Significance
Synthetic [4+2] cycloaddition reactions are prevalent and useful transformations employed in syntheses of valuable products. Coincidentally, nature also takes advantage of this transformation, and several cases of devoted enzymes have been found to facilitate this reaction in the production of small-molecule natural products. Prior examples have involved either carbocyclic or oxygen heterocycle products. Recently, an enzymatic, formal [4+2] aza-cycloaddition has been confirmed, featuring a nitrogen atom in the final pyridine structure central to a subset of thiopeptide natural products. The alternative synthetic route to substituted pyridines via cycloaddition has proven energetically challenging, highlighting the significance of these biocatalysts. Here, we report a detailed structural and mechanistic analysis of two such pyridine synthases from thiomuracin and GE2270 biosynthesis.
Keywords: thiopeptide, antibiotic, biosynthesis, RiPP, [4+2] cycloaddition
Abstract
The [4+2] cycloaddition reaction is an enabling transformation in modern synthetic organic chemistry, but there are only limited examples of dedicated natural enzymes that can catalyze this transformation. Thiopeptides (or more formally thiazolyl peptides) are a class of thiazole-containing, highly modified, macrocyclic secondary metabolites made from ribosomally synthesized precursor peptides. The characteristic feature of these natural products is a six-membered nitrogenous heterocycle that is assembled via a formal [4+2] cycloaddition between two dehydroalanine (Dha) residues. This heteroannulation is entirely contingent on enzyme activity, although the mechanism of the requisite pyridine/dehydropiperidine synthase remains to be elucidated. The unusual aza-cylic product is distinct from the more common carbocyclic products of synthetic and biosynthetic [4+2] cycloaddition reactions. To elucidate the mechanism of cycloaddition, we have determined atomic resolution structures of the pyridine synthases involved in the biosynthesis of the thiopeptides thiomuracin (TbtD) and GE2270A (PbtD), in complex with substrates and product analogs. Structure-guided biochemical, mutational, computational, and binding studies elucidate active-site features that explain how orthologs can generate rigid macrocyclic scaffolds of different sizes. Notably, the pyridine synthases show structural similarity to the elimination domain of lanthipeptide dehydratases, wherein insertions of secondary structural elements result in the formation of a distinct active site that catalyzes different chemistry. Comparative analysis identifies other catalysts that contain a shared core protein fold but whose active sites are located in entirely different regions, illustrating a principle predicted from efforts in de novo protein design.
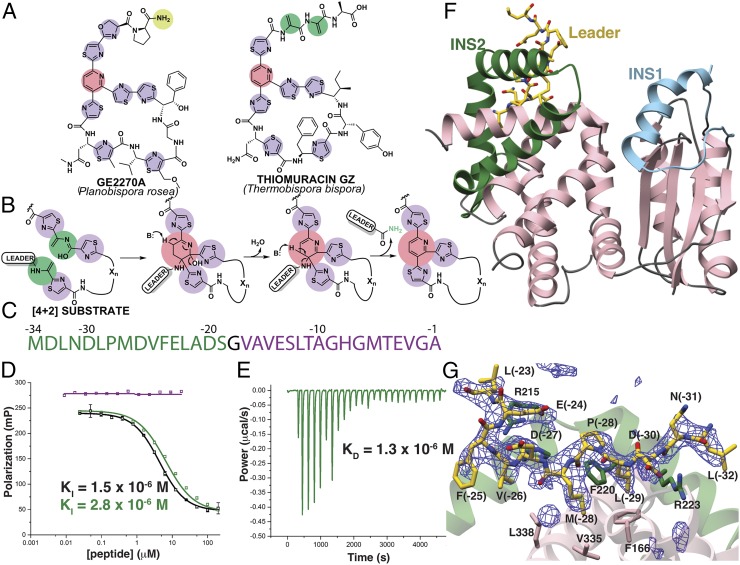
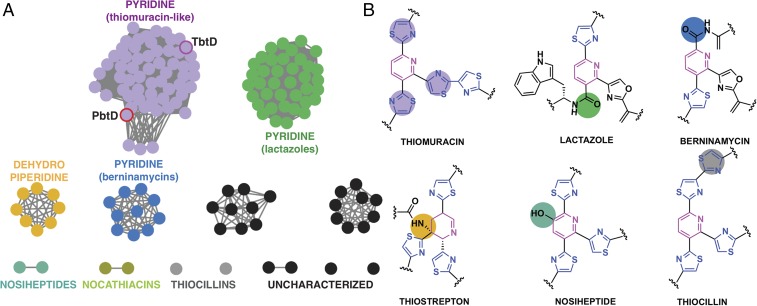
Thiopeptides are a class of ribosomally synthesized and posttranslationally modified peptides (RiPPs) (1) that display a range of biological activities (2–4). The identification of thiopeptides with impressive antibiotic activity toward drug-resistant Gram-positive bacteria, such as methicillin-resistant Staphylococcus aureus, as well as others with antiproliferative and immunosuppressive activities, has spurred a resurgence in their research (2). The antibacterial activity of thiopeptides is exerted through the inhibition of ribosomal protein synthesis. Curiously, the biological target of thiopeptides correlates with the size of the macrocyclic ring that embeds the heterocyclic core; thiostrepton and thiocillins (26-membered rings) inhibit the 50S ribosome, whereas GE2270 and thiomuracin (29-membered rings) (Fig. 1A) bind to elongation factor thermal unstable (EF-Tu) (5, 6). To date, dozens of different types of thiopeptides have been identified, all of which share the class-defining six-membered nitrogenous heterocycle that is at the center of a framework (pink circles) decorated with multiple thiazol(in)es (purple circles), and dehydro amino acids (green circles). The precursor peptide is of ribosomal origin and consists of a leader sequence that directs enzyme recruitment and a core sequence where the modifications occur (7–9). Access to many thiopeptide variants was facilitated owing to the precursor peptide being a direct gene product (4, 10, 11).
Fig. 1.
Scheme of enzyme-catalyzed pyridine formation and leader peptide binding of the pyridine synthase, TbtD. (A) Chemical structures of representative thiopeptides with residues color-coded by posttranslational modification. (B) Proposed enzymatic route for the installation of the central nitrogenous six-membered ring from two dehydroalanine residues, followed by the elimination of water and the leader peptide. (C) Primary sequence and numbering scheme of the TbtA leader peptide with the fragment that is engaged by TbtD colored in green. (D) Competition fluorescence polarization binding experiments where TbtD saturated with fluorescently labeled full-length leader peptide is challenged with either unlabeled full-length leader peptide (black curves), or fragments corresponding to the N-terminal (green curve and sequence) or C-terminal (purple curve and sequence) regions of the leader peptide. The N-terminal fragment is sufficient to compete with avidity similar to that of the full-length leader peptide. (E) ITC shows that the N-terminal leader peptide fragment binds to TbtD. (F) Overall structure of TbtD in complex with the N-terminal leader fragment, showing insertions 1 (cyan) and 2 (green) that distinguish the [4+2] enzymes from the LanB elimination domains. (G) Simulated annealing difference Fourier map (Fobs – Fcalc) contoured at 2.5σ showing residues from the leader peptide that bind to a region near insertion 2 (green) in TbtD.
Reconstitution studies using purified enzymes have established both the nature and the timing of the modifications installed during the biosynthesis of thiomuracin from Thermobispora bispora (12). Specifically, TbtG, a YcaO-type cyclodehydratase (13), converts Cys residues in the core sequence of the TbtA precursor peptide to thiazolines, which are then dehydrogenated to the corresponding thiazoles. A split class I (LanB) lanthipeptide dehydratase (14) modifies the resultant hexathiazole-containing peptide by first O-glutamylating Ser residues in the core sequence, followed by glutamate β-elimination, to yield the corresponding α,β-unsaturated amino acids termed dehydroalanine (Dha). Finally, TbtD forms the central six-membered nitrogenous ring, through the heteroannulation of two Dha residues in the modified core, possibly after tautomerization of one Dha to a conjugated iminol (Fig. 1B). Subsequent aromatization of the central pyridine is established with a dehydration and β-elimination of the leader peptide as a carboxamide (Fig. 1B). Recent in vitro studies using recombinant pyridine synthases demonstrated that a single enzyme plays a direct role in annulation via a formal [4+2] aza-cycloaddition (12, 15, 16). Ala-scanning mutational analysis of TbtA suggested the recognition by TbtD to be most influenced by N-terminal residues of the leader peptide (12).
While other enzymes have been identified as [4+2] cycloaddition catalysts (17, 18), they frequently serve only to enhance a reaction that can occur spontaneously and/or favor product stereochemistry via conformational restriction of the reactant(s) (17). In contrast, the reaction catalyzed by the thiopeptide pyridine synthase is not spontaneous, and synthetically equivalent reactions require either prolonged reaction (∼1 wk) at high temperatures or microwave irradiation for nonenzymatic catalysis, often with competing side reactions (19). The thiopeptide-forming pyridine synthases are dedicated [4+2] cycloaddition catalysts whose only function is catalyzing the cyclization of two Dha residues within the precursor peptide (20). Notably, pyridine synthases catalyze the unique formation of an aza-cyclic product, in contrast to the carbocyclic and dihydropyran products formed in other biosynthetic pathways, such as those for spinosin A (21, 22), pyrroindomycins (23), trans-decalin–containing polyketides (24), and leporin B (25).
Results and Discussion
TbtD Binds the N Terminus of Its Leader Peptide.
We initially focused our studies on TbtD, a pyridine synthase from the thiomuracin biosynthetic cluster from T. bispora. We previously established TbtD activity in vitro utilizing the TbtA precursor substrate with six thiazoles and four Dha residues enzymatically preinstalled (12, 15). Proteolytic removal of the N-terminal 31 residues (out of 34) of the leader peptide from this substrate abolished pyridine formation by TbtD. A similar leader peptide dependency was observed for TclM, the orthologous enzyme from the thiocillin biosynthetic cluster (26). Maltose-binding protein (MBP)-tagged TbtD bound to a fluorescently labeled TbtA leader peptide with a dissociation constant (Kd) of 1.4 × 10−7 M (12). However, sequence analysis of the pyridine synthases failed to identify a precursor peptide recognition element (RRE) that mediates leader peptide binding in many other RiPP enzymes (9) (see Fig. 1C for leader peptide numbering scheme). We carried out fluorescence polarization competition assays to identify the minimal leader peptide-binding motif and provide a quantitative affinity assessment for TbtD independent of any catalytic events (Fig. 1D). Competition studies using tag-free TbtD and unlabeled full-length TbtA leader peptide yielded an inhibition constant (KI) value of 1.5 × 10−6 M. Prior studies demonstrated that engagement by TbtD is compromised by mutations at residues at the N terminus of the leader peptide (12). Accordingly, a 17-residue peptide corresponding to the C-terminal portion of the leader peptide [from Val(-17) to Ala(-1)] failed to compete against the full-length leader peptide, while a 16-residue N-terminal portion [from Met(-34) to Ser(-19)] yielded a KI of 2.8 × 10−6 M. The binding of this peptide by TbtD was corroborated using isothermal titration calorimetry (ITC), which yielded a Kd of 1.3 × 10−6 M (Fig. 1E), while the C-terminal region of the leader peptide did not produce an observable signal.
Overall Fold and New Leader Peptide Binding Mode of the Pyridine Synthase.
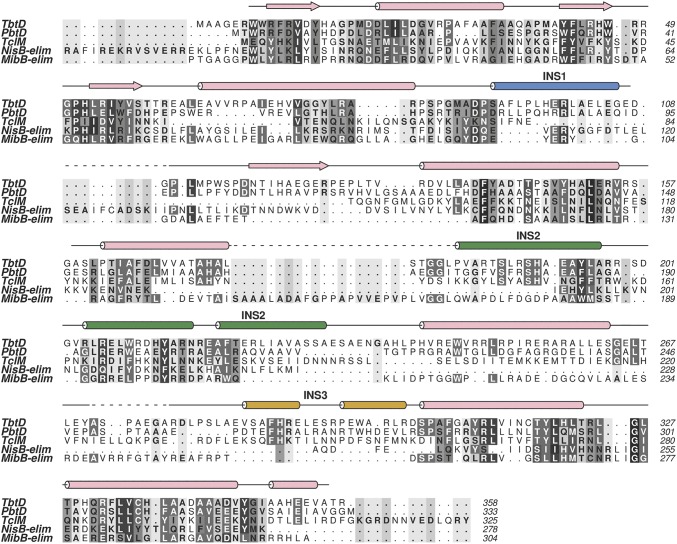
Given the lack of a RRE, we determined crystal structures of TbtD both alone and in complex with the N-terminal region of the leader peptide to define its binding site. TbtD is structurally homologous to the core of the lanthipeptide dehydratase elimination domain (Pfam identifier PF14028; abbreviated as Lant_Dehydr_C) elucidated in NisB (27) and MibB (28), but contains numerous secondary structural insertions (Figs. 1F and 2). Notable examples include a helical insertion in the region spanning Pro86 through Asp116 (insertion region 1), a reorganization of the region encompassing Pro183 through Pro241 (insertion region 2), and a large insertion spanning Gly264 through Arg301 (insertion region 3), which is a short connection loop in the Lant_Dehydr_C domains. Importantly, each of the insertions help to configure the binding sites for the substrates specific to the pyridine synthases, and insertion 3 is only visible in structures with bound pyridine core (see PbtD Binds a Trisubstituted Pyridine Product Analog of GE2270A; Fig. 3C). The cocrystal structure with the N-terminal 16 residues of the TbtA leader peptide shows electron density corresponding to residues Leu(-32) through Leu(-22) in the vicinity of insertion region 2 (Fig. 1G). Residues Ser200 through Ala234 of TbtD form a large, bent helical insert that abuts the enzyme core and form the binding pocket for the leader peptide. The equivalent region in both MibB (Gly990 through Glu1035) and NisB (Asp882 through Gly919) is largely devoid of secondary structure (Fig. 2).
Fig. 2.
Secondary-structure–based multiple sequence alignment (MSA). Included in the MSA are the pyridine synthases TbtD, PbtD, and TclM in addition to the class I LanB dehydratase elimination domains (-elim) NisB-elim, and MibB-elim generated using ALINE software (45). A majority of secondary structural elements are conserved in these functionally divergent enzymes and notable differences are observed in the insertion regions (INS 1, INS 2, and INS 3) found mainly in the pyridine synthases.
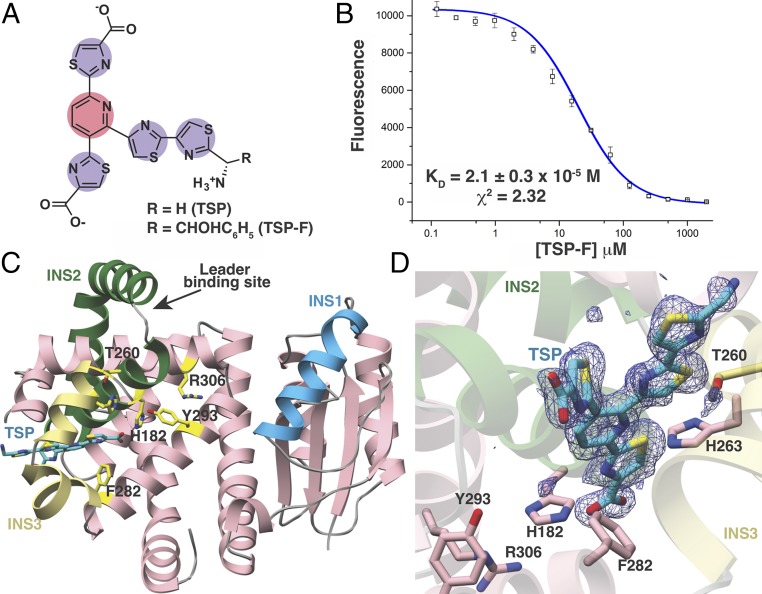
Fig. 3.
Structural and binding studies of the PbtD–TSP complex. (A) Chemical structure of the trisubstituted pyridine (TSP) core produced by acid hydrolysis of GE2270A. (B) Binding of TSP to PbtD as measured by the changes in FRET upon complex formation. (C) Overall structure of PbtD in complex with TSP shows that residues in insertion 3 (yellow) become ordered upon ligand binding. (D) Simulated annealing difference Fourier map (Fobs – Fcalc) contoured at 2.5σ showing the TSP core binds to a region near insertion 3 (yellow) in PbtD. Active-site residues implicated in catalysis, without influencing leader peptide binding, are shown in stick figures.
Mutagenesis and Leader Peptide Binding Studies.
Previous binding experiments between TbtD and site-directed alanine variants of TbtA identified several residues within the leader peptide that are important for binding (12), which can now be rationalized in light of the structural data. For example, an Ala substitution at Leu(-29) severely impairs TbtA binding, and in the cocrystal structure, this residue is docked into a hydrophobic pocket that is ensconced by residues from insertion 3. Similar hydrophobic docking interactions are important for leader peptide engagement by enzymes involved in the biosynthesis of lanthipeptides (27) and cyanobactins (29). Additionally, binding by TbtD is compromised by substitutions at one of several acidic residues in TbtA, including Asp(-30), Asp(-26), and Glu(-23) (12), and the structural data reveal that TbtD engages some of these residues through electrostatic complementarity. We carried out mutational analysis of the binding site in TbtD (SI Appendix, Fig. S2 and Table S3) to establish the importance of residues in mediating interactions with TbtA, using a shorter peptide corresponding to the N-terminal 12 residues visualized in the structure. Binding of TbtA by the pyridine synthase is compromised upon substitutions at enzyme residues that define the hydrophobic pocket for Leu(-29), including Phe166, Phe220, and Val335. An Ala mutation at Arg223, which is involved in additional hydrophobic contacts with Leu(-29) and hydrogen-bonding interactions with main-chain amides of the leader peptide, also diminishes TbtA binding (SI Appendix, Fig. S2 and Table S3). These structural and mutational data establish the validity of a leader peptide-binding site in TbtD and demonstrate that the C-terminal region of the leader peptide is dispensable for precursor binding to the enzyme.
PbtD Binds a Trisubstituted Pyridine Product Analog of GE2270A.
Repeated attempts to crystallize a complex of hexazole-containing tetradehydrated TbtA (designated as the [4+2] substrate in Fig. 1B) bound to catalytically deficient variants of TbtD proved recalcitrant. Binding measurements using fluorescently tagged, modified TbtA hexazole demonstrated that TbtD binds with nanomolar affinity (SI Appendix, Fig. S3), ruling out weak binding as the cause of crystallization issues. TbtD also binds with micromolar affinity a fluorescently tagged and modified TbtA hexazole treated with endoproteinase GluC, containing only three leader peptide residues, Val(-3) through Ala(-1) (SI Appendix, Fig. S5), but crystallization attempts using this substrate were not fruitful. Likewise, attempts at cocrystallization of PbtD with intact GE2270A failed. Rationalizing that flexible regions of the modified peptide substrate may interfere with lattice packing, we focused efforts on studies using analogs that retained the central pyridine moiety (Fig. 3A). Prior studies have shown that acid hydrolysis of GE2270A produced a trithiazole-substituted pyridine (TSP) (5); therefore, we acid-treated GE2270A and purified two resultant TSP variants to near homogeneity (Fig. 3A and SI Appendix, Fig. S8). We exploited the overlap in the absorption spectrum of TSP-F (λex ∼ 333 nm) and the emission spectrum for the corresponding PbtD pyridine synthase from its biosynthetic cluster (λex = 275 nm; λem = 333 nm) to directly measure binding using Förster resonance energy transfer (FRET), yielding a Kd of 2.1 × 10−5 M (Fig. 3B and SI Appendix, Fig. S6).
Structure-Guided Active-Site Mutagenesis.
To discern active-site details of the pyridine synthases, we determined the structure of PbtD both alone and in complex with the GE2270A TSP. The overall structure of PbtD recapitulates the architecture observed in the homologous TbtD, including the lack of interpretable electron density for residues Glu248 through His273 within insertion 3, despite the much higher resolution of this structure. In contrast, the structure with bound TSP reveals that these residues organize and form two short α-helices that define a periphery of the binding pocket for the ligand (Fig. 3C). The helix from insertion 2 forms a base at the bottom of the binding groove. Previous Ala-scanning analysis of TbtD implicated several residues in catalysis. Indeed, substitutions at His191 (equivalent PbtD numbering is given in parentheses; His182), Ser287 (Thr260), His290 (His263), Tyr319 (Tyr293), and Arg332 (Arg306) diminish activity (15). Following a structure-guided mutagenesis approach, this study has revealed an additional residue, Phe308 (Phe282), to be important for product formation after a panel of conserved residues proximal to TSP were tested using similar Ala-scanning mutational approaches (SI Appendix, Fig. S11). Each of these residues is near the bound TSP but are located ∼5–7 Å from each other and are not all spatially clustered (Fig. 3D). Rather, these residues line alternate sides of a shallow cleft that is located at the juncture of the leader peptide-binding site (on TbtD) and bound TSP. It should be noted that some of these variants may carry out the [4+2] cyclization and only be deficient in dehydration activity, but are refractory to identification as there would be no net change in mass.
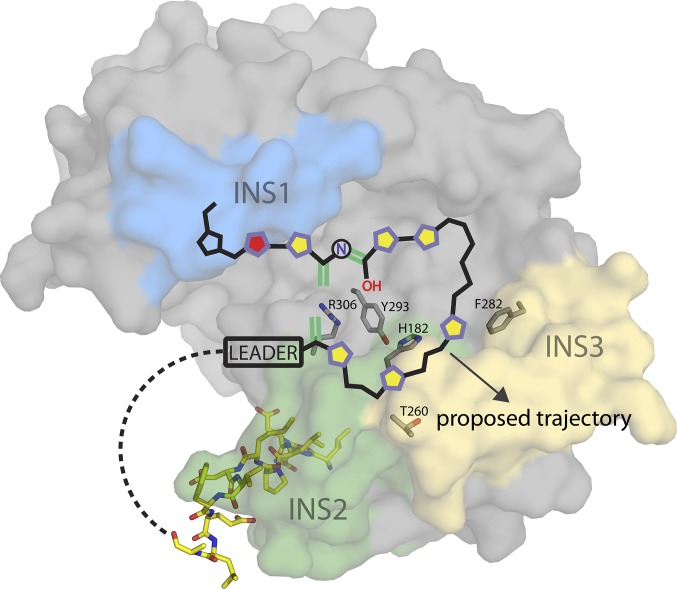
Model of Pyridine Formation.
A feasible model of pyridine formation involves a looped substrate embedded within the shallow cleft in PbtD, where the aforementioned residues may play roles in catalyzing iminol formation, water elimination, and/or leader peptide ejection as the peptide transverses along the binding cleft. In this model, the anchored N terminus of the leader peptide would allow a flexibly tethered core peptide to form this looped structure within the cleft, orienting the C terminus of the substrate away from its N terminus and at the interface of insertion 1 and the periphery of the cleft (Fig. 4). To test this hypothesis, we carried out docking of the [4+2] substrate, as well as various intermediates, and evaluated the stability of each state over a 1-ns time course molecular-dynamics simulation (SI Appendix, Figs. S12–S16). Among the lowest energy poses obtained, we observed the maturing substrate and product docking results to be consistent with the predicted substrate-binding site based on the TSP cocrystal structure (i.e., C terminus and macrocycle orientations). Residues within insertion 3 that are specific to the [4+2] enzymes are largely disordered in the absence of substrate, and the proximity of this region to the active-site cleft suggests that these residues may serve a role in active-site capping and/or modified core binding. Indeed, ordering of insertion 3 upon binding of TSP is consistent with this proposal. Recruitment of the true substrate may lead to additional ordering of insertion 3 to facilitate substrate threading, whereupon the resultant macrocycle becomes solvent exposed and is directed away from the binding cleft. The central nitrogenous ring in thiopeptides varies both in the pattern of substitution, as well as in the oxidation state (13). We generated a sequence similarity network (30) using the available sequences for thiopeptide [4+2] cycloaddition enzymes. This network revealed clear clustering into clades that correlate directly with the substitution pattern and oxidation state of the six-membered heterocycle, even at a fairly low primary sequence similarity cutoff of ∼30% (Fig. 5). Sequence analysis reveals large differences in the composition and length of residues in insertion 3, consistent with our proposed role of this region in directing the path of the core region of the modified peptide substrate, and thus the size of the macrocycle.
Fig. 4.
Model of pyridine formation catalyzed by PbtD. The PbtA [4+2] substrate is superimposed onto the structure of PbtD, with active-site residues drawn as gray sticks (SI Appendix, Figs. S12–S14; state 1). Five-membered heterocycles along the substrate backbone are represented as pentagons (purple outline/yellow center, thiazole; purple outline/red center, oxazoline; black outline/blue center, proline). Dehydroalanine and iminol double bonds involved in a formal [4+2] cycloaddition are drawn in green. The N-terminal leader peptide fragment (N12-mer: LNDLPMDVFELA) observed in the TbtD cocrystal structure has also been superimposed and is represented as yellow sticks.
Fig. 5.
(A) Sequence similarity network of [4+2] cycloaddition proteins found across various thiopeptide biosynthetic clusters showing that the sequences cluster based on the nature of the central nitrogenous ring. Nodes represent protein sequences while edges denote sequences with at least ∼30% amino acid identity. PbtD and TbtD are highlighted. The macrocycle compositions are as follows: thiostrepton-like, 26-membered dehydropiperidines; thiocillin/nosiheptide-like, 26-membered pyridines; thiomuracin/GE2270-like, 29-membered pyridine; lactazole-like, 32-membered pyridine; berninamycin-like, 35-membered pyridine. (B) Structures of the central ring found in different thiopeptide natural products with colored circles to indicate changes in the substitution pattern. The colors correspond with the clusters shown in A. It is noted that the nocathiacin and nosiheptide [4+2] enzymes form two unique clusters, despite these thiopeptides bearing high structural homology and an identical pattern of pyridine substitution. This is likely due to limited sample size and some variation at the N terminus between sequences in these clusters.
A Conserved Scaffold Catalyzes New Chemistry by Active-Site Shifting.
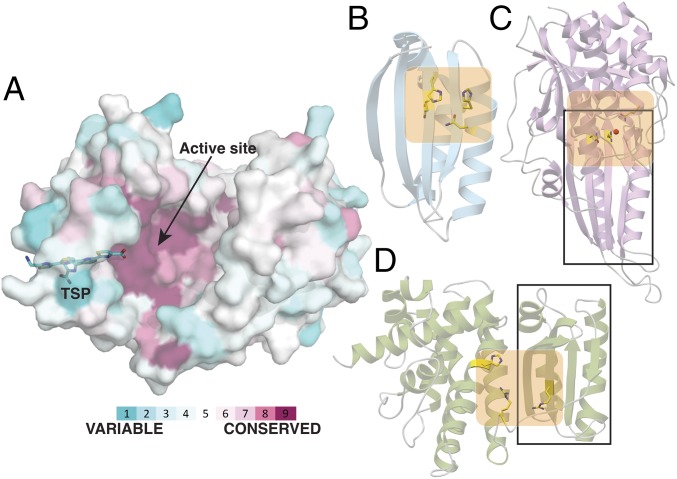
Mapping the primary sequences of nine pyridine synthases onto the structure of the PbtD–TSP cocrystal structure reveals a pattern of conservation at the active site (Fig. 6A). As noted, this active site of the [4+2] enzyme is distinct from that of other Lant_Dehydr_C domains, which are built upon a common scaffold observed in the LsrG epimerase (31), involved in quorum sensing. LsrG contains a ferredoxin-like domain, a prevalent α+β fold observed in numerous metabolic enzymes (32) (Fig. 6B). For example, the architecture of allantoate amidohydrolase, an enzyme involved in purine degradation in both plants and bacteria (33), is formed through the fusion of the LsrG scaffold with a second domain containing a metal-binding active site (34) (Fig. 6C). Likewise, the elimination active site of NisB is formed by the insertion of an additional subdomain onto the LsrG ferredoxin-like fold (Fig. 6D). Here, we demonstrate that the thiopeptide [4+2] cycloaddition enzymes amend the Lant_Dehydr_C fold with functionally relevant insertion regions to achieve gain of function.
Fig. 6.
Pyridine synthase scaffold is conserved in enzymes with divergent functions. (A) PbtD surface rendering with the bound TSP shown as a stick figure in cyan. Using the ConSurf server, the surface has been color-coded according to conservation in primary sequence based upon the alignment of nine sequences of pyridine synthases and colored according to regions of most variability (cyan), modest (white) or highest conservation (magenta) (see SI Appendix, SI Materials and Methods for a list of proteins and accession numbers used in alignment). The strongest sequence conservation occurs near the active site where the [4+2] cycloaddition occurs. (B) Ribbon diagram of the LsrG epimerase (PDB entry 3QMQ) with active-site residues shown as yellow sticks. (C) Ribbon diagram of the allantoate amidohydrolase AAH (PDB entry 4PXD) with active-site residues shown as yellow sticks and a manganese ion shown as a red sphere. (D) Ribbon diagram of the elimination domain of the lanthipeptide dehydratase MibB (PDB entry 5EHK) with active-site residues shown as yellow sticks. The ferredoxin-like α+β folds of AAH and MibB are demarcated within the rectangle, and the active sites are colored in orange.
A comparison of the TbtD/PbtD structures with that of MibB, and LsrG also provides an unexpected occurrence of proteins that contain active sites located at different regions of the polypeptide and catalyze different reactions, illustrating how a common scaffold can accommodate new chemistry at a new active-site region (Fig. 6). Recent de novo protein design efforts by Baker and coworkers (35) describe methodologies for engineering proteins with entirely new primary sequences through considerations of long-range interactions observed in prevalent folds such as the ferredoxin-like domain. These studies predict the possibility of engineering custom-designed large assemblies from such robust building blocks. The structure of the pyridine synthase provides a conspicuous example of how nature has already employed this strategy to yield new catalysts, based on an ancestral ferredoxin-like fold.
Materials and Methods
Peptide Purification.
Peptides used for cocrystallization and fluorescence polarization (FP) experiments were synthesized by GenScript and purified by HPLC using a Grace VisionHT C18 HL column with 5-μm particle size and 250-mm length (Part No. 5151988). A gradient of 5–55% aqueous acetonitrile (MeCN) containing 0.1% formic acid (vol/vol) was used to separate peptides. Peptide identity was verified using a Bruker UltrafleXtreme MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) in reflector positive mode at the University of Illinois School of Chemical Sciences Mass Spectrometry Laboratory.
Purification of Wild-Type and Site-Directed Variants of TbtD and PbtD.
All protein expression constructs were generated using the following procedure with the exception of N-terminal hexahistidine maltose-binding protein (His6-MBP)-tagged TbtD, TbtD-R332A, and PbtD, which were cloned according to methods previously described (15). Details of protein production and purification are provided in SI Appendix, SI Materials and Methods.
ITC.
ITC data were collected on a Nano ITC calorimeter (TA Instruments) using the low-volume sample cell (190 µL) maintained at 25 °C with stirring at 220 rpm. All protein and peptide dilutions were prepared using the same binding buffer as used in FP experiments. The sample cell contained TbtD at 40 μM and peptides at 0.5 mM were injected into the sample cell in 2-µL increments for 26 total injections (Fig. 1E). Data processing was carried out using the AFFINImeter online data analysis tool (https://www.affinimeter.com). The first and sixth injections from the Left in Fig. 1E were omitted in the calculations.
FRET Binding Assay.
Binding experiments were initially carried out using TSP; however, due to insolubility, we opted to use the more soluble TSP-F (SI Appendix, Fig. S8). To ensure a spectral overlap between PbtD native protein fluorescence and TSP-F absorbance, emission and absorbance scans were performed on a Synergy H4 Hybrid plate reader (BioTek) and an Epoch microplate spectrophotometer (BioTek), respectively. PbtD (5 μM) was excited at 275 nm before collecting an emission scan from 300 to 600 nm. An absorbance spectrum of TSP-F (0.5 mM) was collected over a similar range (SI Appendix, Fig. S6). A λex of 275 nm and λem of 333 nm were selected to satisfy spectral overlap. Emission spectra were also taken of TSP-F in water and methanol by exciting at 333 nm and monitoring emission from 333 to 750 nm. Fluorescence of TSP-F was only observed in methanol, preventing detection of fluorescence in aqueous binding buffer. Instead, quenching of PbtD fluorescence (observed in aqueous binding buffer) at 333 nm was monitored as a function of TSP-F concentration. To ensure adequate fluorescence signal above the instrument background emission, PbtD was maintained at 5 μM, while TSP-F was serially diluted from 2 mM to 122 nM. To correct for background emission quenched by TSP-F, the following formula was applied:
where Fl(x) is the total fluorescence in the presence of PbtD and some ligand concentration (x), Flb is the total background fluorescence in the absence of protein or ligand, and Flb(x) is the total background fluorescence in absence of protein and presence of some concentration of ligand (x). To obtain values for Flb and Flb(x), a blank solution containing only binding buffer was measured in addition to serially diluted TSP-F in the absence of PbtD. Measurements of Kd were approximated by fitting the curves to a dose–response curve using OriginPro 2016 (OriginLab) and taking Kd to be equal to the ligand concentration at 50% Flmax (36, 37) (Fig. 3B and SI Appendix, Fig. S7).
Crystallography Data Collection, Structure Determination, and Refinement.
Before data collection, crystals were cryoprotected in similar crystallization conditions supplemented with 20% ethylene glycol and flash-frozen in liquid nitrogen. Data were collected at the Advanced Photon Source at Argonne National Laboratory using the Life-Science Collaborative Access Team (LS-CAT) 21-ID-D, 21-ID-F, and 21-ID-G beamlines. Initial diffraction data were processed using autoPROC (38). Experimental phases for PbtD were determined by using diffraction data from SeMet-PbtD and running Phenix AutoSol (39). The initial structure was built using Phenix AutoBuild and further refined using REFMAC5 (40) in combination with manual rebuilding using COOT (41). Solvent molecules were incorporated using Phenix Refine (42). Before fitting ligands into electron density difference maps, geometry optimization and restraint parameters for TSPs were produced using Phenix eLBOW (43). Crystallographic phases for TbtD were determined by molecular replacement using PbtD coordinates as a search model for Phaser (44). Calculated phases were used to build an initial model, followed by manual and automated refinements as described for PbtD.
Fluorescence Polarization, Modified TbtA and TSP/TSP-F Preparation and Isolation, Protein Crystallization, Bioinformatics, Docking, and Molecular Dynamics.
Details are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Keith Brister and the staff at LS-CAT at the Advanced Photon Source (Argonne National Laboratory) for facilitating data collection, and Jennifer A. Leeds and Matthew J. LaMarche (Novartis) for providing GE2270A. This work was supported in part by grants from the National Institutes of Health (GM097142, to D.A.M.) and the Howard Hughes Medical Institute (to W.A.v.d.D.). G.A.H. is supported by a Pines Fellowship from the Department of Chemistry, and D.P.C. is supported in part by a Westcott Fellowship from the Department of Biochemistry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB), www.wwpdb.org [PDB ID codes 5W98 (PbtD), 5W99 (PbtD–TSP), 5WA3 (TbtD), and 5WA4 (TbtD-Leader16mer)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716035114/-/DCSupplemental.
References
- 1.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Just-Baringo X, Albericio F, Álvarez M. Thiopeptide engineering: A multidisciplinary effort towards future drugs. Angew Chem Int Ed Engl. 2014;53:6602–6616. doi: 10.1002/anie.201307288. [DOI] [PubMed] [Google Scholar]
- 3.Li C, Kelly WL. Recent advances in thiopeptide antibiotic biosynthesis. Nat Prod Rep. 2010;27:153–164. doi: 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, Liu W. Biosynthesis of thiopeptide antibiotics and their pathway engineering. Nat Prod Rep. 2013;30:218–226. doi: 10.1039/c2np20107k. [DOI] [PubMed] [Google Scholar]
- 5.Kettenring J, et al. Antibiotic GE2270 a: A novel inhibitor of bacterial protein synthesis. II. Structure elucidation. J Antibiot (Tokyo) 1991;44:702–715. doi: 10.7164/antibiotics.44.702. [DOI] [PubMed] [Google Scholar]
- 6.Morris RP, et al. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 7.Oman TJ, van der Donk WA. Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X, van der Donk WA. Ribosomally synthesized and post-translationally modified peptide natural products: New insights into the role of leader and core peptides during biosynthesis. Chemistry. 2013;19:7662–7677. doi: 10.1002/chem.201300401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burkhart BJ, Hudson GA, Dunbar KL, Mitchell DA. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat Chem Biol. 2015;11:564–570. doi: 10.1038/nchembio.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tran HL, et al. Structure-activity relationship and molecular mechanics reveal the importance of ring entropy in the biosynthesis and activity of a natural product. J Am Chem Soc. 2017;139:2541–2544. doi: 10.1021/jacs.6b10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Kelly WL. In vivo production of thiopeptide variants. Methods Enzymol. 2012;516:3–24. doi: 10.1016/B978-0-12-394291-3.00022-8. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Biosynthetic timing and substrate specificity for the thiopeptide thiomuracin. J Am Chem Soc. 2016;138:15511–15514. doi: 10.1021/jacs.6b08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkhart BJ, Schwalen CJ, Mann G, Naismith JH, Mitchell DA. YcaO-dependent posttranslational amide activation: Biosynthesis, structure, and function. Chem Rev. 2017;117:5389–5456. doi: 10.1021/acs.chemrev.6b00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Repka LM, Chekan JR, Nair SK, van der Donk WA. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem Rev. 2017;117:5457–5520. doi: 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson GA, Zhang Z, Tietz JI, Mitchell DA, van der Donk WA. In vitro biosynthesis of the core scaffold of the thiopeptide thiomuracin. J Am Chem Soc. 2015;137:16012–16015. doi: 10.1021/jacs.5b10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wever WJ, et al. Chemoenzymatic synthesis of thiazolyl peptide natural products featuring an enzyme-catalyzed formal [4 + 2] cycloaddition. J Am Chem Soc. 2015;137:3494–3497. doi: 10.1021/jacs.5b00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon BS, Wang SA, Ruszczycky MW, Liu HW. Natural [4 + 2]-cyclases. Chem Rev. 2017;117:5367–5388. doi: 10.1021/acs.chemrev.6b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HJ, Ruszczycky MW, Liu HW. Current developments and challenges in the search for a naturally selected Diels-Alderase. Curr Opin Chem Biol. 2012;16:124–131. doi: 10.1016/j.cbpa.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes RA, Thompson SP, Alcaraz L, Moody CJ. Total synthesis of the thiopeptide antibiotic amythiamicin D. J Am Chem Soc. 2005;127:15644–15651. doi: 10.1021/ja0547937. [DOI] [PubMed] [Google Scholar]
- 20.Bowers AA, Walsh CT, Acker MG. Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J Am Chem Soc. 2010;132:12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fage CD, et al. The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition. Nat Chem Biol. 2015;11:256–258. doi: 10.1038/nchembio.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HJ, Ruszczycky MW, Choi SH, Liu YN, Liu HW. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian Z, et al. An enzymatic [4+2] cyclization cascade creates the pentacyclic core of pyrroindomycins. Nat Chem Biol. 2015;11:259–265. doi: 10.1038/nchembio.1769. [DOI] [PubMed] [Google Scholar]
- 24.Li L, et al. Biochemical characterization of a eukaryotic decalin-forming Diels-Alderase. J Am Chem Soc. 2016;138:15837–15840. doi: 10.1021/jacs.6b10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohashi M, et al. SAM-dependent enzyme-catalysed pericyclic reactions in natural product biosynthesis. Nature. 2017;549:502–506. doi: 10.1038/nature23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wever WJ, Bogart JW, Bowers AA. Identification of pyridine synthase recognition sequences allows a modular solid-phase route to thiopeptide variants. J Am Chem Soc. 2016;138:13461–13464. doi: 10.1021/jacs.6b05389. [DOI] [PubMed] [Google Scholar]
- 27.Ortega MA, et al. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature. 2015;517:509–512. doi: 10.1038/nature13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortega MA, et al. Structure and tRNA specificity of MibB, a lantibiotic dehydratase from Actinobacteria involved in NAI-107 biosynthesis. Cell Chem Biol. 2016;23:370–380. doi: 10.1016/j.chembiol.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koehnke J, et al. Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat Chem Biol. 2015;11:558–563. doi: 10.1038/nchembio.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerlt JA, et al. Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 2015;1854:1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marques JC, et al. Processing the interspecies quorum-sensing signal autoinducer-2 (AI-2): Characterization of phospho-(S)-4,5-dihydroxy-2,3-pentanedione isomerization by LsrG protein. J Biol Chem. 2011;286:18331–18343. doi: 10.1074/jbc.M111.230227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma BG, et al. Characters of very ancient proteins. Biochem Biophys Res Commun. 2008;366:607–611. doi: 10.1016/j.bbrc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Werner AK, Romeis T, Witte CP. Ureide catabolism in Arabidopsis thaliana and Escherichia coli. Nat Chem Biol. 2010;6:19–21. doi: 10.1038/nchembio.265. [DOI] [PubMed] [Google Scholar]
- 34.Shin I, Percudani R, Rhee S. Structural and functional insights into (S)-ureidoglycine aminohydrolase, key enzyme of purine catabolism in Arabidopsis thaliana. J Biol Chem. 2012;287:18796–18805. doi: 10.1074/jbc.M111.331819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koga N, et al. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolovska-Coleska Z, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 37.Martin SF, Tatham MH, Hay RT, Samuel ID. Quantitative analysis of multi-protein interactions using FRET: Application to the SUMO pathway. Protein Sci. 2008;17:777–784. doi: 10.1110/ps.073369608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moriarty NW, Grosse-Kunstleve RW, Adams PD. electronic Ligand Builder and Optimization Workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr D Biol Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.