Significance
Associations between genes and languages occur even with sustained migration among communities. By comparing phylogenies of genes and languages, we identify one source of this association. In traditional tribal societies, marriage customs channel language transmission. When women remain in their natal community and men disperse (matrilocality), children learn their mothers’ language, and language correlates with maternally inherited mitochondrial DNA. For the converse kinship practice (patrilocality), language instead correlates with paternally inherited Y chromosome. Kinship rules dictating postmarital residence can persist for many generations and determine population genetic structure at the community scale. The long-term association of languages with genetic clades created by kinship systems provides information about language transmission, and about the structure and persistence of social groups.
Keywords: language, kinship, coevolution, cultural evolution, population genetics
Abstract
Languages are transmitted through channels created by kinship systems. Given sufficient time, these kinship channels can change the genetic and linguistic structure of populations. In traditional societies of eastern Indonesia, finely resolved cophylogenies of languages and genes reveal persistent movements between stable speech communities facilitated by kinship rules. When multiple languages are present in a region and postmarital residence rules encourage sustained directional movement between speech communities, then languages should be channeled along uniparental lines. We find strong evidence for this pattern in 982 individuals from 25 villages on two adjacent islands, where different kinship rules have been followed. Core groups of close relatives have stayed together for generations, while remaining in contact with, and marrying into, surrounding groups. Over time, these kinship systems shaped their gene and language phylogenies: Consistently following a postmarital residence rule turned social communities into speech communities.
Language moves down time in a current of its own making, as Edward Sapir famously observed (1). Here, we suggest that these currents flow in channels created by kinship systems (2–4). To discover how durable such channels are, and how long languages remain within them, we investigated language transmission in 25 communities on the remote eastern Indonesian islands of Sumba and Timor, where language diversity and traditional settlement structures are still largely intact (5). In these traditional societies, movements among communities are mostly motivated by marriage (6–8). Broadly, there are four possibilities: (i) All individuals marry and reside within the same community (endogamy); (ii) individuals marry and live outside their natal community (ambilocality or neolocality); (iii) men remain in their natal community and women disperse (patrilocality or virilocality); or (iv) women remain in their natal community and men disperse (matrilocality or uxorilocality) (9). In tribal societies like Sumba and Timor, where language diversity is high, these dispersal practices have different consequences (10–13). When women remain in their natal communities and men disperse (matrilocality), language transmission is channeled through women, and children will learn the community language of their mothers. In this case, if men often marry outside the radius of their mother’s speech community, language might be expected to correlate with the maternally inherited mitochondrial DNA (mtDNA), but not with the paternally inherited Y chromosome (Y). Conversely, if men stay in their natal community and women disperse (patrilocality), the opposite pattern should hold (14).
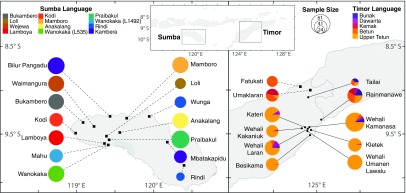
To examine these mechanisms of language transmission, we analyzed linguistic and genetic data from two Indonesian islands where postmarital residence rules are still largely observed and dispersals are mostly local. These communities mimic how the rest of the world once looked and provide a model system to investigate language transmission without many of the repercussions of globalization. On Sumba, we surveyed 14 patrilocal villages where 12 Austronesian languages are spoken (Fig. 1). Many communities on neighboring Timor are also patrilocal, but there is also a centuries-old cluster of matrilocal villages in the region of Wehali near the center of the island (15). We surveyed nine matrilocal villages in Wehali, along with two patrilocal villages located just to the north. The languages spoken in these villages include a non-Austronesian language (Bunak) and four Austronesian languages. Thus, our study system included 25 villages on two islands, representing both matrilocal and patrilocal communities, speaking 17 languages belonging to two language families. How have these languages traveled through time, within and between communities?
Fig. 1.
Languages in sampled villages on Sumba and Timor, eastern Indonesia. Pie charts show the languages spoken, scaled by sample size. (Left) The 14 patrilocal villages (■) on Sumba and the Austronesian languages spoken by the 505 sampled men. (Right) The 11 communities on Timor, including 9 matrilocal villages (•) and 2 patrilocal villages (■). Each of the 477 men sampled on Timor speaks one or more of five local languages belonging to two language families, Austronesian (Dawanta, Kemak, Betun, and Upper Tetun) and non-Austronesian Bunak.
To find out, we began by collecting data on language and kinship from representative samples of men in each village. Our goal was to compare matrilineal and patrilineal pathways of language transmission for each individual. Although matrilineal descent can be inferred for both men and women from their maternally inherited mtDNA, patrilineal descent must be traced with the Y chromosome, carried only by men. Therefore, only men were sampled for this study. We recorded the languages spoken by each individual, as well as genealogical records (including birthplace) extending back to great-grandparents. The goal was to discover which ancestors were the source of the language(s) each individual learned as a child. Genetic distances were inferred between individuals for both mtDNA and Y, and this information was used to trace genealogical relationships much deeper into the past.
Results
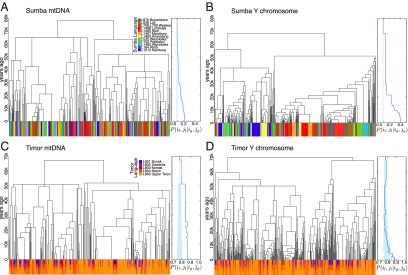
The resulting associations between groups of related men on Sumba and Timor, and the languages they speak, are shown in Fig. 2. Matrilineal relatedness is shown in Fig. 2 A and C, and patrilineal relatedness is shown in Fig. 2 B and D. The color bands beneath each phylogeny indicate the languages spoken by each individual, including multiple colors if the individual is bilingual or multilingual. The horizontal thickness of each color band segment indicates the group sizes of individuals who speak a common language and are closely related genetically. Both the Y chromosome tree for patrilocal Sumba communities (Fig. 2B) and the mtDNA tree for mostly matrilocal Timor communities (Fig. 2C) contain larger clades of related individuals who speak a common language (25 and 47 individuals in the largest clades of the Sumba Y and Timor mtDNA trees), compared with the trees of the dispersing sex (12 and 26 individuals in the largest clades of the Sumba mtDNA and Timor Y trees). Larger groups of individuals who have close genetic relationships and speak a common language are therefore found in the lineages of the nondispersing sex.
Fig. 2.
Language sharing in the phylogenies of Sumba (A and B) and Timor (C and D). (A and C) mtDNA. (B and D) Y chromosome. Color bands beneath the phylogenies show the languages spoken by each individual (monolingual on Sumba; sometimes multilingual on Timor). Plots to the right of each phylogeny show the probability of sharing a language given that each pair of individuals are in the same genetic clade at a given time in the past. Solid lines represent the observed metric, with shaded bands indicating the result of random permutations of the linguistic data. Higher probabilities that close genetic relatives share a language, compared with random expectations, were observed for Sumba Y (B) and Timor mtDNA (C) at all time periods.
The hypothesis that kinship practices create durable channels for language transmission was tested by comparing the topologies of the mtDNA and Y chromosome trees. We surmised that the statistically significant difference in the group sizes of closely related individuals who speak a common language reflects the influence of community structure on language transmission. Specifically, they represent persistent speech communities created by the vertical transmission of languages along genetic clades, as mediated by kinship rules. To test this hypothesis, we defined a probability that a pair of individuals share a common language , given that they belong to the same genetic clade at some time in the past . This metric indicated greater than expected language sharing, even at decreasing degrees of genetic relatedness backward in time. We show this in the plots to the right of the trees in Fig. 2, with solid lines representing the observed data, and shaded regions indicating the range of probabilities [∼0.1 in monolingual Sumba and (0.7, 0.9) in multilingual Timor] seen when languages are shuffled randomly among samples.
As we tracked back through time (i.e., deeper along branches in the trees), men in the patrilocal villages of Sumba were consistently more likely to speak a common language compared with random cases. This tendency was stronger along patrilines (Fig. 2B) than matrilines (Fig. 2A). Conversely, in mostly matrilocal Timor, men were more likely to share a common language with their close matrilineal kin (Fig. 2 C and D). We therefore found evidence for the persistence of speech communities, where probabilities of sharing a common language with close kin were stronger and distinguishable from random chance. These speech communities appeared to have formed as genes and languages followed the nondispersing sex—on the father’s side (Y) on patrilocal Sumba and on the mother’s side (mtDNA) on mostly matrilocal Timor. These results suggested two further hypotheses, which we explore below:
-
1.
Kinship rules concerning marriage and postmarital residence can persist for many generations and predict population genetic structure at the community scale; and
-
2.
The association between language and genetic clades, as created by kinship structures, provides information not only about language transmission, but also about the structure and persistence of social groups.
These arguments turn on the answers to two questions. First, how do kinship rules relate to population genetic structure? And second, what is the relationship between the channels created by kinship practices and the transmission of languages? In other words, how long do such channels persist, and when do languages shift between them? We began with a simple model to explore these scenarios, and then compared the results to the empirical data.
Kinship and Population Genetic Structure.
We began by exploring how kinship rules affect the movements of men and women between villages. For this purpose, we adapted an isolation with migration (IM) coalescent model (16) to capture the genetic consequences of different male and female migration patterns (SI Appendix). This model can be used to directly assess evidence for the four postmarital residence practices: (i) village endogamy, (ii) ambilocality or neolocality, (iii) patrilocality, and (iv) matrilocality.
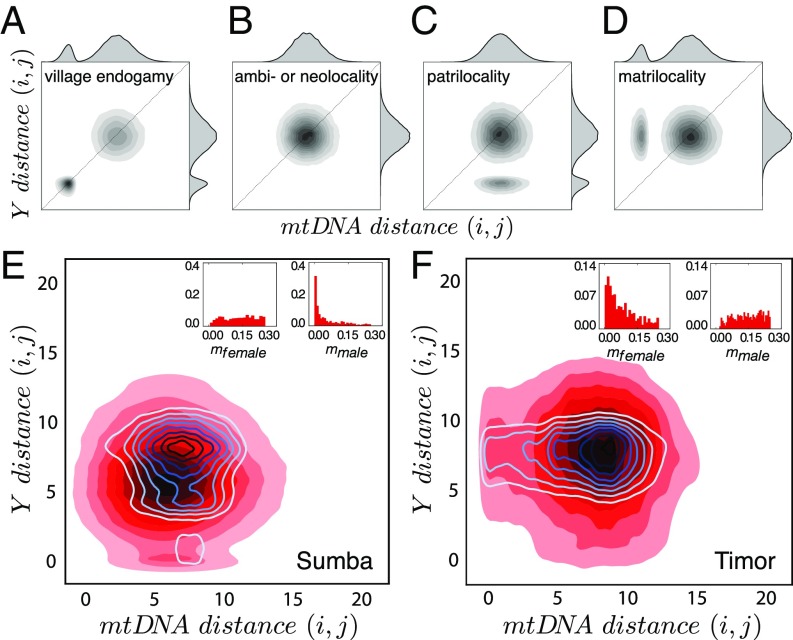
Fig. 3 A–D shows typical outputs from this model in the form of paired genetic distances. Here, each individual is paired with every other individual, and the pairs are represented by points corresponding to how closely they are related on both mtDNA (matriline) and Y (patriline). In the simplest cases (endogamy and ambilocality or neolocality), there is no bias toward matrilineal or patrilineal relatedness (Fig. 3 A and B). Consequently, given equal mutation rates, the distribution of pairwise mtDNA and Y distances lies on the one-to-one correlation line, at a distance from the origin that depends on demographic parameters. If both females and males marry and reside within their natal villages (Fig. 3A), we see two clusters of pairwise genetic distances: (i) near the origin, a cluster of closely related kin who reside in the same village, and (ii) a larger cluster consisting of individuals who live in different villages and are thus less closely related. When there is no gender bias in dispersal and both sexes move frequently, the distributions of genetic distances for mtDNA and Y are similar (Fig. 3B). Introducing a matrilocal or patrilocal bias in marriage customs shifts the cluster of closely related kin toward one or the other axis (Fig. 3 C and D). In patrilocal villages, where men remain in their natal villages while women may move to marry, pairs of men remain closely related on their Y, but not their mtDNA (Fig. 3C). The opposite holds for matrilocal villages where women remain in their natal villages and men may move to marry (Fig. 3D). These are idealized patterns. In reality, we expect a tendency toward endogamy in both matrilocal and patrilocal systems—for example, when a man in a matrilocal village marries a woman from the same village.
Fig. 3.
Genetic structure from an IM model with migration influenced by kinship practices. (A–D) The theoretical role of kinship practices on genetic diversity is shown for populations that are endogamous (A), ambilocal or neolocal (B), patrilocal (C), and matrilocal (D). (E) Close correspondence of the IM model (red shading) with observed data (blue contours) for patrilocal Sumba using (, , , , , ). (F) Close correspondence of the IM model with only matrilocal villages on Timor, using (, , , , , ). Insets in E and F show the posterior distributions of migration rates for the Sumba and Timor kinship systems based on 3 million samples drawn from prior distributions of all IM model parameters.
These predictions were borne out in the genetic data from Sumba (Fig. 3E) and the matrilocal villages of Timor (Fig. 3F). By plotting pairwise genetic distances, we could see how closely individuals are related on their mtDNA (matriline) and Y (patriline). On Sumba, the cluster of small Y distances (close patrilineal kin: of pairs) is more distinct, compared with the faint cluster of small mtDNA distances (close matrilineal kin: of pairs). Conversely, on Timor, the cluster of small mtDNA distances ( of pairs) is more pronounced, in contrast to the cluster of small Y distances ( of pairs). Comparing the two islands (with more detail shown in Fig. 4 A and B), there is a tendency for closer patrilineal relatedness on Sumba, while on Timor, we see the opposite pattern of closer matrilineal relatedness.
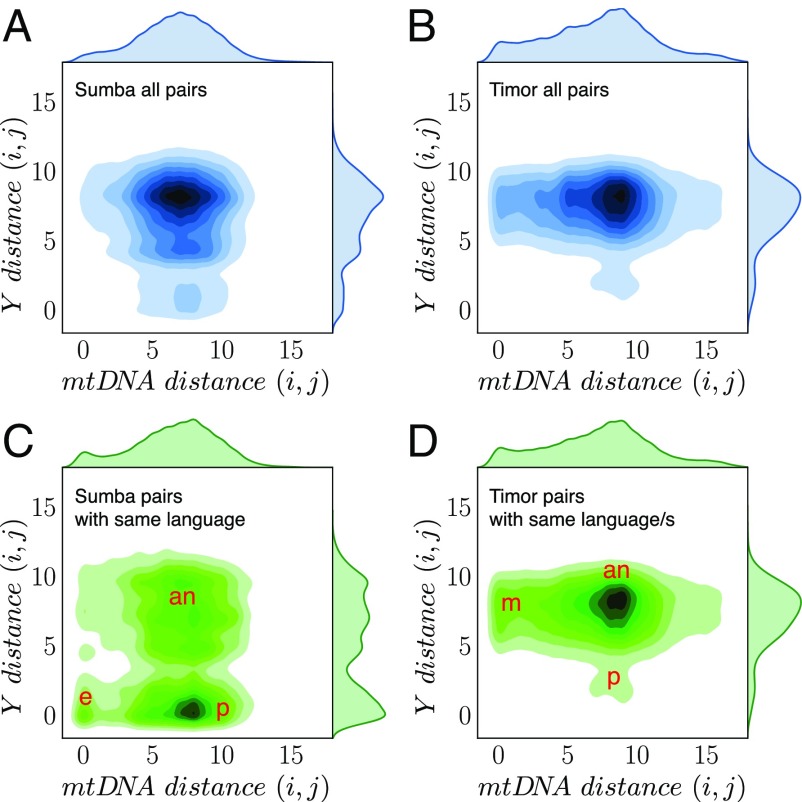
Fig. 4.
Genetic distances on Sumba (A and C) and Timor (B and D), between all pairs of individuals (A and B) and only between individuals who speak a common language (C and D). Conditioning on language sharing reveals three distinct clusters in the Sumba data (C), showing evidence of village endogamy (e), ambilocality or neolocality (an), and patrilocality (p). For Timor, the high degree of multilinguality means that most pairs of individuals in B are also included in D. B and D are dominated by matrilocality (m) and ambilocality or neolocality (an), with a small patrilocal cluster (p).
How do these patterns emerge, assuming a constant sex bias in migration rates? To find out, we used approximate Bayesian computation (ABC) rejection sampling to assess which IM model parameters closely matched the data. This returns the posterior distribution of female and male migration rates that best fit the observed data (Fig. 3 E and F, Insets). The inferred migration rate of the rarely dispersing sex was substantially lower than the migration rate of the other sex, in Sumba especially.
Kinship and Language Transmission.
Thus, the genetic evidence suggests that sex-biased migration rates on both islands are consistent with the observed kinship rules and have persisted for many generations. Do these kinship practices sustain the association between language and genes, and, in so doing, create persistent speech communities? To find out, we analyzed pairwise distance plots, weighted by the degree of language sharing between individuals. If languages are transmitted along uniparental clades, then genetic distances between pairs of individuals who speak a common language should enhance, and further reveal, clusters that reflect the expected sex-biased migration patterns. To test this expectation, we compared the pairwise distance plots weighted by degree of language sharing (Fig. 4 C and D) with the unweighted plots for all pairs of individuals (Fig. 4 A and B).
In Sumba, the signal of patrilocality was strongly enhanced ( of sampled pairs) when only individuals who share a language were considered (compare Fig. 4C with 4A). All of these villages are monolingual, and all but one language is found in only one village, the exception being Kambera, which is spoken in the two villages of Bilur Prangadu and Mbatakapidu. Consequently, most paired individuals who speak the same language come from the same village. While this may seem to be a limitation in the data, it was actually ideal for the purpose of distinguishing whether each man inherits his language from his patriline, matriline, or both. Comparing Fig. 4A (all pairs of Sumba men) with Fig. 4C (only men who share the same language), there is a strong trend for the male children of women who marry into a community to learn the language of that community. In other words, on Sumba, language is transmitted along male lines.
Timor differed from Sumba in two key ways: Our sample included a mix of nine matrilocal and two patrilocal villages, and multilinguality is common. Consequently, the resulting pairwise distances showed a more complex pattern of three clusters (Fig. 4D):
-
1.
A matrilocal cluster (m), comprising pairs of men closely related on the matriline;
-
2.
A weak patrilocal cluster (p), comprising pairs of men closely related on their patriline due to the two patrilocal villages in the sample; and
-
3.
A large ambilocal or neolocal cluster (an), comprising pairs of men less closely related on both the matriline and patriline. This likely reflects a real tendency to effectively ambilocal marriage, consistent with Fig. 3F ABC results.
Importantly, conditioning on the number of languages shared enlarged the matrilocal cluster from to (compare Fig. 4D with Fig. 4B), the converse pattern to Sumba.
We found that the extent of language sharing enhances the expected sex-biased migration patterns observed on genetic distances. To further test whether channels of kinship have guided the current of language evolution itself, we calculated the overall statistical cophylogenetic association between languages and either matrilineal or patrilineal genetic clades (Table 1). In patrilocal Sumba, a far stronger association was seen between genes and languages for Y (; bold type in Table 1). A similar pattern (stronger for Y) existed for the two patrilocal villages in the Timor sample, while the opposite pattern (stronger for mtDNA) was found for the matrilocal villages of Timor.
Table 1.
Z scores of gene–language associations
| Sumba | Timor | |||||
| Genetic locus | All | Matrilocal | Patrilocal | All | Matrilocal | Patrilocal |
| mtDNA | 8.32 | — | 8.32 | 5.43 | 6.62 | 1.01 |
| Y | 67.7 | — | 67.7 | 3.85 | 4.04 | 2.67 |
Comparing within each group of villages (columns), stronger gene–language associations (bold type) are found for the Y chromosome in all Sumba villages, mtDNA in all Timor villages, mtDNA in matrilocal Timor villages, and Y in patrilocal Timor villages.
Language Switching Between Genetic Clades.
We have seen that kinship creates channels for language transmission along uniparental clades and that this occurs over time scales that can structure language sharing between related individuals. Yet it remains unclear how long these associations persist. The phylogenetic trees for mtDNA and Y chromosome (Fig. 2) have roots in the very distant past, long before any conceivable relationship with the languages spoken today could have existed. Many of these genetic lineages are from the first settlers in the region, before Austronesian languages were introduced ∼5,000 y ago. Intriguingly, however, some information about the association between languages and genetic clades in the deep past is preserved by the branching points where languages either become attached to or leave genetic clades.
Using this information, we could estimate “host switching” probabilities for the observed gene–language tree associations (Fig. 5). We adapted this idea from cophylogenies in ecology, where host switching refers to movements of parasites between host species. Here, the hosts are people and languages are the parasites (17, 18). To estimate host-switching probabilities, we generated a stochastic model of language transmission along the branches of the gene tree (SI Appendix). We then performed a standard cophylogeny statistical test to examine the extent of congruence, if any, between the gene and language trees given the mappings simulated from the model.
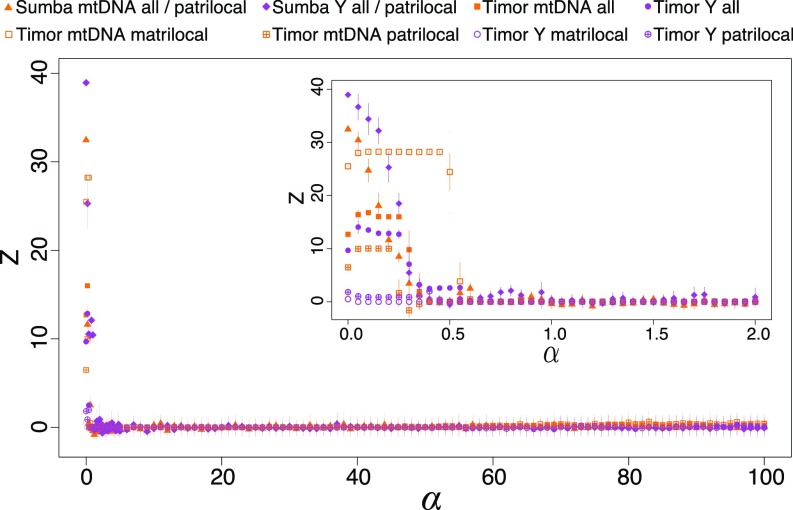
Fig. 5.
The Z score measure of association between gene and language phylogenies on Sumba and Timor for different language switching rates . Language switching rates below ∼0.5% per generation are required to generate the type of association between languages and clades observed in the empirical data (Table 1). All cases independently converge and abruptly lose gene–language associations, behaving similarly to randomized cases, when the language switch rate exceeds ∼0.5% per generation. Inset zooms in to show the fine scale of inferred host-switching rates.
We investigated eight cophylogenies separately, as indicated in Fig. 5. In each case, we found that low rates of host switching () were necessary to generate cophylogenies with strengths of association like those seen in the real data (Table 1). Language-switching rates were lower for the nondispersing sex. This was expected, as fewer opportunities exist for the nondispersing sex to be exposed to new languages, so the rate of language switching is lower. These low language-switching rates strengthen the association between languages and genes. Note, however, that neighboring villages often speak the same language, such that movement between villages may not trigger host switching. For the case of multilingual Timor, host switching means learning a new language while continuing to speak their original language with some probability.
Discussion
The mtDNA and Y chromosome phylogenies told us how closely the individuals in our sample are related, but provided no information about how those relationships came to be. However, because these individuals were sampled at the community scale, pairwise distances revealed sex-biased migration rates from which we could test inferences about their respective kinship systems.
Language added a further dimension. For each individual in our sample, we reconstructed two cophylogenies of language and genetic inheritance—for matrilineal and patrilineal inheritance, respectively. If there were no migration between villages, the signal of association in these cophylogenies would be identical because individuals would learn a single language spoken by both parents. However, if people sometimes marry into villages where a different language is spoken, the gene and language phylogenies will diverge. If many languages are spoken within a geographical region and rules of postmarital residence encourage sustained, directional, and biased population movement between speech communities, then languages will be channeled along uniparental lines.
This channeling creates distinctive patterns. Specifically, kinship practices channel both mtDNA lineages and languages in matrilocal communities, and Y chromosome lineages and languages in patrilocal communities. We found strong evidence for these patterns on both Sumba and Timor. In contrast, lineages on the autosomes—and, to a lesser extent, on the X chromosome—move into and out of both men and women at each generation. Thus, autosomes move between communities relatively independently of the kinship channels (19). Nuclear genes can flow unfettered through cultures: Cultures, not genes, are the stable systems.
The question of how long language transmission might have persisted along these uniparental lines can be addressed by comparing gene–language cophylogenies. In each generation, an opportunity exists to weaken or scramble the correlation between genetics and language through host switching, which occurs when children learn a different language than one of their parents. This can lead to three outcomes. In the absence of host switching, the emergence of new languages (branching in the language tree) can introduce persistent clades of related individuals who speak a common language, and hence extensive correlation between the gene and language trees. When host switching occurs only rarely, some correlation remains; however, frequent host switching and language losses quickly break down the correlation.
Intriguingly, host-switching rates converged to 0.3– per generation for all of the trees examined here. This translated to probability that a single host switch event would occur within a clade (i.e., the “half-life” of a language on a lineage) every 1,700–5,750 y, with near certainty of a host switch event occurring over much longer time frames. This raises the possibility that genetic clades could retain a shared language longer than any single language exists, by related individuals replacing one language with another together as a group.
Overall, we can infer that low rates of host switching captured the observed gene–language tree associations. To interpret this result, it was useful to distinguish between social communities and speech communities. For social communities, kinship rules persist long enough to leave clear traces in population genetic structure. In the 25 communities included in our study, core groups of close relatives must often have stayed together for generations, while simultaneously maintaining contact with neighboring groups with whom they intermarried. In this way, kinship systems directly shaped the language phylogeny over time: Consistently following a postmarital residence rule turned social communities into speech communities.
This was particularly clear on Sumba, where each village became a monolingual speech community. Conversely, most villages on Timor are not monolingual, but instead form multilingual speech communities. The likely explanation for multilinguality in central Timor is the political turmoil of the past century, which led to extensive local migration, as documented by Therik (20). It is noteworthy that the association of languages with genetic clades persists on Timor despite these recent population movements.
The low rates of past host switching were surprisingly similar for both islands, suggesting that kin-structured speech communities remain stable over long time scales. However, we also saw genetic evidence of ongoing contact between social communities that practice exogamous matrilocal or patrilocal marriage (as in ref. 21). In the past, contact between social communities like these would often have been synonymous with contact between speech communities.
Previous correlational studies of gene and language trees have focused on the effects of drift, geography (isolation by distance), and large-scale population movements to explain patterns of correlation at different scales of space and time (5, 22, 23). Our cophylogenetic analyses at the community scale clarify how kinship practices actively channel and continually renew language transmission, creating the observed patterns of language diversity and leaving a strong signal in population genetic structure. Analysis of gene–language cophylogenies and pairwise distances suggested that this channeling process usually persists long enough to be regarded as the norm. The Timor data made this point particularly clearly—beneath the surface variation of language diversity in communities created by recent population movements, enduring associations of language with uniparental genetic clades still persist.
Although our focus has been on the role of kinship in channeling language transmission, clearly, language also protects those channels—shared language helps connect and define matrilocal kin in the Wehali region of Timor, just as it connects and defines patrilocal kin on Sumba. Thus, while language and kinship are typically treated as unrelated subjects, their dynamic interaction in these villages has been fundamental to the structure of social life.
Materials and Methods
Genetic and Linguistic Data.
The assemblage of published genetic data (5, 7, 24–26) used in this study consists of mtDNA HVS-I sequences (positions 16,001–16,540 of the revised Cambridge Reference Sequence) and hierarchically screened SNPs on the Y chromosome, together with 14 Y chromosome STRs. The language(s) spoken by each individual were determined by asking their level of comprehension for each language recorded in their village.
We followed protocols for the protection of human subjects approved by the institutional review boards of the Eijkman Institute, Nanyang Technological University, and the University of Arizona. Informed consent was obtained from all study participants.
Probability of Shared Gene–Language Heritage.
The probability that a pair of individuals share a common language given that they belong to the same genetic clade at generation is given by
| [1] |
ABC Fitting of the IM Model with Kinship.
We adapted the IM model (16) by assuming that the evolution of the mtDNA and Y chromosome can be modeled independently, allowing male and female migration rates to vary. We fit the predictions of this model to the observed distribution of pairwise distances between mtDNA and Y chromosome samples using ABC. Further details are provided in SI Appendix.
Cophylogeny of Genes and Languages.
To test whether a significant association exists between the evolution of genes and languages, we used statistics that are functions of binary association link matrix that indicates which of the local languages each individual speaks, and the principal coordinates of the genetic and linguistic distances ( and ) (27). If genes and languages occupied corresponding positions in both phylogenetic trees with a significant degree of congruence, we rejected the global null hypothesis that their evolution has been independent. The fourth-corner statistics matrix (27) was used to calculate the congruence of the gene and language trees. From , we defined the global association . The score metric of the observed association between genes and languages was then calculated from the values of the original and permuted data.
Language Switching on Genetic Trees.
A plausible stochastic model of language transmission predicting the languages spoken at each gene tree branch was run forward in time over the trees by using two rules. First, where the language tree branches, all genetic clades speaking the ancestral language were randomly assigned one of the daughter languages. Second, a proportion of lineages switched to a new language at each generation. For multilingual Timor, a language switching event alternatively appended a language to the list with probability . Variants of this gene–language coevolution model yielded qualitatively similar results (see SI Appendix for details).
Supplementary Material
Acknowledgments
This study was supported by National Science Foundation Grant SES 0725470 and Singapore Ministry of Education Tier II Grant MOE2015-T2-1-127 (to J.S.L.). Indonesian samples were obtained by J.S.L. and H.S., with assistance from Golfiani Malik, Wuryantari Setiadi, Loa Helena Suryadi, Isabel Andayani, Gludhug Ariyo Purnomo, and Meryanne Tumonggor (Eijkman Institute for Molecular Biology, Jakarta), with the assistance of Indonesian Public Health clinic staff. Permission to conduct this research was granted to J.S.L. by the Indonesian Institute of Sciences and the Ministry for Research, Education and Technology. Additional Swadesh word lists for Sumba languages were provided by the National Language Center of the Indonesian Department of Education. M.P.C. was supported by Te Pūnaha Matatini and a fellowship from the Alexander von Humboldt Foundation. The Santa Fe Institute provided vital support via a working group dedicated to this project. Finally, we thank our anonymous reviewers for their thoughtful comments, including a particularly lovely turn of phrase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706416114/-/DCSupplemental.
References
- 1.Sapir E. Language: An Introduction to the Study of Speech. Harcourt, Brace & Co.; New York: 1949. [Google Scholar]
- 2.Fitch WT. Kin selection and ‘mother tongues’: A neglected component in language evolution. In: Oller DK, Griebel U, editors. Evolution of Communication Systems: A Comparative Approach. MIT Press; Cambridge, MA: 2004. pp. 275–296. [Google Scholar]
- 3.Tosi A. The notion of “community” in language maintenance. In: Extra G, Verhoeven L, editors. Bilingualism and Migration. Mouton de Gruyter; Berlin: 1999. pp. 325–343. [Google Scholar]
- 4.Barnard A. 2008. The co-evolution of language and kinship in Early Human Kinship: From Sex to Social Reproduction, eds Allen NJ, Callan H, Dunbar R, James W Speech (Blackwell Publishing Ltd., Oxford, UK), pp 232–243.
- 5.Lansing J, et al. Coevolution of languages and genes on the island of Sumba, eastern Indonesia. Proc Natl Acad Sci USA. 2007;104:16022–16026. doi: 10.1073/pnas.0704451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lansing J, et al. An ongoing Austronesian expansion in island Southeast Asia. J Anthropol Archaeol. 2011;30:262–272. [Google Scholar]
- 7.Guillot E, et al. Relaxed observance of traditional marriage rules allows social connectivity without loss of genetic diversity. Mol Biol Evol. 2015;32:2254–2262. doi: 10.1093/molbev/msv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balaresque P, Jobling MA. Human populations: Houses for spouses. Curr Biol. 2007;17:R14–R16. doi: 10.1016/j.cub.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 9.Murdock GP. Social Structure. Macmillan; London: 1949. [Google Scholar]
- 10.Barnard A. Kinship, language and production: A conjectural history of Khoisan social structure. Africa. 1988;58:29–50. [Google Scholar]
- 11.Cavalli-Sforza LL. Genes, peoples, and languages. Proc Natl Acad Sci USA. 1997;94:7719–7724. doi: 10.1073/pnas.94.15.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balanovsky O, et al. Parallel evolution of genes and languages in the Caucasus region. Mol Biol Evol. 2011;28:2905–2920. doi: 10.1093/molbev/msr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyer E, et al. Genetic diversity and the emergence of ethnic groups in Central Asia. BMC Genet. 2009;10:49. doi: 10.1186/1471-2156-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes SM, et al. Lack of gene–language correlation due to reciprocal female but directional male admixture in Austronesians and non-Austronesians of East Timor. Eur J Hum Genet. 2017;25:246–252. doi: 10.1038/ejhg.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forshee J. Review: Wehali – the female land: Traditions of a Timorese Ritual Centre, by Tom Therik. Indonesia. 2006;82:133–137. [Google Scholar]
- 16.Wilkinson-Herbots HM. The distribution of the coalescence time and the number of pairwise nucleotide differences in the “Isolation with Migration” model. Theor Popul Biol. 2008;73:277–288. doi: 10.1016/j.tpb.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Legendre P, Desdevises Y, Bazin E, Page RDM. A statistical test for host–parasite coevolution. Syst Biol. 2002;51:217–234. doi: 10.1080/10635150252899734. [DOI] [PubMed] [Google Scholar]
- 18.Hunley K. Reassessment of global gene–language coevolution. Proc Natl Acad Sci USA. 2015;112:1919–1920. doi: 10.1073/pnas.1425000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudjashov G, et al. Complex patterns of admixture across the Indonesian archipelago. Mol Biol Evol. 2017;34:2439–2452. doi: 10.1093/molbev/msx196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Therik T. Wehali – The Female Land: Traditions of a Timorese Ritual Centre. Pandanus Books: ANU research School of Pacific and Asian studies; Canberra, Australia: 2004. [Google Scholar]
- 21.Vallée F, Luciani A, Cox M. Reconstructing demography and social behavior during the Neolithic expansion from genomic diversity across island Southeast Asia. Genetics. 2016;204:1495–1506. doi: 10.1534/genetics.116.191379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creanza N, et al. A comparison of worldwide phonemic and genetic variation in human populations. Proc Natl Acad Sci USA. 2015;112:1265–1272. doi: 10.1073/pnas.1424033112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan FM, Gray RD, Greenhill SJ, Mace R. Matrilocal residence is ancestral in Austronesian societies. Proc R Soc B. 2009;276:1957–1964. doi: 10.1098/rspb.2009.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karafet T, et al. Major east-west division underlies Y chromosome stratification across Indonesia. Mol Biol Evol. 2010;27:1833–1844. doi: 10.1093/molbev/msq063. [DOI] [PubMed] [Google Scholar]
- 25.Tumonggor MK, et al. The Indonesian archipelago: An ancient genetic highway linking Asia and the Pacific. J Hum Genet. 2013;58:165–173. doi: 10.1038/jhg.2012.154. [DOI] [PubMed] [Google Scholar]
- 26.Tumonggor MK, et al. Isolation, contact and social behavior shaped genetic diversity in West Timor. J Hum Genet. 2014;59:494–503. doi: 10.1038/jhg.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dray S, Legendre P. Testing the species traits–environment relationships: The fourth-corner problem revisited. Ecology. 2008;89:3400–3412. doi: 10.1890/08-0349.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.