Significance
Nitric oxide synthase-2 (NOS2) and cyclooxygenase-2 (COX2) are inflammation-associated enzymes with oncogenic function in breast cancer. We show that crosstalk between NOS2/COX2 promotes aggressive phenotypes and that elevated coexpression of NOS2/COX2 in tumors predict significantly reduced patient survival (33%) when compared with 95% survival of estrogen receptor-negative patients with low NOS2/COX2 tumor expression. In addition, we have identified a tumor subtype-specific mechanism showing involvement of TNFα and/or endoplasmic reticulum stress as key players in this autocrine loop. Importantly, the simultaneous inhibition of NOS2/COX2 significantly reduced tumor growth in a xenograft murine model, suggesting that targeted inhibition of NOS2/COX2 may be therapeutically beneficial.
Keywords: breast cancer, nitric oxide, PGE2, NOS2, COX2
Abstract
Proinflammatory signaling pathways are commonly up-regulated in breast cancer. In estrogen receptor-negative (ER−) and triple-negative breast cancer (TNBC), nitric oxide synthase-2 (NOS2) and cyclooxygenase-2 (COX2) have been described as independent predictors of disease outcome. We further explore these findings by investigating the impact of their coexpression on breast cancer survival. Elevated coexpression of NOS2/COX2 proteins is a strong predictor of poor survival among ER− patients (hazard ratio: 21). Furthermore, we found that the key products of NOS2 and COX2, NO and prostaglandin E2 (PGE2), respectively, promote feed-forward NOS2/COX2 crosstalk in both MDA-MB-468 (basal-like) and MDA-MB-231 (mesenchymal-like) TNBC cell lines in which NO induced COX2 and PGE2 induced NOS2 proteins. COX2 induction by NO involved TRAF2 activation that occurred in a TNFα-dependent manner in MDA-MB-468 cells. In contrast, NO-mediated TRAF2 activation in the more aggressive MDA-MB-231 cells was TNFα independent but involved the endoplasmic reticulum stress response. Inhibition of NOS2 and COX2 using amino-guanidine and aspirin/indomethacin yielded an additive reduction in the growth of MDA-MB-231 tumor xenografts. These findings support a role of NOS2/COX2 crosstalk during disease progression of aggressive cancer phenotypes and offer insight into therapeutic applications for better survival of patients with ER− and TNBC disease.
Unresolved inflammation characterized by chronic activation of humoral immunity and infiltration of protumor polarized M2 macrophages and Th2 cells is associated with the promotion and disease progression of breast and other forms of cancer (1, 2). Chronic inflammation within the tumor microenvironment (TME) favors aberrant wound repair characterized by dysregulated release of cytokines, chemokines, and growth factors that the tumor exploits for survival and invasion (1, 3). This inflammation in the TME promotes cancer.
Earlier studies identified inducible nitric oxide synthase (NOS2) as a predictor of poor survival among ER-negative (ER−) breast cancer patients with no predictive effect among ER-positive (ER+) patients (4, 5). In ER− patients, NOS2 correlated with unfavorable prognostic biomarkers including mutant P53, CD31, CD44, basal-like breast cancer markers cytokeratin 5/6 and P-cadherin, and the Toll-receptor agonist S100A8 (4). These data suggest a role of elevated tumor NOS2 expression in tissue remodeling and neovascularization. Aberrant NOS2 has been associated with unique mechanisms to promote oncogenic pathways by redox modulation in a ligand-independent manner (6). Thus, NOS2, via NO signaling, is a unique multifunctional oncoprotein.
Another enzyme involved in cancer inflammation is cyclooxygenase-2 (COX2), which catalyzes the conversion of arachidonic acid to prostaglandins, including prostaglandin E2 (PGE2), which in turn can enhance the metastatic phenotype of breast tumors (7). Increased COX2 expression occurs early in breast cancer development and is detectable in ductal carcinoma in situ, invasive breast carcinoma, and metastatic lesions (8, 9). High expression of COX2 is associated with poor clinical survival in ER− as well as triple-negative breast cancer (TNBC) patients (10, 11). These findings strengthen the association between inflammation and ER− breast cancer through COX2 and NOS2, warranting a deeper understanding of the impact of coexpression on cancer progression and patient survival.
Here we show that high NOS2/COX2 in tumors is a strong predictor of poor survival. Furthermore, we show NOS2/COX2 crosstalk involves TNFα and endoplasmic reticulum stress (ERS) as key players in signaling pathways. Lastly, we demonstrate significant reductions in tumor growth by COX2/NOS2 inhibition using commercially available inhibitors in murine models of ER− breast cancer, suggesting that some patients may benefit from targeted NOS2/COX2 therapy.
Results
NOS2/COX2 Coexpression in ER− Tumors Predicts Poor Outcome.
Expression of NOS2/COX2 was determined immunohistochemically in 248 surgically resected tumors (4, 10). Demographic and clinicopathological features of patients were stratified by ER status (Table S1). Patients with ER− disease had significantly higher-grade tumors and more frequent p53 mutations. Neither COX2 nor NOS2 is differentially expressed between ER− and ER+ tumors. ER− disease is predictive of reduced survival, and patients are more often of African American descent, diagnosed at younger age, and have marked macrophage infiltration (CD68). Elevated tumor COX2/NOS2 expression is observed in 36% and 73% of ER− patients, respectively (Table S1). High COX2 in the absence of NOS2 is rare (3/91 tumors). In contrast, NOS2 was commonly overexpressed as a single marker (36/91 tumors).
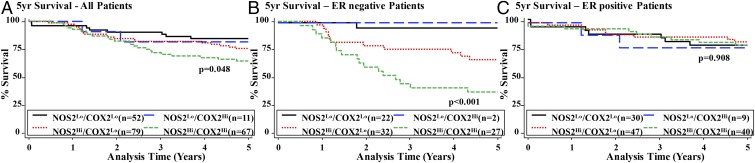
Table S2 shows that univariate analysis of both NOS2 and COX2 predicts poor outcome in ER− patients. NOS2 was a stronger predictor of 5-y survival than COX2. Table S3 shows that NOS2 and COX2 remain independent predictors of 5-y survival in multivariate Cox regression analysis, although the strength of association of NOS2 with outcome decreases, indicating a possible influence of COX2 on NOS2-related outcome. Accordingly, interaction analysis reveals a statistically significant interaction between NOS2 and COX2 on patient survival (P < 0.001). Therefore, the impact of NOS2/COX2 coexpression on patient outcome was explored. Fig. 1A shows the correlation of NOS2/COX2 expression with the 5-y survival rates of breast cancer patients. Stratification based on ER status showed that ER− patients with high levels of both NOS2 and COX2 had substantially poorer survival than those expressing only high NOS2, and the best outcome was seen in patients with low levels of both NOS2 and COX2 (Fig. 1B and Table S4). While no deaths occurred in ER− patients with NOS2lo/COX2hi-expressing tumors within the study interval, the number of patients in this group was limited (n = 3), making it difficult to infer the impact on patient outcome. However, data suggest that high expression of COX2 in the absence of NOS2 is rare.
Fig. 1.
Association between COX2 and NOS2 expression and breast cancer survival by ER status. (A) Kaplan–Meier cumulative breast cancer-specific 5-y survival curves of all breast cancer patients by COX2 and NOS2 status (n = 209); log-rank test: P = 0.048. (B) Kaplan–Meier cumulative breast cancer-specific 5-y survival curves of ER− patients by COX2 and NOS2 status (n = 83); log-rank test: P < 0.001. (C) Kaplan–Meier cumulative breast cancer-specific 5-y survival curves of ER+ patients by COX2 and NOS2 status (n = 126); log-rank test: P = 0.908.
Multivariate Cox regression analysis adjusting for age at diagnosis, race, tumor grade, tumor, node, and metastasis (TNM) stage, chemotherapy, and tumor p53 mutation status identified NOS2hi/COX2hi tumor expression as a predictor of a dismal survival [hazard ratio (HR) = 21.2, 95% CI 2.78–161.9, P = 0.003] in ER− disease (Table S4), with only 33% of patients surviving at 5 y. In contrast, the 5-y survival rate of ER− patients with NOS2lo/COX2lo tumor expression was >95%. Last, NOS2hi/COX2hi did not predict poor survival in ER+ patients (Fig. 1C and Table S4).
NOS2/COX2 Coexpression and Tumor Characteristics.
Earlier studies correlated high tumor NOS2 expression with disease grade (4). In the same cohort, COX2 coexpression status had no association with histologic grade in patients with NOS2hi/COX2lo tumors (91%) versus NOS2hi/COX2hi (83%), as summarized in Table S5. In contrast, increased p53 mutation frequency was observed in patients with a NOS2hi/COX2hi tumor signature (40%) compared with those with NOS2lo/COX2lo-expressing tumors (18%). Likewise expression of the p53 regulatory protein MDM2 was highest in NOS2hi/COX2hi (77%) and NOS2hi/COX2lo (58%) tumors but was lower in NOS2lo/COX2lo (32%) tumors. Also, NOS2hi/COX2hi and NOS2hi/COX2lo tumors tended to be high grade. NOS2hi/COX2hi patients also had elevated phosphorylated epidermal growth factor receptor levels (Table S5). These results suggest that histologic grade and MDM2 expression are most likely affected by NOS2 levels in the tumor, while COX2 may enhance NOS2-associated p53 mutation accumulation. Expression of CD68 (macrophage infiltration) and CD31 (microvessel density) markers showed similar effects, with NOS2 being the key inducer (Table S5).
NOS2/COX2 Coexpression and Akt/Caspase-9/BAD Signaling.
Positive correlations between COX2 expression and signaling via the Akt, caspase-9 (casp9), and BAD pathways were recently identified in ER− breast cancers (10). Casp9 and BAD are downstream targets of Akt, and phosphorylation blocks their apoptotic function. Table S5 shows that phosphorylated Akt (pAkt)-S473 is equally abundant in both NOS2hi/COX2lo (89%) and NOS2hi/COX2hi (87%) ER− tumors compared with NOS2lo/COX2lo lesions (64%), while pAkt-T308 is more abundant in NOS2hi/COX2hi (73%) than in NOS2hi/COX2lo (49%) tumors, with the lowest levels of staining in NOS2lo/COX2lo (33%) tumors. Downstream effectors of Akt signaling, pBAD and pCasp9, were more abundant in NOS2hi/COX2hi than in NOS2hi/COX2lo ER− tumors (Table S5). Multivariate logistic regression supported these results, showing a strong association between NOS2hi/COX2hi expression and Akt pathway activation independent of age at diagnosis and race (Table S5). The direction and magnitude of the adjusted odds ratios point to coexpression of COX2 enhancing the association of NOS2 with pAkt and downstream targets. Furthermore, statistically significant interactions between NOS2hi/COX2lo and pAkt-S473 (P < 0.001), and NOS2hi/COX2hi and pBAD-S136 (P < 0.001) predicted poor survival, further indicating a significant role of Akt signaling in the poor survival associated with NOS2hi/COX2hi-expressing ER− tumors. The NO-mediated role of pBAD was also verified in vitro (Fig. S1).
NOS2/COX2 Coregulation in ER− Breast Cancer.
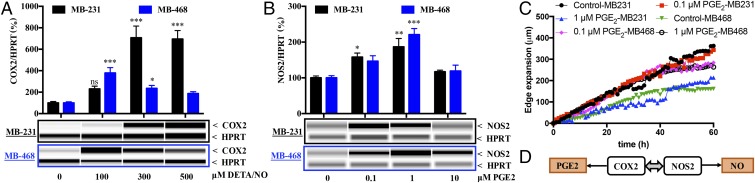
Multivariate analysis of ER− disease implicates a NOS2hi/COX2hi tumor signature as a strong predictor of poor survival. Breast cancer is a heterogeneous disease, with 15–20% of patients having TNBC. Among TNBC tumors, 79% were basal-like (12). Therefore, the NOS2/COX2 relationship was further examined in vitro in two widely used TNBC cells: (i) MDA-MB-468 (MB-468), an epithelium-like basal A breast cancer cell line, and (ii) MDA-MB-231 (MB-231), a more aggressive mesenchymal (claudin-low) basal B breast cancer cell line (13). These cell lines were chosen because they represent different breast cancer phenotypes and thus may provide mechanistic information regarding different tumor subtypes (13). MB-231 and MB-468 cells were treated with the NO donor diethylenetriamine-NO (DETA/NO) or PGE2. WES capillary electrophoresis and RT-PCR analysis revealed COX2 induction in both MB-231 and MB-468 cells after 24-h treatment with 100–500 μM DETA/NO (Fig. 2A and Fig. S2A). Elevated PGE2, reflective of increased COX2 enzymatic activity, was observed in the media of MB-231 cells treated with 300 μM DETA/NO, which was blocked by the COX inhibitor indomethacin (Fig. S2B). These results support NO induction of COX2 as a key regulatory pathway in the more aggressive MB-231 cells.
Fig. 2.
DETA/NO and PGE2 regulate COX2 and NOS2, respectively. MB-231 and MB-468 cells were treated for 24 h with different doses of (A) DETA/NO to determine effect on COX2 protein levels; or (B) PGE2 to determine effect on NOS2 protein levels. In A and B, error bars show SEM; *P < 0.05, **P < 0.01, ***P < 0.001; n ≥ 3; one-way ANOVA. (C) The effect of PGE2 on the migration of MB-468 and MB-231 cells. (D) Summary of data.
While elevated COX2 protein was also observed in MB-468 cells, there was no significant change in PGE2 level with DETA/NO even though the basal PGE2 production decreased in the presence of indomethacin (Fig. S2B). A role of the NO–cGMP pathway in COX2 expression was assessed using the soluble guanylyl cyclase (sGC) inhibitor 1H-(1,2,4)oxadiazolo(4,3-a)quinoxalin-1-one (ODQ). The induction of COX2 by DETA/NO was not affected by ODQ (Fig. S2C) and was thus NO-cGMP independent. Conversely, PGE2 induced NOS2 expression in both cell types (Fig. 2B). Previous studies indicated exogenous PGE2 increased in vitro cancer growth (14); the role of PGE2 in MB-231 and MB-468 cell migration in serum-free medium was examined. PGE2 increased the motility of MB-468 but not of MB-231 cells (Fig. 2C). These data demonstrate NOS2/COX2 crosstalk consistent with our clinical observations and suggest that COX2 expression is likely associated with greater tumor cell motility (Fig. 2D).
NOS2/COX2 Regulation and Cytokine Expression.
Several cytokines have been implicated in cancer progression. Increased levels of IL-6 and IL-8 in patient serum correlate with poor prognosis (15, 16). IL-8 is associated with enhanced tumor stem cell maintenance, survival, proliferation, and invasion as well as with increased angiogenesis and neutrophil infiltration within the TME (15, 16). Another important regulator of the inflammatory response is IL-6, which activates STAT-1 and STAT-3 during tumor progression (15, 16). STATs are elevated in multiple tumors, including breast cancer. Therefore, cytokine levels in cells treated with DETA/NO or PGE2 were examined. Both NO and PGE2 dose-dependently increased IL-6 production in MB-231 cells (Fig. S3 A and B). Consistent with earlier reports, NO induced IL-8 production after 24 h in MB-231 cells (4), while PGE2 had little or no effect (Fig. S3 C and D). A similar trend was observed for IL-6 and IL-8 in MB-468 cells; however, in MB-468 cells the effect of NO on IL-6 was significant only at 100 μM DETA/NO (Fig. S3 A–D). Further examination of NO and PGE2 on GM-CSF, an important immune modulator responsible for recruitment of myeloid cells to the TME (17), exhibited opposing effects (Fig. S3 E and F), suggesting a tight control of cytokine production by tumor cells.
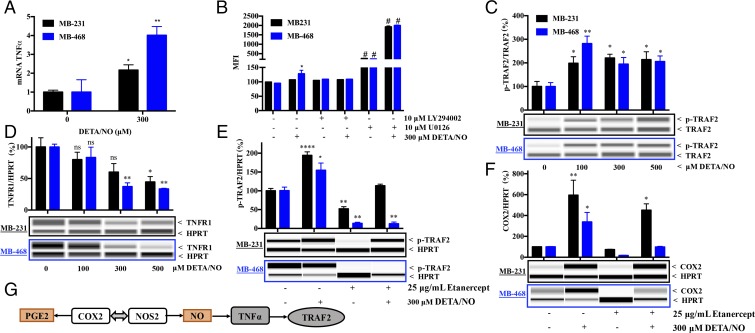
TNFα is an important inflammatory cytokine implicated in breast cancer progression (15, 16). Exposure to DETA/NO increased TNFα mRNA in both MB-231 and MB-468 cells (Fig. 3A). While there was no detectable TNFα in the medium, DETA/NO induced a mild increase in cell-surface–bound TNFα in MB-468 but not in MB-231 cells (Fig. 3B). Since the downstream effects of TNFα are mediated through PI3K and MEK signaling cascades, the NO-mediated effect on membrane-bound TNFα was further examined in the presence of the PI3K and MEK inhibitors LY294002 and U0126, respectively. In the absence of DETA/NO, MEK inhibition led to significant elevations in cell-surface–bound TNFα in both cell types, while PI3K inhibition had no effect. The addition of DETA/NO in the presence of U0126 further elevated cell-surface–bound TNFα, thereby suggesting a role for MEK signaling in the inhibition of NO-induced TNFα in both MB-231 and MB-468 cells (Fig. 3B).
Fig. 3.
NO-mediated signaling of COX2 is TNFα dependent. (A) TNFα mRNA levels induced in MB-231 and MB-468 cells by 300 μM DETA/NO. (B) Change in surface TNFα in MB-231 and MB-468 cells with 300 μM DETA/NO with or without the MEK inhibitor U0126 and the PI3K inhibitor LY294002. (C–F) The effect in MB-231 and MB-468 cells of NO on pTRAF2 (C) and TNFR1 (D) levels and on pTRAF2 (E) and COX2 (F) levels in the presence and absence of the TNFα antagonist etanercept. Error bars show SEM; ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and #P < 0.0001; n ≥ 3; one-way ANOVA. (G) Summary of data.
TRAF2 Mediates NOS2/COX2 Signaling.
Since TNFα signaling activates TNFα receptor-2 (TRAF2), an adaptor protein that regulates the activation of the c-Jun N-terminal kinase and IκB kinase (18), a role for NOS2/COX2 in TNFα-effector signaling was examined. Treatment of MB-231 and MB-468 cells with 100–500 μM DETA/NO increased intracellular pTRAF2 (Fig. 3C). Phosphorylation of TRAF2 was further examined in the presence or absence of the TNFα inhibitor etanercept. Etanercept abated NO-induced pTRAF2 and COX2 expression in MB-468 cells but not in MB-231 cells, suggesting a subtype-specific role of TNFα in COX2 regulation (Fig. 3 E and F).
TNFα signaling is mediated by two cell-surface receptors: TNFR1 mediates proinflammatory signaling, and TNFR2 mediates antiinflammatory pathways. Potential NO effects on TNFR1 and TNFR2 expression were examined. While TNFR2 was not detected, TNFR1 levels decreased with NO treatment in both MB-231 and MB-468 cells (Fig. 3D). This may explain the peak of pTRAF2 and COX2 at 100 µM DETA/NO in MB-468, as higher NO concentrations led to decreased TNFR1, lower pTRAF2, and less COX2 (Figs. 2A and 3C). While high NO concentrations also reduced TNFR1 expression in MB-231 cells, NO-mediated pTRAF2 and COX2 occur independently of TNFα interactions with its receptor (Fig. 3 E and F). Given that NO-induced COX2 in MB-231 cells is TNFα independent, exogenous TNFα is still capable of inducing COX2 in these cells (Fig. S4). These results suggest that in MB-468 cells NO induces autologous TNFα, which in turn activates pTRAF2, leading to COX2 expression (Fig. 3G). In contrast, in more aggressive MB-231 cells, COX2 expression is associated with TRAF2 activation in a TNFα-independent manner.
NO Increases TRAF2 Activation via ERS in MB-231 Cells.
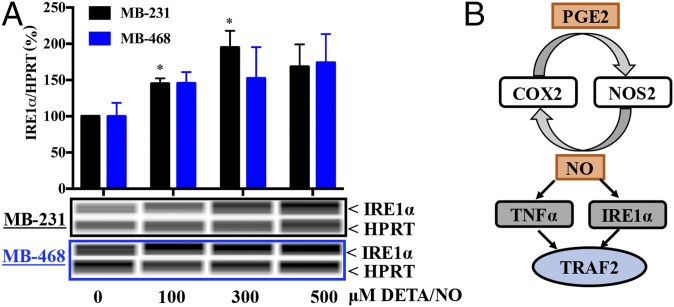
An earlier report showed NO-induced inositol-requiring protein-1 (IRE1α) in MB-231 cells that was abated by NOS2 inhibition (5). TRAF2 activation is induced by IRE1α in response to ERS. To extend these findings, we showed that NO induced IRE1α more effectively in MB-231 cells and less effectively in MB-468 cells. (Fig. 4). These results suggest two distinct TRAF2-activation pathways by NO: a TNFα-dependent mechanism in MB-468 cells and an ERS-dependent pathway in MB-231 cells.
Fig. 4.
The NO-mediated increase in pTRAF2 is mediated by ERS in the MB-231 cell line. (A) Cells were treated with different doses of DETA/NO, and IRE1α protein was measured in MB-231 and MB-468 cells. Error bars indicate SEM; *P < 0.05; n ≥ 3; one-way ANOVA. (B) Summary of data.
NO-Induced Activation of NFκB and AP1.
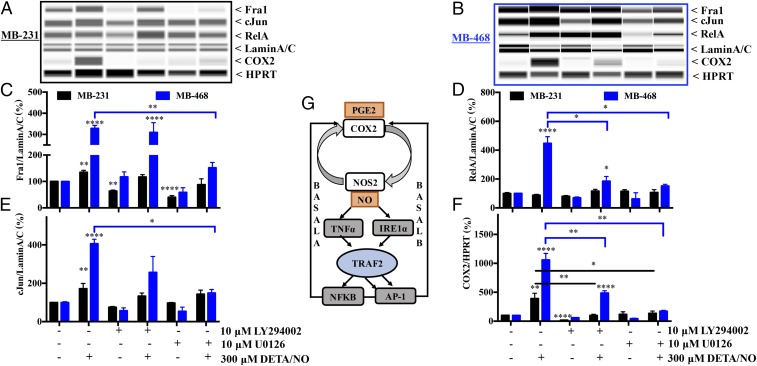
Classical TRAF2 signaling leads to the activation of AP1 and/or NFκB. COX2 and NOS2 promoters contain both NFκB- and AP1-binding sites, suggesting potential collaboration of these transcription factors. Canonical NFκB signaling involves a heterodimeric complex between RelA and p50. Next, nuclear RelA levels were examined in MB-231 and MB-468 cells exposed to NO. RelA was induced by 300 μM NO in MB-468 cells but not in MB-231 cells (Fig. 5 A, B, and D). Serum starvation also induces NOS2-derived NO; modulation of RelA by NO was verified in serum-starved MB-468 cells in the presence and absence of both the NOS inhibitor l-NAME (NG-nitro-l-arginine methyl ester) and DETA/NO (Fig. S5A). Exogenous TNFα also increased nuclear RelA in both MB-231 and MB-468 cells (Fig. S5B). These data suggest that NO-mediated TRAF2 is subtype specific, involving NFκB in the epithelial-like MB-468 cells but not in MB-231 cells.
Fig. 5.
NO-induced COX2 signaling is mediated by the PI3K and MEK pathways. (A–F) The expression of Fra1, cJun, and RelA proteins in the nucleus and COX2 expression in whole-cell lysate was measured after 24 h treatment with 300 μM DETA/NO alone or in the presence of 10 μM LY294002 or U0126 in MB-231 cells (A) and MB-468 cells (B), and their levels after 24 h of treatment were quantified (C–F). Error bars show SEM; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; n ≥ 3; one-way ANOVA. (G) Summary of data.
The AP1 transcription factor, a dimeric complex of members of Jun, Fos, ATF, and MAF family proteins, is also considered tumorigenic in some settings (19). Fra1, a member of the Fos family, is elevated in metastatic breast cancer (20). We found that NO increased cJun and Fra1 levels, suggesting a role for AP1 (Fig. 5 A–C and E). These results again emphasize the distinction of subtype-specific signaling responses within TME: AP1 is dominant in MB-231 cells, while both AP-1 and NFκB are involved in MB-468 cells.
NO-Mediated COX2 Expression Is MAPK and PI3K Dependent.
Specific NO levels activate the ERK/MAPK and/or PI3K/Akt pathways during tumor progression and promote poor disease-specific survival (4, 10, 21, 22). The role of these pathways was examined during the induction of COX2. Inhibition of these pathways abated COX2 induction by NO in MB-231 and MB-468 cells (Fig. 5 A, B, and F). Inhibition of the PI3K and MAPK pathways reduced basal levels but not NO induction of Fra1 in MB-231 cells. In MB-468 cells, abated ERK/MAPK signaling limited NO-induced Fra1 and cJun expression (Fig. 5 A–C and E). Similarly, blockade of either ERK/MAPK or PI3K/Akt signaling suppressed NO-mediated nuclear RelA translocation (Fig. 5 B and D).
Taken together, our in vitro studies bolster our immunohistochemistry results by directly establishing the cross-regulation between NOS2 and COX2. Furthermore, these data define critical roles for TNFα signaling and ERS in the regulation of COX2 and suggest AP-1 and NFκB as important components in this tumor subtype-specific process (Fig. 5G).
NOS2/COX2 Promotes Tumor Growth.
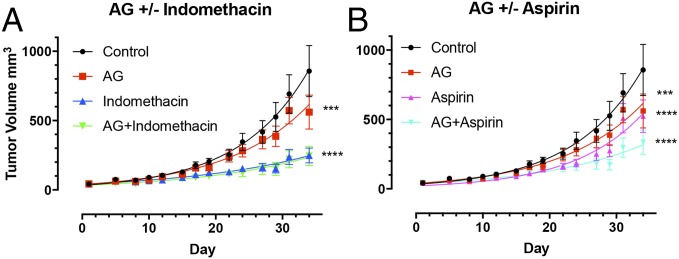
Next, the effect of NOS2/COX2 inhibition on tumor growth in MB-231-GFP xenografts was examined. Fig. 6 demonstrates significant reductions in tumor growth upon amino-guanidine (AG), aspirin, or indomethacin treatment and combination therapy. While the addition of AG to indomethacin had no additional benefit, an additive effect is clearly visible when AG is combined with aspirin therapy. After 34 d of treatment, AG+indomethacin and AG+aspirin suppressed tumor volume by 71% and 60%, respectively, compared with control. Examination of pTRAF2 in tumor xenografts revealed a decreasing trend in mice treated with both NOS2 and COX2 inhibitor (Fig. S6), thus supporting our in vitro data. Together these data demonstrate that simultaneous inhibition of NOS2/COX2 may be therapeutically beneficial in the treatment of ER− breast cancer subtypes.
Fig. 6.
NOS2/COX2 promotes tumor progression. Inhibition of NOS2 and COX2 in MB-231 xenograft model using AG and (A) indomethacin and (B) aspirin, respectively. Error bars show SEM; ***P < 0.001, ****P < 0.0001; two-way ANOVA, n ≥ 3.
Discussion
Breast cancer is a heterogeneous disease with distinct tumor subtypes; TNBC accounts for 15% of patients, is highly metastatic, and is resistant to therapy. NOS2 and COX2 are independent predictors of poor survival in ER− patients (4, 10). Recent evidence suggests a strong association between these two inflammation-associated enzymes in various epithelial cancers (23). Understanding their crosstalk through the elucidation of key molecular mechanisms is an important step toward the development of targeted therapeutics. The impact of coexpression of high NOS2/COX2 on ER− patient survival was investigated.
Our results indicate that ER− patients with NOS2hi/COX2hi tumors have a significantly greater risk of breast cancer-specific death than patients with NOS2lo/COX2lo tumors. The number of patients with NOS2lo/COX2hi tumors was very low, suggesting a critical role of NO in maintaining COX2 regulation in breast cancer. The NOS2/COX2 autocrine loop suggested by our histochemical analysis was further examined in vitro in mesenchymal-like basal B MB-231 and epithelium-like basal A MB-468 TNBC cells to better understand the cellular mechanisms in different progressive tumor subtypes. We found that NO-induced COX2 was sGC independent and that PGE2 induced NOS2 expression. The detrimental effects of PGE2 we define here may even be exacerbated by the known role of PGE2 in immunosuppression in the TME, making the environment more conducive for tumor growth. We suggest that this feed-forward amplification loop is critical in driving the severity of ER− disease. Substantial support for this notion comes from our finding that pharmacological inhibitors of NOS2 and COX2 significantly reduced tumor growth in a xenograft murine model.
The role of TME in establishing immunosuppressive conditions is well established. Tumor cells play a critical role in shaping the TME by secreting various cytokines and growth factors, thus continuously modulating its resistance to therapy and immune response. Proinflammatory cytokines and growth factors such as IL-6 and IL-8 have been established as important NO-induced biomarkers that are up-regulated in breast cancer patients’ serum and correlate with poor prognosis (15, 16). NO induced IL-8 production, a survival- and angiogenesis-promoting chemokine, and GM-CSF, a cytokine responsible for leukocyte recruitment and survival (15–17), while PGE2 was found to be an effective inducer of IL-6, an activator of MAPK, PI3K, and STAT3 associated with poor prognosis and cancer stemness (24). NO also induced TNFα in MB-468 cells. Several reports have shown elevated levels of TNFα in cancer patients (15, 16). The ability of NO and PGE2 to regulate different cytokines in TME further elucidates their crosstalk in the promotion of tumor growth and metastasis as well as in the escape from immune surveillance.
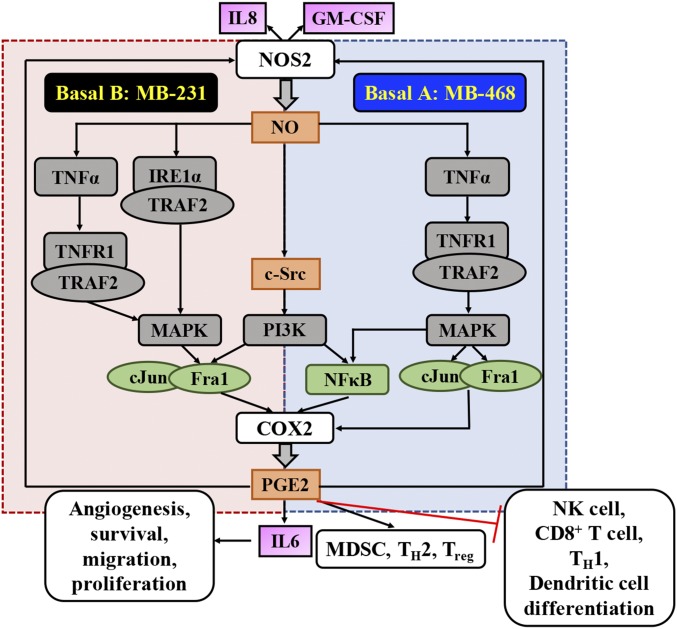
The cellular heterogeneity within and among different breast tumors poses a significant challenge in developing effective therapeutics, thus making it important to understand subtype-specific mechanisms. Thus, subtype-specific differences in cellular signaling were investigated. Our current data, when considered in context with our previous findings, support two distinct but overlapping signaling modules (Fig. 7). In MB-468 cells, NO induced TNFα, which in turn increased pTRAF2, resulting in the activation of both NFκB and AP1 and COX2 up-regulation. In contrast, in MB-231 cells, NO induced TNFα expression, but this cytokine was not required for the accumulation of pTRAF2. Instead, in agreement with earlier reports showing that NOS2 inhibition significantly reduces IRE1α and other ERS markers (5), we found that in MB-231 cells NO potently induced IRE1α, permitting TRAF2 phosphorylation even in the absence of TNFα. Notably, exogenous TNFα can directly induce COX2 in both cell types, demonstrating the power of TMEs rich in TNFα to bypass cell-subtype differences in these pathways. Lastly, we find a complex interaction between the PI3K and MEK signaling pathways in the regulation of COX2. NO is known to activate c-Src, resulting in PI3K activation (25), but its inhibition suppressed COX2 via different mechanisms in MB-231 and MB-468 cells. In MB-468 cells, PI3K inhibition suppresses NFκB activation with no effect on Fra1, whereas in MB-231 cells NFκB activation is absent, but abated PI3K signaling prevents Fra1 nuclear accumulation. The importance of NO in these regulatory pathways is underscored by previous reports of increased levels of Fra1 in breast cancers and its association with high metastatic potential (20). In addition to its role in the regulation of COX2, tumor PI3K likely plays a key role in the high levels of pBAD we detect in patients. Patients with NOS2hi/COX2hi tumors showed greater levels of both pAkt and pBAD than patients with NOS2hi/COX2lo tumors. Moreover, our in vitro data confirm the ability of NO to induce pBAD accumulation directly, regardless of tumor subtype. Interestingly, although PGE2 did not directly induce pBAD in vitro, our patient data suggest that COX2-mediated PGE2 may support pBAD accumulation in tumors with low levels of NO.
Fig. 7.
Summary of NOS2/COX2 crosstalk in ER− breast cancer.
Conclusions
Here we have identified a collaborative interaction between NOS2 and COX2 that leads to poor survival in ER− patients. NO induction of COX2 and PGE2 induction of NOS2 promote feed-forward signaling that involves numerous oncogenic pathways such as ERK, PI3K, NFκB, and AP1. These findings intrinsic to the tumor suggest that the participation of inflammatory factors such as TNFα and other cytokines that are found in the TME can have a major effect on survival. The complexity introduced by the heterogeneity of breast cancer is also shown here, with different signaling pathways as key players based on subtype. In basal A MB-468 cells NO induced TNFα, while in basal B MB-231 cells ERS was a key player; both led to TRAF2-mediated induction of COX2, thus offering insight into mechanisms relevant to different patient subtypes. Moreover, simultaneous inhibition of NOS2 and COX2 using inexpensive pharmaceutics significantly reduced tumor growth, thus suggesting NOS2 and COX2 as potential therapeutic targets in the treatment of aggressive breast cancer.
Materials and Methods
Tumor specimens were obtained from breast cancer patients recruited at the University of Maryland (UMD) Medical Center, the Baltimore Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and the Sinai Hospital in Baltimore between 1993 and 2003. Informed consent was obtained from all patients. The collection of tumor specimens, survey data, and clinical and pathological information (UMD protocol no. 0298229) was reviewed and approved by the UMD Institutional Review Board (IRB) for the participating institutions. The research was also reviewed and approved by the NIH Office of Human Subjects Research (OHSR no. 2248). Animal care was provided in accordance with the procedures outlined in the Guide for Care and Use of Laboratory Animals (26). The approval was obtained from NCI-Frederick IRB. A detailed description of materials and methods, including the assays and instrumentation used, animal model, and analysis methods, is provided in the SI Materials and Methods file.
Supplementary Material
Acknowledgments
This work was supported by the NIH Intramural Research Programs Cancer and Inflammation Program (D.A.W., D.B., M.G., V.S., J.H.N., R.Y.S.C., L.A.R., J.M.W., and D.W.M.), Optical Microscopy and Image Analysis Laboratory (D.A.S., W.F.H., and S.J.L.), and Laboratory of Human Carcinogenesis (S.A.); and by the National University of Ireland Galway and Breast Cancer Now Grant 2013MayPR019 (S.A.G., P.G., and A.E.R.). This project was funded in whole or in part with Federal funds from the National Cancer Institute, NIH, under Contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709119114/-/DCSupplemental.
References
- 1.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: Crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glynn SA, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest. 2010;120:3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granados-Principal S, et al. Inhibition of iNOS as a novel effective targeted therapy against triple-negative breast cancer. Breast Cancer Res. 2015;17:25. doi: 10.1186/s13058-015-0527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wink DA, et al. Nitric oxide and redox mechanisms in the immune response. J Leukoc Biol. 2011;89:873–891. doi: 10.1189/jlb.1010550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howe LR, et al. HER2/neu-induced mammary tumorigenesis and angiogenesis are reduced in cyclooxygenase-2 knockout mice. Cancer Res. 2005;65:10113–10119. doi: 10.1158/0008-5472.CAN-05-1524. [DOI] [PubMed] [Google Scholar]
- 8.Costa C, et al. Cyclo-oxygenase 2 expression is associated with angiogenesis and lymph node metastasis in human breast cancer. J Clin Pathol. 2002;55:429–434. doi: 10.1136/jcp.55.6.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeshita E, Osanai T, Higuchi T, Soumaoro LT, Sugihara K. Elevated cyclooxygenase-2 expression is associated with histological grade in invasive ductal breast carcinoma. J Med Dent Sci. 2005;52:189–193. [PubMed] [Google Scholar]
- 10.Glynn SA, et al. COX-2 activation is associated with Akt phosphorylation and poor survival in ER-negative, HER2-positive breast cancer. BMC Cancer. 2010;10:626. doi: 10.1186/1471-2407-10-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chikman B, et al. COX2 expression in high-grade breast cancer: Evidence for prognostic significance in the subset of triple-negative breast cancer patients. Med Oncol. 2014;31:989. doi: 10.1007/s12032-014-0989-1. [DOI] [PubMed] [Google Scholar]
- 12.Prat A, et al. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18:123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Zhang Y, Kim WJ, Daaka Y. PGE2 promotes renal carcinoma cell invasion through activated RalA. Oncogene. 2013;32:1408–1415. doi: 10.1038/onc.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vendramini-Costa DB, Carvalho JE. Molecular link mechanisms between inflammation and cancer. Curr Pharm Des. 2012;18:3831–3852. doi: 10.2174/138161212802083707. [DOI] [PubMed] [Google Scholar]
- 16.Esquivel-Velázquez M, et al. The role of cytokines in breast cancer development and progression. J Interferon Cytokine Res. 2015;35:1–16. doi: 10.1089/jir.2014.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong IS. Stimulatory versus suppressive effects of GM-CSF on tumor progression in multiple cancer types. Exp Mol Med. 2016;48:e242. doi: 10.1038/emm.2016.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackwell K, et al. TRAF2 phosphorylation modulates tumor necrosis factor alpha-induced gene expression and cell resistance to apoptosis. Mol Cell Biol. 2009;29:303–314. doi: 10.1128/MCB.00699-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eferl R, Wagner EF. AP-1: A double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 20.Dhillon AS, Tulchinsky E. FRA-1 as a driver of tumour heterogeneity: A nexus between oncogenes and embryonic signalling pathways in cancer. Oncogene. 2015;34:4421–4428. doi: 10.1038/onc.2014.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prueitt RL, et al. Inflammation and IGF-I activate the Akt pathway in breast cancer. Int J Cancer. 2007;120:796–805. doi: 10.1002/ijc.22336. [DOI] [PubMed] [Google Scholar]
- 22.Ridnour LA, et al. Nitric oxide synthase and breast cancer: Role of TIMP-1 in NO-mediated Akt activation. PLoS One. 2012;7:e44081. doi: 10.1371/journal.pone.0044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorokin A. Nitric oxide synthase and cyclooxygenase pathways: A complex interplay in cellular signaling. Curr Med Chem. 2016;23:2559–2578. doi: 10.2174/0929867323666160729105312. [DOI] [PubMed] [Google Scholar]
- 24.Kim SY, et al. Role of the IL-6-JAK1-STAT3-Oct-4 pathway in the conversion of non-stem cancer cells into cancer stem-like cells. Cell Signal. 2013;25:961–969. doi: 10.1016/j.cellsig.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Switzer CH, et al. S-nitrosylation of EGFR and Src activates an oncogenic signaling network in human basal-like breast cancer. Mol Cancer Res. 2012;10:1203–1215. doi: 10.1158/1541-7786.MCR-12-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Research Council . Guide for Care and Use of Laboratory Animals. National Academy Press; Washington, DC: 2011. [Google Scholar]
- 27.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis — Correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 28.Boersma BJ, et al. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J Natl Cancer Inst. 2006;98:911–919. doi: 10.1093/jnci/djj245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.