Significance
Captive rearing is known to impact the fitness of individuals released in the wild, but the relative role of genetic vs. nongenetic underlying processes is still debated. We measured genome-wide methylation profiles to document epigenetic differences between Pacific salmon originating from a hatchery and their natural-born congeners in two geographically distant rivers. Our results provide evidence that the epigenetic modifications induced by hatchery rearing provide a potential explanatory mechanism for reduced fitness of hatchery-reared salmon once released in the wild.
Keywords: epigenetics, methylation, coho salmon, hatchery, RAD sequencing
Abstract
Wild stocks of Pacific salmonids have experienced sharp declines in abundance over the past century. Consequently, billions of fish are released each year for enhancing abundance and sustaining fisheries. However, the beneficial role of this widely used management practice is highly debated since fitness decrease of hatchery-origin fish in the wild has been documented. Artificial selection in hatcheries has often been invoked as the most likely explanation for reduced fitness, and most studies to date have focused on finding signatures of hatchery-induced selection at the DNA level. We tested an alternative hypothesis, that captive rearing induces epigenetic reprogramming, by comparing genome-wide patterns of methylation and variation at the DNA level in hatchery-reared coho salmon (Oncorhynchus kisutch) with those of their wild counterparts in two geographically distant rivers. We found a highly significant proportion of epigenetic variation explained by the rearing environment that was as high as the one explained by the river of origin. The differentially methylated regions show enrichment for biological functions that may affect the capacity of hatchery-born smolts to migrate successfully in the ocean. Shared epigenetic variation between hatchery-reared salmon provides evidence for parallel epigenetic modifications induced by hatchery rearing in the absence of genetic differentiation between hatchery and natural-origin fish for each river. This study highlights epigenetic modifications induced by captive rearing as a potential explanatory mechanism for reduced fitness in hatchery-reared salmon.
A major question in captive breeding of plants and animals for conservation efforts is how to maintain the fitness of captive-bred individuals upon release into the wild (1–3). This question is central with respect to the objective of rehabilitating declining or threatened species (4–6). For salmonid species, change in fitness-related traits and gene expression has been reported to occur in a single generation of captivity in a hatchery environment (7–9). Such rapid changes may in turn lead to maladaptation in the natural environment (8). Most studies investigating the molecular basis for rapid change in fitness-related traits occurring in hatcheries have focused on finding signatures of selection at the genome level by identifying loci with a large effect (7, 10–13). Consequently, it still remains to be elucidated if such rapid selection on complex phenotypic traits would rather induce subtle changes in allele frequency over multiple loci (5, 14, 15). Similarly, the relative roles of the genetic vs. nongenetic underlying processes responsible for such phenotypic changes are also still debated.
Numerous wild stocks of anadromous salmon and trout (genus Oncorhynchus and Salmo) have experienced fluctuating abundance over the past century, with a series of sharp declines in abundance (16–18). As a consequence, conservation hatcheries have been flourishing, with the goal of preserving ecosystem integrity, enhancing declining populations, and sustaining fisheries. This situation is common along the North American Pacific coast where billions of salmonids, all species included, are released from hatcheries each year. Despite substantial improvement in production practices (see Supporting Information for details), the beneficial role of hatcheries in enhancing and restoring wild stocks is still debated because many studies have provided evidence for reduced fitness and maladaptation of hatchery fish when released in the wild (7, 9, 19–24). While some discrepancies may be observed between salmonid species (25), studies of coho salmon are concordant in showing that survival of hatchery-born fish compared with their wild counterparts is significantly reduced (20–22, 24). It has also been shown that the hatchery environment may affect a wide range of fitness-related traits, including reproductive success (represented by the number of eggs and the number of eggs surviving to hatch), swimming endurance (swimming time to fatigue), and predator avoidance (20). Although some studies have shown that selection induced by the hatchery environment was involved in such fitness impairment, they also have reported that different environmental conditions (e.g., fish density) may significantly modulate the extent of physiological acclimation to the hatchery environment (8, 20, 23, 26).
In the current study, we used a genome-wide sequencing approach to compare global patterns of genetic variation and methylation in white muscle tissue of hatchery-reared juvenile (smolt) coho salmon with those of their wild counterparts in two geographically distant rivers in British Columbia, Canada. Our results show that, despite a nonsignificant genetic difference between hatchery and wild salmon originating from the same river drainage, the hatchery environment induces hypermethylation for regions associated with ion homeostasis, synaptic and neuromuscular regulation, and immune and stress response, as well as control of swimming functions. This study highlights epigenetic modifications induced by captive rearing as a potential explanatory mechanism for reduced fitness previously reported in hatchery-produced coho salmon.
Results
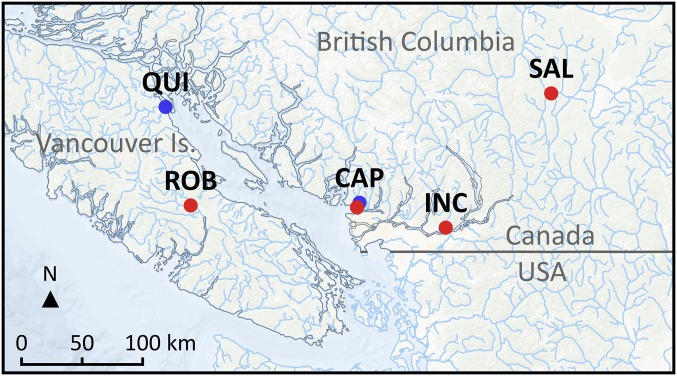
The white dorsal muscle was sampled at the same exact place on each fish to measure variation both at the genetic and epigenetic level between hatchery (HOR) and natural (NOR) origin fish on 40 juvenile (smolt) coho salmon originating from two geographically distant rivers in British Columbia (Canada): the Quinsam and the Capilano rivers (Fig. 1). HOR fish were produced from Salmon Enhancement Program (SEP) hatcheries following a primary production strategy (see Supporting Information for details). Fish were sampled either before release (May 15, 2014) or in a reservoir (May 23, 2014) for the Capilano HOR and NOR fish, respectively, and in the estuary (June 19, 2014) for both NOR and HOR individuals from the Quinsam River, ∼2 to 6 wk following the last production release (see Methods for more details).
Fig. 1.
Samples locations. Samples for epigenetic (methylation profiling) and population genomics analyses are shown in blue: CAP, Capilano River (n = 20), QUI, Quinsam River (n = 19). Samples for whole-genome resequencing (n = 20) are shown in red. CAP, Capilano (n = 5); INC, Inch Creek (n = 5); ROB, Robertson River (n = 5); SAL, Salmon River (n = 5). Resequencing samples were obtained from a hatchery located downstream of each river of origin, except Salmon River, for which the river of origin is represented while the fish were reared at Spius Creek hatchery, Meritt (British Columbia, BC, Canada).
Evidence for Parallel Epigenetic Modifications in Hatchery Environment.
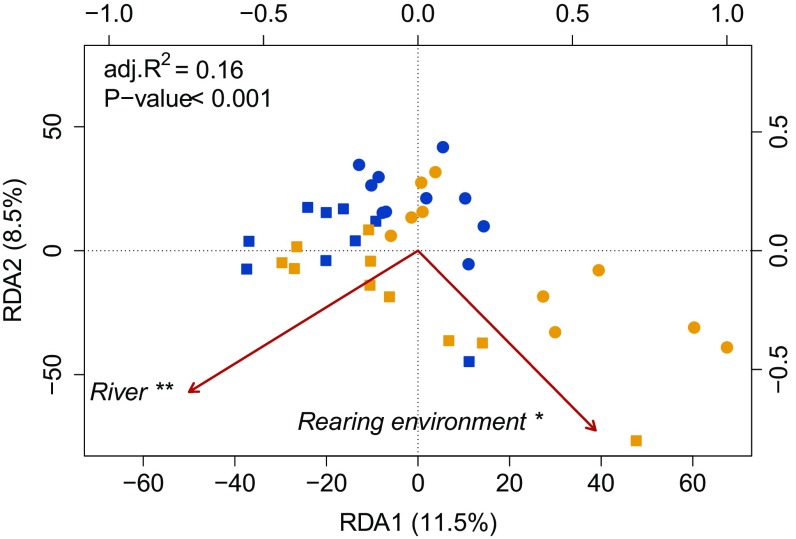
We used a tiling window approach to quantify the percentage of methylation over 1,000-bp regions throughout the genome masked for cytosine to thymine (C-T) polymorphism and retained only cytosines in a cytosine-phosphate-guanine (CpG) context for downstream analyses, as these regions represent the responsive methylation context in vertebrates. We used a distance-based redundancy analysis (db-RDA) to document methylation variation among HOR and NOR fish from both rivers, with river of origin, rearing environment, and sex as explanatory variables. The model was highly significant (P < 0.001) with an adjusted R2 of 0.16 (Fig. 2). Both river of origin and rearing environment were significant whereas no significant effect was detected for sex (Fig. 2). Partial db-RDAs revealed that the net variation explained by rearing environment [adjusted (adj.) R2 = 0.08; F = 4.34; P < 0.05] was identical to that explained by river of origin (adj. R2 = 0.08; F = 4.66; P < 0.01). This shared variation between HOR salmon from both rivers relative to their NOR congeners provides evidence for similar (parallel) epigenetic modifications induced by hatchery rearing.
Fig. 2.
Distance-base redundancy analysis (db-RDA) performed on the methylation data. A db-RDA for DNA methylation levels based on 131,807 1,000-bp sliding window regions for each individual. Symbols represent rivers: circle, Capilano; square, Quinsam. Colors represent rearing environment: blue, hatchery; yellow, wild. The db-RDA was globally significant and explained 16% of all DNA methylation regions variation (adj. R2 = 0.16). River of origin and rearing environment both significantly explained 8% of the variation after controlling for each other with subsequent partial db-RDAs. **P value < 0.01 and *P value < 0.05, related to the explanatory factors.
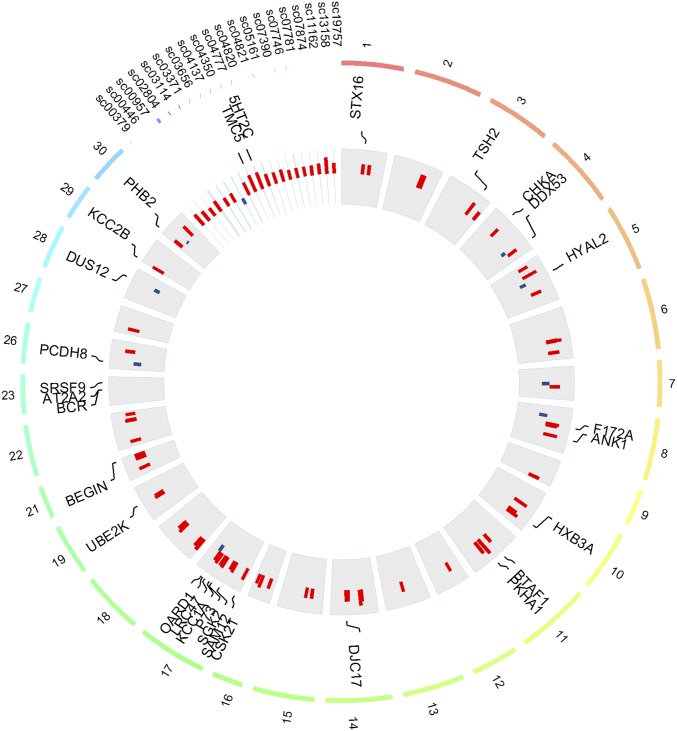
We identified differentially methylated regions (DMRs) (defined as having >15% overall difference; q-value < 0.001) (Supporting Information) between rearing environments, using a logistic regression, with sex and river of origin as covariates. We identified a total of 100 DMRs that were distributed among 27 chromosomes and 20 unmapped scaffolds (Fig. 3). The proportion of hypermethylated DMRs observed in both rivers was eight times greater in HOR relative to NOR salmon (89 vs. 11; χ2 = 60.84, df = 1, P < 0.001), suggesting a global pattern of down-regulation of genes associated with these DMRs in HOR salmon.
Fig. 3.
Circos plot of differentially methylated regions between hatchery and wild fish. Only the chromosomes (n = 27) and scaffolds (sc) (n =20) containing differentially methylated regions are plotted. Bar plots show the difference of methylation levels between hatchery and wild fish. Red bar plots represent hypermethylated regions in hatchery fish, and blue bar plots represent hypomethylated regions in hatchery fish. Only annotated regions (blastx e-value < 10−6) are represented.
Functional Annotation and Gene Ontology of DMRs.
We identified 37 DMRs out of 100 that overlapped 52 unique transcripts and regions comprising 5 kb upstream and downstream of these transcripts. A blastx approach successfully identified 29 unique Uniprot IDs, which again revealed an excess of hypermethylation in HOR relative to NOR fish (25 hypermethylated vs. 4 hypomethylated; χ2 = 15.21, df = 1, P < 0.001) (Fig. 3 and Table S1). These regions were mostly located within a gene body or in UTR regions, supporting their functional roles in gene expression regulation and/or splicing events (Table S1). Gene ontology (GO) analysis revealed an overrepresentation (P value < 0.05 and at least three genes by GO term) of modules associated with ion homeostasis (GO:0055080, cation homeostasis; GO:0042592, homeostatic process; GO:0043167, ion binding; GO:0055065, metal ion homeostasis). A previous study in the closely related rainbow trout (Oncorhynchus mykiss) showed that hatchery rearing negatively affects acclimation to seawater by reducing the specific activity of NA+ K+ ATPase, which resulted in lower survival following seawater transfer (27). We also observed a significant enrichment for functions associated with the immune response (GO:0031347, regulation of defense response; GO:0050727, regulation of inflammatory response; GO:0045321, leukocyte activation), as well as synaptic signal modulation and locomotion functions (GO:0099572, postsynaptic specialization; GO:0050885, neuromuscular process controlling balance). The neuromuscular process controlling balance includes the calcium/calmodulin-dependent protein kinase type II subunit beta (CAMK2B), a main actor of the neuromuscular communication and regulating Ca2+ signaling in skeletal muscle tissue (28), which was hypermethylated in HOR salmon. Its activation has also been associated, together with the Ca2+ signaling, with sustained and endurance muscle exercise in humans and the control of muscle development and excitation (29, 30). Lower critical swimming performance (Uct) has been documented in hatchery-reared coho salmon compared with their wild counterparts following transfer to seawater, and reduced average swimming speed has been documented in F1-hatchery smolts relative to wild smolts of Atlantic salmon (Salmo salar) and brown trout (Salmo trutta) (31, 32). The serotonin receptor 2C (HTR2C), which regulates appetite and feeding behavior (33), was also hypermethylated in HOR salmon. Finally, we observed a GO enrichment for transcription factors (GO:0006357, regulation of transcription from RNA polymerase II promoter), which comprised the TATA-binding protein-associated factor 172, also hypermethylated in HOR fish, which is involved in global transcription regulation. Genes under TATA box regulation are more able to respond rapidly to environmental stress, they show more variability in their expression range (phenotypic plasticity) compared with non-TATA regulated genes, and they account for the appearance of stress-induced phenotypes (34).
No Evidence for Genome-Wide Genetic Differentiation Between HOR and NOR Salmon.
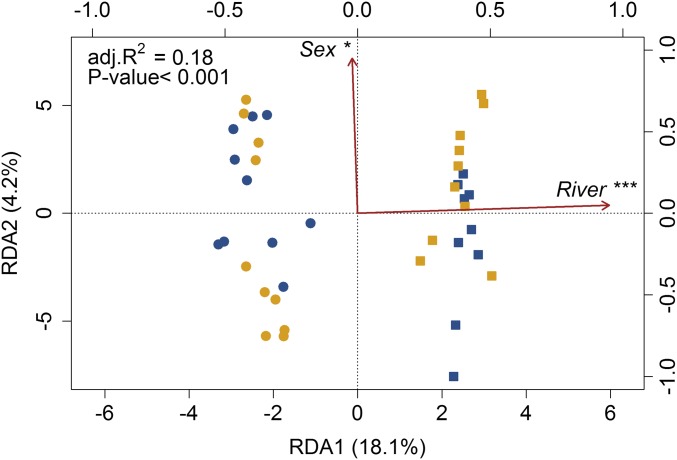
The principal coordinates analysis (PCoA) was performed on a Euclidean distance matrix of the 15,044 markers, and a db-RDA was produced on the genetic variation explained by these PCoA factors (response matrix), with river of origin, rearing environment, and sex as explaining variables. The model was highly significant (P < 0.001) with an adjusted R2 of 0.18 (Fig. 4). Both river of origin and sex were significant whereas no significant effect was detected for rearing environment (Fig. 4). No significant outlier with a genome-scan approach (Bayescan v2.0) (35) was detected between sexes (Fig. S1). Moreover, an analysis of molecular variance (AMOVA) revealed no significant genome-wide difference between HOR and NOR salmon [genetic differentiation (Fst) = 0.005 and 0.002, for Capilano River and Quinsam River populations, respectively; P > 0.05] while the net difference between rivers was highly significant (36) (mean Fst = 0.038 ± 0.003; P < 0.001) (Table S2). Additionally, heterozygosity and inbreeding values (Gis) were not significantly different between rivers or between HOR and NOR salmon (Table S3). No outlier [false discovery rate (FDR) > 0.05] was detected between HOR and NOR fish using Bayescan v2.0 (Fig. S2) whereas random forest identified 114 covarying markers, distributed over the 30 chromosomes. Nevertheless, permutations revealed that a similar pattern of apparent polygenic selection according to the distributions of the out-of-bag (OOB) errors could indeed be obtained by chance alone (Fig. S3). Population genomics analyses confirmed the prediction that HOR and NOR salmon belong to a single panmictic population within a given river. Our results cannot rule out that selection within one generation has caused changes in allele frequencies between HOR and NOR fish in genome regions that were not screened. Nevertheless, they indicate that such an effect would be modest relative to parallel differences observed at the epigenetic level.
Fig. 4.
Distance-base redundancy analysis (db-RDA) performed on the genetic data. The db-RDA performed on the total filtered 15,044 SNPs identified. Symbols represent rivers: circle, Capilano; square, Quinsam. Colors represent captivity treatment: blue, hatchery; yellow, wild. The db-RDA was globally significant and explained 18% of all SNPs variation (adj. R2 = 0.18). River of origin and sex explained significantly 16% and 2%, respectively, of the variation after controlling for each other with subsequent partial db-RDAs. ***P value < 0.001 and *P value < 0.5, related to the explanatory factors.
Discussion
The decline of many wild stocks of Pacific salmon encouraged the development of conservation hatcheries for enhancement. However, the hatchery environment during early life stages induces significant physiological and behavioral changes that may ultimately reduce the fitness of hatchery-born fish (25, 37). Hatchery fish have been shown to have higher reproductive success than their wild counterparts in hatchery conditions, but lower success when released in the wild with an accumulative impact over a generation, indicating inadvertent selection occurring after a single generation of hatchery rearing (8, 9, 37). Recent work provided evidence for a pronounced difference in gene expression between wild and hatchery fish after 1 y of captivity, despite no significant differences at the genome level (7). Similarly, a differential pattern of gene expression between domesticated and wild Atlantic salmon evolved in parallel in North America and Europe within five generations (10). Here, our results support the hypothesis that epigenetic modifications induced by hatchery rearing during early developmental stages may represent a potential explanatory mechanism for rapid change in fitness-related traits in juvenile salmon during their seaward migration. As such, our results are in line with accumulating evidence for epigenetic reprogramming caused by environmental conditions at a specific time that may induce phenotypic changes which may persist in subsequent life stages (38–41).
Strikingly, these parallel epigenetic modifications, mainly in the form of hypermethylation, were induced independently in two genetically distinct populations and in the apparent absence of overall neutral and adaptive variation between HOR and NOR salmon in these systems. As such, our results confirm that HOR and NOR coho salmon within a given river belong to a single panmictic population, as predicted based on the hatchery programs applied in these rivers. These so-called “integrated programs” are based on local populations and involve spawning in hatchery and natural environments. HOR and NOR fish in each river are not kept separate; thus HOR fish spawn in both the hatchery and the natural habitat as do NOR fish, which can maintain high gene flow in the whole system. Furthermore, no difference in genetic diversity (heterozygosity or inbreeding coefficient) was observed between HOR and NOR salmon, hence not supporting that there is no evidence for increased inbreeding depression in hatchery fish for the populations we studied. Finally, we found no evidence of either large effect or polygenic selection acting between hatchery and wild samples when using either a standard genome scan approach or a statistical framework appropriate for investigating the effect of weak selection in multiple regions of the genome. Therefore, our work corroborates a recent study on juvenile steelhead trout (Oncorhynchus mykiss), which showed that a single generation in captivity induced differences in the expression of hundreds of genes in offspring reared in identical environments, but produced from parents that experienced different rearing environments, in the absence of significant genetic differentiation (7). However, comparisons between both studies should be interpreted cautiously given the differences in experimental setting.
In contrast to the absence of significant genetic differences, our results revealed highly significant epigenetic differences between HOR and NOR salmon that were as pronounced as those observed between populations from different rivers. Our results differ from a study that compared hatchery-born and wild steelhead trout where no significant difference in methylation profiles was observed (38). However, rather than biological differences (differences among species and/or life stages), it is most likely that the absence of difference in levels of methylation originate from the lower resolution of the molecular technique available at that time. Indeed, the authors noted that they were only able to detect “all-or-nothing” changes (near 100% methylation) and thus could not rule out that moderate levels of differences could exist, which the reduced-representation bisulfite sequencing (RRBS) protocol allowed us to detect in our study. With an approach offering a substantial increase in genomic resolution, we found evidence for a highly significant effect of hatchery rearing on DNA methylation profiles across several regions of the coho salmon epigenome. Moreover, our results revealed that the same epigenetic modifications developed in parallel between the two independent study systems, mainly in the form of hypermethylation.
In Atlantic salmon, it has been shown that hatchery fish are not as efficient as wild fish in rapid seawater acclimation (27). In addition, acclimation to seawater induces profound, yet transient, changes in methylation levels in brown trout (Salmo trutta L.) (39). Here, we showed that genomic regions with differential methylation profiles between HOR and NOR salmon in both rivers were enriched for ion homeostasis and control of body fluid level functions, adding to growing evidence that hatchery rearing may affect the osmoregulatory process during smoltification. For instance, serine/threonine-protein kinase (SGK2), which was hypermethylated in HOR salmon, is a potent stimulator of epithelial Na+ channels (40). Similarly, seawater acclimation in the killifish (Fundulus heteroclitus) is associated with the level of SGK1 expression (no SGK2 or SGK3 ortholog present in the killifish genome) (41). Considering the fundamental role of these biological functions during smoltification (physiological acclimation to seawater) and seaward migration of juvenile salmonids (42), we propose that hatchery-induced epigenetic modifications during early developmental stages could be partly responsible for the saltwater acclimation deficiency reported in coho salmon (43). Moreover, neuromuscular communication, through regulation of Ca2+ levels, was among the biological functions showing the most pronounced pattern of hypermethylation in HOR salmon, including a major regulator of motoneuron signal transmission through Ca2+levels (CAMK2). This strongly suggests an alteration of the neuromuscular communication that could reduce swimming performance as documented in hatchery-reared coho salmon (31). It is noteworthy that a previous study in rainbow trout revealed differential levels of methylation in key genes responsible for muscle function, which could possibly impact swimming performance in this species (44).
In addition to functions that could impact swimming performance, it is noteworthy that, while we investigated white muscle tissue only, we observed an enrichment for overall synaptic signal control functions, which raises the hypothesis that the hatchery environment causes epigenetic modifications that could cause important physiological and endocrinal change in HOR salmon. In particular, the patterns of hypermethylation we observed at some major neurological regulators, such as HTR2C in HOR salmon, may play a role in the commonly reported behavioral differences between captive-reared and wild fish, such as increased aggressiveness, foraging, and boldness (30, 45–50). This hypothesis could be tested by comparing methylation profiles in the brain of fish with different aggressiveness, foraging, and boldness characteristics (50).
One methodological factor that may warrant a cautious interpretation of our results is that white muscle represents a mixed-cell tissue. Therefore, variable proportions of different cell types among individuals and/or different cell status could introduce biases in measures of methylation levels (51). However, the fact that we were still able to detect pronounced parallel changes in the form of hypermethylation between two independent systems makes our results conservative and suggests that possible biases would be inherent to biological differences between hatchery and wild individuals. Finally, because of the tissue specificity of the methylation patterns (52–54), further studies extending the experimentation on a broader range of tissues will be necessary for a more comprehensive characterization of the impact of hatchery rearing on salmon smolts.
The reduced genome representation method used here and the fact that we investigated only one tissue (yet representing 80% of young salmon body weight) resulted in only a partial coverage of all possible epigenetic differences that may exist between HOR and NOR coho salmon. Our results most likely underestimate the magnitude of epigenetic modifications incurred in the hatchery environment. Nevertheless, our results showed that hatchery-induced methylation changes happened in a similar manner in independent hatcheries, that these differences are much more pronounced than differences at the genome level, and that shared methylation changes persisted in the smolts of both rivers, despite the fact that they were exposed to different environments for several weeks after their release. It is therefore plausible that the hypermethylation of important physiological functions could have an immediate impact on the out-migrating smolt’s capability to acclimate to the freshwater–saltwater transition and the short-term survival at sea, which could ultimately impact the fitness of HOR fish. Moreover, based on previous studies, it is reasonable to hypothesize that hatchery-induced epigenetic modifications during early developmental stages (postfertilization and germ cell differentiation) could impact on lifelong phenotypic changes (55, 56). However, whether or not the epigenetic modifications that persisted for several weeks in smolts are maintained in returning adults and/or are inherited and acted upon by natural selection cannot be answered from our results.
Different practices in hatchery rearing are currently evaluated to circumvent the general observation that captive rearing reduces fitness in the wild. Alternative rearing practices may differ in environmental conditions (e.g., hatchery facilities or open lake), age at release (fry or smolt), or nutrition (supplemented or not by commercial food), which may significantly affect fish survival (25, 26, 39, 57, 58). The effect of such factors could also be detected at the epigenetic level (39). Clearly, improving our understanding of the dual role of genetic and nongenetic variation induced by captive rearing will contribute to the development of the best practices for the management and conservation of salmonids and numerous other species that are managed through supplementation worldwide (1).
Methods
Hatchery Procedures and Sampling.
The Salmon Enhancement Program (SEP) hatcheries have standard operating procedures employed across hatcheries, with the primary production strategy (PPS) being used for coho salmon at both Capilano and Quinsam hatcheries, British Columbia, Canada (see details in Supporting Information). Coho yearling smolts, defined as 1+ year after hatching, are released over a month. In this study, the progeny of fall-run 2012 Capilano and Quinsam River adult coho salmon were released in each respective river as yearling smolts in 2014. Capilano River coho salmon juveniles were collected in fresh water before production releases; the hatchery fish were collected at the hatchery on May 15, 2014 while the wild samples were caught via trap nets in the reservoir on May 23, 2014. These freshwater fish were classified as smolts as all physiological changes in preparation for saltwater had occurred, with minimal size differences in fork length or weight between the two groups: 116 mm and 16.9 g for hatchery and 111 mm and 13.7 g for wild individuals on average. Quinsam River smolts were collected via beach seine nets inside the Campbell River estuary, where the Quinsam River outflows to the sea, on June 19, 2014, ∼2 to 6 wk following the last production coho release from the hatchery. Hatchery fish were identified by their “marked” or clipped adipose fin while the wild samples were initially collected as “unmarked” coho and later confirmed as wild due to their lack of coded wire tag (CWT) detection and lack of an adipose fin clip. We collected a total of 40 coho salmon, including 10 juveniles from each river (smolt stage) reared in captivity in a local hatchery and 10 smolts born in the wild. Whole smolts were anesthetized, frozen on dry ice, transported to the Molecular Genetics Laboratory [Fisheries and Oceans Canada (DFO)] in Nanaimo, BC, and held at −80 °C until subsampled for analysis. Frozen white muscle sections were taken from whole smolts, ∼4 mm above to 4 mm below the lateral line, shipped on dry ice to Laval University, and subsampled for analysis. White muscle tissue was preferred because of its importance in both migration and homeostasis in fish (making up to 80% of the body weight) and because previous studies identified key markers linked to muscle development and activity as differentially methylated between migratory and nonmigratory ecotypes of rainbow trout (Oncorhynchus mykiss) (44, 59–62).
DNA Extraction and Reduced-Representation Bisulfite Sequencing Library Preparation.
The RRBS library preparation was adapted from a previously published protocol (63). Libraries were sequenced on a HiSEq. 2000 platform (five individuals by lane) at the McGill University and Génome Québec Innovation Centre (Montréal, QC) using a 100-bp single-end reads approach. In parallel, sex information was inferred by PCR using a method previously described for salmonids (sdY_E2S2 5′-GTGGAGTACTGCGAAGAGGAGGT-3′ and sdY_E2AS4 5′-CTTAAAACCACTCCACCCTCCAT-3′ primers) (64). Sex information for each individual is available in Table S4. Detailed methods are provided in Supporting Information.
Methylation Calling.
To avoid the possibility of falsely interpreting existing C-T DNA polymorphism as epigenetic variation, we masked these SNPs from the genome of the coho salmon (GenBank assembly accession no. GCA_002021735.1). We used Bismark v0.14.5 (65) and extracted only CpGs with sufficient coverage (≥10×). CpGs were assembled in 1,000-pb regions, and a logistic regression, with the river of origin and sex as covariates, was conducted to identify differentially methylated regions (DMRs) with the MethylKit R package (66). The DMRs were retained when showing at least 15% of difference between treatment, q-value < 0.001, and when a given 1,000-bp region comprised at least three CpGs. For functional annotation, we mapped the coho salmon transcriptome (67) to the genome (65) and annotated the DMRs overlapping genes location (5 kb up- and downstream) according to ref. 68. We added more information to DMRs relative position (3′ and 5′ UTRs, gene body, and CpG islands, shores, and shelves) based on a previous paper on rainbow trout (44). Detailed methods are provided in Supporting Information.
Population and Rearing Environment Effect on DMR Analysis.
We first computed a Euclidian distance matrix on the 131,807 regions and performed a principal coordinates analysis (PCoA). A distance-based redundancy analysis (db-RDA) was then produced with the retained PCo factors (n = 6) as the response matrix and the variables population, rearing environment, and sex as the explanatory matrix using a stepwise model selection. Partial db-RDAs were produced to test for the effect of the selected variables after controlling for the remaining variables. The effect of a given factor was considered significant when the P value was < 0.05. Detailed methods are provided in Supporting Information.
Genotyping for Genetic Data.
For population genomics analysis, mapping and genotyping were conducted with the BISulfite-seq CUI Toolkit (69). Only biallelic markers with minimum and maximum depth of coverage between 5× and 100×, minor allele frequency (maf) of >0.05, minimum quality of 5, maximum missing of 20%, and in Hardy–Weinberg equilibrium (P value > 0.05) were conserved. Markers with statistical linkage disequilibrium (LD) above R2 0.8 were also orphaned (one SNP dropped) (70). From the initial 12,375,758 SNPs, only 15,044 were retained for subsequent population genomics analysis after applying these filtering criteria. Detailed methods are provided in Supporting Information.
Genomic Differentiation Between Hatchery and Wild Origin Fish from Each River.
Similarly to DMR analysis, we computed a Euclidian distance matrix using the 15,044 filtered SNPs to perform a principal coordinates analysis (PCoA). A db-RDA was then produced with the retained PCo factors (n = 10) as the response matrix and the same explanatory variables, using a stepwise model selection. Partial db-RDAs were produced to test for the effect of the selected variables, after controlling for the other variables. The effect of a given factor was considered significant when the P value was <0.05. Pairwise genetic differentiation (Fst), individual coefficients of inbreeding (Gis), and observed and expected heterozygosity within samples were estimated using GENODIVE v2.0b27 (36) (Tables S2 and S3). Detailed methods are provided in Supporting Information. To detect outlier loci between sexes (Fig. S1) and test for possible selective effect within a single generation between HOR and NOR fish within each river (Fig. S2), we first conducted a standard genome scan approach using Bayescan v1.2 (35) on the 15,044 filtered markers. We also tested for polygenic selection using a multilocus analysis with Random Forest while accounting for population structure (rivers). We used permutations (n = 1,000) to assess whether a signal of polygenic selection similar to the one that was detected (Results) could be obtained by chance (e.g., due to genetic drift or sampling error). We compiled the final out-of-bag (OOB) error statistics for each run of simulation and compared it to the final OOB statistics in our empirical dataset (Fig. S3). Detailed methods are provided in Supporting Information.
Supplementary Material
Acknowledgments
We thank L. Benestan, O. Bichet, B. Bougas, B. Boyle, A.-M. Dion Côté, C. Hernandez, M. Krick, and E. Normandeau for laboratory and bioinformatics support. We also thank two anonymous referees for their constructive comments on a previous version of this manuscript. Computations were carried out on the supercomputer Colosse, Université Laval, managed by Calcul Québec and Compute Canada and on local servers (Katak). This research was carried out in conjunction with EPIC4 (Enhanced Production in Coho: Culture, Community, Catch), a project supported by the government of Canada through Genome Canada, Genome British Columbia, and Genome Quebec.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra/ (BioProject accession no. PRJNA389610).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711229114/-/DCSupplemental.
References
- 1.Laikre L, Schwartz MK, Waples RS, Ryman N. GeM Working Group Compromising genetic diversity in the wild: Unmonitored large-scale release of plants and animals. Trends Ecol Evol. 2010;25:520–529. doi: 10.1016/j.tree.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Stockwell CA, Hendry AP, Kinnison MT. Contemporary evolution meets conservation biology. Trends Ecol Evol. 2003;18:94–101. [Google Scholar]
- 3.Ford MJ. Selection in captivity during supportive breeding may reduce fitness in the wild. Conserv Biol. 2002;16:815–825. [Google Scholar]
- 4.Fraser DJ. How well can captive breeding programs conserve biodiversity? A review of salmonids. Evol Appl. 2008;1:535–586. doi: 10.1111/j.1752-4571.2008.00036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeaman S. Local adaptation by alleles of small effect. Am Nat. 2015;186(Suppl 1):S74–S89. doi: 10.1086/682405. [DOI] [PubMed] [Google Scholar]
- 6.Snyder NFR, et al. Limitations of captive breeding in endangered species recovery. Conserv Biol. 1996;10:338–348. [Google Scholar]
- 7.Christie MR, Marine ML, Fox SE, French RA, Blouin MS. A single generation of domestication heritably alters the expression of hundreds of genes. Nat Commun. 2016;7:10676. doi: 10.1038/ncomms10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie MR, Marine ML, French RA, Blouin MS. Genetic adaptation to captivity can occur in a single generation. Proc Natl Acad Sci USA. 2012;109:238–242. doi: 10.1073/pnas.1111073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araki H, Cooper B, Blouin MS. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science. 2007;318:100–103. doi: 10.1126/science.1145621. [DOI] [PubMed] [Google Scholar]
- 10.Mäkinen H, Vasemägi A, McGinnity P, Cross TF, Primmer CR. Population genomic analyses of early-phase Atlantic Salmon (Salmo salar) domestication/captive breeding. Evol Appl. 2015;8:93–107. doi: 10.1111/eva.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia JH, et al. Signatures of selection in tilapia revealed by whole genome resequencing. Sci Rep. 2015;5:14168. doi: 10.1038/srep14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, et al. A genome scan for selection signatures comparing farmed Atlantic salmon with two wild populations: Testing colocalization among outlier markers, candidate genes, and quantitative trait loci for production traits. Evol Appl. 2016;10:276–296. doi: 10.1111/eva.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai H-Y, et al. Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics. 2015;16:969. doi: 10.1186/s12864-015-2117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laporte M, et al. RAD sequencing reveals within-generation polygenic selection in response to anthropogenic organic and metal contamination in North Atlantic Eels. Mol Ecol. 2016;25:219–237. doi: 10.1111/mec.13466. [DOI] [PubMed] [Google Scholar]
- 15.Bourret V, Dionne M, Bernatchez L. Detecting genotypic changes associated with selective mortality at sea in Atlantic salmon: Polygenic multilocus analysis surpasses genome scan. Mol Ecol. 2014;23:4444–4457. doi: 10.1111/mec.12798. [DOI] [PubMed] [Google Scholar]
- 16.Noakes DJ, Beamish RJ, Kent ML. On the decline of Pacific salmon and speculative links to salmon farming in British Columbia. Aquaculture. 2000;183:363–386. [Google Scholar]
- 17.Irvine JR, Fukuwaka M-A. Pacific salmon abundance trends and climate change. ICES J Mar Sci. 2011;68:1122–1130. [Google Scholar]
- 18.Krkošek M, et al. Declining wild salmon populations in relation to parasites from farm salmon. Science. 2007;318:1772–1775. doi: 10.1126/science.1148744. [DOI] [PubMed] [Google Scholar]
- 19.Araki H, Schmid C. Is hatchery stocking a help or harm? Evidence, limitations and future directions in ecological and genetic surveys. Aquaculture. 2010;308(Suppl 1):S2–S11. [Google Scholar]
- 20.Chittenden CM, et al. Genetic versus rearing-environment effects on phenotype: Hatchery and natural rearing effects on hatchery- and wild-born coho salmon. PLoS One. 2010;5:e12261. doi: 10.1371/journal.pone.0012261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chittenden CM, et al. Riverine, estuarine and marine migratory behaviour and physiology of wild and hatchery-reared coho salmon Oncorhynchus kisutch (Walbaum) smolts descending the Campbell River, BC, Canada. J Fish Biol. 2008;72:614–628. [Google Scholar]
- 22.Irvine JR, O’Neill M, Godbout L, Schnute J. Effects of smolt release timing and size on the survival of hatchery-origin coho salmon in the Strait of Georgia. Prog Oceanogr. 2013;115:111–118. [Google Scholar]
- 23.Evans ML, Wilke NF, O’Reilly PT, Fleming IA. Transgenerational effects of parental rearing environment influence the survivorship of captive-born offspring in the wild. Conserv Lett. 2014;7:371–379. [Google Scholar]
- 24.Zimmerman MS, Irvine JR, O’Neill M, Anderson JH, Greene CM. Spatial and temporal patterns in smolt survival of wild and hatchery coho salmon in the Salish sea. Mar Coast Fish. 2015;7:116–134. [Google Scholar]
- 25.Christie MR, Ford MJ, Blouin MS. On the reproductive success of early-generation hatchery fish in the wild. Evol Appl. 2014;7:883–896. doi: 10.1111/eva.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berejikian BA, et al. Rearing strategies alter patterns of size-selective mortality and heritable size variation in steelhead trout (Oncorhynchus mykiss) Can J Fish Aquat Sci. 2016;74:273–283. [Google Scholar]
- 27.McDonald DG, et al. Condition and performance of juvenile Atlantic salmon (Salmo salar): Effects of rearing practices on hatchery fish and comparison with wild fish. Can J Fish Aquat Sci. 1998;55:1208–1219. [Google Scholar]
- 28.Martinez-Pena y Valenzuela I, Mouslim C, Akaaboune M. Calcium/calmodulin kinase II-dependent acetylcholine receptor cycling at the mammalian neuromuscular junction in vivo. J Neurosci. 2010;30:12455–12465. doi: 10.1523/JNEUROSCI.3309-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose AJ, Kiens B, Richter EA. Ca2+-calmodulin-dependent protein kinase expression and signalling in skeletal muscle during exercise. J Physiol. 2006;574:889–903. doi: 10.1113/jphysiol.2006.111757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose AJ, Frøsig C, Kiens B, Wojtaszewski JFP, Richter EA. Effect of endurance exercise training on Ca2+ calmodulin-dependent protein kinase II expression and signalling in skeletal muscle of humans. J Physiol. 2007;583:785–795. doi: 10.1113/jphysiol.2007.138529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brauner CJ, Iwama GK, Randall DJ. The effect of short-duration seawater exposure on the swimming performance of wild and hatchery-reared juvenile coho salmon (Oncorhynchus kisutch) during smoltification. Can J Fish Aquat Sci. 1994;51:2188–2194. [Google Scholar]
- 32.Pedersen L-F, Koed A, Malte H. Swimming performance of wild and F1‐hatchery‐reared Atlantic salmon (Salmo salar) and brown trout (Salmo trutta) smolts. Ecol Freshwat Fish. 2008;17:425–431. [Google Scholar]
- 33.Pérez Maceira JJ, Mancebo MJ, Aldegunde M. The involvement of 5-HT-like receptors in the regulation of food intake in rainbow trout (Oncorhynchus mykiss) Comp Biochem Physiol C Toxicol Pharmacol. 2014;161:1–6. doi: 10.1016/j.cbpc.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Roelofs D, Morgan J, Stürzenbaum S. The significance of genome-wide transcriptional regulation in the evolution of stress tolerance. Evol Ecol. 2010;24:527–539. [Google Scholar]
- 35.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meirmans PG, Van Tienderen PH. Genotype and genodive: Two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes. 2004;4:792–794. [Google Scholar]
- 37.Araki H, Berejikian BA, Ford MJ, Blouin MS. Fitness of hatchery-reared salmonids in the wild. Evol Appl. 2008;1:342–355. doi: 10.1111/j.1752-4571.2008.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blouin MS, et al. No evidence for large differences in genomic methylation between wild and hatchery steelhead (Oncorhynchus mykiss) Can J Fish Aquat Sci. 2010;67:217–224. [Google Scholar]
- 39.Morán P, Marco-Rius F, Megías M, Covelo-Soto L, Pérez-Figueroa A. Environmental induced methylation changes associated with seawater adaptation in brown trout. Aquaculture. 2013;392–395:77–83. [Google Scholar]
- 40.Friedrich B, et al. The serine/threonine kinases SGK2 and SGK3 are potent stimulators of the epithelial Na+ channel αhe serine. Pflugers Arch. 2003;445:693–696. doi: 10.1007/s00424-002-0993-8. [DOI] [PubMed] [Google Scholar]
- 41.Shaw J, et al. The role of SGK and CFTR in acute adaptation to seawater in Fundulus heteroclitus. Cell Physiol Biochem. 2008;22:69–78. doi: 10.1159/000149784. [DOI] [PubMed] [Google Scholar]
- 42.Boeuf G. 1993. Salmonid smolting: A pre-adaptation to the oceanic environment. Fish Ecophysiology, Fish and Fisheries Series, eds Rankin JC, Jensen FB (Chapman & Hall, London), pp 105–135.
- 43.Shrimpton JM, Bernier NJ, Iwama GK, Randall DJ. Differences in measurements of smolt development between wild and hatchery-reared juvenile coho salmon (Oncorhynchus kisutch) before and after saltwater exposure. Can J Fish Aquat Sci. 1994;51:2170–2178. [Google Scholar]
- 44.Baerwald MR, et al. Migration-related phenotypic divergence is associated with epigenetic modifications in rainbow trout. Mol Ecol. 2016;25:1785–1800. doi: 10.1111/mec.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olla BL, Davis MW, Ryer CH. Understanding how the hatchery environment represses or promotes the development of behavioral survival skills. Bull Mar Sci. 1998;62:531–550. [Google Scholar]
- 46.Brown C, Laland K. Social learning and life skills training for hatchery reared fish. J Fish Biol. 2001;59:471–493. [Google Scholar]
- 47.Álvarez D, Nicieza AG. Predator avoidance behaviour in wild and hatchery-reared brown trout: The role of experience and domestication. J Fish Biol. 2003;63:1565–1577. [Google Scholar]
- 48.Metcalfe NB, Valdimarsson SK, Morgan IJ. The relative roles of domestication, rearing environment, prior residence and body size in deciding territorial contests between hatchery and wild juvenile salmon. J Appl Ecol. 2003;40:535–544. [Google Scholar]
- 49.Sundström LF, Johnsson JI. Experience and social environment influence the ability of young brown trout to forage on live novel prey. Anim Behav. 2001;61:249–255. doi: 10.1006/anbe.2000.1593. [DOI] [PubMed] [Google Scholar]
- 50.Aubin-Horth N, Landry CR, Letcher BH, Hofmann HA. Alternative life histories shape brain gene expression profiles in males of the same population. Proc Biol Sci. 2005;272:1655–1662. doi: 10.1098/rspb.2005.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston IA, Bower NI, Macqueen DJ. Growth and the regulation of myotomal muscle mass in teleost fish. J Exp Biol. 2011;214:1617–1628. doi: 10.1242/jeb.038620. [DOI] [PubMed] [Google Scholar]
- 52.Morán P, Pérez-Figueroa A. Methylation changes associated with early maturation stages in the Atlantic salmon. BMC Genet. 2011;12:86. doi: 10.1186/1471-2156-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venney CJ, Johansson ML, Heath DD. Inbreeding effects on gene-specific DNA methylation among tissues of Chinook salmon. Mol Ecol. 2016;25:4521–4533. doi: 10.1111/mec.13777. [DOI] [PubMed] [Google Scholar]
- 54.Feil R, Fraga MF. Epigenetics and the environment: Emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 55.Johnson LJ, Tricker PJ. Epigenomic plasticity within populations: Its evolutionary significance and potential. Heredity (Edinb) 2010;105:113–121. doi: 10.1038/hdy.2010.25. [DOI] [PubMed] [Google Scholar]
- 56.Faulk C, Dolinoy DC. Timing is everything: The when and how of environmentally induced changes in the epigenome of animals. Epigenetics. 2011;6:791–797. doi: 10.4161/epi.6.7.16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fast DE, et al. A synthesis of findings from an integrated hatchery program after three generations of spawning in the natural environment. N Am J Aquac. 2015;77:377–395. [Google Scholar]
- 58.Tatara CP, et al. Age and method of release affect migratory performance of hatchery Steelhead. N Am J Fish Manag. 2017;37:700–713. [Google Scholar]
- 59.Parry G. Osmotic and ionic changes in blood and muscle of migrating salmonids. J Exp Biol. 1961;38:411–427. [Google Scholar]
- 60.Björnsson BT, Stefansson SO, McCormick SD. Environmental endocrinology of salmon smoltification. Gen Comp Endocrinol. 2011;170:290–298. doi: 10.1016/j.ygcen.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 61.Stefansson SO, et al. Growth, osmoregulation and endocrine changes in wild Atlantic salmon smolts and post-smolts during marine migration. Aquaculture. 2012;362:127–136. [Google Scholar]
- 62.Wilke NF, O’Reilly PT, MacDonald D, Fleming IA. Can conservation-oriented, captive breeding limit behavioural and growth divergence between offspring of wild and captive origin Atlantic salmon (Salmo salar)? Ecol Freshwat Fish. 2015;24:293–304. [Google Scholar]
- 63.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–481. doi: 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- 64.Yano A, et al. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol Appl. 2013;6:486–496. doi: 10.1111/eva.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akalin A, et al. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J-H, Leong JS, Koop BF, Devlin RH. Multi-tissue transcriptome profiles for coho salmon (Oncorhynchus kisutch), a species undergoing rediploidization following whole-genome duplication. Mar Genomics. 2016;25:33–37. doi: 10.1016/j.margen.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 68.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou W. 2016 biscuit-0.1.3. Zenodo. Available at doi.org/10.5281/zenodo.48262. Accessed October 3, 2016.
- 70.Larson WA, et al. Genotyping by sequencing resolves shallow population structure to inform conservation of Chinook salmon (Oncorhynchus tshawytscha) Evol Appl. 2014;7:355–369. doi: 10.1111/eva.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.