Abstract
Background
A recently constructed cellulolytic Yarrowia lipolytica is able to grow efficiently on an industrial organosolv cellulose pulp, but shows limited ability to degrade crystalline cellulose. In this work, we have further engineered this strain, adding accessory proteins xylanase II (XYNII), lytic polysaccharide monooxygenase (LPMO), and swollenin (SWO) from Trichoderma reesei in order to enhance the degradation of recalcitrant substrate.
Results
The production of EG I was enhanced using a promoter engineering strategy. This provided a new cellulolytic Y. lipolytica strain, which compared to the parent strain, exhibited higher hydrolytic activity on different cellulosic substrates. Furthermore, three accessory proteins, TrXYNII, TrLPMOA and TrSWO, were individually expressed in cellulolytic and non-cellulolytic Y. lipolytica. The amount of rhTrXYNII and rhTrLPMOA secreted by non-cellulolytic Y. lipolytica in YTD medium during batch cultivation in flasks was approximately 62 and 52 mg/L, respectively. The purified rhTrXYNII showed a specific activity of 532 U/mg-protein on beechwood xylan, while rhTrLPMOA exhibited a specific activity of 14.4 U/g-protein when using the Amplex Red/horseradish peroxidase assay. Characterization of rhTrLPMOA revealed that this protein displays broad specificity against β-(1,4)-linked glucans, but is inactive on xylan. Further studies showed that the presence of TrLPMOA synergistically enhanced enzymatic hydrolysis of cellulose by cellulases, while TrSWO1 boosted cellulose hydrolysis only when it was applied before the action of cellulases. The presence of rTrXYNII enhanced enzymatic hydrolysis of an industrial cellulose pulp and of wheat straw. Co-expressing TrXYNII and TrLPMOA in cellulolytic Y. lipolytica with enhanced EG I production procured a novel engineered Y. lipolytica strain that displayed enhanced ability to degrade both amorphous (CIMV-cellulose) and recalcitrant crystalline cellulose in complex biomass (wheat straw) by 16 and 90%, respectively.
Conclusions
This study has provided a potent cellulose-degrading Y. lipolytica strain that co-expresses a core set of cellulolytic enzymes and some accessory proteins. Results reveal that the tuning of cellulase production and the production of accessory proteins leads to optimized performance. Accordingly, the beneficial effect of accessory proteins for cellulase-mediated degradation of cellulose is underlined, especially when crystalline cellulose and complex biomass are used as substrates. Findings specifically underline the benefits and specific properties of swollenin. Although in our study swollenin clearly promoted cellulase action, its use requires process redesign to accommodate its specific mode of action.
Electronic supplementary material
The online version of this article (10.1186/s13068-017-0990-y) contains supplementary material, which is available to authorized users.
Keywords: Yarrowia lipolytica, Cellulolytic biocatalyst, Consolidated bioprocessing, Accessory proteins, Xylanase, Lytic polysaccharide monooxygenase, Swollenin
Background
The use of lignocellulosic biomass (LCB) as a manufacturing raw material is regarded as a key feature of the bioeconomy, because it will allow industry to transit to a sustainable, low carbon future [1, 2]. However, processing lignocellulosic biomass in an economically viable way remains a challenge, despite the considerable worldwide efforts.
Cellulose, the main component of lignocellulosic biomass, is built from linear β-glucan chains containing several hundreds of β-1,4-linked glucosyl units. In nature, cellulose exists in both ordered crystalline forms, in which multiple polysaccharide chains are densely packed into microfibrils, and to a lesser extent in disordered amorphous forms. One of the aims of biomass transformation processes is to increase the proportion of amorphous cellulose, since this form is then more amenable for further processing [3]. However, when processing plant cell walls, cellulose crystallinity is not the only hurdle to overcome, since rather like reinforced concrete, cellulose ‘rods’ (bundles of microfibrils) are embedded in a matrix, which in this case is composed of hemicellulose and lignin [4]. Together, these features render lignocellulosic biomass particularly recalcitrant to all but the most severe processing strategies [5].
Biochemical conversion of lignocellulose into target products usually occurs in three macro-operations. Biomass pretreatment often involves a combination of physical and chemical strategies that are deployed to modify the structure of lignocellulosic biomass, separating cellulose from lignin and hemicellulose, reducing complexity and increasing the accessible surface area [6]. Pretreatment is followed by enzymatic depolymerization of the polysaccharides, generating fermentable sugars using cellulases and hemicellulases. Finally, fermentation using a suitable microorganism is employed to transform sugars into the desired product(s) [7]. Among these steps, pretreatment and enzymatic hydrolysis are still significant cost drivers, with enzyme loadings and thus costs still exceeding those that have been targeted for many years [8].
One way to reduce cost in biomass processing is to integrate some of the steps and reduce or eliminate the need for external commercial enzymes. This is known as consolidated bioprocessing (CBP) and to achieve this, it is necessary to employ a cellulolytic microorganism (or a microbial consortium) that can both hydrolyse biomass polysaccharides and convert sugars into a target product [9]. However, despite the attractiveness of this concept, it is difficult to achieve because very few naturally occurring microorganisms can perform these two functions in an economically viable and industrially compatible manner [10]. Therefore, it is pertinent to look towards microbial strain engineering strategies in order to confer to an engineered microorganism the attributes required for CBP.
To engineer a microorganism for CBP purposes, it is most common to first select a microorganism that displays a suitable metabolism for target product manufacture and then confer it with cellulolytic capability, using heterologous expression of cellulases of fungal or bacterial origin [9]. Using this approach, several engineered CBP microorganisms have been successfully produced (reviewed in [9, 11]). However, these have mostly been tested using model cellulose substrates that are excessively amenable to enzyme hydrolysis and/or produce low product titers [7]. As a result, when such engineered CBP strains are confronted with industrial cellulose pulps, the addition of exogenous enzymes is necessary to achieve complete hydrolysis [11].
The extensively studied cellulolytic secretome of the soft-rot fungus T. reesei (syn. Hypocrea jecorina) contains several cellulases that synergistically act on complex substrates [12]. In the T. reesei cellulolytic secretome, EG I and EG II are the main endo-acting enzymes, representing approximately 15 and 10% (w/w) of the total protein content, respectively [13], while CBH I and CBH II (Cel7A and Cel6A) are the major exo-acting components, representing 50 and 20% of the total protein content, respectively [1]. The minimal requirement for a cellulase cocktail composed of free enzymes includes at least one endoglucanase (EG, EC 3.2.1.4), one cellobiohydrolase (CBH, EC 3.2.1.91) and a β-glucosidase (BGL, EC 3.2.1.21) [14], the latter being necessary to ensure the production of glucose from cellodextrins. Nevertheless, most commercial cocktails are more complex and are completed with a range of accessory enzymes, including hemicellulases, cinnamic acid and acetyl esterases that together remove hemicellulose and thus increase cellulose accessibility and decrease the risk of cellulase inhibition by xylo-oligosaccharides [15, 16]. Until recently, assembling a sufficiently comprehensive array of polysaccharide hydrolases in a cocktail constituted the main strategy to achieve the hydrolysis of cellulose pulps. However, the recent discovery of lytic polysaccharide monooxygenases (LPMOs) has dramatically changed this view and opened up new options for the engineering of efficient cellulolytic microbial cell factories [8].
Belonging to the so-called Auxiliary Activity family 9 (AA9, formerly GH61) of the CAZy classification [17], LPMOs have been shown to significantly boost the overall efficiency of cellulose hydrolysis when using canonical hydrolytic cellulases. Studies reveal that the LPMOs catalyze the oxidative cleavage of insoluble polysaccharides using molecular oxygen or peroxide and an electron donor [18, 19]. Although most characterized LPMOs oxidize polysaccharides at the C1 position (type 1 PMOs), yielding lactone [19–21], some perform oxidation at the C4 position (type 2 PMOs) or at both the C1 and C4 positions (type 3 PMOs), or at C6. In all cases, the action of LPMOs leads to the formation of a ketoaldose [20, 22, 23]. Considering the clear benefits of LPMOs for cellulase-mediated hydrolysis of cellulose, enzyme manufacturers have incorporated these into the latest generation of commercial cellulase cocktails [24].
In addition to LPMOs, it has been known for some time that fungal cellulolytic secretomes and plant cell walls contain non-enzymatic proteins that possess the ability to disrupt the ordered hydrogen-bonding network in crystalline cellulose. Although the exact role of proteins such as swollenins (fungal origin) and expansins (plant origin) has not been fully elucidated, research reveals that they enhance cellulose-mediated hydrolysis of cellulose, probably by disrupting the cellulose surface, making it more amenable to attack by cellulases [25–27].
Recently, we conferred cellulolytic properties to Yarrowia lipolytica, an oleaginous yeast that is recognized for its industrial usefulness and safety (it is classified by the FDA as a Generally Recognized As Safe strain) [28, 29]. To achieve this, we expressed in a Y. lipolytica strain a range of enzymes including BGLs, EGs, and CBHs, taking care to control the relative proportions of each of these in order to obtain a combination that is quantitatively similar to that of the native cellulase system of T. reesei. The engineered cellulolytic Y. lipolytica was shown to grow efficiently on industrial cellulose pulp (CIMV-cellulose, mostly amorphous), but displayed some difficulty to degrade recalcitrant crystalline cellulose [30]. Therefore, in the present work, we describe how the cellulolytic Y. lipolytica strain has been further manipulated to increase the hydrolysis of crystalline cellulose and complex substrates. This has been achieved by altering the proportions of the expressed cellulases and adding accessory proteins XYNII, LPMO, and swollenin.
Results and discussion
Enhancing the production of T. reesei endoglucanase I in cellulolytic Y. lipolytica
Previous studies have revealed that the efficient hydrolysis of pretreated LCB requires the presence of 25–35% (w/w) EG I in the cellulase cocktail, and that EG I cannot be replaced by EG II [31–33]. Additionally, our results showed that recombinant EG I exhibits a twofold higher specific activity on insoluble substrates, such as Avicel, β-1,3 and β-1,4 glucans, than EG II. Together, these findings underline the important role of EG I for the hydrolysis of recalcitrant biomass. Unfortunately, in the previously described engineered cellulolytic Y. lipolytica strain (YLC6), the secretion yield of T. reesei EG I was approximately 60% lower than that of EG II [30]. Therefore, to alter this ratio, a hybrid promoter strategy [34] was employed, modifying the core TEF promoter element controlling EG I expression. Following successful promoter modification, analyses revealed that all of the previously introduced genes were present (Additional file 1: Figure S1) and that the new strain, YLC7, was able to hydrolyze CMC, PASC, and Avicel cellulose (Fig. 1). Comparing the performance of YLC7 to the parental strain YLC6 revealed that HTEF promoter-controlled expression of rTrEG I led to 18, 14, 10, and 17% increases in hydrolytic activity on CMC, PASC, Avicel, and CIMV-Cellulose, respectively. Using anti-His Western blot analysis, it was possible to correlate these increases with the improved expression level of rhTrEG I in the secretome of YLC6 (Additional file 1: Figure S2). Since our previous results showed that His6-tagging did not influence rTrEG I production [30], it is reasonable to attribute the increase in hydrolytic activity of YLC7 to its higher rTrEG I expression, although the reported cellulolytic activity is the composite result of all secreted cellulase activity.
Fig. 1.
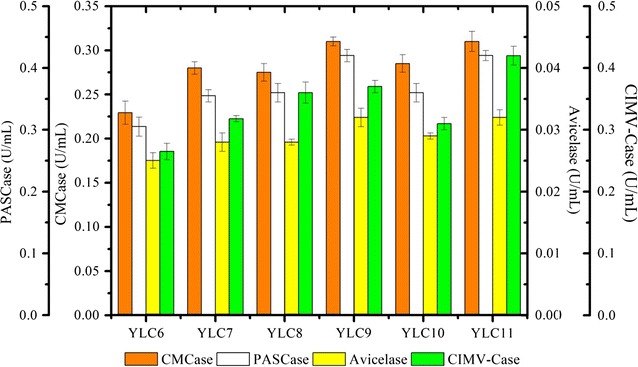
Comparison of the hydrolytic activity (mg reducing sugar/min/mL supernatant) on various cellulosic substrates of the secretomes produced by cellulolytic Y. lipolytica strains YLC6 (∆poxB12 strain expressing EG I, EG II, CBH I, and CBH II), YLC7 (YLC6 + enhanced EG I expression), YLC8 (YLC7 + XYN II), YLC9 (YLC7 + LPMOA), YLC10 (YLC7 + SWO1), and YLC11 (YLC7 + XYNII + LPMOA)
Expression and characterization of T. reesei xylanase II in Y. lipolytica
The results of previous studies focused on the design of optimal cellulase formulations for complex biomass hydrolysis have emphasized the requirement for xylanase, which acts synergistically with cellulases [31–33]. Accordingly, we expressed the T. reesei xylanase II (rTrXYNII) in Y. lipolytica JMY1212 under TEF promoter control and using the lipase 2 pre-pro region to facilitate protein secretion, characterized the resultant recombinant enzyme, and then investigated how rTrXYNII could be expressed in the genetic background of YLC7. Screening on solid medium containing AZCL-arabinoxylan of Y. lipolytica transformants readily revealed the presence of clones producing either rTrXYNII or its His-tagged variant rhTrXYNII (Fig. 2a). To further confirm successful expression of TrXYNII, positive clones were grown in liquid YTD medium and xylanase activity was assayed in the culture supernatant. Accordingly, it was possible to demonstrate that during cultivation xylanase activity steadily increased over a 96-h period, reaching a final activity (in the case of rhTrXYNII) of 32.0 U/mL (Fig. 2b). Furthermore, using SDS-PAGE the comparison of the rhTrXYNII-containing culture supernatant to that of a control culture (Fig. 2c) revealed the presence of a discrete species migrating to a position corresponding to an approximate Mw of 22 kDa, which is consistent with the theoretical Mw of TrXYNII (22.0 kDa).
Fig. 2.
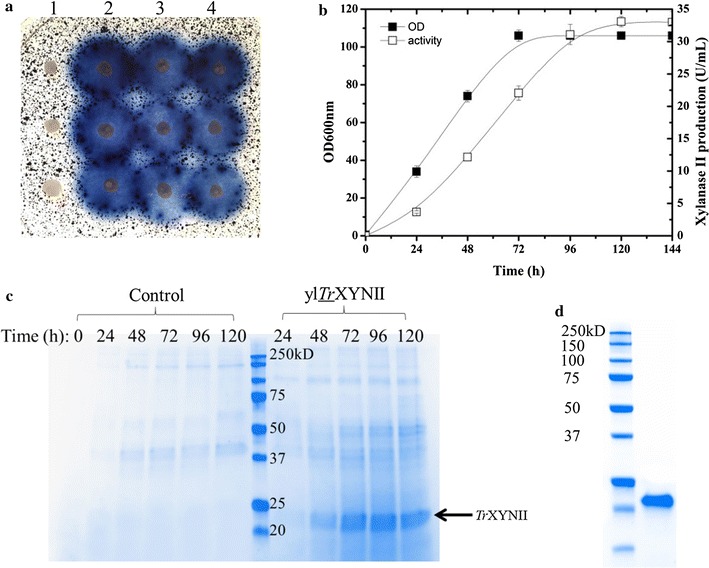
Expression of rTrXYNII in Y. lipolytica. a Screening of Y. lipolytica transformants on agar plates containing 0.2% w/v AZCL-arabinoxylan: lane 1, Y. lipolytica-control; lane 2 to 3, Y. lipolytica expressing rTrXYNII and rhTrXYNII respectively; L4, YLC8; b rTrXYNII production on YTD medium versus time; c SDS-PAGE analysis of the culture supernatant of Y. lipolytica control strain expressing empty plasmid and the recombinant strain expressing rhTrXYNII; d SDS-PAGE analysis of the rhTrXYNII after purification, protein marker (lane 1) and rhTrXYNII (lane 2)
In order to further investigate the properties of rhTrXYNII, the protein was purified from the culture supernatant (Fig. 2d) and its activity was measured on beechwood xylan in different conditions of pH and temperature. This revealed that rhTrXYNII reached optimal activity (532 U/mg-protein) at pH 6.0 and 60 °C (Additional file 1: Figure S3), and that higher temperatures led to rapid enzyme inactivation (data not shown). These findings are consistent with previous data obtained for rTrXYNII expressed in Pichia pastoris [35]. Knowing the specific activity of rhTrXYNII, it was also possible to calculate that 62 mg/L of rhTrXYNII was secreted by Y. lipolytica, when growing in batch mode in YTD medium contained in flasks.
Expression of T. reesei lytic polysaccharide monooxygenase A in Y. lipolytica
In the T. reesei secretome LPMOs form a very minor component [36]. However, recent in vitro studies have shown that the inclusion of these enzymes in cellulase cocktails can reduce overall enzyme loadings, while maintaining the efficiency of cellulose conversion [37, 38]. Therefore, considering that it would be useful to include TrLPMOA in the cellulase cocktail expressed by YLC7, we cloned and expressed a His-tagged rhTrLPMOA in Y. lipolytica JMY1212 under TEF promoter control and using the lipase 2 pre-pro region to facilitate protein secretion. Successful secretion of rhTrLPMOA into the culture supernatant was first established by SDS-PAGE and anti-His Western blot analyses. These revealed a smear in the Mw range 60–200 kDa, suggesting that the expected 34.4 kDa protein species was glycosylated (Fig. 3), which is unsurprising since yeasts are well known to perform hyper N-mannosylation [39]. To investigate this, rhTrLPMOA was submitted to Endo H-mediated deglycosylation and further SDS-PAGE/Western blot analysis, which revealed a better defined protein population that migrated to a position correlating to a median Mw of 55 kDa (Fig. 3a). This result suggests that even after the removal of N-glycosyl moieties, rhTrLPMOA still bears additional post-translational modifications, possibly O-glycosylation of serine/threonine-rich linker region [40].
Fig. 3.
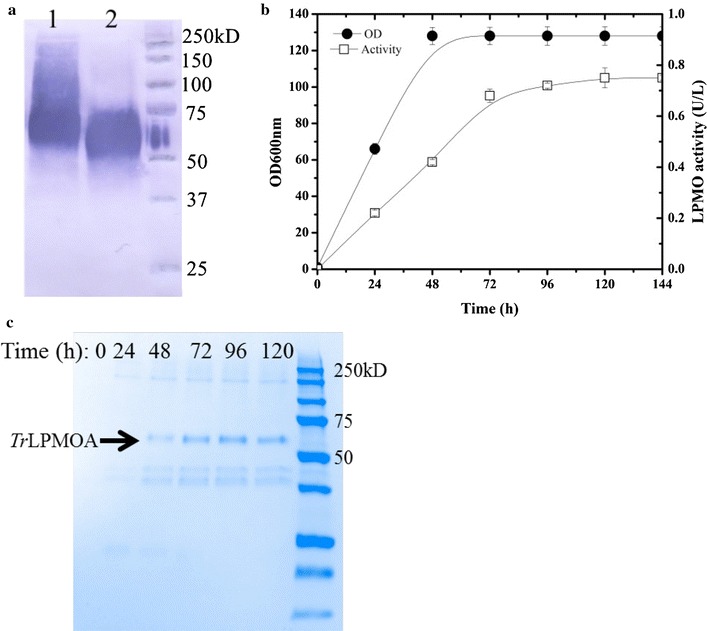
Expression of TrLPMOA in Y. lipolytica. a Western blot analysis of the heterologous LPMOA protein secreted by Y. lipolytica: lane 1, rhTrLPMOA; lane 2, Endo H-treated rhTrLPMOA; b time-related enzyme production by yeast growing in liquid YTD; c SDS-PAGE analysis of the culture supernatant of Y. lipolytica transformants
To confirm that the expressed recombinant protein was active, the ability of rhTrLPMOA to mediate the reduction of O2 to H2O2 in the presence of ascorbate was established. Moreover, using this test, it was shown that the expression level of rhTrLPMOA steadily increased over a 72-h period, reaching a final activity in the culture supernatant of 0.75 U/L (Fig. 3b).
Characterization of the rhTrLPMOA expressed in Y. lipolytica
To further assess the functionality of rhTrLPMOA, the protein was purified in a single step, with an overall yield > 60% (Fig. 4a) and characterized. The purified rhTrLPMOA exhibited a specific activity of 14.4 U/g. This value is similar to that of the Neurospora crassa LPMO9F (15 U/g) expressed in P. pastoris, which is the most active LPMO reported so far using the same activity assay [8]. The amount of rhTrLPMOA secreted by Y. lipolytica in YTD medium during batch cultivation in flasks was approximately 52 mg/L, a yield that is moderate compared to other examples of LPMO heterologous expression [8, 41].
Fig. 4.
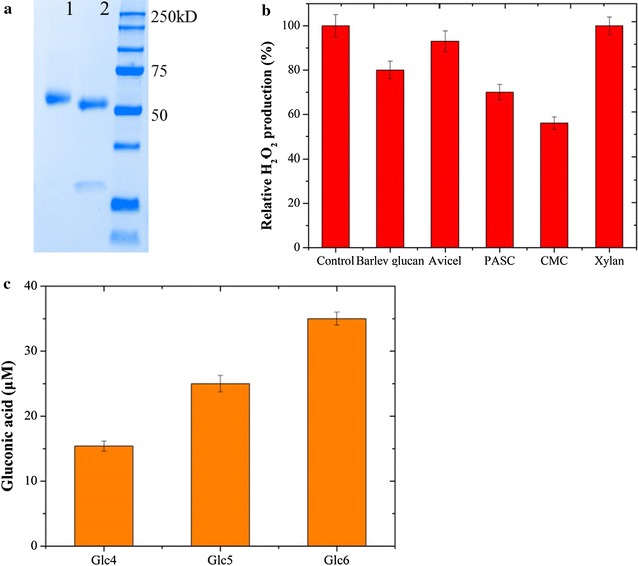
Characterization of TrLPMOA expressed in Y. lipolytica. a SDS-PAGE analysis of the purified rhTrLPMOA (lane 1) and rhTrLPMOA treated by Endo H (lane 2); b H2O2 generation by rhTrLPMOA in the presence and absence of various oligosaccharides substrates; c gluconic acid produced from cello-oligosaccharides (DP4-DP6) by action of rhTrLPMOA in the presence of ascorbic acid
To further investigate the substrate specificities of rhTrLPMOA, we measured the production of H2O2 in the presence of 1.0% (w/v) PASC, CMC, Avicel, barley glucan (β-1,3; 1,4), and beechwood xylan. The repression in H2O2 production was observed for rhTrLPMOA in the presence of the four glucose-polymers (Fig. 4b) while xylan had no effect on H2O2 production. These results suggest that rhTrLPMOA displays broad specificity on β-(1,4)-linked glucans, but is inactive on xylan.
To further investigate the regioselectivity of rhTrLPMOA, tests were performed using cellodextrins (DP4-6). These revealed the production of d-gluconic acid, confirming that oxidation occurred at C1 (Fig. 4c). Moreover, HPAEC analysis failed to reveal any C4-oxidized species (data not shown). Taken together, these results indicate that rhTrLPMOA displays type 1-like LPMO activity, a conclusion that contradicts previous sequence and phylogeny analyses that suggest it is a type 3 LPMO (i.e., one that oxidizes at both the C1 and C4 positions) [40]. However, these apparently conflictual findings can be resolved by attributing TrLPMOA to a recently described type 3 subgroup (PMO3*) that contains LPMOs such as the one from Myceliophthora thermophila AA9 LPMO (MYCTH_92668) that also only performs oxidation at C1 [40].
Expression and characterization of T. reesei swollenin 1 in Y. lipolytica
In this work, swollenin 1 (SWO1), the major expansin-like protein produced by T. reesei [36], was first expressed independently in Y. lipolytica JMY1212 under TEF promoter control and using the lipase 2 pre-pro region to facilitate protein secretion. Compared to a control culture, SDS-PAGE analysis of culture supernatants of Y. lipolytica transformants expressing rhTrSWO1 revealed the presence of a smear (150–200 kDa), which is much higher than the expected Mw of rhTrSWO1 (50 kDa) (Fig. 5a). However, Western blot analysis using anti-His antibody confirmed that this large protein species was His-tagged (Fig. 5b), indicating that it is a highly glycosylated variant of rhTrSWO1, a finding that concurs with data in the literature [26, 27]. Using NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) to analyze the amino acid sequence of rhTrSWO1 has predicted that SWO1 bears three potential N-glycosylation sites. Therefore, to explore this possibility the protein was treated with Endo H and analyzed by Western blot. This revealed a newly formed discrete protein species that migrates to position corresponding to a Mw of approximately 75 kDa (Fig. 5b), a result consistent with the analysis of native SWO1 isolated from the T. reesei secretome [27]. Presumably the discrepancy between the predicted Mw for the polypeptide sequence of SWO1 and that measured (75 KDa) is due to other post-translational modifications, such as O-glycosylation of serine/threonine-rich linker region [26]. Finally, to further investigate the functionality of rhTrSWO1, the protein was purified (Fig. 5c).
Fig. 5.
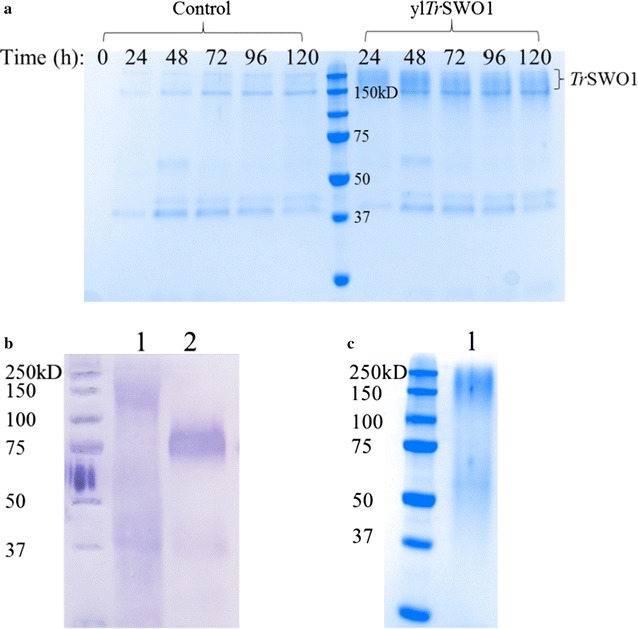
Expression of TrSWO1 in Y. lipolytica. a SDS-PAGE analysis of the culture supernatant of Y. lipolytica control strain expressing empty plasmid and the recombinant strain expressing rhTrSWO1; b Western blot analysis of the heterologous SWO1 protein produced by Y. lipolytica: lane 1, rhTrSWO1; lane 2, rhTrSWO1 treated by Endo H; c SDS-PAGE analysis of the purified rhTrSWO1
Combining the action of LPMOA or SWO1 with cellulases for Avicel hydrolysis
To investigate the synergy of rhTrLPMOA and rhTrSWO1 with cellulases, tests were performed on microcrystalline cellulose (Avicel) using a commercial cellulase cocktail (Celluclast) supplemented with each recombinant protein at different concentrations. Celluclast was used because, unlike Cellic™ CTEC2 [37], it is devoid of LPMOs and SWO. Adding rhTrLPMOA alone significantly accelerated cellulose hydrolysis (Fig. 6a), with rate being dependant on the concentration of rhTrLPMOA. When using 15 mg rhTrLPMOA per g cellulose, the conversion of Avicel (20 g/L) to soluble reducing sugars reached 60% within 72 h, which constitutes a 40% increase compared with the control (Fig. 6a). In contrast, adding rhTrSWO1 alone failed to produce any measurable effect on cellulose hydrolysis, a result that is consistent with previous data and suggests that swollenin does not display synergy with EG and CBH [42]. Nevertheless, the incubation of Avicel with rhTrSWO1 (5–15 mg rhTrSWO1/g cellulose) for a 24-h period prior to the reaction enhanced Avicel hydrolysis, with up to 70% conversion to reducing sugars occurring after 72 h when using 15 mg rhTrSWO1/g cellulose (Fig. 6b), although higher loadings did not procure further improvements (data not shown). Overall, these data indicate that swollenin adsorption is a prerequisite for amorphogenesis and that this is governed by a time-dependant equilibrium. For optimal Avicel amorphogenesis, it is necessary to leave sufficient time for several cycles of adsorption/desorption to occur [26, 42]. Finally, it is noteworthy that when using a similar protein load, the effect of rhTrSWO1 on cellulase-mediated Avicel hydrolysis was more potent than that of rhTrLPMOA, indicating that the use of SWO instead of LPMO in a biorefinery process might be more cost-effective. However, for this to be feasible, it would be necessary to propose a redesigned process and account for the cost burden associated with the time needed for the action of SWO.
Fig. 6.
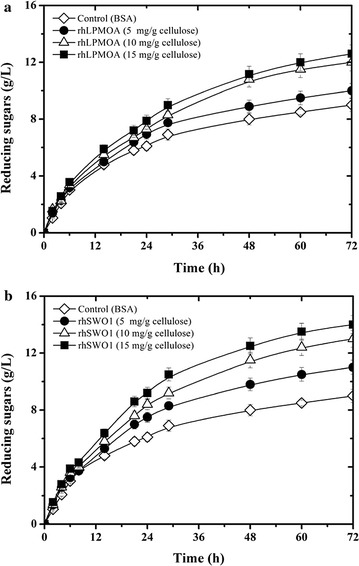
Hydrolysis of Avicel (a) in the presence of rhLPMOA at different concentrations; b after pretreatment with swollenin at different concentrations
Construction of recombinant Y. lipolytica strains expressing multiple cellulases and accessory enzymes
To further enhance the potency of cellulose hydrolysis by an engineered Y. lipolytica, the genes encoding TrXYNII, TrLPMOA, and TrSWO1 were singly expressed in the cellulase-producing strain YLC7 (Table 1). The presence of all of the exogenous genes was established using PCR (Additional file 1: Figure S1). In case of YLC8 (i.e., expressing rTrXYNII), subsequent screening on two different solid culture media containing AZCL-arabinoxylan and Azo-CMC respectively, revealed that clones displayed both xylanase and cellulase activity (Additional file 1: Figure S4; Fig. 2). Testing the ability of the supernatants of each of the daughter strains, YLC8 (rTrXYNII), YLC9 (rTrLPMOA), and YLC10 (rTrSWO1), respectively, to hydrolyze CMC, PASC, Avicel, and CIMV-cellulose (containing hemicellulose), revealed contrasting results (Fig. 1). A 12% increase in glucose production compared to YLC7 was procured by YLC8 on CIMV-cellulose hydrolysis, indicating that the presence of a xylanase is specifically beneficial to hydrolyze industrial cellulose pulp. This suggests that the xylanase works synergistically with the cellulases probably by removing hemicelluloses wrapped around glucan microfibrils and thus increasing the cellulose accessibility [43]. Indeed, the action of the xylanase was further confirmed by HPAEC analysis (data not shown), which revealed the presence of xylodextrins among the hydrolysis products. The presence of TrLPMOA (YLC9) enhanced the hydrolysis of all of the substrates tested, which is consistent with its ability to oxidize cellulose (Fig. 1). In contrast, the co-expression of rTrSWO1 with the cellulases (YLC10) did not procure a significant improvement of hydrolysis, consistent with the observation that rTrSWO1 needs sufficient contact time and confirming that the mode of action of this protein is incompatible with a co-expression strategy. For this reason, and in view of the other results, only rTrXNYII and TrLPMOA were retained for co-expression in the genetic background of YLC7, thus producing YLC11. When using the latter strain for hydrolysis tests on the different substrates, it became clear that the highest enhancement was procured on CIMV-cellulose, while the hydrolysis of pure cellulose (CMC and Avicel) did not change when compared to YLC9 (Fig. 1). We further compared the hydrolytic efficiency of the YLC11 secretome with that of the commercial cocktail Cellic CTec2, using CIMV-cellulose, Avicel and wheat straw as substrates. The results showed that when using equivalent enzyme loadings (10 FPU/g-cellulose) the YLC11 secretome degraded all three substrates faster than Cellic CTec2 (Fig. 7). The CIMV-cellulose is the most amenable substrate, since the YLC11 secretome achieved 95% conversion of this substrate to glucose in 24 h, while Cellic CTec2 achieved 86% over the same time period (Fig. 7a). However, the superiority of the YLC11 secretome is better illustrated by its action on Avicel and wheat straw. On these substrates, the secretome achieved 75 and 30% conversion, respectively, whereas the action of Cellic CTec2 yielded 63% conversion of Avicel and 24% conversion of cellulose in wheat straw (Fig. 7b, c). When compared to the commercial cocktail, the higher potency of the YLC11 secretome can probably be attributed to several factors, the first being the presence of a more efficient CBH I [30] Secondly, the ratio of the different enzymes is optimized in the YLC11 secretome and, thirdly, the amount of LPMO present is probably higher. Additionally, the presence of xylanase in the YLC11 secretome is not doubt useful for the hydrolysis of wheat straw. Therefore, we conclude that YLC11 is a particularly relevant strain for use with industrial cellulose pulp and recalcitrance biomass.
Table 1.
Microbial strains used in the present study
| Strains | Relevant genotype | Source of reference |
|---|---|---|
| T. reesei QM9414 | Wildtype | DSMZ |
| E. coli DH5 | Φ80dlacZΔm15, recA1, endA1, gyrA96, thi-1, hsdR17 (rk−, mk+), supE44, relA1, deoR, Δ(lacZYA-argF) U169 | Invitrogen |
| Y. lipolytica JMY1212 (Zeta) | MATA, ura3-302, leu2-270-LEU2-zeta, xpr2-322 ∆lip2, ∆lip7, ∆lip8 | [52] |
| ylTrXYNII | Zeta, pTEF-XYNII-His6 | This investigation |
| ylTrLPMOA | Zeta, pTEF-LPMOA-His6 | This investigation |
| ylTrSWO1 | Zeta, pTEF-SWO1-His6 | This investigation |
| Y. lipolytica YLC6 | ∆poxB12, pTEF-EGI, pTEF-EGII; pHTEF-NcCBHI, pHTEF-TrCBHII | [30] |
| YLC6b | ∆poxB12, pTEF-EGI-His6, pTEF-EGII; pHTEF-NcCBHI, pHTEF-TrCBHII | This investigation |
| YLC7 | ∆poxB12, pHTEF-EGI, pTEF-EGII; pHTEF-NcCBHI, pHTEF-TrCBHII | This investigation |
| YLC7b | ∆poxB12, pHTEF-EGI-His6, pTEF-EGII; pHTEF-NcCBHI, pHTEF-TrCBHII | This investigation |
| YLC8 | YLC7, pTEF-XYN II | This investigation |
| YLC9 | YLC7, pTEF-LPMOA | This investigation |
| YLC10 | YLC7, pTEF-SWO1 | This investigation |
| YLC11 | YLC7, pTEF-XYNII, pTEF-LPMOA | This investigation |
Fig. 7.
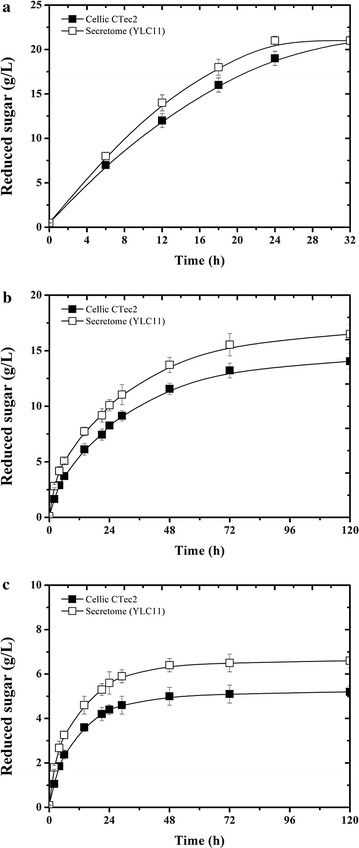
Comparison of hydrolytic efficiency of the secretome of YLC11 and Cellic CTec2 in enzymatic hydrolysis of a CIMV-cellulose, b Avicel, and c wheat straw at the enzyme loading of 10 FPU/g-cellulose
The growth of recombinant Y. lipolytica strains on cellulosic feedstocks
To investigate the suitability of the different strains described in this study for use in a consolidated bioprocessing concept, each of them (YLC6 to 11) was grown in liquid defined medium containing either CIMV-cellulose, Avicel, or milled wheat straw as the carbon source. Results revealed that all of the strains were able to use CIMV-cellulose, with conversion yields of more than 50% being achieved. In contrast, Avicel and wheat straw were less amenable to hydrolysis (Table 2). In terms of microbial biomass yields, these reached approximately 0.4 g-DCW/g-cellulose on CIMV-cellulose, but only (average) 0.3 and 0.1 g-DCW/g-cellulose on Avicel and wheat straw, respectively. It is noteworthy that the increased expression of rTrEGI (i.e., comparing YLC7 to YLC6) resulted in a slight increase in the conversion of all the cellulosic substrates, and predictably the expression of rTrXYNII (YLC8) enhanced the conversion of CIMV-cellulose (64.0 ± 0.4%), while not affecting Avicel conversion. Interestingly, the expression of rTrXYNII also enhanced the conversion of wheat straw (12.3 ± 0.5%), consistent with the fact that significant amounts of xylan are present in this substrate. Strain YLC9 expressing TrLPMOA consumed 65% of CIMV-cellulose, 36% of Avicel, and 16% of cellulose in wheat straw, results that are comparable or better than those obtained with YLC7 and 8 (producing rTrXYNII). When both rTrXYNII and rTrLPMOA (YLC11) were present, the conversion of CIMV-cellulose and wheat straw were further enhanced (68 and 18%, respectively). Finally, while the treatment with rTrSWO1 (15 mg-swollenin/g-cellulose) before strain cultivation failed to affect CIMV-cellulose conversion, it did prove to be beneficial for Avicel and wheat straw, with YLC11-mediated conversion rates being 42% (Avicel) and 22% (wheat straw), respectively, after 5 days of growth. However, it is also noteworthy that CIMV-cellulose is obtained using organosolv technology, which is known to produce relatively pure cellulose [44]. In further work, it will be interesting to confront YLC11 with cellulose pulps produced using less costly, but more ‘quick and dirty’ technologies [45].
Table 2.
Comparison of cellulose utilization and biomass yield of cellulolytic Y. lipolytica grown on different cellulosic substrates for 120 h in aerobic cultivation
| Strains | CIMV-cellulose consumed % | Biomass yield % | Avicel consumed % | Biomass yield % | Cellulose consumed % (wheat straw) | Biomass yield % |
|---|---|---|---|---|---|---|
| YLC6 | 58.6 ± 0.5 | 0.41 ± 0.03 | 30.2 ± 0.3 | 0.32 ± 0.03 | 9.7 ± 0.6 | 0.12 ± 0.01 |
| YLC7 | 61.5 ± 0.6 | 0.40 ± 0.05 | 32.0 ± 0.5 | 0.31 ± 0.02 | 10.5 ± 0.7 | 0.11 ± 0.01 |
| YLC8 | 64.0 ± 0.4 | 0.40 ± 0.02 | 31.8 ± 1.0 | 0.31 ± 0.02 | 12.3 ± 0.5 | 0.12 ± 0.01 |
| YLC9 | 65.4 ± 0.7 | 0.39 ± 0.03 | 36.3 ± 0.9 | 0.29 ± 0.03 | 15.8 ± 0.9 | 0.15 ± 0.01 |
| YLC10 | 59.0 ± 1.5 | 0.40 ± 0.04 | 31.0 ± 1.2 | 0.31 ± 0.04 | 10.3 ± 0.9 | 0.11 ± 0.02 |
| YLC11 | 67.8 ± 1.0 | 0.39 ± 0.02 | 36.5 ± 0.7 | 0.29 ± 0.02 | 18.4 ± 0.8 | 0.15 ± 0.02 |
| YLC11 + Sa | 68.3 ± 1.3 | 0.38 ± 0.01 | 42.0 ± 1.1 | 0.30 ± 0.04 | 21.6 ± 0.5 | 0.17 ± 0.01 |
The results were calculated from at least three biological replicates, and are given as the mean value ± standard deviation. The initial cellulose content was 25 g/L for all the substrates
aThe cellulose was treated by SWO1 at the dosage of 15 mg/g cellulose for 24 h before enzymatic hydrolysis
Among the different accessory proteins that have been used in this study, LPMOA stands out, because it requires an electron donor and oxygen to function. Therefore, when tests were performed, ascorbic acid was included in the reactions. However, for CIMV-cellulose and wheat straw, this was unnecessary as similar yields of cellulose conversion and biomass production were achieved for YLC9 and YLC11 without the supplementation of ascorbic acid (Additional file 1: Table S2), probably because lignin present in the substrates acts as an electron donor [46]. Additionally, it has been shown that when using conventional cellulase cocktails supplemented with LPMOs, enhancement of cellulose hydrolysis will only occur under aerobic conditions [47]. In this respect, it is noteworthy that Y. lipolytica is strictly aerobic, thus the production of LPMOA in CBP-mode should be quite advantageous [29]. Finally, it is also important to note that Y. lipolytica can assimilate gluconic acid, an important attribute when LPMOA is present (Additional file 1: Figure S5).
Regarding swollenins, our study clearly confirms the great potential for these proteins, but underlines their incompatibility with a straightforward CBP concept. To draw benefit from swollenins, it will be necessary to devise a hybrid process in which exogenously produced swollenins are used at some point during biomass pretreatment, taking into account their specific operating requirements (i.e., contact time, temperature, and protein loading).
Finally, this study confirmed the usefulness of xylanases in the context of cellulose hydrolysis. Based on our findings and those of previous studies [48], we postulate that further engineering of Y. lipolytica to confer additional β-d-xylosidase activity and the ability to metabolize xylose efficiently [49] will procure a quite potent CBP strain. If successful, this could pave the way for the use of Y. lipolytica in the biorefinery industry.
Conclusion
In the present study, a Y. lipolytica strain co-expressing core cellulolytic enzymes and accessory proteins has been successfully developed. The results reveal that accessory proteins can significantly enhance cellulose hydrolysis and confer cellulolytic Y. lipolytica with the ability on one hand to better handle industrial pulps containing xylan and on the other to hydrolyze more recalcitrant crystalline substrates. Overall, the data presented here confirm the feasibility of a Y. lipolytica-based consolidated bioprocess concept and reveal further ways to improve performance. In particular, the results obtained with swollenin provide insight into how this type of protein could be used in an advanced biorefinery concept involving a swollenin-mediated biomass pretreatment step and a CBP unit operation. Further strain engineering combined with appropriate process design will undoubtedly lead to much better performances in the future.
Methods
Strains and media
The genotypes of the microbial strains used in the present study are summarized in Table 1. E. coli DH5 was purchased from Invitrogen (Paisley, UK) and used for plasmid construction. The Y. lipolytica strains were routinely cultivated in a medium composed of 1% w/v yeast extract, 1% w/v Bacto peptone, and 1% w/v glucose (YPD), solid media contained 1.5% agar. Transformants were selected on solid YNB medium (0.17% w/v YNB, 1% glucose or cellobiose w/v, 0.5% w/v ammonium chloride, with (for Ura+) or without (for Leu+) 0.2% w/v casamino acids and 50 mM sodium–potassium phosphate buffer, pH 6.8), supplemented with uracil (440 mg/L) or leucine (440 mg/L) depending on the auxotrophic requirements. The detection of xylanase activity in solid YNB medium was achieved by incorporating 2.0% w/v AZCL-arabinoxylan (Megazyme). For cellulase characterization, enzymes were produced in YTD medium (1% w/v yeast extract, 2% w/v tryptone, 5% w/v glucose and 100 mM phosphate buffer, pH 6.8). To evaluate the growth of engineered cellulolytic Y. lipolytica on cellulose, transformants were aerobically cultivated in defined medium containing vitamins, trace elements [50], and salts, including 3.5 g/L (NH4)2SO4, 3.0 g/L K2HPO4, 3.0 g/L NaH2PO4, and 1.0 g/L MgSO4·7H2O with 27.5 g/L CIMV-cellulose (91% w/v cellulose, provided by CIMV S.A.) [44], or 25 g/L Avicel PH-101 (Sigma) or 46 g/L wheat straw (particle size ~ 0.5 mm, 27% w/w arabinoxylan, and 44% w/w cellulose) [51]. In addition, ascorbate (1 mM) was added into the cultures as the reducing agent to reactivate LPMOA.
Plasmid constructions
The plasmids constructed in the present study are summarized in Table 3 and all primers are listed in Table 4. Briefly, the total RNA from 5-day cultured T. reesei QM9414 was isolated using RNeasy Plus Mini Kit (QIAGEN) and reverse transcription was performed with iScript™ cDNA Synthesis Kit (BIO-RAD) according to the manufacturer’s instructions. LPMOA gene (formerly known as GH61A, GenBank accession code: Y11113.1) was amplified from the cDNA of T. reesei by PCR using F (1) as forward primer and R (1) as reverse primer. A 15-base pair homologous sequence of the target plasmid was introduced into the upstream and downstream of each gene during PCR amplification. After that, the gene encoding LPMOA was fused with the PCR fragment (primers JMP1F/JMP1R) of secretion vector JMP62UraTEF under the control of TEF promoter and the pre-pro sequence of lipase 2 of Y. lipolytica by In-Fusion Cloning (Clontech). XYN II gene (GenBank accession code: XM_006968885.1), encoding T. reesei xylanase II, was amplified from the plasmid JMP63UraXYNII (kindly provided by Dr. Cédric Montanier, INSA-Toulouse) by PCR using primers F (2) and R (2) and fused with secretion vector JMP62UraTEF as described above. The SWO1 gene (GenBank accession code: AJ245918.1), encoding T. reesei swollenin, was synthesized by GenScript (USA) after codon optimization based on the codon bias of Y. lipolytica (Additional file 2), and cloned into the plasmid JMP62UraTEF.
Table 3.
Plasmids used or created in the present study
| Plasmids | Description | Source of reference |
|---|---|---|
| JMP62UraTEF | URA3, pTEF | [52] |
| JMP62LeuTEF | LEU2, pTEF | [52] |
| JMP62UraTB1his | URA3, pTEF-BGL1-His6 | [57] |
| PUB4-CRE | hph, hp4d-CRE | [53] |
| JMP62UraHTEF | URA3, pHTEF | [30] |
| JMP62LeuHTEF | LEU2, pHTEF | [30] |
| JMP62hphTEF | Hph, pTEF | This investigation |
| JMP62LeuHTEF-EG1 | LEU2, pHTEF-EG I | This investigation |
| JMP62UraXYNIIhis | URA3, pTEF-XYNII-His6 | This investigation |
| JMP62UraLPMOAhis | URA3, pTEF-LPMOA-His6 | This investigation |
| JMP62UraSWO1his | URA3, pTEF-SWO1-His6 | This investigation |
| JMP62UraXYNII | URA3, pTEF-XYNII | This investigation |
| JMP62LeuXYNII | LEU2, pTEF-XYNII | This investigation |
| JMP62hphXYNII | hph, pTEF-XYNII | This investigation |
| JMP62UraLPMOA | URA3, pTEF-LPMOA | This investigation |
| JMP62LeuLPMOA | LEU2, pTEF-LPMOA | This investigation |
| JMP62hphLPMOA | hph, pTEF-LPMOA | This investigation |
| JMP62hphLPMOA/XYNII | hph, pTEF-LPMOA, pTEF-XYNII | This investigation |
| JMP62UraSWO1 | LEU2, pTEF-SWO1 | This investigation |
| JMP62Leu SWO1 | LEU2, pTEF-SWO1 | This investigation |
| JMP62hph SWO1 | hph, pTEF-SWO1 | This investigation |
Table 4.
Sequences of the oligonucleotide primers used in this study
| Primer names | Sequence (5′–3′), 15-bp homologous sequence for infusion is underlined |
|---|---|
| F1 | GTTCTCCAGAAGCGACATGGACATATTAATGACATTGTCATCAACG |
| R1 | CACAGACACCCTAGGCTAGTTAAGGCACTGGGCGTAGTAGGG |
| F2 | GTTCTCCAGAAGCGACAGACCATCCAGCCCGGCACC |
| R2 | CACAGACACCCTAGGTTAGCTCACGGTGATAGAGGCAGAGCCA |
| JMP1F | CCTAGGGTGTCTGTGGTATCTAAGCTATT |
| JMP1R | TCGCTTCTGGAGAACTGCGG |
| F3 | ACACCCGAAGGATCCCACAATGAAGCTTTCCACCATCC |
| R3 | ATGGTGATGATGGTGGCTCACGGTGATAGAGGCAGAG |
| R4 | ATGGTGATGATGGTGGTTAAGGCACTGGGCGTAGTAGGG |
| R5 | ATGGTGATGATGGTGGTTCTGGGAAAACTGGACGCC |
| JMP2F | CACCATCATCACCATCATTAAAACT |
| JMP2R | GGATCCTTCGGGTGTGAGTTG |
| HphF | CCACACACATCCACAATGAAAAAGCCTGAACTCACCGCGA |
| HphR | TAGCAGGGCAGGGCCCTATTCCTTTGCCCTCGGACGAGTG |
| LHFF | GGCCCTGCCCTGCTAATGAAATG |
| LHFR | TGTGGATGTGTGTGGTTGTATGTGTGATG |
| XLF | GCTCTAGACGATGCCGCCGCAAGGAATG (XbaI) |
| XLR | CGTCTAGATGGAATTCGATTTGTCTTAGAGGAACGCA (XbaI) |
For the expression of His-tagged proteins, XYNII, LPMOA, and SWO1 were cloned by PCR with F3 as forward primer and R (3–5) as reverse primers using the expression vectors constructed in last step as template, and fused with PCR fragment (primers JMP2F/JMP2R) of the vector JMP62UraTB1his [30]. This enables the addition of a sequence encoding 6-histidine at the C-terminus of the proteins.
To improve the expression level of EG I, the plasmid JMP62UraEG1 was digested using BamHI/AvrII, and the EG I gene was recovered and then inserted into the corresponding sites of the previously constructed plasmid JMP62UraHTEF under the control of TEF promoter with an enhancer comprising 4 tandem copies of upstream activation sequences (4UASs) [30].
After construction, all expression vectors were verified by DNA sequencing (GATC Biotech, Konstanz, Germany). For Y. lipolytica transformation, vectors were digested using NotI, thus generating a linear DNA with Zeta sequences at both extremities. Then the gel purified expression cassettes were introduced into the Zeta docking platform of Y. lipolytica JMY1212 Zeta for the expression of single recombinant protein, or randomly into the genome of ∆poxB12 strain, for co-expression of multiple cellulases, using the lithium acetate method [52]. For the latter case, the LoxP-Cre recombination system was used for marker rescue and to ensure the multistep insertion of the target genes [53]. However, after the integration of six cellulase genes (BGL1, BGL2, EGI, EGII, CBHI, and CBHII), the removal of the selection markers (URA3 and LEU2) from cellulolytic Y. lipolytica caused loss of the previously introduced genes, probably due to the presence of multiple LoxP sites. In order to further introduce accessory proteins into cellulolytic Y. lipolytica, the hygromycin phosphotransferase coding gene hph, conferring hygromycin resistance to Y. lipolytica, was amplified from the vector pUB4-Cre by PCR using primers HphF and HphR. Then, the hph gene was fused with DNA fragment amplified from the vectors JMP62LeuTEFLPMOA, JMP62LeuTEFXYNII, and JMP62LeuTEFSWO1 using primers LHFF and LHFR, respectively. The plasmid for co-expression of TrLPMOA and TrXYNII was constructed by insertion of the expressing cassette containing TrLPMOA under the control of TEF promoter, obtained by PCR amplification from the vector JMP62UraTEFLPMOA using primers XLF and XLR, into the XbaI site of the vector JMP62hphTEFXYNII. The successful multiple integration of the heterologous genes into the genome of Y. lipolytica was verified by PCR using gene-specific primers (Additional file 1: Table S1). In addition, transformants expressing multiple enzymes were tested for growth on cellobiose, and for degradation of Azo-CM-Cellulose and AZCL-arabinoxylan. Clones displaying both activities were retained for further analysis. Table 3 summarizes the expressed cellulase genes and their corresponding Y. lipolytica transformants.
Protein production and hydrolytic activity assay
Recombinant protein production by Y. lipolytica was carried out in liquid YTD medium in shake flask at 28 °C and 120 rpm for 5 days. PASC was prepared from Avicel PH-101 as previously described [54]. The overall cellulolytic activities were measured on CMC, PASC, CIMV-cellulose, and Avicel PH-101 using a previously described method with slight modifications [55]. Briefly, the reaction mixture contained 1% (w/v) cellulosic substrate, 50 mM citrate buffer (pH 4.8), and proper volume of diluted enzyme solution.
Xylanase activity was measured on 2% (w/v) beechwood xylan (Megazyme) in 50 mM citrate buffer (pH 6.0). The reaction was conducted at 50 °C for 10 min.
The reducing sugars were quantified using the dinitrosalicylic acid (DNS) reagent [30]. One unit of activity (U) was defined as the amount of enzyme or culture supernatant required to release 1 μmol of reducing sugars per min for xylanase activity, or 1 mg of reducing sugars per min for cellulolytic activity.
In addition, glucose and xylose were measured using an Aminex HPX87-H column (Bio-Rad Laboratories, Germany), operating at 50 °C using a mobile phase (5 mM H2SO4) flowing at a rate of 0.5 mL/min. Glucose and xylose were detected using a Shodex RI-101 refractive index detector (Showa Denko, New York, NY, USA).
All protein concentrations were measured using the Bradford method and bovine serum albumin as a standard [56]. All enzymatic activity measurements were performed in triplicate unless otherwise stated.
SDS-PAGE and Western blot analysis
SDS-PAGE was conducted using Mini-PROTEAN TGX Stain-Free precast gels (Biorad) according to the manufacturer’s instructions. Fifteen microlitre of culture supernatant or enzyme solution was loaded into each well. Western blotting of proteins was performed as described previously [57]. Crude supernatant of Y. lipolytica JMY1212 expressing LPMOA and SWO1 fused with the His6 tag were concentrated tenfold by ultrafiltration on an Omega™ membrane disc filter with a 10 kDa cut off (Pall, France). Blots were sequentially treated with mouse non-position-specific His-Tag antibody 1:2500 (THE™ from Genscript, Piscataway, NJ, US) and the alkaline phosphatase-conjugated goat anti-mouse IgG. Phosphatase activity was detected by NBT/BCIP as substrate.
Purification and deglycosylation of recombinant proteins
Yarrowia lipolytica JMY1212 expressing His6-tagged XYNII, LPMOA and SWO1, respectively, was grown in 200 mL YTD medium at 120 rpm, 28 °C for 48 h. After centrifugation (8000×g for 5 min at 4 °C), the supernatant was concentrated tenfold by ultrafiltration with an Omega™ membrane disc filter at 10 kDa cut off (Pall, France), and applied to 2 mL of TALON Metal Affinity Resin (Clontech, Takara-Bio, Kyoto, Japan). Subsequently, protein was eluted using buffer containing imidazole according to the manufacturer’s instructions. Deglycosylation was carried out by treating the purified proteins with endoglycosidase H (New England Biolabs, Beverly, MA, USA) to remove N-linked carbohydrates at 37 °C for 1 h. Protein samples were analyzed by SDS-PAGE and visualized with colloidal Coomassie blue staining.
Amplex Red/horseradish peroxidase assay
The oxygen reactivity of TrLPMOA was measured in a quantitative time resolved assay of H2O2 using Amplex Red and horseradish peroxidase as described previously [8, 22]. The reaction mixture (100 μL) contained 100 mM sodium acetate buffer (pH 6.0), 50 μM Amplex Red reagent (Invitrogen, France), 7.14 U/mL horseradish peroxidase, 0.1–0.5 μM LPMOA and 30 μM ascorbate as reductant, and was assembled in the well of a 96-well microtiter plate and incubated at 30 °C. The fluorescence (excitation and emission wavelengths of 560 and 595 nm, respectively) was measured using a Tecan Infinite M200Pro plate reader (Tecan, Männedorf, Switzerland). The activity of TrLPMOA was derived from the data using a standard H2O2 calibration curve. The inhibition of H2O2 production was used to test the activity of TrLPMOA on various β-glucan and xylan substrates as described previously [41]. A final concentration of 1.0% (w/v) PASC, CMC, Avicel PH-101, barley glucan (β-1,3; 1,4), and xylan was added into the above reaction mixture. All measurements were performed in triplicates.
Cello-oligosaccharide cleavage assay
The cleavage of cello-oligosaccharides by LPMOA was assayed by incubating 16 mg of TrLPMOA, 1 mM of ascorbate, and 5 mM cello-oligosaccharides (DP4-DP6) in 50 mM sodium acetate buffer (pH 4.8) at 50 °C for 24 h under shaking (1000 rpm). To stop the reaction, samples were boiled (100 °C for 10 min) and then cooled before adding an aliquot of Novozyme 188 β-glucosidase (gift from Novozyme, Denmark) (12.0 IU/g cello-oligosaccharides, 810 IU/mL). After that, the reaction was incubated for a further 12 h and then, the oxidized mono-saccharides in the supernatant were measured using a d-gluconic acid/d-glucono-δ-Lactone assay kit (Megazyme) according to manufacturer’s instructions. All assays were carried out in triplicate.
Study of the synergy of LPMOA and SWO1 on enzymatic hydrolysis of Avicel
Enzymatic hydrolysis of cellulose was carried out in 50 mM citrate buffer (pH 4.8) containing 2.0% (w/v) microcrystalline cellulose (Avicel PH-101, Sigma). Celluclast 1.5 L (60 FPU/mL) and β-glucosidase (810 IU/mL, Novozyme 188) were added at 5.0 FPU/g-cellulose and 12.0 IU/g-cellulose, respectively. The reaction (1 mL) was conducted in Eppendorf tubes (2 mL) using a thermomixer at 50 °C and 1000 rpm. To study the synergistic effect of TrLPMOA on the enzymatic hydrolysis of cellulose, different amounts of this protein (5, 10, and 15 mg-protein/g-cellulose, respectively) were added to the reaction mixtures containing Avicel (2.0% w/v). In addition, ascorbate (1 mM) was added into the reactions as the reducing agent to reactivate LPMOA. To study the synergistic effect of swollenin on the enzymatic hydrolysis of cellulose, this protein was either added into reaction mixture together with cellulases at different concentrations (5, 10, and 15 mg-swollenin/g-cellulose respectively) or, incubated with the Avicel at 50 °C under shaking for 24 h prior to the addition of cellulases. Control experiments were conducted under the same conditions, with or without, BSA (at 5, 10, and 15 mg-protein/g-cellulose). Samples were taken at regular intervals to determine the concentration of reducing sugars using DNS.
Enzymatic hydrolysis of cellulosic feedstocks
Enzymatic hydrolysis of different cellulosic feedstocks was carried out in 50 mM citrate buffer (pH 4.8). CIMV-cellulose, Avicel and wheat straw, respectively, was added into the reaction to yield a final cellulose concentration of 2.0% (w/v). Cellic® CTec2 (a gift from Novozyme) or concentrated secretome of Y. lipolytica YLC11 was added at 10.0 FPU/g-cellulose. The reaction mixture was incubated at 50 °C under shaking for 120 h. Samples were taken at regular intervals to determine the concentration of glucose using HPLC.
Growth of yeast expressing multiple cellulases on cellulosic feedstocks
Yeast growth on different cellulosic feedstock was performed in 50 mL defined medium containing CIMV-cellulose, Avicel cellulose or wheat straw in 250-mL Erlenmeyer flasks. Yeasts were pre-cultivated in defined media on glucose until mid-exponential phase and the cells were collected by centrifugation. After washing with deionized water, the cells were used to inoculate the defined medium containing cellulose to yield an initial biomass concentration of 1.0 g-DCW/L. In addition, CIMV-cellulose, Avicel cellulose, and wheat straw were incubated with swollenin at 50 °C under shaking for 24 h prior to the inoculation of strain YLC11. All the cultivations were conducted at 28 °C and under shaking at 120 rpm. Samples were taken at the end of 5 days to determine concentrations of biomass and residual cellulose (see below). The data are presented as mean values of at least three biological replicates ± standard deviation.
Analysis of cellulose residues and determination of dry cell weight
The quantification of residual cellulose and dry cell matter was conducted as previously described with slight modifications [30]. Briefly, the cell pellets mixed with cellulose from the above cultures were harvested by centrifugation at 8000×g for 10 min at 4 °C. After two washes with distilled water, the collected cellulose-cell pellet was freeze-dried and weighed. The amount of remaining cellulose and hemicellulose was calculated from the total glucose and xylose released from diluted acid hydrolysis of the residues with 2.5% sulfuric acid at 121 °C for 1 h. Dry cell weight was deduced by subtracting the amount of cellulose or lignocellulose from the weight of cellulose or lignocellulose-cell pellet. Glucose and xylose were measured by HPLC as described before [57]. The biomass yield was calculated as the ratio of the amount of biomass obtained divided by the amount of carbon source consumed.
Additional files
Additional file 1: Figure S1. PCR verification of Y. lipolytica transformants expressing multiple cellulases and accessory proteins (A) YLC7, Lane 1 to 6: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII; (B) YLC8, Lane 1 to 6: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrXYNII; (C) YLC9, Lane 1 to 7: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrLPMOA; (D) YLC10, Lane 1 to 7: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrSWO1; (D) YLC11, Lane 1 to 8: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrXYNII, TrLPMOA. Figure S2. Western blot analysis of the heterologous rhTrEGI protein secreted by the engineered Y. lipolytica strains: lane 1, Endo H-treated secretome of YLC6b (20 μL); lane 2, Endo H-treated secretome of YLC7b (20 μL). Figure S3. Characterization of the recombinant XYNII expressed in Y. lipolytica. (a) Effect of pH on the activity of rhXYNII; (B) Effect of temperature on the activity of rhXYNII. Figure S4. Screening of Y. lipolytica expressing cellulases and accessory enzymes on YNB indication plate containing supplemented with 0.2% w/vAzo-CM-Cellulose. Lane 1, Y. lipolytica control; Lane 2 to 4, YLC8, YLC9 and YLC10. Figure S5. The growth of Y. lipolytica in defined medium containing 10 g/L gluconic acid or 10 g/L glucose. Table S1. The sequences of the oligonucleotide primers used for PCR verification of Y. lipolytica-transformants. Table S2. Comparison of cellulose utilization and biomass yield of cellulolytic Y. lipolytica grown on different cellulosic substrates for 120 h in aerobic cultivation without the addition of ascorbic acid.
Additional file 2. Nucleotide sequence of codon-optimized SWO1.
Authors’ contributions
ZPG, SB, SD, AM, and MJOD conceived the study and participated in its design. ZPG designed the constructs, carried out all the experiments, and drafted the manuscript. SB, SD, JMN, AM, and MJOD revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to express their gratitude to Nelly Monties for her help with chromatographic analyses. We thank Dr. Cédric Montanier for kingly providing us with the plasmid JMP63UraXYNII. We thank CIMV S.A. and Françoise Ouarne for kindly providing us with CIMV-cellulose. We thank the ICEO facility dedicated to enzyme screening and discovery, and part of the Integrated Screening Platform of Toulouse (PICT, IBiSA) for providing access to its screening, protein purification and analytical facilities.
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
All data generated or analyzed in the present study are included in this published article and a supporting material “Additional file 1.”
Consent for publication
All authors consent for publication.
Ethical approval and consent to participate
Not applicable.
Funding
This work was funded by the Agence Nationale de la Recherche (Investissements d’Avenir program; reference ANR-11-BTBR-0003). Dr. Guo is a recipient of a postdoctoral fellowship from the Institut National de la Recherche Agronomique.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- BGL
β-glucosidase
- CBH
cellobiohydrolase
- CBP
consolidated bioprocessing
- CMC
carboxymethyl cellulose
- DCW
dry cell weight
- EG
endoglucanase
- Endo H
endoglycosidase
- LCB
lignocellulosic biomass
- LPMO
lytic polysaccharide monooxygenase
- Mw
molecular weight
- NBT
nitroblue tetrazolium
- Nc
Neurospora crassa
- PAGE
polyacrylamide gel electrophoresis
- PASC
phosphoric acid swollen cellulose
- r
recombinant proteins
- rh
His6-tagged recombinant proteins
- HPAEC
high performance anion exchange chromatography
- SWO
swollenin
- Tr
Trichoderma reesei
- yl
Y. lipolytica
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13068-017-0990-y) contains supplementary material, which is available to authorized users.
Contributor Information
Zhong-peng Guo, Email: zguo@insa-toulouse.fr.
Sophie Duquesne, Email: sbozonne@insa-toulouse.fr.
Sophie Bozonnet, Email: duquesne@insa-toulouse.fr.
Jean-Marc Nicaud, Email: jean-marc.nicaud@inra.fr.
Alain Marty, Email: marty@insa-toulouse.fr.
Michael Joseph O’Donohue, Email: michael.odonohue@insa-toulouse.fr.
References
- 1.Zhang YHP. A sweet out-of-the-box solution to the hydrogen economy: is the sugar-powered car science fiction? Energy Environ Sci. 2009;2:272–282. doi: 10.1039/b818694d. [DOI] [Google Scholar]
- 2.Sheldon RA. Green and sustainable manufacture of chemicals from biomass: state of the art. Green Chem. 2014;16:950. doi: 10.1039/C3GC41935E. [DOI] [Google Scholar]
- 3.Chundawat SP, Beckham GT, Himmel ME, Dale BE. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng. 2011;2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 4.Kumar M, Campbell L, Turner S. Secondary cell walls: biosynthesis and manipulation. J Exp Bot. 2016;67(2):515–531. doi: 10.1093/jxb/erv533. [DOI] [PubMed] [Google Scholar]
- 5.Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng. 2008;10:295–304. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Kumar P, Barrett DM, Delwiche MJ, Stroeve P. Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res. 2009;48(8):3713–3729. doi: 10.1021/ie801542g. [DOI] [Google Scholar]
- 7.Brethauer S, Studer MH. Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ Sci. 2014;7:1446–1453. doi: 10.1039/c3ee41753k. [DOI] [Google Scholar]
- 8.Kittl R, Kracher D, Burgstaller D, Haltrich D, Ludwig R. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol Biofuels. 2012;5(1):79. doi: 10.1186/1754-6834-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasunuma T, Kondo A. Development of yeast cell factories for consolidated bioprocessing of lignocellulose to bioethanol through cell surface engineering. Biotechnol Adv. 2012;30(6):1207–1218. doi: 10.1016/j.biotechadv.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Mazzoli R. Development of microorganisms for cellulose-biofuel consolidated bioprocessings: metabolic engineers’ tricks. Comput Struct Biotechnol J. 2012;3:e201210007. doi: 10.5936/csbj.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Haan R, van Rensburg E, Rose SH, Görgens JF, van Zyl WH. Progress and challenges in the engineering of non-cellulolytic microorganisms for consolidated bioprocessing. Curr Opin Biotechnol. 2015;33:32–38. doi: 10.1016/j.copbio.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Sticker AR, Mach RL, de Graaff LH. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl Microbiol Biotechnol. 2008;78:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- 13.Chokhawala HA, Roche CM, Kim TW, Atreya ME, Vegesna N, Dana CM, Blanch HW, Clark DS. Mutagenesis of Trichoderma reesei endoglucanase I: impact of expression host on activity and stability at elevated temperatures. BMC Biotechnol. 2015;15:11. doi: 10.1186/s12896-015-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Álvarez C, Reyes-Sosa FM, Díez B. Enzymatic hydrolysis of biomass from wood. Microb Biotechnol. 2016;9(2):149–156. doi: 10.1111/1751-7915.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selig MJ, Knoshaug EP, Adney WS, Himmel ME, Decker SR. Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresour Technol. 2008;99(11):4997–5005. doi: 10.1016/j.biortech.2007.09.064. [DOI] [PubMed] [Google Scholar]
- 17.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37(Database issue):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VG. Novel enzymes for the degradation of cellulose. Biotechnol Biofuels. 2012;5(1):45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sørlie M, Eijsink VG. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 20.Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem Biol. 2011;6:1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 21.Hemsworth GR, Taylor EJ, Kim RQ, Gregory RC, Lewis SJ, Turkenburg JP, Parkin A, Davies GJ, Walton PH. The copper active site of CBM33 polysaccharide oxygenases. J Am Chem Soc. 2013;135:6069–6077. doi: 10.1021/ja402106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaksen T, Westereng B, Aachmann FL, Agger JW, Kracher D, Kittl R, Ludwig R, Haltrich D, Eijsink VG, Horn SJ. A C4-oxidizing lytic polysaccharide monooxygenase cleaving both cellulose and cello-oligosaccharides. J Biol Chem. 2014;289(5):2632–2642. doi: 10.1074/jbc.M113.530196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bey M, Zhou S, Poidevin L, Henrissat B, Coutinho PM, Berrin JG, Sigoillot JC. Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (FamilyGH61) from Podospora anserina. Appl Environ Microbiol. 2013;79:488–496. doi: 10.1128/AEM.02942-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cannella D, Jørgensen H. Do new cellulolytic enzyme preparations affect the industrial strategies for high solids lignocellulosic ethanol production? Biotechnol Bioeng. 2014;111(1):59–68. doi: 10.1002/bit.25098. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert HJ. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 2010;153(2):444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jäger G, Girfoglio M, Dollo F, Rinaldi R, Bongard H, Commandeur U, Fischer R, Spiess AC, Büchs J. How recombinant swollenin from Kluyveromyces lactis affects cellulosic substrates and accelerates their hydrolysis. Biotechnol Biofuels. 2011;4(1):33. doi: 10.1186/1754-6834-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eibinger M, Sigl K, Sattelkow J, Ganner T, Ramoni J, Seiboth B, Plank H, Nidetzky B. Functional characterization of the native swollenin from Trichoderma reesei: study of its possible role as C1 factor of enzymatic lignocellulose conversion. Biotechnol Biofuels. 2016;9(1):178. doi: 10.1186/s13068-016-0590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, Otoupal P, Alper HS. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. 2014;5:3131. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 29.Groenewald M, Boekhout T, Neuvéglise C, Gaillardin C, van Dijck PW, Wyss M. Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit Rev Microbiol. 2014;40(3):187–206. doi: 10.3109/1040841X.2013.770386. [DOI] [PubMed] [Google Scholar]
- 30.Guo ZP, Duquesne S, Bozonnet S, Cioci G, Nicaud JM, Marty A, O’Donohue MJ. Conferring cellulose-degrading ability to Yarrowia lipolytica to facilitate a consolidated bioprocessing approach. Biotechnol Biofuels. 2017;10:132. doi: 10.1186/s13068-017-0819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerjee G, Car S, Scott-Craig JS, Borrusch MS, Aslam N, Walton JD. Synthetic enzyme mixtures for biomass deconstruction: production and optimization of a core set. Biotechnol Bioeng. 2010;106(5):707–720. doi: 10.1002/bit.22741. [DOI] [PubMed] [Google Scholar]
- 32.Billard H, Faraj A, Lopes Ferreira N, Menir S, Heiss-Blanquet S. Optimization of a synthetic mixture composed of major Trichoderma reesei enzymes for the hydrolysis of steam-exploded wheat straw. Biotechnol Biofuels. 2012;5(1):9. doi: 10.1186/1754-6834-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao DH, Chundawat SPS, Krishnan C, Balan V, Dale BE. Mixture optimization of six core glycosyl hydrolases for maximizing saccharification of ammonia fiber expansion (AFEX) pretreated corn stover. Bioresour Technol. 2010;101(8):2770–2781. doi: 10.1016/j.biortech.2009.10.056. [DOI] [PubMed] [Google Scholar]
- 34.Blazeck J, Liu L, Redden H, Alper H. Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol. 2011;77(22):7905–7914. doi: 10.1128/AEM.05763-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J, Yu B, Zhang K, Ding X, Chen D. Expression of endo-1, 4-beta-xylanase from Trichoderma reesei in Pichia pastoris and functional characterization of the produced enzyme. BMC Biotechnol. 2009;9:56. doi: 10.1186/1472-6750-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adav SS, Chao LT, Sze SK. Quantitative secretomic analysis of Trichoderma reesei strains reveals enzymatic composition for lignocellulosic biomass degradation. Mol Cell Proteom. 2012 doi: 10.1074/mcp.M111.012419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannella D, Hsieh CW, Felby C, Jørgensen H. Production and effect of aldonic acids during enzymatic hydrolysis of lignocellulose at high dry matter content. Biotechnol Biofuels. 2012;5(1):26. doi: 10.1186/1754-6834-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu J, Chandra R, Arantes V, Gourlay K, van Dyk JS, Saddler JN. The addition of accessory enzymes enhances the hydrolytic performance of cellulase enzymes at high solid loadings. Bioresour Technol. 2015;186:149–153. doi: 10.1016/j.biortech.2015.03.055. [DOI] [PubMed] [Google Scholar]
- 39.Hamilton SR, Zha D. Progress in yeast glycosylation engineering. Methods Mol Biol. 2015;1321:73–90. doi: 10.1007/978-1-4939-2760-9_6. [DOI] [PubMed] [Google Scholar]
- 40.Vu VV, Beeson WT, Phillips CM, Cate JH, Marletta MA. Determinants of regioselective hydroxylation in the fungal polysaccharide monooxygenases. J Am Chem Soc. 2014;136(2):562–565. doi: 10.1021/ja409384b. [DOI] [PubMed] [Google Scholar]
- 41.Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou S, Fanuel M, Ropartz D, Rogniaux H, Gimbert I, Record E, Berrin JG. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol Biofuels. 2015;8:90. doi: 10.1186/s13068-015-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gourlay K, Hu J, Arantes V, Andberg M, Saloheimo M, Penttilä M, Saddler J. Swollenin aids in the amorphogenesis step during the enzymatic hydrolysis of pretreated biomass. Bioresour Technol. 2013;142:498–503. doi: 10.1016/j.biortech.2013.05.053. [DOI] [PubMed] [Google Scholar]
- 43.Jeoh T, Ishizawa CI, Davis MF, Himmel ME, Adney WS, Johnson DK. Cellulase digestibility of pretreated biomass is limited by cellulose accessibility. Biotechnol Bioeng. 2007;98:112–122. doi: 10.1002/bit.21408. [DOI] [PubMed] [Google Scholar]
- 44.Snelders J, Dornez E, Benjelloun-Mlayah B, Huijgen WJ, de Wild PJ, Gosselink RJ, Gerritsma J, Courtin CM. Biorefining of wheat straw using an acetic and formic acid based organosolv fractionation process. Bioresour Technol. 2014;156:275–282. doi: 10.1016/j.biortech.2014.01.069. [DOI] [PubMed] [Google Scholar]
- 45.Lantz SE, Goedegebuur F, Hommes R, Kaper T, Kelemen BR, Mitchinson C, Wallace L, Ståhlberg J, Larenas EA. Hypocrea jecorina CEL6A protein engineering. Biotechnol Biofuels. 2010;3:20. doi: 10.1186/1754-6834-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westereng B, Cannella D, Wittrup Agger J, Jørgensen H, Larsen Andersen M, Eijsink VG, Felby C. Enzymatic cellulose oxidation is linked to lignin by long-range electron transfer. Sci Rep. 2015;5:18561. doi: 10.1038/srep18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller G, Várnai A, Johansen KS, Eijsink VG, Horn SJ. Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnol Biofuels. 2015;8:187. doi: 10.1186/s13068-015-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, Wei H, Alahuhta M, Chen X, Hyman D, Johnson DK, Zhang M, Himmel ME. Heterologous expression of xylanase enzymes in lipogenic yeast Yarrowia lipolytica. PLoS ONE. 2014;9(12):e111443. doi: 10.1371/journal.pone.0111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ledesma-Amaro R, Lazar Z, Rakicka M, Guo Z, Fouchard F, Coq AC, Nicaud JM. Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab Eng. 2016;38:115–124. doi: 10.1016/j.ymben.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Verduyn C, Postma E, Scheffers WA, van Dijken JP. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 51.Song L, Siguier B, Dumon C, Bozonnet S, O’Donohue MJ. Engineering better biomass-degrading ability into a GH11 xylanase using a directed evolution strategy. Biotechnol Biofuels. 2012;5(1):3. doi: 10.1186/1754-6834-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duquesne S, Bordes F, Fudalej F, Nicaud JM, Marty A. The yeast Yarrowia lipolytica as a generic tool for molecular evolution of enzymes. Methods Mol Biol. 2012;861:301–312. doi: 10.1007/978-1-61779-600-5_18. [DOI] [PubMed] [Google Scholar]
- 53.Fickers P, Le Dall MT, Gaillardin C, Thonart P, Nicaud J-M. New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J Microbiol Methods. 2003;55(3):727–737. doi: 10.1016/j.mimet.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Zhang YHP, Cui J, Lynd LR, Kuang LR. A transition from cellulose swelling to cellulose dissolution by o-phosphoric acid: evidence from enzymatic hydrolysis and supramolecular structure. Biomacromol. 2006;7:644–648. doi: 10.1021/bm050799c. [DOI] [PubMed] [Google Scholar]
- 55.Ghose TK. Measurement of cellulase activities. Pure Appl Chem. 1987;59:257–268. [Google Scholar]
- 56.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 57.Guo Z, Duquesne S, Bozonnet S, Cioci G, Nicaud JM, Marty A, O’Donohue MJ. Development of cellobiose-degrading ability in Yarrowia lipolytica strain by overexpression of endogenous genes. Biotechnol Biofuels. 2015;8:109. doi: 10.1186/s13068-015-0289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. PCR verification of Y. lipolytica transformants expressing multiple cellulases and accessory proteins (A) YLC7, Lane 1 to 6: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII; (B) YLC8, Lane 1 to 6: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrXYNII; (C) YLC9, Lane 1 to 7: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrLPMOA; (D) YLC10, Lane 1 to 7: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrSWO1; (D) YLC11, Lane 1 to 8: BGL1, BGL2, 4UASTrEGI, TrEGII, 4UASNcCBHI, 4UASTrCBHII, TrXYNII, TrLPMOA. Figure S2. Western blot analysis of the heterologous rhTrEGI protein secreted by the engineered Y. lipolytica strains: lane 1, Endo H-treated secretome of YLC6b (20 μL); lane 2, Endo H-treated secretome of YLC7b (20 μL). Figure S3. Characterization of the recombinant XYNII expressed in Y. lipolytica. (a) Effect of pH on the activity of rhXYNII; (B) Effect of temperature on the activity of rhXYNII. Figure S4. Screening of Y. lipolytica expressing cellulases and accessory enzymes on YNB indication plate containing supplemented with 0.2% w/vAzo-CM-Cellulose. Lane 1, Y. lipolytica control; Lane 2 to 4, YLC8, YLC9 and YLC10. Figure S5. The growth of Y. lipolytica in defined medium containing 10 g/L gluconic acid or 10 g/L glucose. Table S1. The sequences of the oligonucleotide primers used for PCR verification of Y. lipolytica-transformants. Table S2. Comparison of cellulose utilization and biomass yield of cellulolytic Y. lipolytica grown on different cellulosic substrates for 120 h in aerobic cultivation without the addition of ascorbic acid.
Additional file 2. Nucleotide sequence of codon-optimized SWO1.


