Significance
IFNγ is a cytokine that induces various downstream genes for host defense against pathogens. How IFNγ induces downstream genes via the classical JAK1-STAT1 pathway is well understood, but how IFNγ induces a subset of downstream genes via the kinase IKKβ remains enigmatic. We identified SNX8 protein as an important component of this IFNγ-triggered noncanonical pathway. SNX8 deficiency impairs IFNγ-triggered induction of a subset of downstream genes, rendering the mouse more susceptible to infection of intracellular bacteria Listeria monocytogenes. Mechanistically, IFNγ induces phosphorylation of SNX8, which promotes the recruitment of IKKβ to the JAK1 complex and subsequent activation of IKKβ. Our findings suggest that SNX8 acts as a link for IFNγ-triggered noncanonical signaling pathway and host defense against intracellular bacteria infection.
Keywords: interferon, SNX8, noncanonical, IKK, phosphorylation
Abstract
IFNγ is a cytokine that plays a key role in host defense against intracellular pathogens. In addition to the canonical JAK-STAT1 pathway, IFNγ also activates an IKKβ-mediated noncanonical signaling pathway that is essential for induction of a subset of downstream effector genes. The molecular mechanisms and functional significance of this IFNγ-triggered noncanonical pathway remains enigmatic. Here, we identified sorting nexin 8 (SNX8) as an important component of the IFNγ-triggered noncanonical signaling pathway. SNX8-deficiency impaired IFNγ-triggered induction of a subset of downstream genes. Snx8−/− mice infected with Listeria monocytogenes exhibited lower serum cytokine levels and higher bacterial loads in the livers and spleens, resulting in higher lethality. Mechanistically, SNX8 interacted with JAK1 and IKKβ and promoted their association. IFNγ induced JAK1-mediated phosphorylation of SNX8 at Tyr95 and Tyr126, which promoted the recruitment of IKKβ to the JAK1 complex. SNX8-deficiency impaired IFNγ-induced oligomerization and autophosphorylation of IKKβ at Ser177, which is critical for selective induction of downstream genes. Our findings suggest that SNX8 acts as a link for IFNγ-triggered noncanonical signaling pathway, which induces a subset of downstream genes important for host defense against L. monocytogenes infection.
IFNγ is a cytokine that plays pivotal roles in host defense to microbial infection. IFNγ activates macrophages and other cell types, leading to production of various cytokines, phagosomal maturation, autophagy, and bactericidal activity (1). Individuals with partial or complete defects in the IFNγ signaling pathways have increased susceptibility to Listeria monocytogenes as well as other bacterial species (2).
The canonical IFNγ-triggered signaling pathway is characterized by JAK-mediated phosphorylation of STAT1 (3). The binding of IFNγ homodimer to IFNGR1 and IFNGR2 results in spatial proximity of JAK1 and JAK2, leading to phosphorylation of IFNGR1 and JAKs. The phosphorylation of IFNGR1 at Tyr440 by JAKs provides a docking site for the recruitment of STAT1, and the phosphorylated JAKs subsequently phosphorylate STAT1 at Tyr701. Phosphorylated STAT1 forms homodimers, which translocate to the nucleus and bind to the conserved IFNγ-activated sites (GASs) on the promoters of the IFN-stimulated genes (ISGs) to initiate the transcription of these downstream target genes (4, 5). In addition to the phosphorylation of Tyr701 of STAT1, IFNγ induces phosphorylation of STAT1 at Ser727, which is mediated by CDK8, PI3K, and the downstream protein kinase C family members PKC-δ and PKC-ε, and this phosphorylation is critical for the full activation of STAT1 (6–10). In addition to the well-known IFNγ-JAK-STAT1 canonical pathway, IFNγ activates additional signal pathways (11, 12). IKKβ, a master activator of inflammatory response, is required to mediate the transcriptional induction of a subset of IFNγ-stimulated genes, such as guanylate binding proteins (GBPs) and several chemokines (CXCL9, CXCL10, and CXCL11) (1, 13–18). The phosphorylation of STAT1 is necessary but not sufficient for the activation of these genes in response to IFNγ. The mechanisms and biological functions of this IFNγ-triggered noncanonical signaling pathway remain enigmatic.
Sorting nexin 8 (SNX8) belongs to the sorting nexin protein family, which is involved in endocytosis, endosomal sorting, and signaling (19). It has been shown that SNX8 is a β-amyloid (Aβ) toxicity enhancer and associated with Alzheimer’s disease (20, 21). A few of sorting nexin family proteins form the retromer complex with VPS26-VPS29-VPS35 heterotrimer that has been implicated in membrane recruitment and formation of recycling tubules (22, 23). In addition, SNXs play an important role in modulating the degradation of receptors through endocytic pathways (19, 24). Whether and how SNX8 is involved in cellular signaling events are unclear.
In this study, we identified SNX8 as a key adaptor in the IFNγ-triggered noncanonical signaling pathway. SNX8 deficiency impaired expression of a subset of downstream genes induced by IFNγ. Snx8−/− mice were more susceptible to lethal infection by Listeria. SNX8 interacted with JAK1 and IKKβ and selectively promoted IKKβ dimerization/oligomerization and autophosphorylation. Moreover, JAK1-mediated phosphorylation of SNX8 at Tyr95 and Tyr126 was required for its recruitment of IKKβ to JAK1. These findings reveal molecular mechanisms of IFNγ-triggered and SNX8-mediated noncanonical signaling pathway that is important for host defense to microbial infection.
Results
SNX8 Positively Regulates IFNγ-Triggered Signaling.
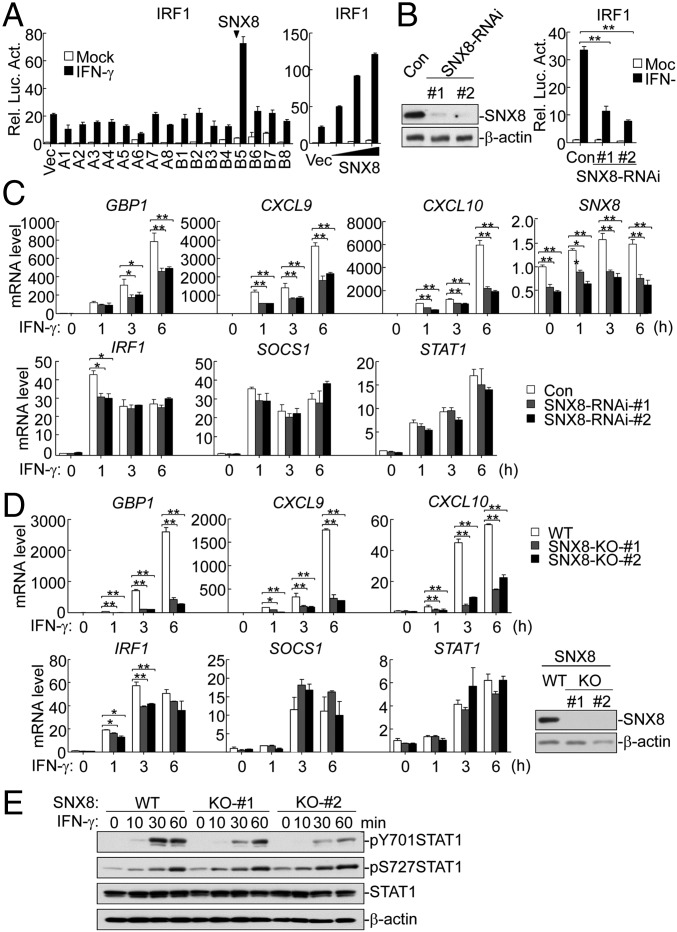
It has been demonstrated that IFNγ stimulation induces the expression of IFN-regulatory factor 1 (IRF1) gene whose promoter contains two conserved STAT1-binding sites (25). To identify additional molecules involved in IFNγ-triggered signaling, we screened ∼10,000 individual human cDNA clones for their ability to activate the IRF1 promoter by reporter assays in 293 cells. These efforts led to the identification of SNX8, which activated the IRF1 promoter and dramatically potentiated IFNγ-triggered activation of the IRF1 promoter (Fig. 1A). Further experiments indicated that SNX8 potentiated IFNγ-triggered activation of the IRF1 promoter in a dose-dependent manner (Fig. 1A).
Fig. 1.
Identification of SNX8 as a positive regulator of IFNγ-triggered signaling. (A) Expression screens for clones that activate the IRF1 promoter. The 293 cells were transfected with the IRF1 reporter plasmids and ∼10,000 individual cDNA clones (Left histograph shows a few representative clones) or increased SNX8 plasmid (Right histograph) for 24 h. Cells were left untreated or treated with IFNγ for 12 h before luciferase assays. (B) Effects of SNX8 knockdown on IFNγ-induced activation of the IRF1 promoter. The 293 cells were transfected with the indicated RNAi plasmids for 36 h before immunoblot analysis with the indicated antibodies (Left blots). Or, the 293 cells were transfected with the IRF1 promoter reporter and the indicated RNAi plasmids. Thirty-six hours after transfection, cells were left untreated or treated with IFNγ for 12 h before luciferase assays. (C) Effects of SNX8 knockdown on IFNγ-induced transcription of downstream genes in THP1 cells. The cells were transduced with control or SNX8-RNAi by retroviral-mediated gene transfer. The cells were then either untreated or treated with IFNγ for the indicated times before qPCR experiments. (D) Effects of SNX8 deficiency on IFNγ-induced transcription of downstream genes. SNX8-deficient HeLa cells were untreated or treated with IFNγ for the indicated times before qPCR experiments. SNX8 levels in the cells were analyzed by immunoblots (Right). (E) Effects of SNX8 deficiency on IFNγ-induced phosphorylation of STAT1 in HeLa cells. The cells were untreated or treated with IFNγ for the indicated times, and cell lysates were analyzed by immunoblots with the indicated antibodies. Graphs show mean ± SD, n = 3. **P < 0.01, *P < 0.05.
We next examined the role of endogenous SNX8 in regulation of IFNγ-triggered signaling. We made two SNX8 RNAi constructs that could markedly inhibit the expression of endogenous SNX8 (Fig. 1B). Knockdown of SNX8 inhibited IFNγ-triggered activation of the IRF1 promoter (Fig. 1B). In addition, knockdown of SNX8 inhibited transcription of a subset of IFNγ-induced downstream genes in THP1 cells, such as GBP1, CXCL9, and CXCL10 (Fig. 1C). In these experiments, knockdown of SNX8 moderately inhibited transcription of IRF1 at an early time point (1 h) but not later time points after IFNγ stimulation, whereas knockdown of SNX8 had no marked effects on transcription of other examined IFNγ-inducible genes, including SOCS1 and STAT1 (Fig. 1C). We then confirmed these observations using SNX8-knockout HeLa. Consistently, SNX8 deficiency inhibited IFNγ-induced transcription of GPB1, CXCL9, CXCL10, and IRF1 but not SOCS1 or STAT1 genes (Fig. 1D). Moreover, SNX8 deficiency reduced IFNγ-triggered phosphorylation of STAT1 at Tyr701 but not Ser727 (Fig. 1E). These data suggest that SNX8 is essential for IFNγ-induced transcription of a subset of downstream genes.
SNX8 Is Essential for IFNγ-Triggered Signaling in Murine Cells.
Human SNX8 consists of 465 amino acid (aa) residues and shares 88.9% sequence identity with its murine ortholog. To investigate the functions of SNX8 in IFNγ-triggered signaling, we generated SNX8-deficient mice (Fig. S1A). Immunoblot analysis confirmed that SNX8 was undetectable in Snx8−/− mouse lung fibroblasts (MLFs) and mouse embryonic fibroblasts (MEFs) (Fig. S1B). Homozygous Snx8−/− mice were born at the Mendelian ratio (Fig. S1C). The numbers and compositions of major immune cells in lymph nodes, spleen, and thymus were similar between Snx8+/+ and Snx8−/− mice (Fig. S1D), suggesting that Snx8 is not essential for development of the examined immune cells.
To determine whether SNX8 is essential for IFNγ-triggered induction of downstream genes in murine cells, we examined expression of downstream genes induced by IFNγ in mouse bone-marrow-derived macrophages (BMDMs) and dendritic cells (BMDCs). As shown in Fig. S2A, SNX8 deficiency markedly inhibited IFNγ-induced transcription of Gbp1, Cxcl9, Cxcl10, and Cxcl11 genes in BMDMs and BMDCs. SNX8 deficiency moderately inhibited IFNγ-induced transcription of Irf1 gene at the early time points of stimulation while having no marked effects on IFNγ-induced transcription of Stat1 and Socs1 genes in BMDMs and BMDCs. The levels of secreted CXCL9 and CXCL10 chemokines induced by IFNγ were markedly lower in Snx8−/− in comparison with Snx8+/+ BMDMs and BMDCs (Fig. S2B). In addition, SNX8 deficiency inhibited IFNγ-triggered phosphorylation of STAT1 at Tyr701 but not Ser727 in both BMDMs and BMDCs (Fig. S2C). Taken together, these results suggest that SNX8 is essential for IFNγ-triggered induction of a subset of downstream genes in mouse immune cells.
SNX8 and IKKβ Mediate Transcription of Overlapping Genes Induced by IFNγ.
It has been reported that IKKβ, a master activator of inflammatory response, is required to activate a subset of IFNγ-induced genes (14). Consistently, transcription of Gbp1, Cxcl9, and Cxcl10 genes induced by IFNγ was decreased in IKKβ−/− in comparison with IKKβ-reconstituted MEFs (Fig. S3A). However, the Y701 and S727 phosphorylation of STAT1 induced by IFNγ was similar in IKKβ−/− in comparison with IKKβ-reconstituted MEFs (Fig. S3B). As shown above, the IKKβ- and SNX8-dependent downstream genes induced by IFNγ overlap well. To further investigate the relationship between IFNγ-induced IKKβ- and SNX8-mediated downstream genes, we constructed JAK1-, SNX8-, and IKKβ-deficient HeLa cells by the CRISPR-Cas9 method (Fig. S3C) and compared transcription levels of downstream genes induced by IFNγ in these cell lines. With a cutoff of twofold changes, we found that transcription levels of 411, 236, and 229 genes from IFNγ-triggered JAK1-, SNX8-, and IKKβ-deficient cells had twofold or greater changes compared with those from IFNγ-treated wild-type cells. Among them, 110 genes were identified in all three groups, whereas 153 genes overlapped between SNX8- and IKKβ-deficient cells (Fig. S3D). A subset of IFNγ-induced downstream genes were down-regulated in both SNX8- and IKKβ-deficient cells, including GBP1, GBP2, CXCL10, and CXCL11 that was identified by the qPCR experiments mentioned in Fig. 1D and Fig. S3A (Fig. S3E). Interestingly, a subset of IFNγ-induced downstream genes was down-regulated in JAK1-deficient cells, but not in SNX8- and IKKβ-deficient cells (Fig. S3F). These results suggest that SNX8 and IKKβ mediate induction of an overlapping set of downstream genes.
SNX8 Interacts with JAK1 and IKKβ After IFNγ Stimulation.
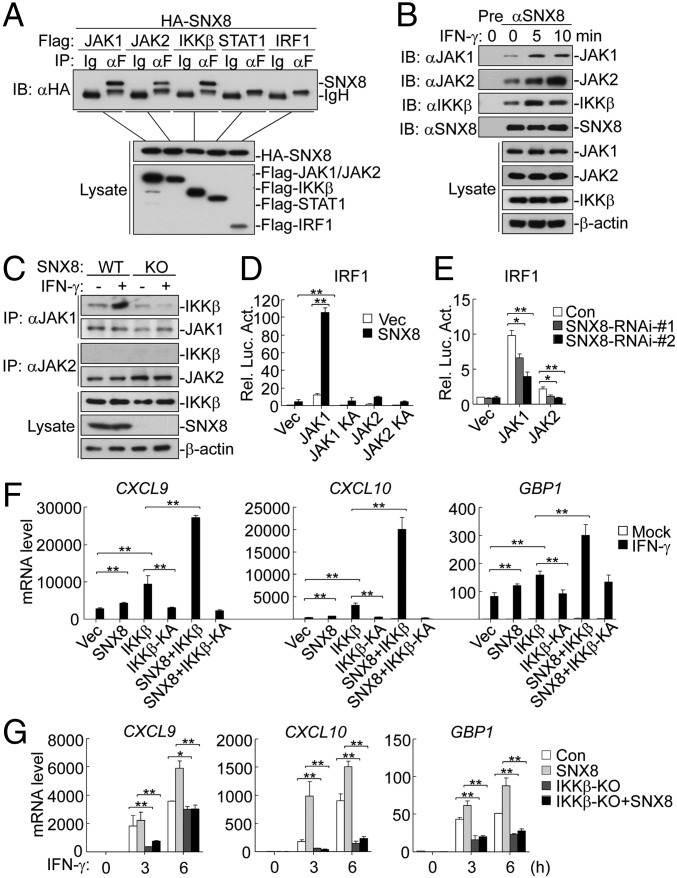
We next determined whether SNX8 is associated with signaling components, including IKKβ in IFNγ-triggered pathways. SNX8 was associated with JAK1, JAK2, and IKKβ, but not with STAT1 or IRF1 in mammalian overexpression system (Fig. 2A). Endogenous SNX8 interacted weakly with JAK1, JAK2, and IKKβ in unstimulated cells and these associations were enhanced after IFNγ stimulation (Fig. 2B). These results suggest that associations of SNX8 with JAK1/2 and IKKβ are increased after IFNγ stimulation.
Fig. 2.
SNX8 interacts with JAK1/2 and IKKβ. (A) SNX8 interacts with JAK1/2 and IKKβ. The 293 cells were transfected with the indicated plasmids for 24 h before coimmunoprecipitation and immunoblot analysis. (B) Endogenous associations between SNX8 and JAK1/2 or IKKβ. The THP1 cells were left untreated or treated with IFNγ for the indicated times before coimmunoprecipitation and immunoblot analysis. (C) SNX8 deficiency impairs association of IKKβ with JAK1. HeLa cells were either untreated or treated with IFNγ for 10 min before coimmunoprecipitation and immunoblot analysis. (D) Effects of SNX8 on JAK1/2-mediated activation of the IRF1 promoter. The 293 cells were transfected with the IRF1 promoter reporter and the indicated plasmids for 20 h before luciferase assays. (E) Effects of SNX8 knockdown on JAK1/2-mediated activation of the IRF1 promoter. The 293 cells were transfected with SNX8-RNAi plasmids for 24 h, followed by further transfection of an IRF1 promoter reporter and the indicated expression plasmids for 20 h before luciferase assays. (F) The synergetic effects of SNX8 on IKKβ-mediated transcription of IFNγ-stimulated genes. HeLa cells were transfected with the indicated plasmids for 24 h and then either untreated or treated with IFNγ for the indicated times before qPCR experiments. (G) Effects of IKKβ deficiency on SNX8-mediated transcription of IFNγ-stimulated genes. HeLa cells were transfected with the indicated plasmids for 24 h and then either untreated or treated with IFNγ for the indicated times before qPCR experiments. Graphs show mean ± SD, n = 3. **P < 0.01, *P < 0.05.
It has been shown that PX-containing SNX family proteins may act as scaffold proteins (26). Because SNX8 interacted with JAK1/2 and IKKβ, we hypothesized that SNX8 might positively regulate IFNγ-triggered signaling by facilitating the associations between IKKβ and JAKs. SNX8 deficiency impaired the association of IKKβ with JAK1 after IFNγ stimulation (Fig. 2C), whereas JAK2-IKKβ association was undetectable (Fig. 2C). These data suggest that SNX8 acts as a scaffolding protein that facilitates JAK1-IKKβ association.
Since SNX8 is associated with JAK1/2 and IKKβ, we next determined whether SNX8 regulates JAK1/2 and IKKβ activity. As shown in Fig. 2D, overexpression of SNX8 activated the IRF1 promoter, which was potentiated by JAK1 or JAK2 and inhibited by dominant negative mutants of JAK1 or JAK2. Conversely, JAK1- or JAK2-mediated activation of the IRF1 promoter was substantially inhibited by knockdown of SNX8 (Fig. 2E). These data suggest that JAK1/2 and SNX8 are mutually required for activation of the IRF1 promoter. In addition, SNX8-mediated induction of CXCL9, CXCL10, and GBP1 genes was potentiated by IKKβ but inhibited by a kinase inactive mutant of IKKβ (IKKβ-K44A) (Fig. 2F). IKKβ deficiency also inhibited SNX8-mediated induction of CXCL9, CXCL10, and GBP1 genes (Fig. 2G). These results suggest that IKKβ acts downstream of SNX8 in regulating a subset of IFNγ-induced downstream genes.
It has been shown that the IFNγ-triggered and IKKβ-dependent noncanonical induction of a subset of downstream genes does not require NF-κB activation (14). We further confirm this observation. Although p65 deficiency impaired TNFα-induced transcription of downstream genes including Tnf, Il6, Cxcl10, and Il1β (Fig. S4A), it did not inhibit IFNγ-induced phosphorylation of STAT1 and transcription of downstream genes, including Gpb1, Cxcl9, and Cxcl10 (Fig. S4 B and C). Our data suggest that IKKβ-mediated induction of a subset of IFNγ responsive genes (Gbp1, Cxcl9, Cxcl10, etc.) is independent of NF-κB activation.
SNX8 Promotes IKKβ Self-Association and Autophosphorylation.
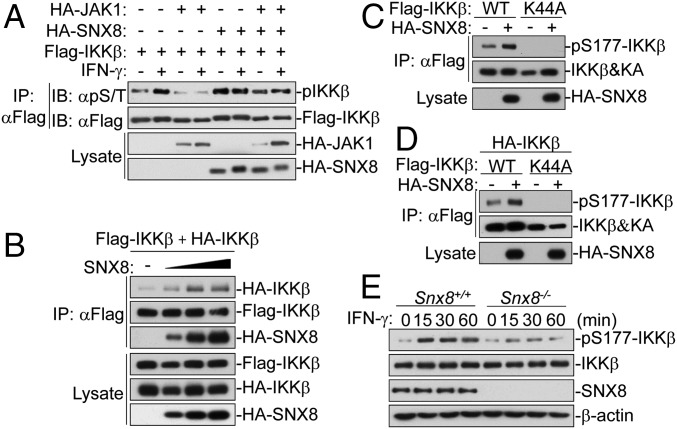
We next investigated how SNX8 regulates IKKβ in IFNγ-triggered signaling. In our earlier experiments, we found that IKKβ-K44A failed to induce Cxcl9, Cxcl10, and Gbp1 genes (Fig. 2F), suggesting that the kinase activity of IKKβ is required for its regulation of IFNγ-triggered signaling. We therefore determined whether IFNγ regulates IKKβ phosphorylation. As shown in Fig. 3A, we found that IFNγ induced serine/threonine phosphorylation of IKKβ. SNX8 markedly increased IKKβ phosphorylation even without IFNγ stimulation (Fig. 3A). We next determined whether SNX8 regulates IKKβ dimerization/oligomerization and autoactivation. SNX8 markedly enhanced IKKβ self-association in a dose-dependent manner in a mammalian overexpression system (Fig. 3B). Because SNX8 has no expected enzymatic activity, we reasoned that SNX8 promotes IKKβ autophosphorylation. Previously, it has been shown that phosphorylation at Ser177 of IKKβ is required for its activation (27–29). As shown in Fig. 3C, overexpression of SNX8 caused phosphorylation of wild-type IKKβ at Ser177 but not IKKβ-K44A. We further determined whether SNX8 promotes IKKβ autophosphorylation or transphosphorylation at Ser177. As shown in Fig. 3D, SNX8 increased the phosphorylation of Flag-tagged wild-type IKKβ but not IKKβ-K44A at Ser177 even in the presence of HA-tagged wild-type IKKβ. These results suggest that SNX8 promotes auto- but not transphosphorylation of IKKβ at Ser177.
Fig. 3.
SNX8 promotes IKKβ self-association and autophosphorylation. (A) SNX8 promotes phosphorylation of IKKβ. The 293 cells were transfected with the indicated plasmids for 24 h and then either untreated or treated with IFNγ before coimmunoprecipitation and immunoblot analysis. (B) SNX8 promotes IKKβ self-association. The experiments were similarly performed as in A. (C) SNX8 does not promote phosphorylation of IKKβ (K44A). The experiments were similarly performed as in A. (D) Effects of SNX8 on phosphorylation of IKKβ and its mutant at Ser177. The experiments were similarly performed as in A. (E) IFNγ-induced phosphorylation of IKKβ at Ser177 in wild-type and SNX8-deficient MLFs. Snx8+/+ and Snx8−/− MLFs were left untreated or treated with IFNγ for the indicated times before immunoblot analysis.
We further examined phosphorylation of endogenous IKKβ in the presence or absence of IFNγ stimulation. As shown in Fig. 3E, IFNγ-induced phosphorylation of IKKβ at Ser177 was impaired in Snx8−/− MLFs. These results suggest that SNX8 mediates IKKβ dimerization/oligomerization and autophoshorylation at Ser177 after IFNγ stimulation.
Phosphorylation of SNX8 at Tyr95 and Tyr126 Is Required for Its Activity.
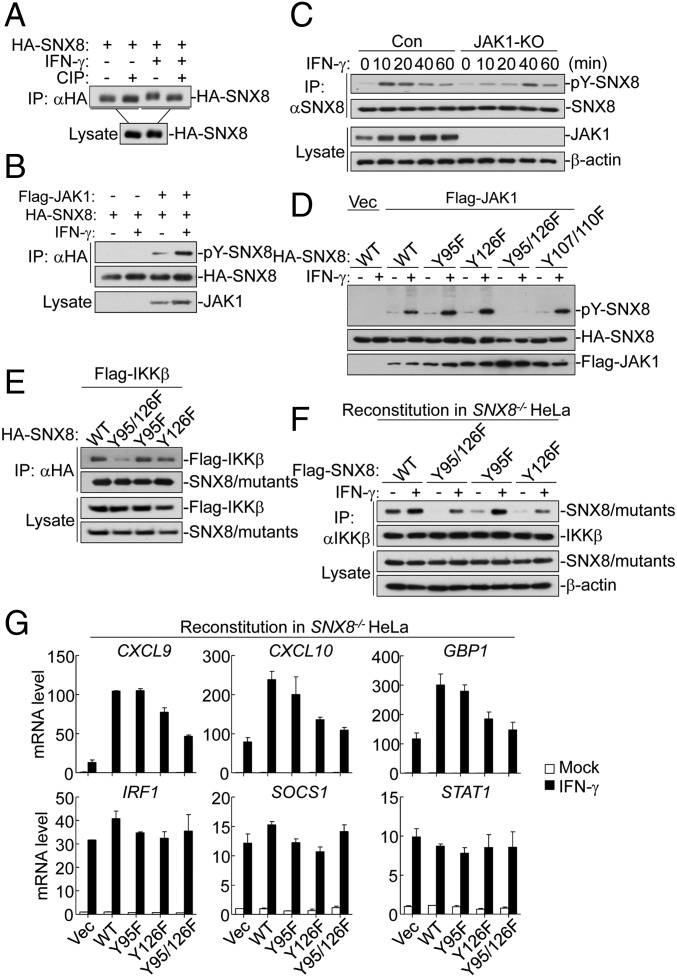
During our coimmunoprecipitation assays, we observed that IFNγ stimulation caused a shift of SNX8 to a higher molecular weight band (Fig. 3A). Such a shift was due to phosphorylation of SNX8 because calf intestine phosphatase (CIP) treatment reversed the shift (Fig. 4A). Interestingly, we found that SNX8 was tyrosine phosphorylated by JAK1, which was further increased by IFNγ stimulation (Fig. 4B). In addition, tyrosine phosphorylation of SNX8 induced by IFNγ was decreased in JAK1-deficient cells in the early phase of stimulation (Fig. 4C). These data suggest that SNX8 was phosphorylated by JAK1 after IFNγ stimulation.
Fig. 4.
Phosphorylation of SNX8 at Tyr95 and Tyr126 is essential for its activity. (A) IFNγ induces phosphorylation of SNX8. The 293 cells were transfected with the indicated plasmids. Cell lysates were immunoprecipitated with anti-HA. The immunoprecipitates were treated with buffer or calf intestine phosphatase (CIP) and analyzed by immunoblots. (B) SNX8 is phosphorylated by JAK1. The 293 cells were transfected with the indicated plasmids for 24 h and then either left untreated or treated with IFNγ before coimmunoprecipitation and immunoblot analysis. (C) Effects of JAK1 deficiency on IFNγ-induced phosphorylation of SNX8. The control or JAK1-deficient HeLa cells were untreated or treated with IFNγ for the indicated times before coimmunoprecipitation and immunoblot analysis. (D) Tyr95 and Tyr126 are the major tyrosine phosphorylation sites of SNX8. The 293 cells were transfected with the indicated plasmids for 24 h and then either left untreated or treated with IFNγ for 10 min before immunoblot analysis. (E) Phosphorylation of SNX8 facilitates its recruitment of IKKβ. The experiments were similarly performed as in D. (F) Endogenous association between IKKβ and SNX8 or its mutants in HeLa cells. SNX8-deficient cells reconstituted with SNX8 or the indicated mutants were untreated or treated with IFNγ for 10 min before endogenous coimmunoprecipitation and immunoblot analysis. (G) Effects of SNX8 mutants on IFNγ-induced transcription of downstream genes in HeLa cells. SNX8-deficient cells reconstituted with SNX8 or the indicated mutants were untreated or treated with IFNγ for 6 h before qPCR experiments.
To identify potential tyrosine phosphorylation residues of SNX8, we performed mutagenesis analysis. The results indicated that Tyr95 and Tyr126 were the major tyrosine phosphorylation residues in human SNX8 (Fig. 4D). Coimmunoprecipitation experiments indicated that double mutation of SNX8 impaired its interaction with IKKβ (Fig. 4E). Consistently, double mutation of SNX8 impaired its interaction with IKKβ after IFNγ stimulation in reconstitution experiments (Fig. 4F). Interestingly, the Y126F mutant of SNX8 had slightly decreased association with IKKβ, but not the Y95F mutant. Consistent with the biochemical results, the Y95F/Y126F double and Y126F single mutants had dramatically or partially reduced abilities to potentiate IFNγ-induced transcription of downstream genes including Cxcl9, Cxcl10, and Gbp1 but not Irf1, Stat1, and Socs1 genes, whereas the Y95F mutant had similar ability as the wild-type SNX8 in promoting IFNγ-induced transcription of downstream genes (Fig. 4G). These results suggest that tyrosine phosphorylation of SNX8 at Tyr95 and Tyr126 is important for its association with IKKβ and activity.
SNX8 Is Essential for Host Defense Against L. monocytogenes Infection in Mice.
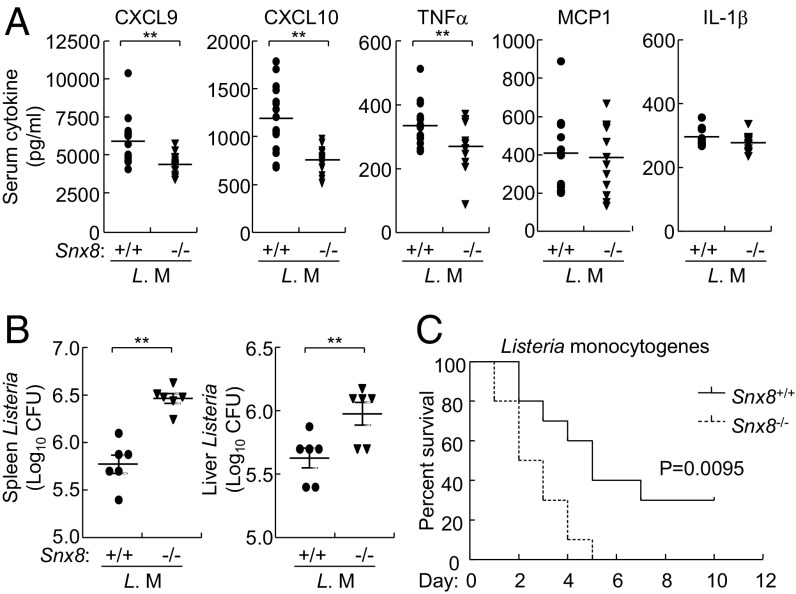
It has been demonstrated that the IFNγ-induced GTPases of the GBP family play pivotal roles in host defense against intracellular pathogens (1, 17, 18). A family of IFNγ-induced chemokines, including CXCL9, CXCL10, and CXCL11, displays direct antimicrobial activity against L. monocytogenes (13, 15). Since SNX8 selectively mediated expression of IFNγ-induced CXCL9, CXCL10, CXCL11, and GBP, we determined whether SNX8 is important for host defense against L. monocytogenes in vivo. We found that serum chemokines and cytokines induced by L. monocytogenes infection, including CXCL9, CXCL10, and TNFα, were severely impaired in Snx8−/− in comparison with wild-type mice (Fig. 5A). In the same experiments, levels of MCP1 and IL-1β induced by L. monocytogenes were similar in Snx8−/− and Snx8+/+ mice (Fig. 5A). In addition, after i.p. infection with L. monocytogenes for 3 d, the bacterial titers in the livers and spleens of Snx8−/− mice were significantly increased in comparison with those of their wild-type littermates (Fig. 5B). We also compared the survival rates of Snx8−/− and Snx8+/+ mice after L. monocytogenes infection intraperitoneally at a high dose (5 × 107 cfu per mouse). The results indicated that Snx8−/− mice were more susceptible to Listeria-triggered death than their wild-type littermates (Fig. 5C). Collectively, these data suggest that SNX8 is essential for host defense against L. monocytogenes in mice.
Fig. 5.
SNX8 is essential for host defense against L. monocytogenes infection in mice. (A) Effects of SNX8 deficiency on serum levels of CXCL9, CXCL10, and TNFα induced by i.p. injection of mice (n = 11 per group) with L. monocytogenes (4 × 106 each mouse) for 6 h (**P < 0.01). (B) Effects of SNX8 deficiency on bacterial loads in mice. Plaque assays of homogenized spleens and livers from mice (n = 6 per group) 3 d after i.p. injection of L. monocytogenes (4 × 106 each mouse) (**P < 0.01). (C) Survival of mice (n = 10 per group) after i.p. injection of L. monocytogenes (5 × 107 each mouse) (**P < 0.01).
Our results demonstrated that SNX8 regulates the IFNγ-triggered noncanonical pathway through IKKβ, which also plays a pivotal role in TLR2 sensing of L. monocytogenes infection. Therefore, we determined whether SNX8 plays a role in TLR2 signaling. SNX8 deficiency had no obvious effects on PGN (ligand for TLR2) induced phosphorylation of IKKβ and IκBα, and transcription of downstream genes in BMDCs (Fig. S5 A and B). In addition, levels of serum cytokines induced by PGN, including TNFα and IL-6, were similar between Snx8−/− and Snx8+/+ mice (Fig. S5C). These results suggest that SNX8 is not required for TLR2-mediated signaling.
Discussion
IFNγ is an important mediator of innate and adaptive immune responses with a key role in clearance of various pathogens. It has been well documented that IFNγ signals through IFNγ receptors (IFNGR1 and IFNGR2), which are associated with tyrosine kinases JAK1 and JAK2. JAKs directly phosphorylate STAT1 at Tyr701 and indirectly lead to its phosphorylation at Ser727. In addition, IKKβ, a master activator of inflammatory response, is required to mediate the induction of a subset of IFNγ-stimulated genes. In this study, we have demonstrated that SNX8 acts as a switch for the IFNγ-triggered noncanonical signaling pathway by selectively mediating JAK1-IKKβ association and plays a critical role in host defense against intracellular bacterial infection.
Our results suggest that a SNX8-IKKβ axis is responsible for induction of a specific subset of IFNγ-responsive genes, such as Gbp1/2, Cxcl9, and Cxcl10. Previously, it has been demonstrated that IFNγ fails to induce expression of CXCL10 and GBP1 with the normal activation of STAT1 in IKKβ-deficient cells (14). Our RNA-sequencing data revealed that SNX8- and IKKβ-mediated genes induced by IFNγ were significantly correlated, including GBP1/2, and CXCL10/11. Interestingly, we found that SNX8 and IKKβ synergistically induced GBP, CXCL9, and CXCL10. Coimmunoprecipitation experiments indicated that SNX8 was constitutively associated with IKKβ, and the association was enhanced by IFNγ stimulation. SNX8 promoted dimerization/oligomerization and autophosphorylation of IKKβ at Ser177. Studies with Snx8−/− MLFs indicated that SNX8 was essential for IFNγ-induced phosphorylation of IKKβ at Ser177. These results suggest that SNX8 recruits IKKβ and mediates IFNγ-induced IKKβ activation as well as transcription of a subset of downstream genes. A previous study and our results suggest that IKKβ-mediated induction of a subset of IFNγ responsive genes (Gbp1, Cxcl9, Cxcl10, etc.) is independent of NF-κB activation. Since IKKβ deficiency does not affect STAT1 Y701 and S727 phosphorylation (Fig. S3B), it is possible that IKK mediates the activation of an unknown transcription factor or STAT1 by an unknown mechanism, which is responsible for the induction of a specific subset of IFNγ-responsive genes.
Our results suggest that IFNγ stimulation induces JAK1/2-mediatd tyrosine phosphorylation of SNX8, which is important for activating IKKβ. The phosphorylation of SNX8 reached peak at the early phase (10–20 min) and then decreased to a lower level at the late phase (40 and 60 min) after IFNγ stimulation. The IFNγ-induced increase of SNX8 phosphorylation was markedly decreased in JAK1-deficient cells. In the late phase of IFNγ stimulation, the phosphorylation level of SNX8 had no obvious difference between wild-type and JAK1-deficient cells. One explanation for this observation is that an additional kinase(s) exists to catalyze SNX8 phosphorylation at the late phase of IFNγ stimulation. Mutagenesis experiments indicated that Tyr95 and Tyr126 were the major phosphorylation residues of SNX8. A mutant in which these two residues were changed to phenylalanines decreased its association with IKKβ and had dramatically reduced ability to potentiate IFNγ-triggered induction of downstream genes, including Gbp1, Cxcl9, and Cxcl10 but not Irf1, Socs1, and Stat1. These results further support the idea that the JAK1/2-SNX8-IKKβ axis mediates the IFNγ-triggered noncanonical pathway.
Our results indicated that SNX8 deficiency partially inhibited tyrosine phosphorylation of STAT1, but inhibited only a subset of downstream genes induced by IFNγ, such as Gbp1 and Cxcl9/10/11. In fact, the effects of SNX8 deficiency on IFNγ-induced downstream genes can be classified into three groups: (i) transcriptional induction is unaffected, such as in Socs1 and Stat1; (ii) transcriptional induction is partially affected, such as in Irf1; and (iii) transcriptional induction is dramatically affected, such as in Gbp1, Cxcl9, and Cxcl10. For the first group of genes (such as Socs1 and Stat1), although SNX8 deficiency partially reduced IFNγ-induced STAT1 Y701 phosphorylation, this might not reach a threshold to affect their transcription. For the second group of genes (such as Irf1), SNX8 deficiency partially affected IFNγ-induced transcription of Irf1 gene at early time points after stimulation, while IKKβ deficiency had no effects on its induction and STAT1 phosphorylation (14) (Fig. S3B). The simplest explanation for this is that Irf1 can be induced by the canonical JAK-STAT1 pathway, independent of IKKβ, and SNX8 may contribute to STAT1 activation and Irf1 induction through an additional unknown mechanism. For the third group of genes (such as Gbp1, Cxcl9, and Cxcl10), their transcription requires both JAK1-STAT1 activation and the JAK1-SNX8-IKKβ axis reported in this paper.
Our results indicated that SNX8 selectively regulates tyrosine but not serine phosphorylation of STAT1. Previous studies have demonstrated that the tyrosine phosphorylation and nuclear translocation are required for STAT1 S727 phosphorylation (30) and the nuclear kinase CDK8 mediates the STAT1 S727 phosphorylation in response to IFN stimulation (7). However, much evidence suggests that the STAT1 S727 phosphorylation is not just a nuclear event. Phosphorylated STAT1 S727 is observed in resting natural killer (NK) cells independent of its tyrosine phosphorylation (31, 32). CDK8 plays an essential role in mediating the basal STAT1 S727 phosphorylation (31). In our experiments, the STAT1 S727 phosphorylation was detected in the cytosol even without its Tyr701 phosphorylation in JAK1-deficient cells, whereas SNX8 deficiency partially inhibited IFNγ-triggered tyrosine phosphorylation and its nuclear localization but not serine phosphorylation (Fig. S6).
Based on these results, we propose a model of the role of SNX8 in IFNγ-triggered signaling and effects. IFNγ stimulation induces activation of JAK1/2, which then recruits SNX8 and phosphorylates it at Tyr95 and Tyr126. Phosphorylated SNX8 acts as a scaffold protein to recruit IKKβ to the JAK1 complex, leading to IKKβ dimerization/oligomerization and autophosphorylation, as well as selective induction of a subset of downstream genes that are important for host defense against intracellular bacterial infection. Although SNX8 is a member of the sorting protein family, it can constitutively associate with JAKs and IKKβ, suggesting that at least a fraction of SNX8 is a component of the JAKs complexes. Our study provides insight into the mechanisms of IFNγ-triggered noncanonical signaling pathways as well as antibacterial effects.
Materials and Methods
All animal experiments were performed in accordance with the Wuhan University Animal Care and Use Committee guidelines. The information on reagents, antibodies, constructs, PCR primers and RNAi target sequences are described in SI Materials and Methods. The methods for expression screens, reporter assays, coimmunoprecipitation and immunoblot analysis, RNAi transduction, CRISPR-Cas9 knockout, RNA-seq, ELISA, flow cytometry, cell fractionation, L. monocytogenes infection and PGN injection in mice, and statistical analysis are previously described (33–35) and the details are presented in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by State Key R&D Program of China Grants 2017YFA0505800, 2016YFA0502102, and 2014CB910103; and National Natural Science Foundation of China Grants 31630045, 31521091, 91429304, and 31671465.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713462114/-/DCSupplemental.
References
- 1.Yamamoto M, et al. A cluster of interferon-γ-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity. 2012;37:302–313. doi: 10.1016/j.immuni.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Kim BH, et al. A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. Science. 2011;332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 3.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden EC, et al. Interferons at age 50: Past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villarino AV, Kanno Y, O’Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. 2017;18:374–384. doi: 10.1038/ni.3691. [DOI] [PubMed] [Google Scholar]
- 6.Decker T, Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 7.Bancerek J, et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity. 2013;38:250–262. doi: 10.1016/j.immuni.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury GG. A linear signal transduction pathway involving phosphatidylinositol 3-kinase, protein kinase Cepsilon, and MAPK in mesangial cells regulates interferon-gamma-induced STAT1alpha transcriptional activation. J Biol Chem. 2004;279:27399–27409. doi: 10.1074/jbc.M403530200. [DOI] [PubMed] [Google Scholar]
- 9.Deb DK, et al. Activation of protein kinase C δ by IFN-γ. J Immunol. 2003;171:267–273. doi: 10.4049/jimmunol.171.1.267. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen H, Ramana CV, Bayes J, Stark GR. Roles of phosphatidylinositol 3-kinase in interferon-gamma-dependent phosphorylation of STAT1 on serine 727 and activation of gene expression. J Biol Chem. 2001;276:33361–33368. doi: 10.1074/jbc.M105070200. [DOI] [PubMed] [Google Scholar]
- 11.Majoros A, et al. Canonical and non-canonical aspects of JAK-STAT signaling: Lessons from interferons for cytokine responses. Front Immunol. 2017;8:29. doi: 10.3389/fimmu.2017.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545–551. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole AM, et al. Cutting edge: IFN-inducible ELR- CXC chemokines display defensin-like antimicrobial activity. J Immunol. 2001;167:623–627. doi: 10.4049/jimmunol.167.2.623. [DOI] [PubMed] [Google Scholar]
- 14.Sizemore N, et al. Inhibitor of kappaB kinase is required to activate a subset of interferon gamma-stimulated genes. Proc Natl Acad Sci USA. 2004;101:7994–7998. doi: 10.1073/pnas.0401593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dussurget O, Bierne H, Cossart P. The bacterial pathogen Listeria monocytogenes and the interferon family: Type I, type II and type III interferons. Front Cell Infect Microbiol. 2014;4:50. doi: 10.3389/fcimb.2014.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shultz DB, Rani MRS, Fuller JD, Ransohoff RM, Stark GR. Roles of IKK-beta, IRF1, and p65 in the activation of chemokine genes by interferon-gamma. J Interferon Cytokine Res. 2009;29:817–824. doi: 10.1089/jir.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. IFN-inducible GTPases in host cell defense. Cell Host Microbe. 2012;12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ireton K, Rigano LA, Dowd GC. Role of host GTPases in infection by Listeria monocytogenes. Cell Microbiol. 2014;16:1311–1320. doi: 10.1111/cmi.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johannes L, Wunder C. The SNXy flavours of endosomal sorting. Nat Cell Biol. 2011;13:884–886. doi: 10.1038/ncb2300. [DOI] [PubMed] [Google Scholar]
- 20.Muirhead G, Dev KK. The expression of neuronal sorting nexin 8 (SNX8) exacerbates abnormal cholesterol levels. J Mol Neurosci. 2014;53:125–134. doi: 10.1007/s12031-013-0209-z. [DOI] [PubMed] [Google Scholar]
- 21.Dyve AB, Bergan J, Utskarpen A, Sandvig K. Sorting nexin 8 regulates endosome-to-Golgi transport. Biochem Biophys Res Commun. 2009;390:109–114. doi: 10.1016/j.bbrc.2009.09.076. [DOI] [PubMed] [Google Scholar]
- 22.Lucas M, et al. Structural mechanism for cargo recognition by the retromer complex. Cell. 2016;167:1623–1635.e14. doi: 10.1016/j.cell.2016.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlton J, et al. Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high- curvature membranes and 3-phosphoinositides. Curr Biol. 2004;14:1791–1800. doi: 10.1016/j.cub.2004.09.077. [DOI] [PubMed] [Google Scholar]
- 24.Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- 25.Sims SH, et al. A novel interferon-inducible domain: Structural and functional analysis of the human interferon regulatory factor 1 gene promoter. Mol Cell Biol. 1993;13:690–702. doi: 10.1128/mcb.13.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. Biochem J. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- 27.Zandi E, Chen Y, Karin M. Direct phosphorylation of IkappaB by IKKalpha and IKKbeta: Discrimination between free and NF-kappaB-bound substrate. Science. 1998;281:1360–1363. doi: 10.1126/science.281.5381.1360. [DOI] [PubMed] [Google Scholar]
- 28.Xu G, et al. Crystal structure of inhibitor of κB kinase β. Nature. 2011;472:325–330. doi: 10.1038/nature09853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mercurio F, et al. IKK-1 and IKK-2: Cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 30.Sadzak I, et al. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc Natl Acad Sci USA. 2008;105:8944–8949. doi: 10.1073/pnas.0801794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putz EM, et al. CDK8-mediated STAT1-S727 phosphorylation restrains NK cell cytotoxicity and tumor surveillance. Cell Reports. 2013;4:437–444. doi: 10.1016/j.celrep.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putz EM, Gotthardt D, Sexl V. STAT1-S727–The license to kill. OncoImmunology. 2014;3:e955441. doi: 10.4161/21624011.2014.955441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei J, Lian H, Zhong B, Shu HB. Parafibromin is a component of IFN-γ-triggered signaling pathways that facilitates JAK1/2-mediated tyrosine phosphorylation of STAT1. J Immunol. 2015;195:2870–2878. doi: 10.4049/jimmunol.1501111. [DOI] [PubMed] [Google Scholar]
- 34.Luo WW, et al. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol. 2016;17:1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 35.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.