Significance
IL-22 has been identified as a cancer-promoting cytokine, but its regulation in cancer tissue has not been addressed. Using both murine and human models, we demonstrate that cancer cells directly induce IL-22 production. We prove that interleukin-1β induced by inflammasome activation is critical for IL-22 production. IL-1β increased the activity of the IL-22 transcription factors in lineage-committed T cells. We show the existence of IL-22–producing Th1, Th17, and Th22 cells in tumor tissue of patients. Use of the clinically approved IL-1 receptor antagonist anakinra in vivo reduced IL-22 production and reduced tumor growth in a breast cancer model. These data provide the basis for therapeutic interventions, particularly using anakinra, aiming at limiting IL-22 production in patients with cancer.
Keywords: interleukin-22, interleukin-1, inflammasome, cancer immunology, anakinra
Abstract
IL-22 has been identified as a cancer-promoting cytokine that is secreted by infiltrating immune cells in several cancer models. We hypothesized that IL-22 regulation would occur at the interface between cancer cells and immune cells. Breast and lung cancer cells of murine and human origin induced IL-22 production from memory CD4+ T cells. In the present study, we found that IL-22 production in humans is dependent on activation of the NLRP3 inflammasome with the subsequent release of IL-1β from both myeloid and T cells. IL-1 receptor signaling via the transcription factors AhR and RORγt in T cells was necessary and sufficient for IL-22 production. In these settings, IL-1 induced IL-22 production from a mixed T helper cell population comprised of Th1, Th17, and Th22 cells, which was abrogated by the addition of anakinra. We confirmed these findings in vitro and in vivo in two murine tumor models, in primary human breast and lung cancer cells, and in deposited expression data. Relevant to ongoing clinical trials in breast cancer, we demonstrate here that the IL-1 receptor antagonist anakinra abrogates IL-22 production and reduces tumor growth in a murine breast cancer model. Thus, we describe here a previously unrecognized mechanism by which cancer cells induce IL-22 production from memory CD4+ T cells via activation of the NLRP3 inflammasome and the release of IL-1β to promote tumor growth. These findings may provide the basis for therapeutic interventions that affect IL-22 production by targeting IL-1 activity.
IL-22 is a cytokine with tumor-promoting properties. It enhances tumor-cell proliferation, protects against apoptosis, and mediates the attraction of immunosuppressive immune cells and the release of pro- and antiinflammatory cytokines (1). IL-22 also promotes neoangiogenesis and epithelial-to-mesenchymal transition, which are hallmarks of cancer (1, 2). Unlike other cytokines, IL-22 is produced only by immune cells and binds to IL-22 receptor-1+ (IL-22-R1+) nonimmune cells (3). Strong evidence links IL-22 to colon cancer pathogenesis in both inflammatory and genetic colon cancer models (4–6). IL-22 drives the progression of hepatocellular carcinoma, potentially via accelerated tumor-cell proliferation (7, 8). The presence of IL-22–producing cells is linked to a more aggressive phenotype in a variety of cancer entities such as lung, breast, gastric, and skin cancer, indicating a more universal function of IL-22 in cancer progression (9–13). The source of IL-22 in these tumor entities varies, including innate immune cells and CD4+ T cells (1, 14). In contrast, the mechanisms by which cancer cells or other cell populations within the tumor environment induce IL-22 production remain unaddressed.
Under physiological conditions and in certain inflammatory diseases such as psoriasis, IL-22 is mainly produced by T cells with smaller contributions from other immune populations (15, 16). IL-22 production is regulated by the transcription factors retinoic acid-related orphan receptor-γ (ROR-γ) and aryl hydrocarbon receptor (AhR) (15). Different cytokines have been reported to induce IL-22 production, but no data are available on how cancer cells regulate IL-22 production (15).
We demonstrate that tumor cells can induce IL-22 production directly from immune cells via IL-1. IL-1 induces the production of IL-22 in a memory CD4+ Th cell population in mice and humans. We show that reducing endogenous IL-1 activity in vivo, by administering anakinra, leads to diminished IL-22 production and subsequently reduced tumor burden in two different tumor models. In mice, dying tumor cells release IL-1α and initiate IL-22 production, but in humans, cancer cells trigger activation of the NLRP3 inflammasome and the release of IL-1β to regulate IL-22 in memory CD4+ T cells. In patients, we found a strong correlation between the expression of IL-22–related genes and inflammasome activation, supporting the in vivo relevance of our findings. Our study unravels a shared mechanism of IL-22 induction across different cancer types. These findings identify IL-1 as a possible target to therapeutically modulate IL-22 production.
Results
IL-22 Production in Splenocytes Is Induced by Soluble Factors Released by Murine Breast and Lung Cancer Cell Lines.
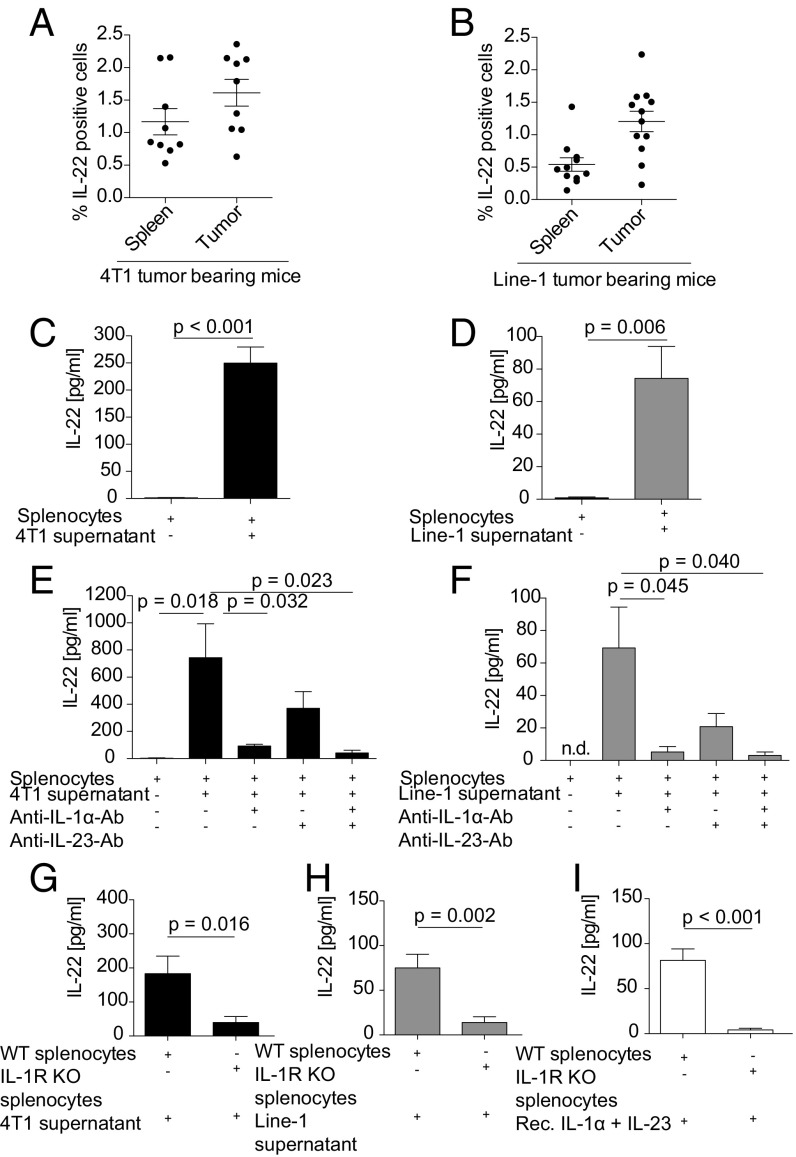
IL-22 is expressed in most cancer tissues studied so far, including breast and lung cancer (9, 10, 17). To investigate the source and the regulation of IL-22 production in these diseases, we first analyzed two murine syngeneic cancer models, 4T1 breast cancer and Line-1 lung cancer, for the presence of IL-22+ cells by flow cytometry. We detected IL-22+ cells with mononuclear cell morphology in spleen and tumor tissue in both models (Fig. 1 A and B). We hypothesized that these IL-22+ cells are nontumor cells, e.g., infiltrating immune cells.
Fig. 1.
Murine lung and breast cancer cell lines induce IL-22 from splenocytes via tumor-derived IL-1α. (A and B) Single-cell suspensions of 4T1 (A) and Line-1 (B) s.c. tumors were analyzed by flow cytometry for total intracellular IL-22 expression. Values in A and B represent pooled data of three independent experiments with three mice per group and four independent experiments with three mice per group, respectively. (C and D) Splenocytes (2 × 106/mL) were stimulated with 50% cell-free 4T1 (C) or Line-1 (D) tumor-cell supernatant for 6 d. Mean values from five independent experiments are shown. (E and F) Splenocytes were stimulated with 4T1 (E) or Line-1 (F) tumor-cell supernatant in the presence or absence of anti–IL-1α or anti–IL-23 blocking antibodies (2.5 µg/mL) or both for 6 d. The mean values of a minimum of four independent experiments are shown. (G–I) Wild-type or IL-1R knock-out splenocytes were stimulated with 4T1 supernatant (G), Line-1 supernatant (H), or 20 ng/mL recombinant IL-1α and IL-23, respectively (I). Values in G–I represent pooled data of two independent experiments with four to seven mice per group. IL-22 production was quantified by ELISA (C–I). Error bars represent the SEM; P values from a two-sided Student’s t test are shown; n.d., not detectable; rec., recombinant.
IL-22 was induced in splenocytes incubated with cell-free tumor cell-conditioned supernatants (Fig. 1 C and D).
Tumor-Derived IL-1α Drives IL-22 Production from Murine Splenocytes.
To identify tumor-derived IL-22–inducing factors, we stimulated splenocytes with 4T1 and Line-1 cell supernatants and found 14 cytokines in the supernatants of the stimulated splenocytes (Fig. S1A). Of these 14 cytokines, IL-1α, IL-6, IL-23, IFN-γ, TNF-α, and G-CSF have been previously described as being involved in IL-22 induction (14, 18–20). IL-1β could not be detected. Testing for the effect of exogenously added cytokines detected only IL-1α and, to a lesser extent, IL-23 induced IL-22 from murine splenocytes (Fig. S1B). IL-1α and IL-23 were found in relevant amounts in both 4T1 and Line-1 cell lysates (Fig. S1 C and D) and in tumor-cell supernatants (Fig. S1 E and F). Stimulation of splenocytes with recombinant IL-1α and IL-23 alone or in combination dose-dependently mimicked stimulation with tumor-cell supernatants in terms of IL-22 induction (Fig. S1G). The addition of anti–IL-1α or anti–IL-23 neutralizing antibodies or both reduced IL-22 induction in splenocytes by tumor-cell supernatants (Fig. 1 E and F). Similarly, the addition of the IL-1 receptor (IL-1R) antagonist anakinra to tumor supernatants abrogated IL-22 induction in splenocytes (Fig. S1H). IL-22 induction was dependent upon IL-1R signaling, as stimulation of IL-1R–KO splenocytes with tumor supernatants or recombinant cytokines did not induce IL-22 production (Fig. 1 G–I).
Tumor Cells Induce IL-22 Production in Splenocytes via AhR and RORγt Signaling.
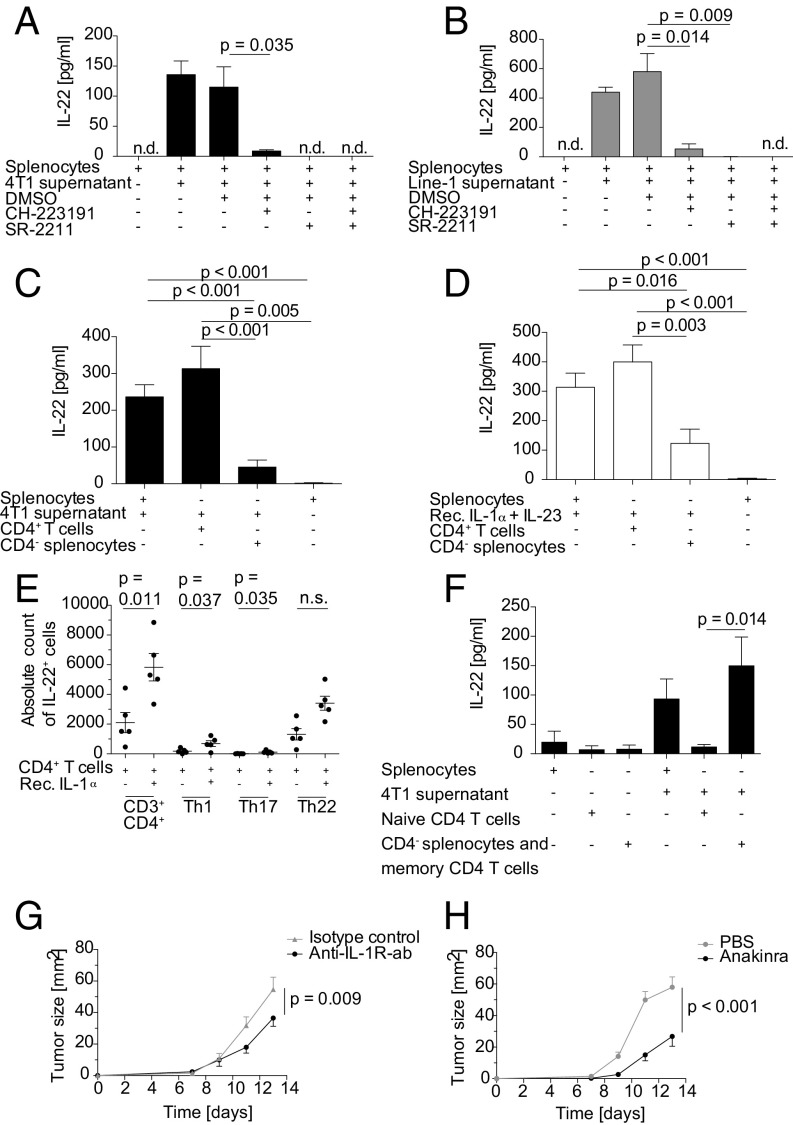
To further dissect the mechanism of IL-22 induction by tumor cells, we sought to identify the transcription factors involved. Both AhR and RORγt have been described as playing a role in IL-22 production by immune cells under physiological conditions (21, 22). The addition of the AhR antagonist CH-223191 or the RORγt antagonist SR-2211 to splenocytes during stimulation with 4T1 and Line-1 cell supernatants significantly reduced IL-22 production. IL-22 production was blocked completely when both antagonists were added (Fig. 2 A and B). Moreover, treatment of tumor-bearing mice with repeated doses of CH-223191 decreased infiltrating IL-22+ immune cells and the amount of IL-22 in tumor tissue in both the 4T1 and the Line-1 tumor models (Fig. S2 A and B), pointing toward a role for AhR signaling in IL-22 production in vivo as well.
Fig. 2.
IL-22 is secreted from memory CD4+ T cells in an AhR- and RORγt-dependent manner, and in vivo neutralization of IL-1 reduces tumor growth. (A and B) Splenocytes (2 × 106/mL) were stimulated with 4T1 supernatant (A) or Line-1 supernatant (B) in the presence or absence of 10 µM CH-223191 (AhR antagonist) or 5 µM SR-2211 (RORγt antagonist) for 6 d. Values in A are the mean of three different experiments performed in triplicate. Values in B are representative of five different experiments performed in triplicate. IL-22 production was quantified by ELISA. (C and D) MACS-enriched CD4+ T cells and CD4− splenocytes or total splenocytes were stimulated with 4T1 tumor supernatant (C) or 100 ng/mL recombinant IL-1α and IL-23 (D) for 6 d. Mean values of a minimum of four independent experiments are shown. (E) MACS-enriched CD4+ T cells were stimulated with 20 ng/mL recombinant IL-1α for 4 d. IL-22 production by Th1 (CD3+CD4+IFN-γ+), Th17 (CD3+CD4+IFN-γ−IL-17+), and Th22 (CD3+CD4+IFN-γ−IL-17−) T cells was analyzed by flow cytometry (data represent two independent experiments with five different mice). (F) MACS-enriched naive CD4+ T cells and CD4− splenocytes including CD4+ CD44+ memory T cells were stimulated with 4T1 supernatant for 6 d. Mean values of three different experiments with eight replicates of supernatants are shown. (G) BALB/c mice bearing 1.25 × 105 4T1 tumor cells s.c. (n = 10 mice per group) were treated i.p. with 300 µg anti-mouse IL-1R antibody or isotype control every second day beginning on day 0. (H) C57BL/6 mice were injected s.c. in the right flank with 2.5 × 105 E0771 tumor cells (n = 15 mice per group). Mice were treated with 1 mg anakinra or PBS i. p. every day beginning on day 0. In A–F error bars represent the SEM, and P values by two-sided Student’s t test are shown. In G and H, statistical significance was analyzed by two-way ANOVA with correction for multiple testing; n.d., not detectable; n.s., not significant; rec., recombinant.
Murine Tumor Cells Induce IL-22 Production from Memory CD4+ T Cells.
Based on the literature, T cells are a major source of IL-22 in mice and humans (14). We hypothesized that, if T cells are the major source of IL-22 in our system, IL-22 production should be conserved in the CD3+ and CD4+ splenocyte fraction while being reduced in the CD3- or CD4-depleted fraction. Purified CD3+ T cells secreted IL-22 in comparable amounts to splenocytes when stimulated with tumor-cell supernatant. In contrast, IL-22 production was significantly lower in the CD3-depleted fraction (Fig. S2 C and D). Similarly, purified CD4+ T cells but not CD4-depleted splenocytes produced IL-22 when stimulated with tumor-cell supernatants or with recombinant IL-1α and IL-23 (Fig. 2 C and D). To further characterize the source of IL-22 within the CD4+ T cell population, we stimulated CD4+ splenocytes with IL-1α and analyzed their phenotype by flow cytometry. Most IL-22+ cells were CD3+CD4+ and consisted of a mixed Th1, Th17, and Th22 cell population (Fig. 2E). These phenotype findings were supported by analyzing the supernatants for the presence of the respective prototypical cytokines, such as IL-17 and IFN-γ. IFN-γ and IL-17 were cosecreted in high amounts (Fig. S2 E and F). The IFN-γ and IL-17 production of T cells was also IL-1α dependent, as the induction of either cytokine could be abrogated by the IL-1R antagonist anakinra (Fig. S2 G and H).
In tumor-bearing mice, we could identify both IL-22–producing CD3+CD4+ and CD3+CD8+ T cells in tumor tissue (Fig. S2 I and J). When mice were treated with the AhR antagonist, the amount of IL-22 production was reduced in these T cell fractions (Fig. S2K).
CD4+CD44+ memory T cells were identified as the target population for IL-1α, as IL-22 induction was detected only in this fraction but not in naive CD4+ T cells or CD4− cells (Fig. 2F).
Blocking of IL-1 Signaling Reduces Tumor Progression and Production of IL-22+ Cells in Vivo.
To confirm our in vitro findings and the relevance of the identified pathway for tumor progression, we tested the impact of IL-1 blockade on tumor growth and IL-22 production in vivo. We used both a neutralizing IL-1R antibody and the soluble IL-1R antagonist anakinra. Tumor progression, as evidenced by tumor growth, was reduced when 4T1 tumor-bearing mice were treated with IL-1R antibody (Fig. 2G). Growth reduction was paralleled by reduced IL-22 production, as analyzed by flow cytometry, confirming the dependence of IL-22 production on IL-1 (Fig. S2L). Similarly, when we treated mice bearing the E0771 breast cancer model with anakinra, we again found a striking retardation of tumor growth (Fig. 2H). As seen in the 4T1 model, IL-22 production was again reduced when IL-1 activity was inhibited (Fig. S2M). These findings highlight the relevance of the IL-1–IL-22 pathway for cancer progression and point toward the potential use of approved IL-1–antagonizing agents such as anakinra for cancer therapy.
Tumor Cell-Derived Factors from Human Breast and Lung Cancer Cells Induce IL-22 Production from Peripheral Blood Mononuclear Cells.
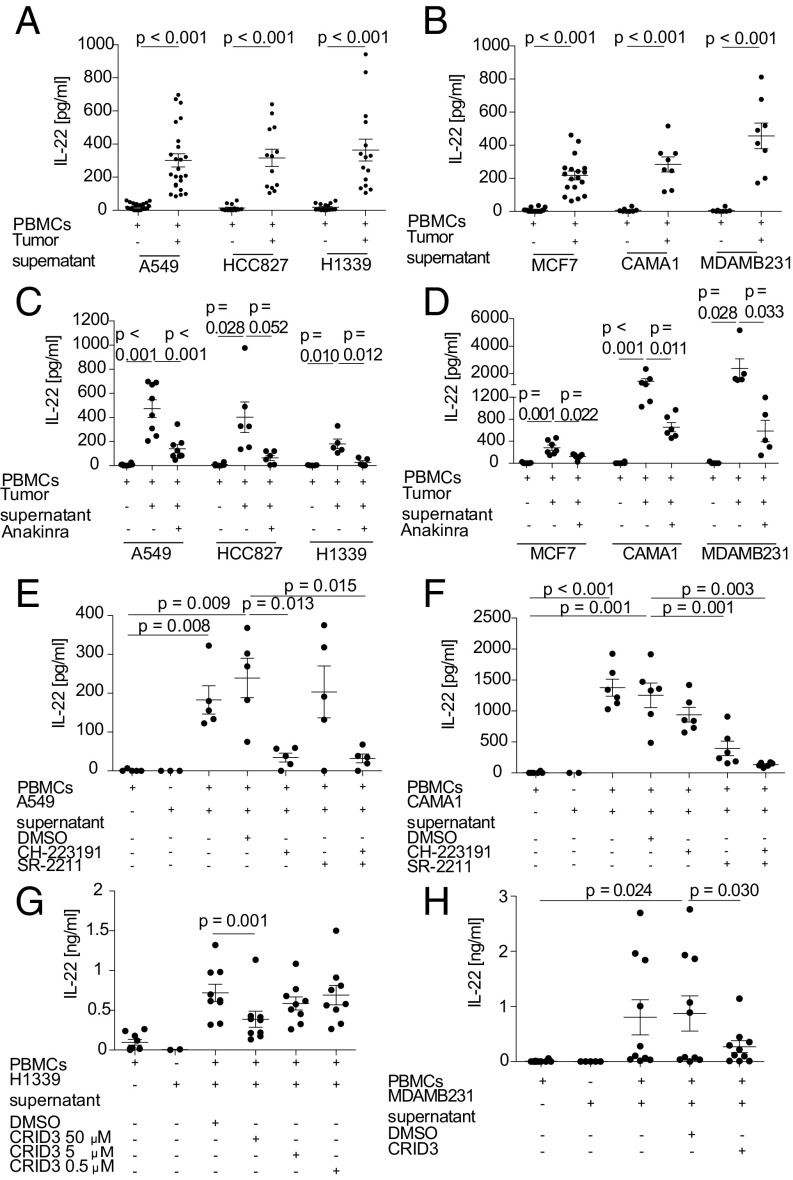
Based on the observations in mice, we next asked whether the observed induction of IL-22 production in immune cells by breast and lung cancer cells would also occur in human cells. Human peripheral blood mononuclear cells (PBMCs) from healthy donors were stimulated with tumor-conditioned supernatants of three human breast cancer (MCF7, CAMA-1, and MDAMB231) and three human lung cancer (A549, HCC827, and H1339) cell lines. Supernatants from both cancer cell types induced IL-22 production in human PBMCs, and this induction was attributable to soluble factors (Fig. 3 A and B). In contrast, stimulation of PBMCs with non–tumor-cell supernatant (from HEK293 cells) failed to lead to IL-22 production (Fig. S3A).
Fig. 3.
Human lung and breast tumor supernatants activate the NLRP3 inflammasome to release IL-1 and induce IL-22 in PBMCs from healthy donors in an AhR- and RORγT-dependent manner. (A–D) PBMCs (2 × 106/mL) were stimulated with 50% cell-free human lung cancer cell (A549, HCC827, H1339) conditioned supernatant (A) or breast cancer cell (MCF7, CAMA-1, MDAMB231) conditioned supernatant (B) for 6 d in the presence or absence of anakinra (500 ng/mL) (C and D). (E and F) PBMCs were stimulated with A549-conditioned (E) or CAMA-1–conditioned (F) tumor-cell supernatant for 6 d in the presence or absence of 10 µM CH-223191 or 5 µM SR-2211. (G and H) PBMCs were stimulated with H1339-conditioned (G) or MDAMB231-conditioned (H) tumor-cell supernatant for 6 d in the presence or absence of 50, 5, or 0.5 µM (H1339) or 50 µM (MDAMB231) CRID3. IL-22 production was quantified by ELISA (A–H). n = 8–24 different donors in A and B, 5–8 different donors in C and D, 5–6 different donors in E and F, 9 different donors in G, and 10 different donors in H. Each dot in the graphs represents one donor. Error bars represent the SEM; P values from a two-sided Student’s t test are shown.
Tumor Cell-Derived IL-1α and Tumor Cell-Induced IL-1β Lead to IL-22 Production in Human PBMCs in an AhR- and RORγt-Dependent Manner.
To further investigate the mechanism of IL-22 induction by cancer cells in human PBMCs, we added the IL-1R antagonist anakinra to the conditioned supernatants of breast and lung cancer cell lines. Anakinra blocked IL-22 induction in PBMCs stimulated with breast and lung cancer cell supernatants in a similar fashion (Fig. 3 C and D). In tumor-cell supernatants, we found IL-1α (but not IL-1β) in two of the human cell lines, H1339 and MDAMB231 cells (Fig. S3 B and C). IL-1α was also induced after stimulation with supernatants except for the H1339 supernatant (Fig. S3 B and C). Incubation of human PBMCs with any of the tumor-cell supernatants tested induced IL-1β production (Fig. S3 D and E). For all human cell lines analyzed, IL-1β was the main driver of IL-22 induction in PMBCs, as anti–IL-1β but not anti–IL-1α antibodies antagonized IL-22 induction in HCC827, H1339, CAMA-1, and MDAMB231 cell lines (Fig. S3 F–I). At the transcription factor level, IL-22 induction was dependent on both AhR and RORγt signaling in PBMCs (Fig. 3 E and F and Fig. S4 A and B).
Human Breast and Lung Cancer Cells Activate the NLRP3 Inflammasome to Induce IL-22 from Memory CD4+ T Cells.
We next addressed the cellular sources of IL-1β within the PBMCs by flow cytometry and detected IL-1β production by myeloid cells and to a lesser extent by CD4+ T cells after stimulation with tumor supernatants (Fig. S4C). To further dissect the mechanism responsible for IL-1β production, we used the specific NLRP3 inhibitor, the cytokine release inhibitory drug CRID3, and the pan-caspase inhibitor Z-Vad. The addition of CRID3 and Z-Vad abolished IL-22 production in a concentration-dependent manner (Fig. 3 G and H and Fig. S5 A and B), suggesting the involvement of inflammasome activation by the tumor cells.
To identify the cell type responsible for IL-22 production upon the addition of tumor cell-conditioned supernatants to PBMCs, we analyzed their phenotype by flow cytometry. Most IL-22+ cells were CD3+CD4+ and consisted of a mixed Th1, Th17, and Th22 cell population (Figs. S4 D and E and S5 C and D). We purified and stimulated CD4+ T cells with lung and breast cancer cell supernatants. Secretion of IL-22 by CD4+ T cells was comparable to that from whole PBMCs, but IL-22 production was almost absent in the CD4-depleted fraction (Figs. S4 F and G and S5 E and F). Similarly, stimulation of CD4+ T cells, but not of CD4-depleted PBMCs, with recombinant IL-1α or IL-1β led to IL-22 production (Fig. S4H), indicating that the CD4+ T cell fraction is the target of IL-1 and the source of IL-22 production. To further characterize the T cell population involved, we isolated naive and memory CD4+ T cells. We found that isolated memory CD4+ T cells were proficient IL-22 producers in our settings. Naive CD4+ T cells did not produce IL-22, whereas the naive CD4+ T cell-depleted fraction retained their ability to induce IL-22 (Fig. S5G). IL-22 induction by A549 tumor supernatant was still seen in the purified CD4+ T cell population and was dependent on the activation of the NLRP3 inflammasome (Fig. S5H).
This mixed phenotype of different Th lineages was further confirmed by analysis of supernatants after stimulation of PBMCs with tumor-conditioned supernatants. IFN-γ and IL-17 cosecretion was found in all cultures analyzed, supporting the presence of Th1, Th17, and Th22 cells in the culture (Fig. S6 A–D). The production of IL-22 appears to be dependent on these subpopulations, as addition of the IL-1R antagonist anakinra to the PBMCs incubated with tumor-cell supernatants blocked the induction of both IFN-γ and IL-17 in addition to blocking IL-22 production (Fig. S6 E–H). These results indicate that tumor cells can induce cytokine production from Th1, Th17, and Th22 cells in an IL-1R–dependent manner via activation of the NLRP3 inflammasome.
Th Cells Are the Main Source of IL-22 in Primary Human Lung and Breast Cancer Tissue.
To confirm the existence of IL-22–producing Th cell populations in primary human breast and lung cancer, we next analyzed tumor samples of patients with lung (n = 23) and breast (n = 11) cancer for the presence of these cells by flow cytometry. In lung cancer samples 0.58% and in breast cancer samples 0.23% of the mononuclear cell fraction expressed IL-22 (Figs. S7 A and B and S8 A and B). Among these IL-22+ cells in lung cancer samples, the main fraction, accounting for 50% of these cells, was of a Th1 phenotype, followed by Th22 and Th17 phenotypes (14% and 6%, respectively) (Fig. S7C). Expression of IL-22 in lung cancer tissue was confirmed in protein lysates of the same tumor samples (Fig. S7D). IL-22 was found at higher levels in lung cancer than in matched nontumor tissue from the same patient (Fig. S7 B and D). To corroborate the link between IL-22 production and IL-1, we next correlated protein levels of both cytokines. We found a significant correlation between IL-22 and IL-1α, compatible with the dependence of IL-22 production on the presence IL-1 in tumor tissue from patients (Fig. S7E). Expression of IL-22 could also be confirmed in breast cancer tissue (n = 7) (Fig. S8D). There, in line with our in vitro findings, we could confirm a predominant expression of IL-22 by CD4+ T cells consisting of Th1 (41%), Th17 (9%), and Th22 (23%) cells (Fig. S8C).
IL-22 Expression and Inflammasome Activation Correlate in Human Lung and Breast Adenocarcinoma.
To further link inflammasome activation with IL-22 production, we next analyzed expression data from two clinical cohorts of patients with lung (n = 80) or breast (n = 45) cancer (23, 24). Thirty-three transcripts related to the IL-22 pathway were arbitrarily selected (Fig. S9A). Lung and breast samples were hierarchically clustered to these 33 transcripts, and transcripts that clustered together and were closely related with IL22 were further analyzed by hierarchical clustering to identify their power to discriminate normal from cancer tissues (Fig. S9 B and D). The inflammasome-related genes enabled differentiation between cancer and noncancer tissue in lung cancer but not in breast cancer (Fig. S9 B and D). When lung and breast cancer tissues were analyzed separately, excluding normal tissues, we found a clear correlation between inflammasome- and IL-22–related genes, further underscoring the relationship between IL-22 production and inflammasome activation (Fig. S9 C and E).
Discussion
Our study describes a mechanism by which cancer cells induce IL-22 production from CD4+ T cells in mice and in humans in an AhR- and RORγt-dependent manner. In mice, IL-22 production is dependent on IL-1α release by cancer cells. IL-22 content in tumor tissue and tumor growth are reduced when IL-1 is neutralized in two different breast cancer models. Human cancer cells induce IL-22 through the activation of the NLRP3 inflammasome, resulting in IL-1β release. In patients, the degree of inflammasome activation correlates with IL-22 content in human breast and lung cancer samples.
A role for IL-22 in cancer development and progression has been recognized in several epithelial cancers, including breast and lung cancer (1, 9). When released by immune cells, IL-22 can act on cancer cells to promote tumor growth, aggressiveness, and treatment resistance (1, 25). However, no study has yet investigated the mechanism by which IL-22 production is induced or which immune cells are able to produce IL-22 in the tumor environment. Our findings provide evidence that cancer cells affect memory CD4+ T cells to express and release IL-22 in an IL-1–dependent manner and that this shared mechanism promotes tumor growth.
IL-1α and IL-1β are two cytokines with shared signaling via IL-1R but with a different biology (26). IL-1β is a driver of IL-22 production by immune cells and by Th17 cells in particular (27–29). In our study, IL-1R signaling was central to cancer cell-driven IL-22 production, but the mediating IL-1 family members differed between species. In mice, IL-1α was the main inducer of IL-22 production and was detected in the supernatants and protein lysates of cancer cells. IL-1α is mostly cell-associated in viable cells but may be released from dying tumor cells (30, 31). In addition, IL-1α can induce its own release from immune cells, further enhancing its effects on IL-1R+ cells, as previously described (32).
IL-1 is detectable in human breast, colon, lung, and head and neck cancers and in melanoma. Its detection is typically associated with a worse prognosis (17). In line with these findings, inhibition of endogenous IL-1 activity by administering anakinra reduces both the extent of metastasis and tumor burden (18). Comparable effects are also known for the NLRP3 inflammasome (33). Our findings link endogenous IL-1 activity to IL-22 induction in two different breast cancer models.
In contrast to the murine system, in human cancer cell lines cocultured with PBMCs, IL-1β is the main inducer of IL-22 and is induced in PBMCs. The specific NLRP3 inhibitor CRID3 abolished both IL-1β and IL-22 production in whole PBMCs and in purified CD4+ memory T cells, indicating that NLRP3 is required for inflammasome activation. A mechanism of NLRP3 inflammasome activation could be the release of IL-1α precursor or uric acid from dying tumor cells. These act on immune cells and activate the NLPR3 inflammasome (34–37). Release of ATP from tumor cells or tumor cell-derived nucleic acids may also result in NLRP3 activation and IL-1β release (38, 39).
We have identified memory CD4+ T cells as a primary target of the released IL-1β for IL-22 induction. Mechanistically, IL-1β could activate a preexisting pool of Th1, Th17, and Th22 cells for IL-22 production (40). Alternatively, there is evidence that IL-1β can drive the differentiation of memory T cells to these Th cell lineages (41, 42).
The transcription factor RORγt is required for Th17 and Th22 polarization, and the transcription factor AhR is additionally required for Th22 differentiation in mice (43–45). Similarly, RORγt and AhR are involved in IL-22 production (46, 47). Our findings show that both transcription factors are also required for IL-22 production in the setting of cancer cell-induced IL-22 production.
The cellular source of IL-22 in the tumor microenvironment varies according to the tumor entity and the species studied (1, 5). In the present study, we demonstrate that murine and human breast and lung cancer cells induce IL-22 production from a mixed population of Th1, Th17, and Th22 cells in an IL-1–dependent manner. Our findings are compatible with studies that have identified Th1 and Th17 cells in breast and lung cancer (48–50). The presence of these cells has been reported to correlate with worsened clinical outcomes (49, 51, 52). We could identify a positive correlation between inflammasome activation and IL-22 production in tumor tissue specimens from two patient cohorts with lung and breast cancer, respectively.
In summary, our study describes a previously unrecognized mechanism by which cancer cells can induce IL-22 in T cells. It provides a link between the widely described expression and function of IL-22 in cancer and its cellular source (Fig. S10). On another line, we provide pathophysiological insights into the effect of clinical IL-1 blockade recently described in lung cancer (53). The availability of clinically approved IL-1–antagonizing agents such anakinra and its favorable safety profile place clinical testing for tumor indications within reach. Clinical trials with anakinra in patients with breast cancer are under way, so far showing promising preliminary data (54). Our findings add to the rationale for developing therapeutic interventions targeting the IL-1–IL-22 axis.
Materials and Methods
Mice.
All animal experiments were approved by the local regulatory agency (Regierung von Oberbayern) or by the Veterinary Administration of the Prefecture of Western Greece (protocol approval no. 118018/578) and adhered to the NIH guidelines for the care and use of laboratory animals.
Mouse Tumor Models and Treatment of Mice.
Five-week-old BALB/c mice were injected s.c. in the right flank with 1.25 × 105 4T1 or 5 × 105 Line-1 tumor cells, and C57BL/6 mice were injected s.c. in the right flank with 2.5 × 105 E0771 tumor cells and were treated as indicated.
Patient Samples.
The tissue samples of non-small cell lung cancer (NSCLC) and corresponding clinical data used in this study were provided by the Biobank under the administration of Human Tissue and Cell Research (HTCR) Foundation at University Hospital, Ludwig Maximilian University, Munich (LMU Munich). The framework of the HTCR includes obtaining written informed consent from all patients with lung cancer and has been approved by the Ethics Committee of the Medical Faculty, LMU Munich (no. 025-12) and by the Bavarian State Medical Association. All operations of Biobank are certified according to ISO 9001:2008. Written informed consent was obtained from all patients with breast cancer before collection of specimens, in line with the respective institutional policies and in accordance with to the Declaration of Helsinki. Tumor specimens were obtained from patients undergoing clinically indicated surgery. Ethical approval was obtained from the Ethics Committee of the Medical Faculty, LMU Munich (reference nos. 220-15 and 249-15).
Cytokine Secretion Assays.
Splenocytes, purified T cells, and PBMCs were cultivated as indicated (see SI Materials and Methods and figure legends for details).
Flow Cytometry.
Flow cytometry was performed according to standard protocols as indicated (see SI Materials and Methods for details).
Statistics.
FlowJo V9.2 software (TreeStar) was used for analysis of FACS datasets. Statistics were calculated with GraphPad Prism software 5.0. Differences between experimental conditions were analyzed using the unpaired two-tailed Student’s t test. The Mann–Whitney U test was used to compare data points from individual mice. A paired two-tailed Student’s t test was used when comparing experimental conditions for individual human donors. Statistical significance was analyzed by two-way ANOVA with correction for multiple testing in case of tumor growth curves. P values < 0.05 were considered significant.
Data Availability.
All data supporting this paper are attached. Raw data and reagents will be made available upon reasonable request to the authors.
Supplementary Material
Acknowledgments
This study was supported by Wilhelm Sander Stiftung Grant 2014.018.1 (to S.E. and S.K.); the International Doctoral Program i-Target: Immunotargeting of Cancer funded by the Elite Network of Bavaria (S.K., S.E., M.S.); Melanoma Research Alliance Grants N269626 (to S.E.) and 409510 (to S.K.); the Marie-Sklodowska-Curie Program Training Network for the Immunotherapy of Cancer funded by the H2020 Program of the European Union (Grant 641549, to S.E., S.K., and M.S.); the Else Kröner-Fresenius-Stiftung (S.K.); the German Cancer Aid (S.K.); the Ernst-Jung-Stiftung (S.K.); LMU Munich’s Institutional Strategy LMUexcellent within the framework of the German Excellence Initiative (S.E. and S.K.); the Bundesministerium für Bildung und Forschung Project Oncoattract (S.E. and S.K.); European Research Council Grant 756017, ARMOR-T (to S.K.); and by the Comprehensive Pneumology Center Research School Program Lung Biology and Disease (C.V.). This study was supported by Human Tissue and Cell Research, a nonprofit foundation under German civil law. G.T.S. was supported by European Research Council 2010 Starting Independent Investigator Grant 260524 and 2015 Proof-of-Concept Grant 679345.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw data and reagents will be made available upon reasonable request to the authors.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705165114/-/DCSupplemental.
References
- 1.Lim C, Savan R. The role of the IL-22/IL-22R1 axis in cancer. Cytokine Growth Factor Rev. 2014;25:257–271. doi: 10.1016/j.cytogfr.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Sonnenberg GF, Fouser LA, Artis D. Border patrol: Regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol. 2011;12:383–390. [Google Scholar]
- 4.Huber S, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259–263. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirchberger S, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kryczek I, et al. IL-22(+)CD4(+) T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity. 2014;40:772–784. doi: 10.1016/j.immuni.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang R, et al. Interleukin-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology. 2011;54:900–909. doi: 10.1002/hep.24486. [DOI] [PubMed] [Google Scholar]
- 8.Kuang DM, et al. B7-H1-expressing antigen-presenting cells mediate polarization of protumorigenic Th22 subsets. J Clin Invest. 2014;124:4657–4667. doi: 10.1172/JCI74381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K, et al. Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinogenesis. 2014;35:1352–1361. doi: 10.1093/carcin/bgu044. [DOI] [PubMed] [Google Scholar]
- 10.Kobold S, et al. Interleukin-22 is frequently expressed in small- and large-cell lung cancer and promotes growth in chemotherapy-resistant cancer cells. J Thorac Oncol. 2013;8:1032–1042. doi: 10.1097/JTO.0b013e31829923c8. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, et al. Increased Tc22 and Treg/CD8 ratio contribute to aggressive growth of transplant associated squamous cell carcinoma. PLoS One. 2013;8:e62154. doi: 10.1371/journal.pone.0062154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, et al. Antiapoptotic activity of autocrine interleukin-22 and therapeutic effects of interleukin-22-small interfering RNA on human lung cancer xenografts. Clin Cancer Res. 2008;14:6432–6439. doi: 10.1158/1078-0432.CCR-07-4401. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang Y, et al. Increased intratumoral IL-22-producing CD4(+) T cells and Th22 cells correlate with gastric cancer progression and predict poor patient survival. Cancer Immunol Immunother. 2012;61:1965–1975. doi: 10.1007/s00262-012-1241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: Immunobiology and pathology. Annu Rev Immunol. 2015;33:747–785. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolk K, Witte E, Witte K, Warszawska K, Sabat R. Biology of interleukin-22. Semin Immunopathol. 2010;32:17–31. doi: 10.1007/s00281-009-0188-x. [DOI] [PubMed] [Google Scholar]
- 16.Sabat R, Ouyang W, Wolk K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov. 2014;13:21–38. doi: 10.1038/nrd4176. [DOI] [PubMed] [Google Scholar]
- 17.Ye ZJ, et al. Interleukin 22-producing CD4+ T cells in malignant pleural effusion. Cancer Lett. 2012;326:23–32. doi: 10.1016/j.canlet.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Savan R, et al. A novel role for IL-22R1 as a driver of inflammation. Blood. 2011;117:575–584. doi: 10.1182/blood-2010-05-285908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Behrends J, Renauld JC, Ehlers S, Hölscher C. IL-22 is mainly produced by IFNγ-secreting cells but is dispensable for host protection against Mycobacterium tuberculosis infection. PLoS One. 2013;8:e57379. doi: 10.1371/journal.pone.0057379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie L, et al. Targeting of MyD88 homodimerization by novel synthetic inhibitor TJ-M2010-5 in preventing colitis-associated colorectal cancer. J Natl Cancer Inst. 2015;108:djv364. doi: 10.1093/jnci/djv364. [DOI] [PubMed] [Google Scholar]
- 21.Sanos SL, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 23.Merdad A, et al. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer: MMP-9 as a potential biomarker for cancer invasion and metastasis. Anticancer Res. 2014;34:1355–1366. [PubMed] [Google Scholar]
- 24.Kabbout M, et al. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin Cancer Res. 2013;19:3383–3395. doi: 10.1158/1078-0432.CCR-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalapour S, Karin M. Immunity, inflammation, and cancer: An eternal fight between good and evil. J Clin Invest. 2015;125:3347–3355. doi: 10.1172/JCI80007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rider P, et al. IL-1α and IL-1β recruit different myeloid cells and promote different stages of sterile inflammation. J Immunol. 2011;187:4835–4843. doi: 10.4049/jimmunol.1102048. [DOI] [PubMed] [Google Scholar]
- 27.Foucher ED, et al. IL-34- and M-CSF-induced macrophages switch memory T cells into Th17 cells via membrane IL-1α. Eur J Immunol. 2015;45:1092–1102. doi: 10.1002/eji.201444606. [DOI] [PubMed] [Google Scholar]
- 28.Mills KH. Induction, function and regulation of IL-17-producing T cells. Eur J Immunol. 2008;38:2636–2649. doi: 10.1002/eji.200838535. [DOI] [PubMed] [Google Scholar]
- 29.Hernández PP, et al. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper AL, Beasley D. Hypoxia stimulates proliferation and interleukin-1alpha production in human vascular smooth muscle cells. Am J Physiol. 1999;277:H1326–H1337. doi: 10.1152/ajpheart.1999.277.4.H1326. [DOI] [PubMed] [Google Scholar]
- 31.Di Mitri D, et al. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature. 2014;515:134–137. doi: 10.1038/nature13638. [DOI] [PubMed] [Google Scholar]
- 32.Ghezzi P, Dinarello CA. IL-1 induces IL-1. III. Specific inhibition of IL-1 production by IFN-gamma. J Immunol. 1988;140:4238–4244. [PubMed] [Google Scholar]
- 33.Weichand B, et al. S1PR1 on tumor-associated macrophages promotes lymphangiogenesis and metastasis via NLRP3/IL-1β. J Exp Med. 2017;214:2695–2713. doi: 10.1084/jem.20160392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voronov E, et al. Unique versus redundant functions of IL-1alpha and IL-1beta in the tumor microenvironment. Front Immunol. 2013;4:177. doi: 10.3389/fimmu.2013.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu DE, Moore AM, Thomsen LL, Brindle KM. Uric acid promotes tumor immune rejection. Cancer Res. 2004;64:5059–5062. doi: 10.1158/0008-5472.CAN-04-1586. [DOI] [PubMed] [Google Scholar]
- 36.Jeha S. Tumor lysis syndrome. Semin Hematol. 2001;38(Suppl 10):4–8. doi: 10.1016/s0037-1963(01)90037-x. [DOI] [PubMed] [Google Scholar]
- 37.Kolb R, Liu GH, Janowski AM, Sutterwala FS, Zhang W. Inflammasomes in cancer: A double-edged sword. Protein Cell. 2014;5:12–20. doi: 10.1007/s13238-013-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolb HJ, Schmid C, Barrett AJ, Schendel DJ. Graft-versus-leukemia reactions in allogeneic chimeras. Blood. 2004;103:767–776. doi: 10.1182/blood-2003-02-0342. [DOI] [PubMed] [Google Scholar]
- 39.Xiao TS. The nucleic acid-sensing inflammasomes. Immunol Rev. 2015;265:103–111. doi: 10.1111/imr.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Sasson SZ, et al. IL-1 acts directly on CD4 T cells to enhance their antigen-driven expansion and differentiation. Proc Natl Acad Sci USA. 2009;106:7119–7124. doi: 10.1073/pnas.0902745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeshita M, et al. Polarization diversity of human CD4+ stem cell memory T cells. Clin Immunol. 2015;159:107–117. doi: 10.1016/j.clim.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Hebel K, et al. IL-1β and TGF-β act antagonistically in induction and differentially in propagation of human proinflammatory precursor CD4+ T cells. J Immunol. 2011;187:5627–5635. doi: 10.4049/jimmunol.1003998. [DOI] [PubMed] [Google Scholar]
- 43.Yang L, et al. IL-21 and TGF-beta are required for differentiation of human T(H)17 cells. Nature. 2008;454:350–352. doi: 10.1038/nature07021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 45.Yeste A, et al. IL-21 induces IL-22 production in CD4+ T cells. Nat Commun. 2014;5:3753. doi: 10.1038/ncomms4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JS, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol. 2011;13:144–151. doi: 10.1038/ni.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawa S, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 48.Gu-Trantien C, et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123:2873–2892. doi: 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goc J, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 2014;74:705–715. doi: 10.1158/0008-5472.CAN-13-1342. [DOI] [PubMed] [Google Scholar]
- 50.Xu B, et al. Promotion of lung tumor growth by interleukin-17. Am J Physiol Lung Cell Mol Physiol. 2014;307:L497–L508. doi: 10.1152/ajplung.00125.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amicarella F, et al. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut. 2015;0:1–13. doi: 10.1136/gutjnl-2015-310016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo B, Fu S, Zhang J, Liu B, Li Z. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Sci Rep. 2016;6:36107. doi: 10.1038/srep36107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridker PM, et al. Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: Exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390:1833–1842. doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
- 54.O’Shaughnessy J, et al. Safety and immunologic activity of anakinra in HER2-negative metastatic breast cancer (MBC) J Clin Oncol. 2016;34(15_Suppl) [Google Scholar]
- 55.McLean M, et al. A BALB/c murine lung alveolar carcinoma used to establish a surgical spontaneous metastasis model. Clin Exp Metastasis. 2004;21:363–369. doi: 10.1023/b:clin.0000046176.33867.c5. [DOI] [PubMed] [Google Scholar]
- 56.Kobold S, et al. Selective bispecific T cell recruiting antibody and antitumor activity of adoptive T cell transfer. J Natl Cancer Inst. 2014;107:364. doi: 10.1093/jnci/dju364. [DOI] [PubMed] [Google Scholar]
- 57.Thasler WE, et al. Charitable state-controlled foundation human tissue and cell research: Ethic and legal aspects in the supply of surgically removed human tissue for research in the academic and commercial sector in germany. Cell Tissue Bank. 2003;4:49–56. doi: 10.1023/A:1026392429112. [DOI] [PubMed] [Google Scholar]
- 58.Kumar N, et al. Identification of SR2211: A potent synthetic RORγ-selective modulator. ACS Chem Biol. 2012;7:672–677. doi: 10.1021/cb200496y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci. 2010;117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting this paper are attached. Raw data and reagents will be made available upon reasonable request to the authors.