Significance
Breast cancer is one the most common cancers and causes of cancer-related death worldwide. Tumor recurrence following therapy is attributed to a subset of tumor-initiating cells (TICs) with stem cell (SC) properties. Similar to normal adult SCs that drive tissue regeneration, TICs regenerate tumors after treatment and thereby enable dissemination throughout the body. Thus, a better appreciation of the mechanisms that induce and maintain SCs in normal and neoplastic tissues is critical for understanding normal tissue regeneration and improving breast cancer therapy. Here we show that key developmental epithelial–mesenchymal transition programs promote stemness of mammary SCs by inducing primary ciliogenesis and Hedgehog signaling. These results provide insights into intraepithelial and intratumoral heterogeneity that have an impact on certain breast cancers.
Keywords: EMT, primary cilia, hedgehog, stemness
Abstract
Tissue regeneration relies on adult stem cells (SCs) that possess the ability to self-renew and produce differentiating progeny. In an analogous manner, the development of certain carcinomas depends on a small subset of tumor cells, called “tumor-initiating cells” (TICs), with SC-like properties. Mammary SCs (MaSCs) reside in the basal compartment of the mammary epithelium, and their neoplastic counterparts, mammary TICs (MaTICs), are thought to serve as the TICs for the claudin-low subtype of breast cancer. MaSCs and MaTICs both use epithelial–mesenchymal transition (EMT) programs to acquire SC properties, but the mechanism(s) connecting EMT programs to stemness remain unclear. Here we show that this depends on primary cilia, which are nonmotile, cell-surface structures that serve as platforms for receiving cues and enable activation of various signaling pathways. We show that MaSC and MaTIC EMT programs induce primary cilia formation and Hedgehog (Hh) signaling, which has previously been implicated in both MaSC and MaTIC function. Moreover, ablation of these primary cilia is sufficient to repress Hh signaling, the stemness of MaSCs, and the tumor-forming potential of MaTICs. Together, our findings establish primary ciliogenesis and consequent Hh signaling as a key mechanism by which MaSC and MaTIC EMT programs promote stemness and thereby support mammary tissue outgrowth and tumors of basal origin.
The adult mammary gland is a regenerative, branching ductal tissue that is formed by a stratified epithelium comprised of luminal and basal cells surrounded by a basement membrane, these being embedded in stroma (1). Mammary gland development begins in midembryogenesis. During puberty, hormonal stimuli trigger the postnatal mammary stem cells (MaSCs) to proliferate and generate the complex ductal structure of the adult mammary gland (2). Previous work combining FACS, mammosphere/organoid assays, and transplantation experiments has shown that adult gland formation is supported by bipotent MaSCs that reside in the basal compartment of the mammary epithelium (3–8). More recently, in vivo lineage-tracing studies confirmed the existence of basal bipotent MaSCs in addition to long-lived unipotent stem/progenitor cells that actively participate in tissue regeneration (1, 9–11).
Molecular analyses have identified Hedgehog (Hh) signaling as a key player regulating MaSC biology (12, 13). Three mammalian Hh ligands, Sonic (Shh), Indian (Ihh), and Desert (Dhh), are known to bind and inhibit the Patched 1 (Ptch1) and/or Patched 2 (Ptch2) receptors. These interactions relieve repression of Smoothened (Smo), allowing activation of the GLI transcription factors (GLI-TFs) GLI1, GLI2, and GLI3 (14). The core Hh signaling pathway components are all expressed at some point in mammary gland morphogenesis (15–20). Accordingly, mouse studies have revealed both specific and redundant requirements for these factors in mammogenesis.
Various observations suggest that Hh signaling acts in both the stromal and epithelial compartments of the mammary gland. Early transplantation studies provided strong evidence for a stromal role. Specifically, a Gli2−/− primitive mammary gland (comprised of both stromal and epithelial components) yielded defective mammary structures when transplanted into cleared mammary fats pads, whereas transplantation of the Gli2−/− epithelial cells alone created normal mammary glands (16), establishing an essential function for Gli2 in the stromal compartment. More recently, the stroma-specific mutation of Gli2 was shown to delay puberty-induced mammogenesis, reflecting a requirement for Gli2 in coordinating the expression of growth factors that support MaSCs (21). Other, equally compelling, data support an epithelial role for Hh signaling. For example, hyperplasia, dysplasia, and/or impaired differentiation of the adult mammary gland are triggered by expression of constitutively active Smo (22, 23), Gli1 (24), or Shh (20) in epithelial/luminal cells. Notably, this Shh study showed that the Hh-responsive cells are located within the basal epithelial compartment and express stem/progenitor markers, consistent with the notion that these are MaSCs (20). However, other work links Hh signaling more directly to the maintenance of adult MaSCs. Thus, Hh pathway components are up-regulated in mammospheres containing human MaSCs. Most importantly, the self-renewal capacity of MaSCs is either activated or suppressed by activation or inhibition of Hh signaling respectively (12, 13).
Unrelated studies show that epithelial–mesenchymal transition (EMT) programs support the stemness of MaSCs (6, 7, 25–27). The EMT transcription factor (EMT-TF) Slug is expressed within populations of basal cells that are enriched for MaSCs (6, 7, 25–27). Moreover, the self-renewal capacity of these cells in organoid and transplantation/reconstitution assays is enhanced or suppressed by Slug overexpression or knockdown, respectively (7, 27). Indeed, Slug inhibition appears to promote luminal epithelial differentiation (26, 28). Consistent with these roles, Slug-knockout mice show a delay in mammary gland development (25). Despite these advances, it remains unclear how EMT programs enable the acquisition of stemness in cells of the mammary gland. When considering this question, it is important to note that EMT programs do not function as a simple binary switch from epithelial to mesenchymal states but instead generate a spectrum of phenotypic E–M states between these two extremes, only a subset of which is thought to enable stemness (29).
Breast cancers have been divided into various molecular subtypes, which are thought to arise from various cell lineages within the mammary epithelial differentiation hierarchy (1). Claudin-low tumors are thought to arise from the MaSCs of the basal compartment, and they display many of the defining characteristics of these cells. Claudin-low tumors are associated with activation of the EMT program (30), and the tumorigenic capacity of their mammary TICs (MaTICs) relies on EMT-TF programs in orthotopic mouse tumor models (6, 7, 26, 31). Additionally, Hh pathway components are up-regulated in poorly differentiated MaTICs of claudin-low and other breast cancer subtypes, and activation of Hh signaling correlates both with MaTIC expansion (12, 32–36) and with the formation of mammary tumors that express markers of the EMT program (37, 38). Most importantly, the self-renewal capacity of MaTICs is either activated or suppressed by activation or inhibition of Hh signaling, respectively, independent of breast cancer subtype (32–36). Collectively, these findings argue that EMT and Hh programs both play key roles in the formation of MaSC and MaTICs. However, the relationship and epistasis of EMT and Hh programs in either population have been obscure. In this study, we show that primary ciliogenesis plays a critical role in linking these two processes.
The primary cilium is a microtubule-based structure that is transiently assembled on the cell surface by the centrosome during the G0/G1 stages of the cell cycle (39). The function of the primary cilium was widely neglected until the discovery that primary ciliogenesis is essential for normal development (40), including development of the mammary gland (41). During embryogenesis, primary cilia coordinate the activation of various core signaling pathways (42, 43). Hh signaling is one of the known cilium-dependent pathways, in part because the GLI-TFs that function as downstream Hh effectors are processed within the primary cilium to yield either cleaved transcriptional repressors or full-length activators (14, 44). In this study, we establish that EMT programs result in the induction of primary ciliogenesis, which in turn enables Hh signaling and consequent acquisition of stem cell (SC) function.
Results
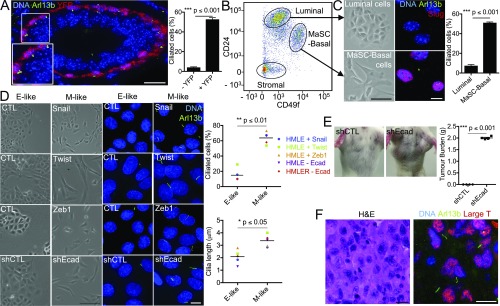
We initiated this study by hypothesizing that EMT programs act to promote the stemness of the MaSCs and MaTICs of claudin-low breast tumors by inducing primary ciliogenesis, thereby enabling active Hh signaling. This hypothesis was based on the prior findings that primary cilia are observed almost exclusively in the basal compartment of the adult mammary epithelium (41) and that epithelium-specific Shh expression induced the formation of hyperplastic ciliated lesions arising from the basal cell lineage (20). To test our hypothesis, we first asked whether primary cilia were observed with any frequency in MaSC-enriched cell populations. Since the Slug EMT-TF is expressed in MaSC-enriched basal cells, albeit in cells that may reside in different states along the E–M axis, we used a Slug-internal ribosome entry site (IRES)-YFP knockin mouse line in which the locus encoding Slug was engineered to express both Slug and a YFP reporter (7). Mammary gland sections from 8- to 10-wk-old virgin females were stained for YFP (indicative of Slug expression), α-smooth muscle actin (αSMA, a basal cell marker), and two primary cilium markers, Arl13b and acetylated tubulin. Remarkably, primary cilia were detected on 51% of the YFP+ cells in the basal compartment of mature ducts but were largely absent from YFP− cells (P ≤ 0.001) (Fig. 1A and Fig. S1 A and B). We also examined remaining terminal end buds, in which the cap cells express Slug (25), and observed primary cilia in the YFP+ cells (Fig. S1C). Consistent with published data (41), we also detected primary cilia in the supporting stroma of both mature ducts and terminal end buds and found that a subset of these ciliated cells were YFP+ (Fig. S1 B and C). Since the stroma is known to express both Slug and, at higher levels, the related EMT-TF Snail (27), we stained for their common downstream target, the EMT-TF Zeb1 (Fig. S1B). We found that Zeb1 was consistently expressed in primary cilia-bearing cells of both the basal layer (lower Zeb1) and stroma (higher Zeb1) of the mature duct. We also assessed ciliogenesis in the mammary epithelium of nontransgenic mice using FACS to isolate stromal-cell–free populations that were enriched for either MaSC-basal (CD24Lo;CD49fHi) or luminal (CD24Hi;CD49fLo) cells, the latter serving as a basal MaSC-deficient control (Fig. 1 B and C). Once again, primary cilia were detected on the majority of MaSC-enriched basal cells but on few of the luminal controls (P ≤ 0.001) (Fig. 1C). Hence, primary cilia were associated with the Slug EMT-TF–expressing, SC-enriched mammary epithelial cell population.
Fig. 1.
EMT programs induce primary ciliogenesis. (A) Normal mammary gland sections from Slug-IRES-YFP animals (8–10 wk old, n = 3) were stained for the indicated proteins (Inset: 1.4× magnification), and the percentage of ciliated cells was quantified (mean ± SEM). Representative results (from three independent experiments) are shown. (B and C) Luminal and MaSC-enriched basal cells from nontransgenic adult females (8–10 wk old, n = 3) were isolated by FACS using the indicated cell-surface markers (B) and were plated and examined for morphology by brightfield microscopy or for ciliated cells by immunofluorescence for the indicated proteins (mean ± SEM) (C). Representative results from three independent experiments are shown. (D) Morphology and percent ciliated cells were determined as described above for E-like, control (CTL), sh (short hairpin)CTL, and M-like (Snail, Twist, Zeb1, shEcad) HMLE cells. Representative results from three independent experiments are shown. (E) Bilateral orthotopic implantations were conducted with shCTL or shEcad HMLER variants; representative mice are shown. Tumor burden per mouse (mean ± SEM) was determined 8 wk postimplantation with two sites of implantation per mouse and four mice per cell type. (F) Sections from the resulting shEcad HMLER tumors were stained with H&E or for large T antigen and Arl13B to identify the tumor cells and cilia, respectively. Representative images are shown. [Scale bars: 100 μm for brightfield images (except in F, where the scale bar: 15 μm) and 15 μm for immunofluorescence images.]
We then asked whether EMT programs actively promote primary ciliogenesis using HMLE cells. These are human mammary epithelial cells harvested from mammoplasty and immortalized by ectopic expression of the hTERT and SV40 Large T proteins (45). HMLE cells are more basal-like, as judged by gene-expression analyses (46). Rare mesenchymal variants exist within the HMLE population (6), but the great majority display epithelial characteristics (6), and thus we refer to HMLE cells as “E-like.” Given the basal-like nature of HMLE cells, we wanted to determine whether primary cilia and Slug expression exist in this population. Thus, we subjected the parental HMLE cells to FACS, employing the same markers used to identify primary MaSC-enriched basal cells, and screened the isolated CD24Lo;CD49fHi population for Slug and Arl13b expression (Fig. S2 A and B). We observed primary cilia on 30.88 ± 5.97% of the SlugHi cells but rarely on the SlugLo cells (P ≤ 0.01). These findings established that primary cilia exist on a subset of HMLE cells and reinforced our conclusion of a correlation between EMT-TF expression and primary ciliogenesis.
In previous work, we generated HMLE variants in which ectopic expression of the Snail, Twist, or Zeb1 EMT-TFs or knockdown of E-cadherin was used to drive EMT and thereby generate more mesenchymal cells (6, 31). These variants display loss of epithelial markers, acquisition of mesenchymal markers, and a more mesenchymal morphology (herein referred to as “M-like”) (Fig. 1D and Fig. S2 C and D). Since primary cilia are assembled specifically during the G0/G1 cell-cycle phases, we serum-starved HMLE lines in either the E-like or M-like state and assessed primary cilia representation by staining for Arl13b. Remarkably, the M-like variants all exhibited dramatic increases in both the number and length of primary cilia relative to their E-like counterparts, indicating that EMT induction promotes primary ciliogenesis (Fig. 1D).
To establish whether cell-cycle phasing contributed to the increased frequency of primary cilia in the EMT-induced cells, we also assessed their representation in the presence of added mitogenic growth factors. Once again, primary cilia were far more prevalent on M-like cells than on their E-like counterparts (Fig. S2D), even though these cell populations contained G0/G1 subpopulations of comparable sizes (Fig. S2E). Analysis of adult mammary gland sections also revealed no difference in proliferative index that could account for the differential representation of cilia on MaSC-enriched basal cells compared with luminal cells (Fig. S2F). We conclude that EMT programs actively induce primary cilia assembly and that this process could not be ascribed simply to promotion of entry into the G0/G1phases of the cell cycle.
HMLE cells have been transformed through H-RASG12V expression to generate a tumorigenic population, termed “HMLER,” which becomes more M-like upon E-cadherin knockdown (Fig. S2C) (31, 45). Of note, this switch causes acquisition of tumor-initiating capacity (31), as evidenced by increased ability to form tumors that model claudin-low human breast cancers following orthotopic transplantation into the mammary fat pad (46) (Fig. 1E). Importantly, we found that E-cadherin knockdown in cultured HMLER (creating M-like cells) also greatly promoted primary ciliogenesis (Fig. 1D and Fig. S2G). Moreover, primary cilia were clearly apparent on a subset (30.4 ± 1.6%) of in vivo mammary tumor cells arising from the orthotopic transplant of the M-like (shEcad expressing) HMLER cells (Fig. 1F). Taken together, these data show that EMT programs operating within both MaSC-enriched basal cells and their neoplastic counterparts are associated with induction of primary ciliogenesis.
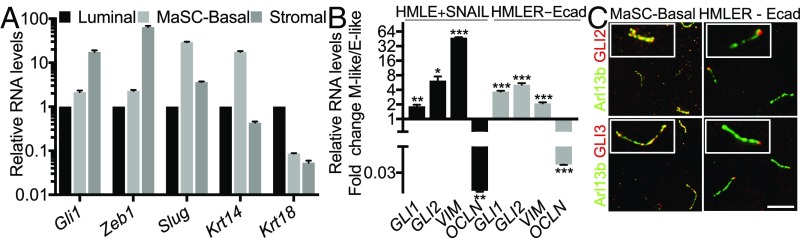
We proceeded to determine whether EMT-TF induction of primary ciliogenesis resulted in Hh pathway activation. As part of the Hh signaling program, active GLI-TFs promote their own transcription, and thus GLI mRNA levels are frequently used as a surrogate marker for Hh pathway activation (12). Accordingly, we isolated by FACS, populations of MaSC-enriched basal and stromal cells (both heterogeneous for EMT-TF expression) and luminal cells (EMT-TF deficient) (Fig. 1B) and conducted real-time qPCR for Gli1 and other markers (Fig. 2A). Gli1 mRNA was detected at higher levels in the MaSC-enriched basal cells (P ≤ 0.01) and at highest levels in stromal cells (P ≤ 0.001), relative to the luminal controls. Notably, the levels of Gli1 correlated well with those of Zeb1, which encodes the key EMT-TF in these three populations (Fig. 2A). Gli2 also showed modestly, but significantly (P ≤ 0.05), higher expression in MaSC-enriched basal cells than in luminal cells (Fig. S3A). Moreover, analysis of two existing gene-expression datasets also showed significantly higher expression of Gli1, Gli2, Snai2 (Slug), and the mesenchymal marker Vim (vimentin) in murine MaSC-enriched basal cells than in luminal or luminal progenitor cells (Fig. S3 B and C).
Fig. 2.
EMT programs induce Hh pathway activation. (A and B) Relative levels of the indicated gene transcripts were determined by real-time qPCR analysis (n = 3, mean ± SEM) of FACS-sorted luminal cells, MaSC-enriched basal cells, and stromal cells (A) and HMLE or HMLER cells in the E-like [CTL, sh (short hairpin)CTL] versus M-like (SNAIL, shEcad) states (B). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Representative results from three independent experiments are shown. (C) Colocalization of GLI2 and GLI3 with the cilium marker Arl13b in MaSC-enriched basal cells and HMLER cells in the M-like state (shEcad). (Scale bar, 5 μm.) Insets: 2× magnification.
To determine whether shifts between epithelial vs. mesenchymal states can modulate Hh pathway signaling, we extended our analyses to the paired HMLE and HMLER cell lines. Here we observed significant up-regulation of GLI1 and GLI2 mRNAs and proteins in the M-like (post-EMT induction) variants compared with the E-like (pre-EMT) controls (Fig. 2B and Fig. S3D). Thus, EMT programs can induce Hh pathway activation in both normal and neoplastic mammary epithelial cells. Gli-TFs can be activated via both Smo-dependent and -independent events. To probe for a potential role of Smo, we treated M-like HMLER cells with the Smo inhibitor erismodegib (Fig. S3E). The drug had opposing and dose-dependent effects on Gli1 (down-regulated) and Gli2 (up-regulated) mRNA levels. These data show that Smo is able to modulate Gli-TF regulation in M-like HMLER cells, but the differential response of Gli1 and Gli2 suggests that both canonical and noncanonical Gli pathways play roles in their activation in M-like cells.
Previous studies in nonmammary tissues had shown that GLI2 and GLI3 accumulate in the primary cilium as part of the process by which Hh signaling mediates GLI-TF activation (14). Accordingly, we used immunofluorescence to determine the localization of GLI2 and GLI3 within both MaSC-enriched basal cells and M-like (shEcad-expressing) populations of HMLER cells. In both cases, these GLI-TFs were detected within the primary cilia (Fig. 2C). Notably, in the M-like HMLER cells, GLI2 and GLI3 were particularly enriched at the cilia tip, a known indicator of potent Hh pathway activation (44). Thus, in the context of both normal and tumorigenic mammary SC populations, EMT programs activate primary ciliogenesis, which then enable engagement of Hh signaling.
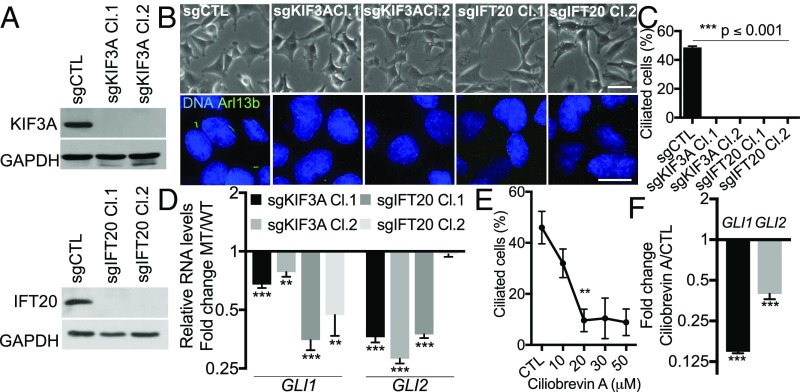
We wished to determine whether primary cilia are actually required for induction of Hh signaling. To address this question, we used CRISPR/Cas9 to mutate the genes encoding two essential ciliogenesis regulators, KIF3A and IFT20, in M-like HMLER cells (Fig. S4A). This yielded cell populations with partial reduction of either KIF3A or IFT20 protein levels and thus partial loss of primary cilia, due to the cell-to-cell variability in the inactivation of KIF3A and IFT20 (Fig. S4 B and C). We then generated single-cell clones to identify two mutant clones for each gene that completely lacked either KIF3A or IFT20 expression due to frameshift mutations (Fig. 3A and Fig. S4D). As anticipated, these clones, but not the sg (small-guide)CTL control cells, lacked primary cilia (Fig. 3 B and C). Notably, the sgKIF3A and sgIFT20 mutant clones all retained their M-like morphologies (Fig. 3B) and displayed no alteration in their proliferative capacity in monolayer (Fig. S4E). Most importantly, we found that all four cilia-deficient clones had significantly lower levels of GLI1 and/or GLI2 mRNA than did the sgCTL control cells (Fig. 3D). As a parallel approach, we also treated M-like HMLER cells with the ciliogenesis inhibitor ciliobrevin A (Fig. 3E and Fig. S4F). Ciliobrevin A (20 µM) reduced the frequency of cilia from 45.95 ± 6.37% to 9.62 ± 4.42%, and this was accompanied by a significant down-regulation (P ≤ 0.001) of GLI1 and GLI2 mRNA (Fig. 3 E and F and Fig. S4F). We also confirmed that GLI1 and/or GLI2 protein levels were reduced by ciliogenesis inhibition (Fig. S3D). Collectively, these data show that primary cilia are dispensable for maintenance of the M-like state and proliferative capacity in monolayers, but they enable induction of Hh signaling within SC-enriched populations. Interestingly, residual levels of GLI transcripts were detected in both the knockout and ciliobrevin A-treated cells, suggesting the existence of cilium-independent Hh/GLI-TF signaling, as previously reported (47).
Fig. 3.
Hh pathway induction relies on primary ciliogenesis. (A) KIF3A and IFT20 knockouts were validated by Western blot of extracts from the indicated cells. (B and C) The impact on morphology and ciliogenesis was assessed as described in the legend of Fig. 1. (Scale bars: brightfield, 50 μm; immunofluorescence, 15 μm.) (D) Relative GLI1 and GLI2 RNA levels (n = 3, mean ± SEM) in indicated control (WT) and mutant (MT) cells were analyzed by real-time qPCR. Representative results from three independent experiments are shown. (E and F) The impact of ciliobrevin A on ciliogenesis and GLI-TF RNA levels in HMLER cells in the M-like state (shEcad) were analyzed as described above. **P ≤ 0.01, ***P ≤ 0.001.
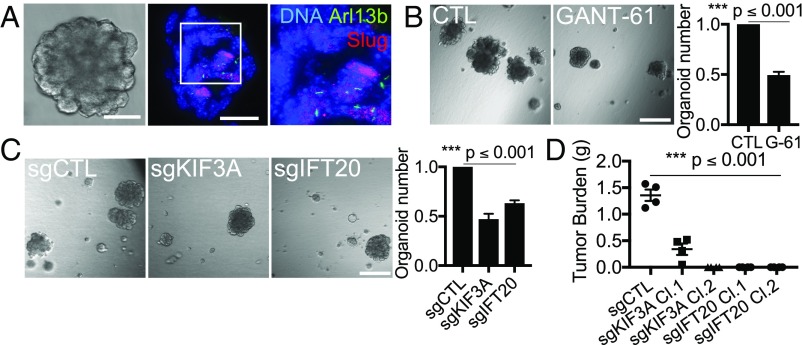
We also wished to learn whether primary cilia and/or induction of Hh signaling play causal roles in the acquisition of stemness. Initially, we addressed this question by assaying the ability of normal basal MaSCs to form organoids using a 3D-Matrigel assay, which has been shown to recapitulate the regenerative capacity of MaSCs faithfully in transplant assays (7). We began by generating organoids from MaSC-enriched basal cells isolated from nontransgenic mice and determining that a subset of the cells displayed primary cilia, which were coincident with Slug expression (Fig. 4A). We then showed that Hh signaling plays a key role in stemness, consistent with prior reports (12, 13). Specifically, we treated isolated MaSC-enriched basal cells with the GLI1/2 inhibitor GANT61 (48) and showed that this significantly reduced their ability to form organoids (P ≤ 0.001) (Fig. 4B). Having validated the contribution of Hh signaling, we also asked whether primary cilia are required for organoid formation. For this, we isolated MaSC-enriched basal cells by FACS and transduced these with lentiviruses expressing the puromycin-resistance gene CAS9 together with CTL, KIF3A, or IFT20 small-guide RNAs to inactivate these genes. Because MaSCs are primary cells, we could not use single-cell cloning to isolate clonal populations of knockout cells. Instead, we cultured the cells briefly in puromycin to enrich for populations of vector-transduced cells, which were heterogeneous in their inactivation of KIF3A or IFT20. We introduced these directly into organoid assays and observed that the sgKIF3A- and sgIFT20-containing populations both yielded approximately half the number of organoids formed by the sgCTL control population (P ≤ 0.001) (Fig. 4C). Importantly, all the organoids arising from the sgKIF3A-containing populations maintained primary cilia, indicating that these arose from nontargeted cells (Fig. S4G). Hence, we concluded that Hh signaling plays a critical role in enabling stemness of basal MaSCs and that primary cilia are essential for this stemness.
Fig. 4.
Primary ciliogenesis promotes stemness of MaSCs and MaTICs. (A) Organoids from MaSC-enriched basal cells were cut and stained for the indicated proteins (Inset: 2× magnification). (Scale bars: brightfield image, 100 μm; and immunofluorescence images, 35 μm.) (B and C) Organoid-forming capacity was determined for sorted MaSC-enriched basal cells after treatment with vehicle (CTL) or GLI1/2 inhibitor (GANT-61/G-61, 20 µM) (B) or transduction with the indicated single-guide (sg) RNAs (C). Organoid numbers were quantified 7 d after plating and normalized to the controls (n = 3, mean ± SEM). (Scale bars in B and C: 500 μm.) Representative results from three independent experiments are shown. (D) Bilateral orthotopic implantations were conducted with the indicated HMLER variants, and tumor burden per mouse was determined as described in the legend of Fig. 1E.
Finally, we asked whether primary cilia also support the stemness of MaTICs using the single-cell KIF3A and IFT20 CRISPR/Cas9 mutant clones that we had generated from the M-like HMLER (shEcad) line. We assayed the tumorigenic capacity of these cells compared with that of the sgCTL HMLER cells using mammary fat pad implantation to do so (Fig. 4D). Transplantation of sgCTL HMLER cells successfully yielded tumors (Fig. 4D) with a frequency similar to that of the parental M-like HMLER cells (Fig. 1E). In stark contrast, although there was no alteration in their proliferative capacity in monolayer (Fig. S4E), the primary cilia-deficient HMLER cells exhibited either greatly reduced (sgKIF3A Cl.1) or completely abolished (sgKIF3A Cl.2, sgIFT20 Cl.1, and sg IFT20 Cl.2) ability to generate tumors (Fig. 4D). Therefore, primary cilia play essential roles in the tumor-initiating capacity of MaTICs.
Discussion
Previous studies have revealed that intraepithelial Hh signaling enables expansion of normal and malignant stem/progenitor cells and promotes tumorigenesis (12, 13, 20, 32–37). Similarly, several EMT-TFs are known to promote the stemness of both MaSCs and MaTICs (6, 7, 25–28). Our data emphasize a clear connection between EMT programs and Hh signaling in basal MaSCs and elucidate an epistatic relationship between these processes. Specifically, we show that EMT programs activate primary ciliogenesis, which then enables Hh signaling. Most importantly, we demonstrate that ablation of primary ciliogenesis abrogates both the stemness of normal MaSCs in organoid assay and the tumor-forming capacity of MaTICs. Collectively, these data establish an ordered pathway of EMT programs → primary ciliogenesis → Hh signaling → stemness, and reveal this as a key mechanism enabling normal and transformed mammary stem cells to maintain their SC properties.
Our data do not exclude the possibility that primary cilia act to promote additional signaling pathways in MaSCs and/or MaTICs; indeed, we think this is entirely likely. Nonetheless, our results clearly highlight the importance of Hh signaling, which, based on the divergent response to Smo inhibitor, seems likely to result from both canonical and noncanonical Hh pathways. There are already extensive existing data supporting both epithelial and stromal roles for Hh signaling in mammogenesis (15–24). Our findings establish a key function for Hh signaling in epithelial cells with activated EMT programs and thus M-like morphology. Interestingly, a recent report also observed that M-like breast cancer cells, including HMLER M-like cells, displayed elevated Hh signaling activation and further showed that this can act in a non–cell-autonomous manner to promote the tumorigenicity of breast cancer cells in the E-like state (49). Notably, this report and our own study demonstrate epithelial roles for Hh, but they do not contradict the idea that stromal Hh signaling also facilitates mammogenesis. However, both prior studies (12, 13, 20) and our current findings are at odds with the recent suggestion that the role of Hh signaling in postnatal mammary gland development is entirely stromal (21). Interestingly, mammary stromal cells are known to exhibit primary cilia (41), and they express high levels of EMT-TFs (27). Our data now establish that cilia-bearing stromal cells express Slug and Zeb1, indicating an association between the EMT programs and primary cilia in the stromal compartment. This raises the intriguing possibility that EMT-TFs might serve to induce primary cilia and thus Hh signaling in stromal cells, in a manner analogous to the Slug → primary ciliogenesis → Hh signaling pathway that we have uncovered in MaSC-enriched populations.
It is important to consider the relevance of our findings to different breast cancer subtypes. Our data reveal a critical role for primary cilia in both MaSC-enriched basal cells and the MaTICs for HMLER M-like cells, which yield tumors that display the hallmarks of basally derived claudin-low subtype. We note that other breast cancer subtypes, including basal-like, HER2+, and luminal A and B, are thought to arise from the luminal lineage at different stages of differentiation (1). As we and others (41) have shown, ciliated cells are rarely observed in the luminal compartment. We hypothesize that the requirement of primary cilia will differ across various breast cancer subtypes, reflecting the presence or absence of cilia in the corresponding cells of origin. Consistent with this model, a number of studies have assessed the representation of primary cilia in mammary hyperplastic lesions and tumors (20, 50–54) and have arrived at differing conclusions about their frequency, ranging from rare (52–54) to elevated (20) levels. Not all of these studies allow conclusions about cilia phenotypes in relation to subtype. However, when the subtype is apparent, the data show that cilia are present at high levels in Shh-dependent hyperplasia arising from the basal layer (20), the setting that is relevant to our study, but are rare in tumors that are derived from the luminal layer (54). Notably, at the time of submission of this manuscript, a study by Hassounah et al. (55) concluded that ciliogenesis acts to suppress breast cancer. While Hassounah et al. apply their findings to breast cancer generally, their data come from the study of MMTV-PyMT–driven tumors, which are luminally derived, and thus do not impact the conclusion that basal-derived tumors are cilia-dependent. In support of this conclusion, a recent study suggested that primary cilia may specifically promote estrogen receptor alpha (ERα)-negative breast cancer metastasis (56). Notably, there is strong reason to believe that subtype-specific dependence on primary cilia in breast cancer is relevant for other tumor types. For example, specific subsets of skin cancer (basal cell carcinomas) and brain cancer (medulloblastomas, SHH subtype) are believed to employ primary cilia, while other subtypes have low levels of cilia (57–59). Additional experiments will be required to establish whether low cilia representation in these other subtypes equates to cilia independence.
Even among basal-derived breast tumors, we anticipate some degree of intratumoral heterogeneity regarding the representation of ciliated cells. First, since basal body maturation and primary ciliogenesis are tightly regulated during the cell cycle, abnormal cancer cell division will likely alter these processes. Second, we still have much to learn about the mechanism(s) that link EMT signaling to primary ciliogenesis, including the degree to which this is a stable or transient response. Since EMT programs appear to generate a spectrum of phenotypic states along the E–M axis (29), it is an open question which of these states enable primary ciliogenesis. It is important to note that only a subset of the EMT-TF–expressing basal MaSCs and MaTICs seems to have SC potential (7, 27). Interestingly, we consistently find that some, but not all, of the EMT-TF–expressing cells display primary cilia, even when arrested in the G0/G1 state. Given these findings, we postulate that stemness is conferred by a particular E/M-like state that depends on and is marked by the primary cilium.
Methods
Cell culture, Western blot experiments, cell-cycle analysis, real-time qPCR, RNA-sequencing (RNA-seq) analysis, and statistical analysis are detailed in Supporting Information.
Animals.
Slug-IRES-YFP mice were generated and selected as previously described (7, 27). Black 6 and NOD SCID animals were obtained from the Jackson laboratory (stock numbers 000664 and 001303, respectively). Mice were housed and handled in accordance with protocols approved by the Animal Care Committee of the Massachusetts Institute of Technology.
Primary Mammary Epithelial Cell Isolation and FACS.
Mammary glands from 8- to 16-wk-old Black 6 females were minced and dissociated using collagenase/hyaluronidase (STEMCELL Technologies) diluted (1:10) in DMEM/F-12 medium at 37 °C for 5 h under constant agitation. The dissociated glands were spun down at 450 × g for 5 min and were resuspended in ammonium chloride solution (STEMCELL Technologies) diluted (4:1) in HBSS buffer supplemented with 10 mM Hepes and 2% FBS (HF buffer). The samples were spun down and resuspended in warm Trypsin-EDTA (STEMCELL Technologies) by pipetting for 3 min. Trypsin was inactivated by the addition of HF buffer. The digested samples were spun down and further digested with dispase solution (STEMCELL Technologies) supplemented with 0.1 mg/mL DNase I for 1 min. Cell suspensions were diluted with HF buffer and filtered through a 40-μm cell strainer to collect single cells. To separate various cell populations, single cells were stained with a Live-Dead fixable violet dye and antibodies against CD24 (PE; BioLegend) and CD49f (APC; BioLegend). Stained cells were sorted on a FACSAria II sorter (Becton Dickinson).
Immunofluorescence and Image Analysis.
Five-micrometer paraffin tissue sections of formalin-fixed mammary glands from 8- to16-wk-old mice, 8-μm frozen sections of organoids, or cells fixed in 4% paraformaldehyde for 10 min on glass coverslips were permeabilized with 0.3% Triton X-100 for 10 min and blocked with 5% goat or donkey serum for 1 h before staining with primary antibodies against Arl13b (NeuroMab 73-287; 1:100), YFP (Cell Signaling 2956; 1:100), Slug (Cell Signaling 9585; 1:100), acetylated tubulin (Cell Signaling 5335; 1:50), γ-tubulin (Sigma T5326; 1:50), SMA (Abcam ab21027; 1:100), Zeb1 (Santa Cruz Biotechnology sc-25388; 1:50), E-cadherin (Cell Signaling 3195; 1:200), Ki-67 (BD Biosciences 550609; 1:50), GLI2 (R&D Systems AF3635; 1:100), and GLI3 (R&D Systems AF3690; 1:100). Secondary antibodies were anti-goat 405 (Abcam ab175664; 1:250), anti-goat 488 (Abcam ab150129; 1:250), anti-mouse 488 (Life Technologies A11001; 1:500), anti-mouse 546 (Life Technologies A11003; 1:500), anti-mouse 555 (Abcam ab150106; 1:250), anti-mouse IgG1 647 (Life Technologies A21240; 1:250), anti-rabbit 488 (Life Technologies A21206; 1:500), anti-rabbit 546 (Life Technologies A11010; 1:500), and anti-rabbit 647 (Life Technologies A31573; 1:500). Mounted coverslips were examined using DeltaVision Olympus IX-71 microscopes. Images were acquired using 40×, 60×, and 100× objectives and a CoolSNAP HQ camera. Z-stacks were deconvolved (Softworx) and processed with ImageJ.
CRISPR Mutations.
KIF3A and IFT20 guide RNAs were selected from the e-crisp-test.dkfz.de/ and crispr.mit.edu/ websites and were cloned into lentiCRIPRV2 plasmids containing, either a puromycin-resistance gene or a blasticidin-resistance gene designed to replace the puromycin one. Lentiviruses were produced in 293FT cells after their transfection with lentiCRISPR (transfer), psPAX2 (packaging), and pMD2.G (envelope) plasmids, using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Supernatants containing lentiviruses were collected 48 and 72 h posttransfection. Primary mammary epithelial cells or HMLER cells growing in a monolayer were transduced with the supernatants-containing viruses in the presence of 8 μg/mL Polybrene (Sigma-Aldrich). Transduced primary mammary epithelial cells and HMLER cells were selected with 2 μg/mL puromycin (GIBCO) or 6 μg/mL blasticidin (GIBCO). HMLER clones were selected by FACS 1 mo after infection. Mutations were validated by PCR amplification and Sanger sequencing of the targeted loci. Oligonucleotides used for CRISPR mutations and sequencing are listed in Table S2. Information.
Organoid Assay and Orthotopic Tumor Cell Implantation.
Matrigel organoid culture was performed as described previously (7). Briefly, freshly isolated mammary epithelial cells or transduced cells were cultured in complete EpiCult-B medium (STEMCELL Technology) containing 5% Matrigel (Corning), 5% heat-inactivated FBS, 10 ng/mL EGF, 10 ng/mL FGF, 4 μg/mL heparin, and 5 μM Y-27632. Cells were seeded at 2,000 cells per well in 96-well ultra-low-attachment plates (Corning). Organoids were counted 7–14 d after seeding. For orthotopic cell implantations, tumor cells were resuspended in a 1:1 mixture of complete MEGM medium (Lonza) with Matrigel. Seven hundred thousand cells in 20 μL of medium + matrigel (1:1 mixture) were injected bilaterally into inguinal mammary fat pads of 8-wk-old NOD SCID females. Tumors were resected 8 wk postimplantation, and their mass was determined to establish the tumor burden per animal.
Supplementary Material
Acknowledgments
We thank Arjun Bhutkar for analysis of gene expression datasets; Guillaume Carmona, Julia Fröse, and Mary Brooks for providing HMLE cells; Ferenc Reinhardt and Joana Liu Donaher for help in development of orthotopic transplantation experiments; Feng Zhang for providing LentiCRISPR v2 (Addgene plasmid no. 52961); and the Koch Institute Swanson Biotechnology Center for technical support. This project was supported by Fondation MIT, the Ludwig Center for Molecular Oncology at MIT, and Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. R.A.W. is an American Cancer Society and Ludwig Foundation Professor. J.A.L. is the Virginia and D. K. Ludwig Professor for Cancer Research at the Koch Institute at MIT. V.J.G. was supported by the Philippe Foundation and by Postdoctoral Fellowships from the Ludwig Foundation and the Koch Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711534114/-/DCSupplemental.
References
- 1.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inman JL, Robertson C, Mott JD, Bissell MJ. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development. 2015;142:1028–1042. doi: 10.1242/dev.087643. [DOI] [PubMed] [Google Scholar]
- 3.Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny from a single cell. Development. 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- 4.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 5.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 6.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 10.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 11.Visvader JE, Clevers H. Tissue-specific designs of stem cell hierarchies. Nat Cell Biol. 2016;18:349–355. doi: 10.1038/ncb3332. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N, et al. Reciprocal intraepithelial interactions between TP63 and hedgehog signaling regulate quiescence and activation of progenitor elaboration by mammary stem cells. Stem Cells. 2008;26:1253–1264. doi: 10.1634/stemcells.2007-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 15.Lewis MT, et al. Defects in mouse mammary gland development caused by conditional haploinsufficiency of Patched-1. Development. 1999;126:5181–5193. doi: 10.1242/dev.126.22.5181. [DOI] [PubMed] [Google Scholar]
- 16.Lewis MT, et al. The Gli2 transcription factor is required for normal mouse mammary gland development. Dev Biol. 2001;238:133–144. doi: 10.1006/dbio.2001.0410. [DOI] [PubMed] [Google Scholar]
- 17.Gallego MI, Beachy PA, Hennighausen L, Robinson GW. Differential requirements for shh in mammary tissue and hair follicle morphogenesis. Dev Biol. 2002;249:131–139. doi: 10.1006/dbio.2002.0761. [DOI] [PubMed] [Google Scholar]
- 18.Michno K, Boras-Granic K, Mill P, Hui CC, Hamel PA. Shh expression is required for embryonic hair follicle but not mammary gland development. Dev Biol. 2003;264:153–165. doi: 10.1016/s0012-1606(03)00401-9. [DOI] [PubMed] [Google Scholar]
- 19.Hatsell SJ, Cowin P. Gli3-mediated repression of Hedgehog targets is required for normal mammary development. Development. 2006;133:3661–3670. doi: 10.1242/dev.02542. [DOI] [PubMed] [Google Scholar]
- 20.García-Zaragoza E, et al. Intraepithelial paracrine Hedgehog signaling induces the expansion of ciliated cells that express diverse progenitor cell markers in the basal epithelium of the mouse mammary gland. Dev Biol. 2012;372:28–44. doi: 10.1016/j.ydbio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Zhao C, et al. Stromal Gli2 activity coordinates a niche signaling program for mammary epithelial stem cells. Science. 2017;356:eaal3485. doi: 10.1126/science.aal3485. [DOI] [PubMed] [Google Scholar]
- 22.Moraes RC, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 23.Visbal AP, et al. Altered differentiation and paracrine stimulation of mammary epithelial cell proliferation by conditionally activated smoothened. Dev Biol. 2011;352:116–127. doi: 10.1016/j.ydbio.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiaschi M, Rozell B, Bergström A, Toftgård R, Kleman MI. Targeted expression of GLI1 in the mammary gland disrupts pregnancy-induced maturation and causes lactation failure. J Biol Chem. 2007;282:36090–36101. doi: 10.1074/jbc.M704280200. [DOI] [PubMed] [Google Scholar]
- 25.Nassour M, et al. Slug controls stem/progenitor cell growth dynamics during mammary gland morphogenesis. PLoS One. 2012;7:e53498. doi: 10.1371/journal.pone.0053498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips S, et al. Cell-state transitions regulated by SLUG are critical for tissue regeneration and tumor initiation. Stem Cell Reports. 2014;2:633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye X, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proia TA, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 30.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kai M, et al. Semi-quantitative evaluation of CD44(+) /CD24(-) tumor cell distribution in breast cancer tissue using a newly developed fluorescence immunohistochemical staining method. Cancer Sci. 2011;102:2132–2138. doi: 10.1111/j.1349-7006.2011.02063.x. [DOI] [PubMed] [Google Scholar]
- 33.Goel HL, et al. GLI1 regulates a novel neuropilin-2/α6β1 integrin based autocrine pathway that contributes to breast cancer initiation. EMBO Mol Med. 2013;5:488–508. doi: 10.1002/emmm.201202078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colavito SA, Zou MR, Yan Q, Nguyen DX, Stern DF. Significance of glioma-associated oncogene homolog 1 (GLI1) expression in claudin-low breast cancer and crosstalk with the nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) pathway. Breast Cancer Res. 2014;16:444. doi: 10.1186/s13058-014-0444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han B, et al. FOXC1 activates smoothened-independent Hedgehog signaling in basal-like breast cancer. Cell Reports. 2015;13:1046–1058. doi: 10.1016/j.celrep.2015.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Memmi EM, et al. p63 sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2015;112:3499–3504. doi: 10.1073/pnas.1500762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiaschi M, Rozell B, Bergström A, Toftgård R. Development of mammary tumors by conditional expression of GLI1. Cancer Res. 2009;69:4810–4817. doi: 10.1158/0008-5472.CAN-08-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou M, et al. LncRNA-Hh strengthen cancer stem cells generation in twist-positive breast cancer via activation of Hedgehog signaling pathway. Stem Cells. 2016;34:55–66. doi: 10.1002/stem.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- 40.Pazour GJ, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDermott KM, Liu BY, Tlsty TD, Pazour GJ. Primary cilia regulate branching morphogenesis during mammary gland development. Curr Biol. 2010;20:731–737. doi: 10.1016/j.cub.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huangfu D, et al. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 44.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prat A, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat. 2013;142:237–255. doi: 10.1007/s10549-013-2743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bishop CL, et al. Primary cilium-dependent and -independent Hedgehog signaling inhibits p16(INK4A) Mol Cell. 2010;40:533–547. doi: 10.1016/j.molcel.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 48.Lauth M, Bergström A, Shimokawa T, Toftgård R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neelakantan D, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8:15773. doi: 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reilova-Velez J, Seiler MW. Abnormal cilia in a breast carcinoma. An ultrastructural study. Arch Pathol Lab Med. 1984;108:795–797. [PubMed] [Google Scholar]
- 51.Yasar B, Linton K, Slater C, Byers R. Primary cilia are increased in number and demonstrate structural abnormalities in human cancer. J Clin Pathol. 2017;70:571–574. doi: 10.1136/jclinpath-2016-204103. [DOI] [PubMed] [Google Scholar]
- 52.Yuan K, et al. Primary cilia are decreased in breast cancer: Analysis of a collection of human breast cancer cell lines and tissues. J Histochem Cytochem. 2010;58:857–870. doi: 10.1369/jhc.2010.955856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nobutani K, et al. Absence of primary cilia in cell cycle-arrested human breast cancer cells. Genes Cells. 2014;19:141–152. doi: 10.1111/gtc.12122. [DOI] [PubMed] [Google Scholar]
- 54.Menzl I, et al. Loss of primary cilia occurs early in breast cancer development. Cilia. 2014;3:7. doi: 10.1186/2046-2530-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hassounah NB, et al. Inhibition of ciliogenesis promotes Hedgehog signaling, tumorigenesis, and metastasis in breast cancer. Mol Cancer Res. 2017;15:1421–1430. doi: 10.1158/1541-7786.MCR-17-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Légaré S, Chabot C, Basik M. SPEN, a new player in primary cilia formation and cell migration in breast cancer. Breast Cancer Res. 2017;19:104. doi: 10.1186/s13058-017-0897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang N, et al. INTU is essential for oncogenic Hh signaling through regulating primary cilia formation in basal cell carcinoma. Oncogene. 2017;36:4997–5005. doi: 10.1038/onc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conduit SE, et al. A compartmentalized phosphoinositide signaling axis at cilia is regulated by INPP5E to maintain cilia and promote Sonic Hedgehog medulloblastoma. Oncogene. 2017;36:5969–5984. doi: 10.1038/onc.2017.208. [DOI] [PubMed] [Google Scholar]
- 59.Cavalli FMG, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–754 e6. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.