureus strain USA300 in Western Australia. Clin Microbiol Infect. 2009;15:770–776. doi: 10.1111/j.1469-0691.2009.02792.x. [DOI] [PubMed] [Google Scholar]
- 25.Coombs GW, et al. Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg Infect Dis. 2006;12:241–247. doi: 10.3201/eid1202.050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyes J, et al. Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin Infect Dis. 2009;49:1861–1867. doi: 10.1086/648426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planet PJ, et al. Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J Infect Dis. 2015;212:1874–1882. doi: 10.1093/infdis/jiv320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monecke S, et al. Population structure of Staphylococcus aureus from Trinidad & Tobago. PLoS One. 2014;9:e89120. doi: 10.1371/journal.pone.0089120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenthal ME, et al. Molecular epidemiology of Staphylococcus aureus in post-earthquake northern Haiti. Int J Infect Dis. 2014;29:146–151. doi: 10.1016/j.ijid.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Schaumburg F, Gast B, Sannon H, Mellmann A, Becker K. Is Africa the origin of major Haitian Staphylococcus aureus lineages? Int J Infect Dis. 2015;34:1–2. doi: 10.1016/j.ijid.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Herrmann M, et al. Staphylococcal disease in Africa: Another neglected ‘tropical’ disease. Future Microbiol. 2013;8:17–26. doi: 10.2217/fmb.12.126. [DOI] [PubMed] [Google Scholar]
- 33.Breurec S, et al. Working Group on Staphylococcus aureus infections Epidemiology of methicillin-resistant Staphylococcus aureus lineages in five major African towns: Emergence and spread of atypical clones. Clin Microbiol Infect. 2011;17:160–165. doi: 10.1111/j.1469-0691.2010.03219.x. [DOI] [PubMed] [Google Scholar]
- 34.Ateba Ngoa U, et al. Epidemiology and population structure of Staphylococcus aureus in various population groups from a rural and semi urban area in Gabon, Central Africa. Acta Trop. 2012;124:42–47. doi: 10.1016/j.actatropica.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Kraef C, et al. Co-detection of Panton-Valentine leukocidin encoding genes and cotrimoxazole resistance in Staphylococcus aureus in Gabon: Implications for HIV-patients’ care. Front Microbiol. 2015;6:60. doi: 10.3389/fmicb.2015.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaumburg F, et al. Transmission of Staphylococcus aureus between mothers and infants in an African setting. Clin Microbiol Infect. 2014;20:O390–O396. doi: 10.1111/1469-0691.12417. [DOI] [PubMed] [Google Scholar]
- 37.Huson MA, et al. Methicillin-resistant Staphylococcus aureus as a cause of invasive infections in Central Africa: A case report and review of the literature. Infection. 2014;42:451–457. doi: 10.1007/s15010-014-0589-1. [DOI] [PubMed] [Google Scholar]
- 38.Egyir B, et al. Molecular epidemiology and antimicrobial susceptibility of clinical Staphylococcus aureus from healthcare institutions in Ghana. PLoS One. 2014;9:e89716. doi: 10.1371/journal.pone.0089716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stegger M, et al. Origin and evolution of European community-acquired methicillin-resistant Staphylococcus aureus. MBio. 2014;5:e01044–e14. doi: 10.1128/mBio.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Botigué LR, et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc Natl Acad Sci USA. 2013;110:11791–11796. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thurlow LR, et al. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe. 2013;13:100–107. doi: 10.1016/j.chom.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen AR, et al. Two distinct clones of methicillin-resistant Staphylococcus aureus (MRSA) with the same USA300 pulsed-field gel electrophoresis profile: A potential pitfall for identification of USA300 community-associated MRSA. J Clin Microbiol. 2009;47:3765–3768. doi: 10.1128/JCM.00934-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jamrozy DM, et al. Pan-genomic perspective on the evolution of the Staphylococcus aureus USA300 epidemic. Microb Genom. 2016;2:e000058. doi: 10.1099/mgen.0.000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holden MTG, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23:653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chadwick SG, et al. Detection of epidemic USA300 community-associated methicillin-resistant Staphylococcus aureus strains by use of a single allele-specific PCR assay targeting a novel polymorphism of Staphylococcus aureus pbp3. J Clin Microbiol. 2013;51:2541–2550. doi: 10.1128/JCM.00417-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaumburg F, et al. Study Group Population dynamics among methicillin-resistant Staphylococcus aureus isolates in Germany during a 6-year period. J Clin Microbiol. 2012;50:3186–3192. doi: 10.1128/JCM.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Köser CU, et al. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med. 2012;366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strommenger B, et al. Evolution of methicillin-resistant Staphylococcus aureus towards increasing resistance. J Antimicrob Chemother. 2014;69:616–622. doi: 10.1093/jac/dkt413. [DOI] [PubMed] [Google Scholar]
- 50.Uhlemann A-C, et al. Molecular tracing of the emergence, diversification, and transmission of S. aureus sequence type 8 in a New York community. Proc Natl Acad Sci USA. 2014;111:6738–6743. doi: 10.1073/pnas.1401006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ofner-Agostini M, et al. Canadian Nosocomial Infection Surveillance Program, Health Canada Methicillin-resistant Staphylococcus aureus in Canadian aboriginal people. Infect Control Hosp Epidemiol. 2006;27:204–207. doi: 10.1086/500628. [DOI] [PubMed] [Google Scholar]
- 52.O’Brien FG, et al. Population dynamics of methicillin-susceptible and -resistant Staphylococcus aureus in remote communities. J Antimicrob Chemother. 2009;64:684–693. doi: 10.1093/jac/dkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauß L, et al. Detecting Staphylococcus aureus virulence and resistance genes: A comparison of whole-genome sequencing and DNA microarray technology. J Clin Microbiol. 2016;54:1008–1016. doi: 10.1128/JCM.03022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol. 2014;52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mellmann A, et al. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol. 2016;54:2874–2881. doi: 10.1128/JCM.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kondo Y, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milheiriço C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex.’. J Antimicrob Chemother. 2007;60:42–48. doi: 10.1093/jac/dkm112. [DOI] [PubMed] [Google Scholar]
- 59.Prabhakara S, et al. Genome sequencing unveils a novel sea enterotoxin-carrying PVL phage in Staphylococcus aureus ST772 from India. PLoS One. 2013;8:e60013. doi: 10.1371/journal.pone.0060013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boakes E, et al. Distinct bacteriophages encoding Panton-Valentine leukocidin (PVL) among international methicillin-resistant Staphylococcus aureus clones harboring PVL. J Clin Microbiol. 2011;49:684–692. doi: 10.1128/JCM.01917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 62.Driebe EM, et al. Using whole genome analysis to examine recombination across diverse sequence types of Staphylococcus aureus. PLoS One. 2015;10:e0130955. doi: 10.1371/journal.pone.0130955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop GCE (Institute of Electrical and Electronics Engineers, Piscataway, NJ), no. 11705685.
- 65.Schaumburg F, et al. Staphylococcus aureus complex from animals and humans in three remote African regions. Clin Microbiol Infect. 2015;21:345.e1–345.e8. doi: 10.1016/j.cmi.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Schaumburg F, et al. Virulence factors and genotypes of Staphylococcus aureus from infection and carriage in Gabon. Clin Microbiol Infect. 2011;17:1507–1513. doi: 10.1111/j.1469-0691.2011.03534.x. [DOI] [PubMed] [Google Scholar]
- 67.Okon KO, et al. Population dynamics of Staphylococcus aureus from Northeastern Nigeria in 2007 and 2012. Epidemiol Infect. 2014;142:1737–1740. doi: 10.1017/S0950268813003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benson MA, et al. Evolution of hypervirulence by a MRSA clone through acquisition of a transposable element. Mol Microbiol. 2014;93:664–681. doi: 10.1111/mmi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diep BA, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 70.Highlander SK, et al. Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 2007;7:99. doi: 10.1186/1471-2180-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.