Significance
Cholesterol is important for membrane protein function, but cholesterol-binding structures of membrane proteins are difficult to determine by X-ray crystallography and electron microscopy due to the small size and dynamic nature of cholesterol. We have developed a solid-state NMR approach to determine the cholesterol-binding structure of membrane proteins in lipid bilayers. Applied to the influenza M2 protein, the measured interatomic distances and cholesterol orientational angles indicate that cholesterol binds M2 in a substoichiometric fashion, flanking methyl-rich transmembrane (TM) residues near an amphipathic helix, without requiring a cholesterol recognition sequence motif, and this substoichiometric binding uniquely correlates with membrane curvature generation. These results give unprecedented insights into how cholesterol clusters M2 to the neck of the budding virus to mediate membrane scission.
Keywords: membrane proteins, 19F-NMR, deuterium NMR, docking, membrane scission
Abstract
The influenza M2 protein not only forms a proton channel but also mediates membrane scission in a cholesterol-dependent manner to cause virus budding and release. The atomic interaction of cholesterol with M2, as with most eukaryotic membrane proteins, has long been elusive. We have now determined the cholesterol-binding site of the M2 protein in phospholipid bilayers using solid-state NMR spectroscopy. Chain-fluorinated cholesterol was used to measure cholesterol proximity to M2 while sterol-deuterated cholesterol was used to measure bound-cholesterol orientation in lipid bilayers. Carbon–fluorine distance measurements show that at a cholesterol concentration of 17 mol%, two cholesterol molecules bind each M2 tetramer. Cholesterol binds the C-terminal transmembrane (TM) residues, near an amphipathic helix, without requiring a cholesterol recognition sequence motif. Deuterium NMR spectra indicate that bound cholesterol is approximately parallel to the bilayer normal, with the rough face of the sterol rings apposed to methyl-rich TM residues. The distance- and orientation-restrained cholesterol-binding site structure shows that cholesterol is stabilized by hydrophobic interactions with the TM helix and polar and aromatic interactions with neighboring amphipathic helices. At the 1:2 binding stoichiometry, lipid 31P spectra show an isotropic peak indicative of high membrane curvature. This M2–cholesterol complex structure, together with previously observed M2 localization at phase boundaries, suggests that cholesterol mediates M2 clustering to the neck of the budding virus to cause the necessary curvature for membrane scission. The solid-state NMR approach developed here is generally applicable for elucidating the structural basis of cholesterol’s effects on membrane protein function.
Cholesterol plays important roles in membrane protein functions such as neurotransmission and ion transport (1–4). Cholesterol can affect protein structure by indirectly modulating the physical properties of the lipid membrane or by direct and site-specific binding. Which mechanism is operative and the details of the mechanism have been elusive due to the scarcity of high-resolution structures of membrane proteins in cholesterol-containing lipid membranes, except for adventitious capturing of cholesterol in crystal structures of membrane proteins (5). A number of cholesterol-binding amino acid sequence motifs have been proposed, including a two-helix cholesterol consensus motif (CCM) in G-protein–coupled receptors (5) and a single-helix cholesterol recognition amino acid consensus (CRAC) motif and its inverted counterpart CARC (6, 7). These motifs share the common feature of a branched hydrophobic residue, an aromatic residue, and a positively charged residue. The spacing between these residues is variable in CRAC and CARC motifs but is strictly defined in the CCM. The CCM additionally includes a second aromatic residue from a neighboring transmembrane (TM) helix. Due to the scarcity of high-resolution structural information, only the CCM has been observed (5), which reduces the predictive power of the loosely defined CRAC and CARC cholesterol-binding motifs (7).
The influenza M2 protein mediates membrane scission in the last step of virus budding in a cholesterol-dependent fashion: in membranes with less than ∼23 mol% cholesterol, M2 causes budding of giant unilamellar vesicles (GUVs) (8). The domain responsible for membrane scission is an amphipathic helix (AH) (8) that is C-terminal to the TM domain. M2 isolated from virus-infected cells copurifies with cholesterol at a ratio of 0.5–0.9 cholesterol per protein monomer (9). Cholesterol also modulates drug binding to the TM pore to block its proton channel activity (10–13). A cholesterol-binding CRAC motif has been proposed for the majority of M2 sequences (14). Located in the amphipathic helix, this CRAC motif consists of a Leu, Tyr, and a cationic Arg or Lys (Fig. 1A). However, 2% of M2 sequences, including the 1968 pandemic flu M2 and the commonly studied Udorn M2, lack the CRAC motif without adverse consequences to virus assembly and replication (15, 16) or proton channel activity (9). In fact, the membrane scission activity was first observed in Udorn M2, which lacks the complete CRAC motif. Deletion of the CRAC motif in the M2 sequence from the 1933 WSN H1N1 strain did not affect in vitro virus replication but attenuated virulence in a mouse model (15). Recently, a solid-state NMR (SSNMR) study detected cholesterol–M2 cross-peaks (11) but was unable to assign the cross-peaks to a specific residue. Thus, whether the CRAC motif is required for M2–cholesterol interaction, and whether cholesterol binds a specific site in the protein, remain unknown.
Fig. 1.
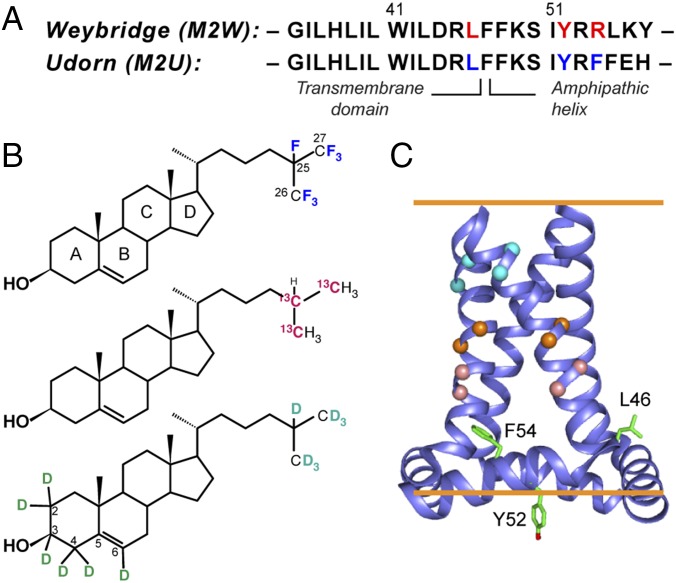
Strategy for determining the cholesterol-binding structure of M2. (A) Amino acid sequences of Weybridge and Udorn M2, showing the CRAC motif residues (red) in M2W and its loss in M2U (blue). (B) Cholesterol labeled with 19F, 13C, and 2H for determining cholesterol binding to M2. (C) SSNMR structure of Udorn M2(22–62) (PDB ID code 2L0J). Cα atoms whose distances to cholesterol were measured are shown as balls, and the putative CRAC residues are shown as sticks.
To elucidate atomic details of cholesterol interaction with M2, we have now determined the cholesterol-binding site of M2 and bound-cholesterol orientation in phospholipid bilayers using magic-angle–spinning (MAS) SSNMR. M2 peptides containing various 13C,15N-labeled residues were reconstituted into 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) bilayers that contain 17 mol% cholesterol, a composition that has been shown by confocal and electron microscopy to permit membrane budding (8). By measuring protein–cholesterol distances, we show that cholesterol binds at a unique site of M2. Comparing M2 sequences with and without the amphipathic helix, and with and without the CRAC motif, we show that cholesterol binding requires the amphipathic helix but not the CRAC motif. Furthermore, we used 2H NMR to measure the orientation of bound cholesterol. The resulting structural model of the cholesterol-binding site of M2 gives molecular insight into how M2 co-opts cholesterol to carry out membrane scission and mediate virus release.
Results
Protein Sequence Requirements for Cholesterol Binding.
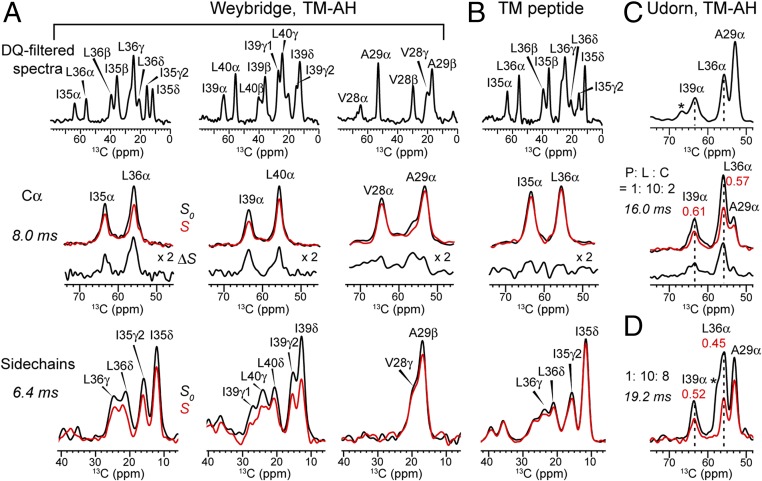
To measure cholesterol–M2 proximity, we combined chain-heptafluorinated cholesterol (F7-cholesterol, Fig. 1B) with 13C-labeled protein and conducted 13C–19F rotational-echo–double-resonance (REDOR) distance measurements (17–19) (Fig. S1). F7-cholesterol has the same interfacial area, molecular orientation, and phospholipid interaction as hydrogenated cholesterol based on pressure-area isotherm data (20). Six residues on the lipid-facing surface and helix–helix interface of the TM domain were 13C-labeled for the distance experiments (Fig. 1C and Table S1). M2 peptides (residues 22–61) corresponding to the CRAC-containing Weybridge strain (M2W) and the CRAC-absent Udorn strain (M2U) were synthesized. The 13C chemical shifts of the labeled residues are well resolved (Fig. 2), thus allowing Cα distances to F7-cholesterol to be measured and compared. In both M2W and M2U TM-AH peptides, the I35, L36, I39, and L40 Cα’s show significant intensity differences between control spectra (S0) measured without 19F pulses and dipolar-dephased spectra (S) measured with 19F pulses (Fig. 2 A and C), indicating that the cholesterol tail lies close to these Cα atoms. Similar dephasing was observed for both M2W and M2U, indicating that the CRAC sequence motif is not required for cholesterol contact to the protein. However, in TM peptides lacking the amphipathic helix, I35 and L36 show negligible intensity differences between the control and dephased spectra, indicating loss of cholesterol binding. This is consistent with previous thiol-disulfide exchange data of the TM peptide that showed limited cholesterol binding (21). Finally, V28 and A29 in the AH-containing M2W peptide show weak dephasing by fluorinated cholesterol. These two residues lie at similar depths in the N-terminal leaflet of the bilayer as the depths of I39 and L40 in the C-terminal leaflet; thus, the weak REDOR dephasing indicates that cholesterol binds only the C-terminal half of the TM helix, near the amphipathic helix. This cholesterol proximity is further supported by significant dephasing of I35, L36, I39, and L40 sidechains, in contrast to the negligible dephasing of V28 and A29 side chains (Fig. 2).
Fig. 2.
Double-quantum (DQ)-filtered 13C spectra and representative REDOR spectra of M2 bound to POPC:POPG (4:1) bilayers containing F7-cholesterol. (A) M2W(22–61) with pairwise 13C-labeled residues at I35/L36, I39/L40, V28/A29. The P:L:C molar ratio is 1:10:2. Top row: 13C spectra. Middle row: Cα J-decoupled REDOR spectra (control S0; dephased S; difference ΔS) at 8.0-ms mixing. Bottom row: Side-chain DQ-filtered REDOR spectra at 6.4-ms mixing. (B) TM peptide (22–46) with 13Cα-labeled I35 and L36. P:L:C = 1:10:2. The rows are the same as in A. (C) M2U(22–61) peptide with 13C-labeled A29, G34, L36, and I39. P:L:C = 1:10:2. Top row: DQ-filtered spectrum. Bottom row: Cα J-decoupled REDOR spectra with 16.0-ms mixing. L36 Cα shows S/S0 = 0.57. (D) REDOR S0 and S spectra of M2U(22–61) at P:L:C = 1:10:8 with 19.2-ms mixing. At this higher cholesterol concentration, L36 Cα shows a lower S/S0 of 0.45, indicating increased cholesterol binding. All REDOR spectra were measured at 235 K under 5-kHz MAS.
The fraction of protein carbons in close proximity to cholesterol is obtained from the lowest REDOR intensity ratios between the 19F-dephased and control spectra. Importantly, in the 17 mol%-cholesterol membrane, the REDOR dephasing curves of both M2W and M2U peptides decayed to a normalized intensity (S/S0) of only 0.5–0.6 instead of 0 (Fig. 3A), indicating that only ∼50% of the protein 13C labels are close to a cholesterol chain. Control experiments on 5-19F-tryptophan gave the expected REDOR dephasing to ∼0 (Fig. S1C), confirming that the incomplete dephasing of M2 results from partial occupancy of cholesterol per M2 monomer. This unexpected half-occupancy in fact lies within the previously reported range of 0.5–0.9 cholesterol per monomer of Weybridge M2 based on tritium-labeling and filipin-staining experiments (9). In addition, electron paramagnetic resonance experiments of M2 peptides with AH spin labels in membranes that contain 20 mol% cholesterol detected the coexistence of a broad, rigid component (∼57%) and a narrow, mobile component, indicating two distinct conformational states of the protein, also consistent with substoichiometric cholesterol binding.
Fig. 3.
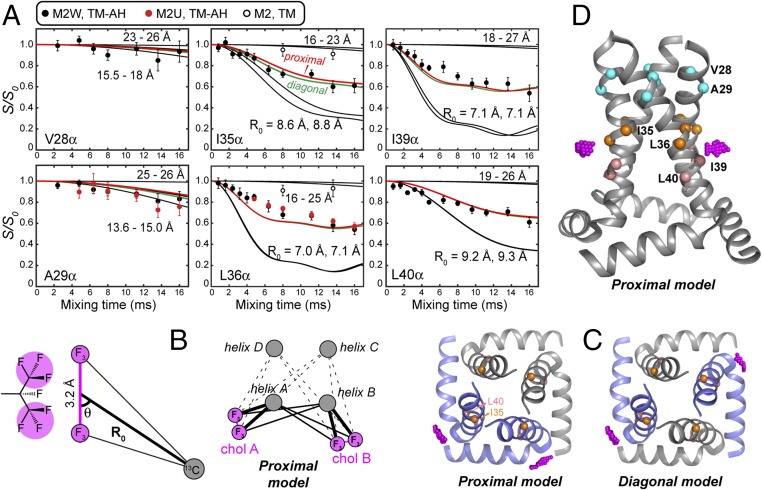
M2–cholesterol distance extraction. (A) Carbon–fluorine REDOR dephasing of Cα sites in M2W, M2U, and M2TM peptides. I35, L36, I39, and L40 Cα in M2(22–61) peptides show significant dipolar dephasing by F7-cholesterol, while V28 and A29 show minimal dephasing. I35 and L36 Cα in M2TM also show negligible dephasing. Best-fit dephasing curves for the proximal (red) and diagonal (green) binding models are both shown. (B) Geometry of a peptide 13C spin near two CF3 groups of cholesterol. Cholesterol can bind in either a proximal mode or a diagonal mode to the tetramer. (C) Top view of the proximal and diagonal binding modes. The CF3 groups are shown in magenta. (D) Side view of the CF3 positions in the proximal binding mode, showing the height of the cholesterol tail relative to the TM helices.
Extraction of Protein–Cholesterol Distances.
Quantitative M2–cholesterol distances were extracted from the mixing-time–dependent REDOR intensity ratios (S/S0) in two steps. We focus on the Cα dephasing, since the backbone structure of the Udorn TM–AH peptide in lipid bilayers has been reported while the side-chain rotameric conformations are not yet known (22). We approximated each CF3 group as a single pseudofluorine (pF) atom, since the dipolar coupling of a 13C atom to a fast-rotating CF3 group can be calculated based on the motional symmetry (Methods). This simplified the consideration of a 13C spin coupled to two CF3 groups to a three-spin system containing one 13C and two pF atoms. Furthermore, when a carbon atom is more than ∼6 Å away from the center of the two methyl groups, the REDOR dephasing mainly depends on the distance R0 (Fig. 3B) and not on the incident angle of the carbon relative to the pF–pF vector (Fig. S2). In the first step, we simulated the REDOR dephasing curves of one Cα coupled to two pF atoms of a single cholesterol molecule, and scaled the resulting curves by 0.5 to reflect the 1:2 binding stoichiometry. This analysis yielded distances of 7–10 Å for the four Cα’s from I35 to L40, while V28 and A29 showed distances longer than 11 Å.
In the second step, we searched a (50 Å)3 cube surrounding the TM–AH structure (PDB ID code 2L0J) at 0.5-Å resolution to find cholesterol tail positions that agree with the four Cα–(pF)2 distances to within ±1 Å. This search restricted the cholesterol isooctyl tail to the exterior of the four-helix bundle, adjacent to the TM helices, while locations inside the pore were readily ruled out by the REDOR data (Fig. S3). Since two cholesterol molecules bind each tetramer (Fig. 3B), we considered simultaneous dephasing of each Cα by four pF atoms. The two cholesterol molecules can bind the tetramer in either a proximal or a diagonal configuration (Fig. 3 C and D). Since the measured 13C signals are the average of four carbons for each residue, we simulated the REDOR curves of all four Cα carbons of each labeled site and averaged these to obtain the final calculated REDOR curves, which were compared with the experimental data. These calculations yielded closest-approach Cα–pF distances of 7.0–7.1 Å for L36 and I39, while I35 and L40 Cα show closest distances of 8.6 and 9.2 Å, respectively (Fig. 3A and Table S2). The proximal and diagonal binding modes gave indistinguishable REDOR curves and are thus both possible from these data.
These protein–cholesterol distances were corroborated by 2D 13C–13C correlation spectra of 25,26,27-13C–labeled cholesterol (Fig. 1B) in M2U-containing membranes. At a mixing time of 1.0 s and with 38-fold sensitivity enhancement by dynamic nuclear polarization (23), the I39 Cα and Cγ2 atoms exhibit cross-peaks with 13C-labeled cholesterol C26 and C27 (Fig. S4). These cross-peaks were absent at short mixing times and absent in samples with unlabeled cholesterol, confirming the assignment. The I39-cholesterol cross-peaks have ∼70% of the intensities of an intramolecular G34–I39 cross-peak, which corresponds to a 9.0-Å distance. Given the 1:2 binding stoichiometry of cholesterol to M2, the cross-peak intensities suggest ∼8-Å distances between cholesterol 26,27-13C and I39 carbons. G34 and A29 do not exhibit cross-peaks to cholesterol, consistent with the 13C–19F REDOR data. The agreement between the 13C–13C cross-peaks and the 13C–19F REDOR data also confirms that fluorination does not affect cholesterol binding to M2. Additionally, at the protein monomer:lipid:cholesterol (P:L:C) molar ratio of 1:10:2, the average distance between the edges of two TM tetramers is ∼20 Å; thus, the cholesterol distances of 7–9 Å to TM residues is not due to confinement of cholesterol to insufficiently solvated M2 tetramers.
Determination of M2-Bound Cholesterol Orientation.
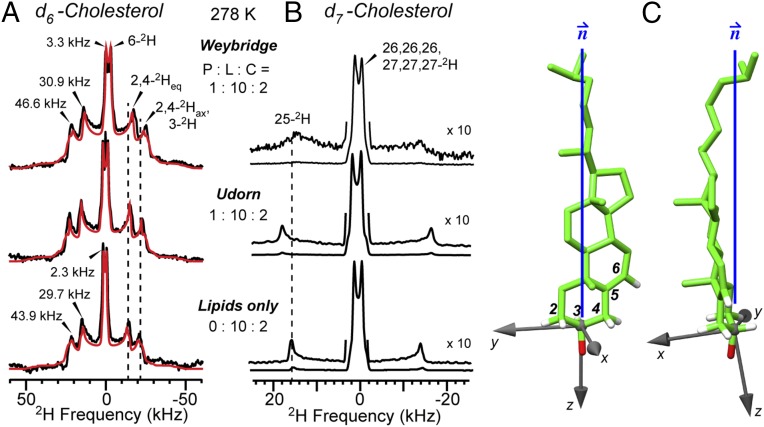
In addition to the distance constraints, we measured the sterol orientation of bound cholesterol using 2H NMR of deuterated cholesterol (Fig. 1B). Cholesterol undergoes fast uniaxial diffusion around the bilayer normal at ambient temperature, giving motionally averaged 2H quadrupolar couplings that reflect the C–D bond orientations from the bilayer normal. The 2H spectra of d6-cholesterol in the presence of M2 show three resolved quadrupolar splittings, whose distribution is similar to spectra of cholesterol in protein-free membranes (Fig. 4A). Since the sterol rings are rigid, these splittings can be assigned based on the fixed relative orientations of the six C–D bonds (24, 25). The observation of only three resolved splittings for the six C–D bonds indicates that some C–D bonds have similar orientations to the bilayer normal. Simulation of the 2H spectra allowed the determination of the sterol ring orientation, which was found to be approximately vertical in the membrane, with a polar angle β of 11° and an azimuthal angle γ of 10° for the bilayer normal in a molecule-fixed coordinate system (Fig. 4C). Deviation from these angles by just a few degrees result in simulated spectra that are clearly incompatible with the experimental spectra (Fig. S5).
Fig. 4.
Orientation of M2-bound cholesterol. (A) Measured (black) and best-fit simulation (red) of 2H spectra of d6-cholesterol at 278 K in the presence and absence of M2U(22–61). (B) 2H spectra of d7-cholesterol. (C) Best-fit orientation of M2-bound cholesterol relative to the bilayer normal, calculated using a C3–OH affixed coordinate system.
The absence of a second set of quadrupolar couplings and the similarity of the 2H NMR line shapes of peptide-containing and peptide-free membranes indicate that bound and free cholesterol adopt similar orientations at the concentration of 17–44 mol% in the membrane. For peptide-free membranes, the best-fit cholesterol orientation has β = 11° and γ = 8° (Table S3). However, the quadrupolar couplings are slightly larger in the presence of M2 than in its absence. This difference is indicated by the molecular order parameter Smol, which describes cholesterol wobbling from the bilayer normal. The Smol value is 0.85 for the peptide-free sample, in good agreement with the literature (25), and increases to 0.89 for M2-containing membranes, indicating that the degree of cholesterol wobbling is restricted by the protein. This is supported by the d7-cholesterol 2H spectra (Fig. 4B), which show that the 25-2H quadrupolar coupling is broadened by M2W and increased by M2U compared with the peptide-free spectrum. Thus, M2 partially immobilizes both the sterol rings and the isooctyl tail.
Distance- and Orientation-Constrained Cholesterol Docking Structure to M2.
To obtain energetically reasonable structures of the M2–cholesterol complex that incorporate these distance and orientational constraints, we docked cholesterol in vacuo onto the Udorn M2 tetramer using Autodock (26). The REDOR distance constraints were implemented by restricting the cholesterol C25 within a steep Gaussian potential imposed about a central point calculated from the (50 Å)3 grid for each helix. The sidechain χ1 and χ2 angles of I39, I42, L43, L46, and F47 were allowed to freely rotate during docking. The docking results were screened based on the calculated (β, γ) angles of the sterol rings; only results that were closest to the experimentally measured orientation were selected.
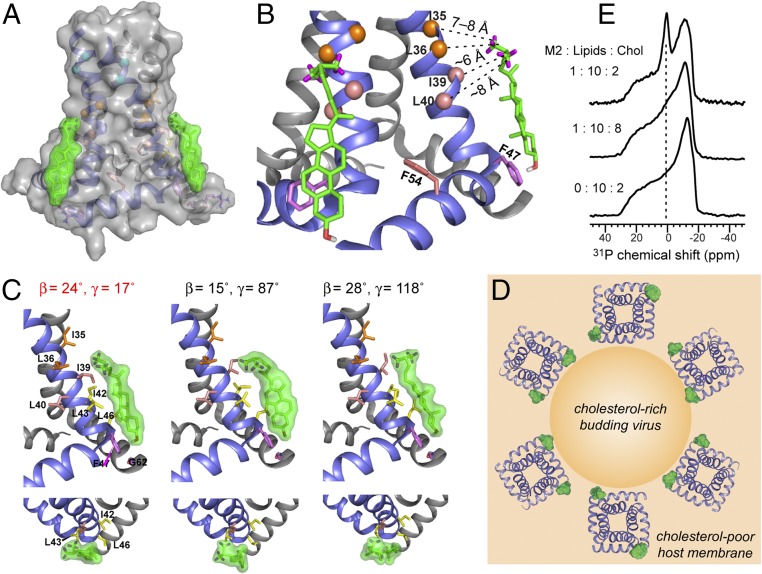
Each docked cholesterol molecule binds at the vertex between two subunits (Fig. 5 A and C), along the C-terminal half of the TM helix. The sterol plane is roughly tangential to the TM helix, with the methyl-rich rough β face apposing the methyl-rich Ile and Leu side chains from residues 35–43, forming favorable van der Waals interactions, while the smooth α face points to membrane phospholipids (Fig. 5B). Many docking results can be ruled out due to sterol plane orientations that are incompatible with the 2H NMR data (Fig. 5C). The CF3 groups lie at the depth of L36 while the hydroxyl group lies near the amphipathic helix, in close proximity to the F47 ring, thus explaining a previously observed cross-peak between cholesterol C3 and a Phe side chain (11). Each cholesterol molecule contacts the same subunit’s TM residues and the beginning of the amphipathic helix, but the sterol head may also be stabilized by polar and aromatic interactions with the neighboring subunit’s R61, and the aromatic residue 57. The side-chain conformations of AH residues are currently unknown, but rotameric flexibility and backbone structural adjustments may facilitate protein interactions with the sterol head. These multiple stabilizing interactions with two neighboring amphipathic helices explain the requirement of the AH for M2–cholesterol interaction and the loss of cholesterol binding to the TM peptide. The bound cholesterol is far from Y52 and residue 54, which is Arg in Weybridge M2 and Phe in Udorn M2, consistent with the CRAC independence of cholesterol binding. The similarity of bound- and free-cholesterol orientations suggests that M2 co-opts the natural tendency of cholesterol, but uses two AHs and the surface of one TM helix to sequester the ligand.
Fig. 5.
Structure of the cholesterol-binding site of M2. (A) Distance- and orientation-constrained docked cholesterol structure to M2. The proximal binding model is shown. (B) Cholesterol contacts methyl-rich TM residues and is stabilized by aromatic and polar interactions with amphipathic helix residues. (C) Representative cholesterol orientations from docking analysis and their (β, γ) angles. The (β, γ) = (24°, 17°) solution agrees with the 2H NMR data, while the other two orientations are incompatible with the 2H NMR data. (D) Proposed model of how the M2–cholesterol complex promotes membrane curvature and scission. M2 tetramers cluster to the neck of the cholesterol-rich budding virus because the proximal complex has a twofold higher statistical probability than the diagonal complex. (E) Static 31P-NMR spectra of POPC:POPG:cholesterol membranes with and without M2 at varying P:L:C ratios. A strong isotopic peak indicative of high curvature is observed in the 17% cholesterol membrane but not in the 44% cholesterol membrane.
Discussion
The above data show that, with 17 mol% cholesterol in the membrane, cholesterol and M2 form a 1:2 complex. This substoichiometric binding provides crucial insights into how cholesterol promotes membrane scission by M2. For a 1 cholesterol:2 protein complex, the proximal binding configuration is statistically twice as likely as diagonal. The higher probability of the proximal configuration attracts the M2–cholesterol complex to the edge of the budding virus, because the virus lipid envelope is far more enriched in cholesterol than the host membrane (27); moreover, the inner leaflet of the host membrane is deficient in cholesterol compared with the outer leaflet while the virus envelope has similar cholesterol levels in both leaflets (28). Thus, in a host membrane with emerging viruses, a significant lateral and longitudinal cholesterol concentration gradient exists, and the proximal M2–cholesterol complex would cluster the M2 tetramers to the boundary between the host membrane and the budding virus (29) (Fig. 5D). Concentrated at this interface, M2 tetramers induce membrane curvature because the wedge-shaped four-helix bundle excludes different amounts of volume in the two lipid leaflets (30), the insertion of the amphipathic helix into the cytoplasmic leaflet of the host membrane can induce stacking defects (31), and cholesterol binding to the inner leaflet on the side of the amphipathic helix can amplify the preexisting line tension at the phase boundary (31, 32) (Fig. S6). Confocal microscopy data of fluorescently tagged GUVs containing sphingomyelin, phosphocholine, and cholesterol showed that M2 clusters at the interface between the cholesterol-rich liquid-ordered (Lo) and cholesterol-poor liquid-disordered (Ld) phases (8). The current data indicate that this interfacial location of M2 is promoted by cholesterol binding to the tetramers in a proximal manner. At the edge of the budding virus, the multiple curvature-inducing effects can act in concert to excise the cholesterol-rich phase from the host membrane, in excellent agreement with the observed outward budding of the Lo phase from GUVs (8, 33).
The 1:2 binding stoichiometry found at 17 mol% membrane cholesterol, where virus budding is active, reflects the equilibrium of cholesterol binding to M2, with an estimated dissociation constant (Kd) of ∼13 mol%. When the cholesterol concentration increased to 44 mol%, a level at which M2 no longer causes budding (8), the REDOR intensity ratio decreased to ∼0.45 (Fig. 2D), indicating a higher binding stoichiometry. This suggests that, when M2 resides in the cholesterol-rich virus envelope, the population of the 1:1 complex increases, whose symmetry would then turn off the curvature-inducing function. To verify that the M2–cholesterol complex indeed causes curvature, we measured static 31P-NMR spectra of the POPC:POPG membranes. Consistent with previous results on virus-mimetic lipid membranes, the spectra show a distinct isotropic peak, which is indicative of high membrane curvature, for the 17% cholesterol membrane (Fig. 5E and Fig. S6), but the isotropic peak is suppressed in the 44% cholesterol membrane. Thus, low cholesterol levels and substoichiometric cholesterol binding to M2 correlate with curvature generation, while high cholesterol levels and increased binding to M2 inhibit curvature induction.
Our data show that cholesterol binding to M2 requires a specific 3D fold instead of a specific primary sequence. The binding-competent 3D fold is an L-shaped structure formed by the TM helix and the amphipathic helix of the same sub unit. The distance- and orientation-constrained structure of the M2–cholesterol complex explains the CRAC independence of cholesterol binding: the aromatic Y52 and the cationic residue 54, which are part of the CRAC motif, are far from cholesterol (Fig. 5). This cholesterol-binding site structure differs from the other cholesterol-docked protein structures associated with the CCM, CRAC, and CARC motifs. While M2 utilizes a TM and a peripheral helix to form the binding pocket, the CCM motif in G-protein–coupled receptors employs multiple TM helices (5), and the CRAC and CARC motifs are proposed for single TM helices. The vertical orientation of M2-bound cholesterol is also qualitatively different from the ∼45° tilted cholesterol proposed to bind α-synuclein (7). Finally, the amyloid precursor protein C99 uses a GXXXG motif to bind cholesterol, in contrast to the large hydrophobic sidechains that constitute the M2–cholesterol interface (3).
M2 is known to have pronounced conformational plasticity in response to pH, drug, and membrane properties (34, 35), and cholesterol is no exception. Cholesterol stabilizes M2 against drug-induced conformational changes (11), and in cholesterol-containing membranes the amphipathic helix adopts two conformations, as shown by double electron–electron resonance EPR measurements (36). Thus, cholesterol affects M2 structure and dynamics in a complex fashion. The SSNMR structure of the TM–AH peptide in cholesterol-free DOPC/DOPE bilayers (22) shows the tetramer as a C4-symmetric bundle. In the presence of cholesterol, the C4 symmetry will likely break, and the protein structure should depend on whether the complex contains proximally or diagonally bound cholesterol. This hypothesized symmetry breaking by substoichiometric cholesterol binding, which is important for virus budding, is unrelated to the symmetry breaking of M2 seen in cholesterol-free diphytanoylphosphocholine membranes (37). Since none of the existing M2 structures were solved in the presence of cholesterol, determining the high-resolution structure of M2 in cholesterol-containing membranes should allow the distinction of the proximal and diagonal binding modes and further elucidate the molecular basis of influenza virus budding.
Methods
M2 peptides were synthesized using custom-designed fast-flow synthesizers and Fmoc solid-phase protocols (38, 39), and were purified using reverse-phase HPLC. Peptide purity and mass were verified using liquid chromatography (LC)-MS. Peptides for 13C–19F REDOR experiments were dialyzed against a 1% acetic acid solution to remove trifluoroacetate ions. Site-specifically 13C,15N-labeled peptides were reconstituted into POPC:POPG (4:1) membranes with varying concentrations of 19F, 13C, or 2H-labeled cholesterol (Table S1). Carbon–fluorine distance experiments and 2H NMR experiments were mainly conducted on a 400-MHz spectrometer, while dynamic nuclear polarization (DNP) spectra were measured on a 600-MHz spectrometer. Methyl-rotation averaged 13C–19F REDOR curves were simulated using the SIMPSON software and analyzed in MATLAB, while orientation calculations were carried out in MATLAB. Cholesterol docking to M2 was performed using Autodock 4.2. Additional details of the experiments and data analysis are given in SI Methods.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant GM088204 (to M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715127114/-/DCSupplemental.
References
- 1.Oates J, Watts A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr Opin Struct Biol. 2011;21:802–807. doi: 10.1016/j.sbi.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Simmonds AC, et al. Annular and non-annular binding sites on the (Ca2+ + Mg2+)-ATPase. Biochim Biophys Acta. 1982;693:398–406. doi: 10.1016/0005-2736(82)90447-3. [DOI] [PubMed] [Google Scholar]
- 3.Barrett PJ, et al. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Scala C, et al. Common molecular mechanism of amyloid pore formation by Alzheimer’s β-amyloid peptide and α-synuclein. Sci Rep. 2016;6:28781. doi: 10.1038/srep28781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanson MA, et al. A specific cholesterol binding site is established by the 2.8 Å structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991–4997. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 7.Fantini J, Barrantes FJ. How cholesterol interacts with membrane proteins: An exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front Physiol. 2013;4:31. doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroeder C, Heider H, Möncke-Buchner E, Lin TI. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur Biophys J. 2005;34:52–66. doi: 10.1007/s00249-004-0424-1. [DOI] [PubMed] [Google Scholar]
- 10.Cady S, Wang T, Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein. J Am Chem Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekanayake EV, Fu R, Cross TA. Structural influences: Cholesterol, drug, and proton binding to full-length influenza A M2 protein. Biophys J. 2016;110:1391–1399. doi: 10.1016/j.bpj.2015.11.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cady SD, et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stouffer AL, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thaa B, Levental I, Herrmann A, Veit M. Intrinsic membrane association of the cytoplasmic tail of influenza virus M2 protein and lateral membrane sorting regulated by cholesterol binding and palmitoylation. Biochem J. 2011;437:389–397. doi: 10.1042/BJ20110706. [DOI] [PubMed] [Google Scholar]
- 15.Stewart SM, Wu WH, Lalime EN, Pekosz A. The cholesterol recognition/interaction amino acid consensus motif of the influenza A virus M2 protein is not required for virus replication but contributes to virulence. Virology. 2010;405:530–538. doi: 10.1016/j.virol.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thaa B, Siche S, Herrmann A, Veit M. Acylation and cholesterol binding are not required for targeting of influenza A virus M2 protein to the hemagglutinin-defined budozone. FEBS Lett. 2014;588:1031–1036. doi: 10.1016/j.febslet.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG. Measurement of 13C-15N distances in uniformaly 13C labeled biomolecules: J-decoupled REDOR. J Am Chem Soc. 1999;121:10237–10238. [Google Scholar]
- 18.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J Magn Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Kim SJ, Cegelski L, Preobrazhenskaya M, Schaefer J. Structures of Staphylococcus aureus cell-wall complexes with vancomycin, eremomycin, and chloroeremomycin derivatives by 13C19F and 15N19F rotational-echo double resonance. Biochemistry. 2006;45:5235–5250. doi: 10.1021/bi052660s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kauffman JM, Westerman PW, Carey MC. Fluorocholesterols, in contrast to hydroxycholesterols, exhibit interfacial properties similar to cholesterol. J Lipid Res. 2000;41:991–1003. [PubMed] [Google Scholar]
- 21.Cristian L, Lear JD, DeGrado WF. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc Natl Acad Sci USA. 2003;100:14772–14777. doi: 10.1073/pnas.2536751100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma M, et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni QZ, et al. High frequency dynamic nuclear polarization. Acc Chem Res. 2013;46:1933–1941. doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarrell HC, Jovall PA, Giziewicz JB, Turner LA, Smith IC. Determination of conformational properties of glycolipid head groups by 2H NMR of oriented multibilayers. Biochemistry. 1987;26:1805–1811. doi: 10.1021/bi00381a003. [DOI] [PubMed] [Google Scholar]
- 25.Marsan MP, et al. Cholesterol orientation and dynamics in dimyristoylphosphatidylcholine bilayers: A solid state deuterium NMR analysis. Biophys J. 1999;76:351–359. doi: 10.1016/S0006-3495(99)77202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerl MJ, et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenard J, Rothman JE. Transbilayer distribution and movement of cholesterol and phospholipid in the membrane of influenza virus. Proc Natl Acad Sci USA. 1976;73:391–395. doi: 10.1073/pnas.73.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb RA, Zebedee SL, Richardson CD. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt NW, Mishra A, Wang J, DeGrado WF, Wong GC. Influenza virus A M2 protein generates negative Gaussian membrane curvature necessary for budding and scission. J Am Chem Soc. 2013;135:13710–13719. doi: 10.1021/ja400146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossman JS, Lamb RA. Viral membrane scission. Annu Rev Cell Dev Biol. 2013;29:551–569. doi: 10.1146/annurev-cellbio-101011-155838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuzmin PI, Akimov SA, Chizmadzhev YA, Zimmerberg J, Cohen FS. Line tension and interaction energies of membrane rafts calculated from lipid splay and tilt. Biophys J. 2005;88:1120–1133. doi: 10.1529/biophysj.104.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong M, DeGrado WF. Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Sci. 2012;21:1620–1633. doi: 10.1002/pro.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim SS, et al. Cholesterol-dependent conformational exchange of the C-terminal domain of the influenza A M2 protein. Biochemistry. 2015;54:7157–7167. doi: 10.1021/acs.biochem.5b01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreas LB, et al. Structure and mechanism of the influenza A M218-60 dimer of dimers. J Am Chem Soc. 2015;137:14877–14886. doi: 10.1021/jacs.5b04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon MD, et al. Rapid flow-based peptide synthesis. ChemBioChem. 2014;15:713–720. doi: 10.1002/cbic.201300796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mijalis AJ, et al. A fully automated flow-based approach for accelerated peptide synthesis. Nat Chem Biol. 2017;13:464–466. doi: 10.1038/nchembio.2318. [DOI] [PubMed] [Google Scholar]
- 40.Acharya A, et al. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc Natl Acad Sci USA. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao SY, Lee M, Wang T, Sergeyev IV, Hong M. Efficient DNP NMR of membrane proteins: Sample preparation protocols, sensitivity, and radical location. J Biomol NMR. 2016;64:223–237. doi: 10.1007/s10858-016-0023-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hohwy M, Rienstra CM, Jaroniec CP, Griffin RG. Fivefold symmetric homonuclear dipolar recoupling in rotating solids: Application to double quantum spectroscopy. J Chem Phys. 1999;110:7983–7992. [Google Scholar]
- 43.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 44.Wang T, Cady SD, Hong M. NMR determination of protein partitioning into membrane domains with different curvatures and application to the influenza M2 peptide. Biophys J. 2012;102:787–794. doi: 10.1016/j.bpj.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T, Hong M. Investigation of the curvature induction and membrane localization of the influenza virus M2 protein using static and off-magic-angle spinning solid-state nuclear magnetic resonance of oriented bicelles. Biochemistry. 2015;54:2214–2226. doi: 10.1021/acs.biochem.5b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bak M, Rasmussen JT, Nielsen NC. SIMPSON: A general simulation program for solid-state NMR spectroscopy. J Magn Reson. 2000;147:296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 47.Sinha N, Schmidt-Rohr K, Hong M. Compensation for pulse imperfections in rotational-echo double-resonance NMR by composite pulses and EXORCYCLE. J Magn Reson. 2004;168:358–365. doi: 10.1016/j.jmr.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 48.Davis JH. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim Biophys Acta. 1983;737:117–171. doi: 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- 49.Dufourc EJ, Parish EJ, Chitrakorn S, Smith ICP. Structural and dynamical details of cholesterol-lipid interaction as revealed by deuterium NMR. Biochemistry. 1984;23:6062–6071. [Google Scholar]
- 50.Burnett LJ, Muller BH. Deuteron quadrupole coupling constants in three solid deuterated paraffin hydrocarbons: C2D6, C4D10, C6D14. J Chem Phys. 1971;55:5829–5831. [Google Scholar]
- 51.Kowalewski J, Lindblom T, Vestin R, Drakenberg T. Deuteron magnetic resonance of monodeuteroethene: Isotropic and anisotropic phase spectra. Mol Phys. 1976;31:1669–1676. [Google Scholar]
- 52.Cady SD, Goodman C, Tatko CD, DeGrado WF, Hong M. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: A 2H, 13C, AND 15N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle. J Am Chem Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 53.Hong M, Doherty T. Orientation determination of membrane-disruptive proteins using powder samples and rotational diffusion: A simple solid-state NMR approach. Chem Phys Lett. 2006;432:296–300. doi: 10.1016/j.cplett.2006.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McMullan RK, Koetzle TF, Fronckowiak MD. Structure of [20-CH3],[20-CD3]-methylpregnene-3, 20-diol methanolate from neutron diffraction at 123 K. Acta Crystallogr C. 1992;48:1509–1512. [Google Scholar]
- 55.Massiot D, et al. Modelling one‐and two‐dimensional solid‐state NMR spectra. Magn Reson Chem. 2002;40:70–76. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.