Abstract
Previously, we discovered and structurally characterized a complex between amyloid β 1–40 and the neuropeptide leucine enkephalin. This work identified leucine enkephalin as a potentially useful starting point for the discovery of peptide-related biotherapeutics for Alzheimer’s disease. In order to better understand such complexes that are formed in vitro, we describe here the analysis of a series of site-directed amino acid substitution variants of both peptides, covering the leucine enkephalin sequence in its entirety and a large number of selected residues of amyloid β 1–40 (residues: D1, E3, F4, R5, H6, Y10, E11, H13, H14, Q15, K16, E22, K28, and V40). Ion mobility-mass spectrometry measurements and molecular dynamics simulations reveal that the hydrophobic C-terminus of leucine enkephalin (Phe-Leu, FL) is crucial for the formation of peptide complexes. As such, we explore here the interaction of the dipeptide FL with both wildtype and variant forms of amyloid β in order to structurally characterize the complexes formed. We find that FL binds preferentially to amyloid β oligomers, and attaches to amyloid β within the region between its N-terminus and its hydrophobic core, most specifically at residues Y10 and Q15. We further show that FL is able to prevent fibril formation.
Graphical abstract
Introduction
The utility of ion mobility-mass spectrometry (IM-MS) as a tool for drug discovery and development, especially in the context of protein misfolding diseases such as Alzheimer’s Disease (AD) and Type II Diabetes, has recently emerged.1–10 While many hypotheses surrounding the causation of AD are proposed, a prominent theory is that the uncontrolled aggregation of Amyloid β (Aβ) peptides, which form small toxic oligomers (dimer to dodecamer) leads ultimately to neuronal cell death.11–15 As Aβ aggregation allows for many oligomeric states simultaneously, studying Aβ oligomers for the purposes of drug discovery is difficult with classical biophysical and structural biology techniques.16 IM-MS allows for the use of low concentrations, small sample volumes, and separates protein oligomers based on mass and collision cross section (CCS)17–19 making it an ideal technology for the analysis of Aβ-small molecule complexes.
We previously reported a set of experiments that identified Aβ-Neuropeptide complexes, with the ultimate goal of identifying a peptide or peptide fragment which might inhibit Aβ fibril formation and provide a scaffold for future potential biotherapeutics20. In these datasets, we found evidence of interactions between Aβ1–40 and both leucine enkephalin and galanin neuropeptides. We characterized the former in detail, providing CCSs, peptide-ligand dissociation constant (KD) values, and molecular dynamics (MD) simulations. In addition, we studied the ability of leucine enkephalin to inhibit Aβ fibril formation using both TEM and gel electrophoresis. Complexes containing Aβ and leucine enkephalin were detected in stoichiometric ratios ranging from 1:1 to 1:3, and from 2:1 to 2:3 Aβ:leucine enkephalin. At increased concentrations of leucine enkephalin, the data indicated that attachment to Aβ dimers is favored over binding to the monomeric form. KD values were measured in the µM range for these complexes, and MD simulations led the determination of the likely binding location of leucine enkephalin along the N-terminus of the Aβ sequence, similar to other natural products.5 Transmission Electron Microscopy (TEM) and gel data reported decreased Aβ fibrillation and an increased amount of lower molecular weight Aβ oligomers when incubated in excess with leucine enkephalin, compared with control.
Here we report IM-MS measurements of complexes formed between Aβ and the site directed amino acid substitutions of the entire sequence of leucine enkephalin, which indicate that the hydrophobic C-terminal residues FL are critical for the formation of Aβ:leucine enkephalin interactions. Furthermore, we study the dipeptide FL and its complexes formed with Aβ sequence variants. For 1:1 Aβ:FL complexes, we identify residues Y10 and Q15 as possessing key interactions with FL, necessary for strong complex formation. This observation matches well with the predicted location of FL binding from MD simulations of Aβ:leucine enkephalin.20 MD simulations of Aβ:FL were conducted and models filtered using IM-MS data. From these, we predict that FL interacts with Aβ primarily through Y10, and that these interactions occur mostly through a series of specific interactions with the C-terminus of FL. Beyond monomeric Aβ complexes, we also study higher-order Aβ complexes (e.g. dimers and trimers) and find that FL interacts with Aβ dimers preferentially. Finally, we explored the downstream effects of FL binding on Aβ fibrillation through TEM analysis, finding strong evidence of the inhibition of fibrilization. We conclude by discussing the potential impact of FL as a scaffold for the future development of AD therapies in the context of previously described small peptide-based amyloid inhibitors.
Results and Discussion
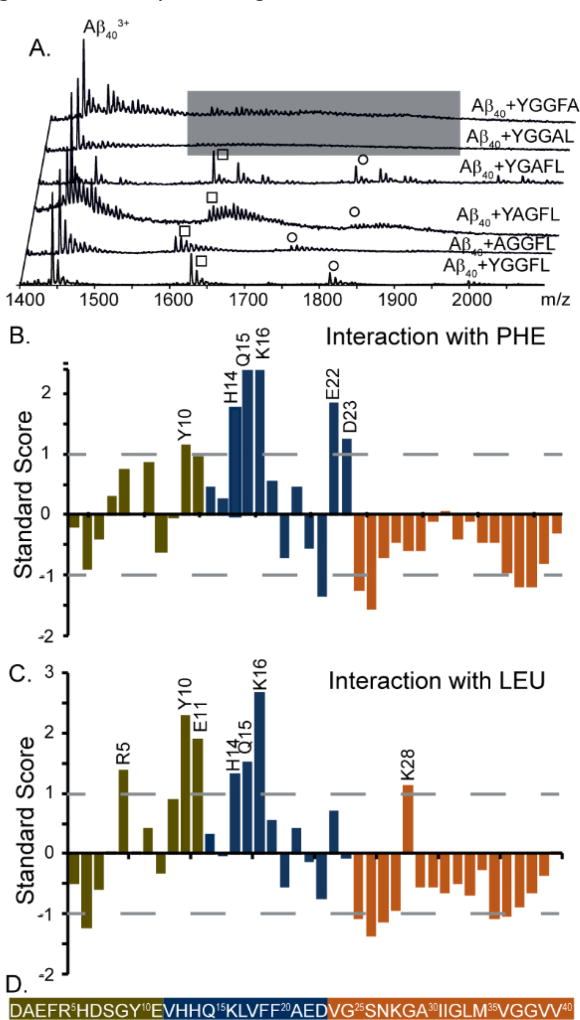
To investigate the specific interactions responsible for Aβ:leucine encephalin binding, we first undertook a series of IM-MS experiments using a comprehensive set of site-directed amino acid substituted forms of leucine enkephalin (sequence: YGGFL). Each of these five leucine enkephalin variants was first evaluated using MS intensity values under controlled conditions as a quantitative measure of Aβ1–40 complex formation. (Figure 1A) Alanine substitution of the first three residues in the leucine enkephalin sequence (tyrosine and both glycine residues) yield complex ion signals similar to control experiments under our experimental conditions. However, when the two hydrophobic C-terminal residues F and L were substituted, neither of these variants were observed to generate signals associated with complex formation, indicating their importance in Aβ binding overall.
Figure 1.
Interactions between Aβ1–40 and leucine enkephalin variants. A) IM-MS analysis of alanine substituted variants of leucine enkephalin reveals complexes formed between Aβ1–40 and AGGFL, YAGFL, and YGAFL in a manner similar to WT leucine enkephalin. Samples containing YGGAL and YGGFA produced no measureable complex signals, indicating F and L to be critical for the interaction with Aβ. Square annotations represent the 1:1 interaction, and circles the 1:2 interaction. B) and C) Plots of the standard score (Z-score) for Aβ residues that are within 4Å of the F and L in leucine enkephalin peptide structure. D) Sequence of Aβ1–40, where the N-terminus (green) is defined as residues 1–11, the hydrophobic core (blue) is residues 12–23, and the C-terminus is residues 24–40. This color-coding for Aβ1–40 applies to all figures in this report.
Using the information provided in Figure 1A as well as CCS information from our previous study,20 we filtered an ensemble of putative Aβ:leucine encephalin structures generated by MD in order to enrich for models where Aβ constituents are within 4 Å of either the F or L residues of the bound leucine enkephalin peptide. This refined population of structures was used to calculate standardized probability values for each residue of Aβ1–40 in terms of their F or L contacts. Our analysis highlights several regions of importance, which are shown in Figures 1B and C, where a standard score greater than 1 indicates an increased significance of the contact observed between the indicated Aβ residue and either the F or L within the leucine enkephalin sequence, respectively. The Aβ1–40 residues which most frequently interact with F within leucine enkephalin are Y10, H14, Q15, K16, E22 and D23, and those interacting most frequently with the L residue within leucine enkephalin are R5, Y10, E11, H14, Q16, K16, and K28. Those interacting significantly with both residues are Y10, H14, Q15, K16. This analysis agrees with our previous models of Aβ:leucine enkephalin complexes, from which we concluded that leucine enkephalin binding likely occurs in the N-terminal hydrophobic core region of Aβ, exhibiting similar behavior to other natural products that act as Aβ amyloid inhibitors.5, 20 As such, R5, Y10, E11, H14, Q15, K16, E22, and K28 alanine substitution variants of Aβ were targeted for downstream experiments. Of the remaining alanine variants, D1A and V40A, the Aβ termini, were used as negative controls. Furthermore, H6A and H13A were selected as additional controls for the H14A while E3 served as an additional control for the E11A and E22A substitutions. Each of these selections were designed to ensure that any detected change in complex KD could be interpreted as related to the disruption of a specific binding motif observed by MD, rather than due to subtle changes in Aβ structure. In addition, the F4A Aβ variant has been identified as a potential binding site in the context of other AD therapeutics6, and so was also used here as a control.
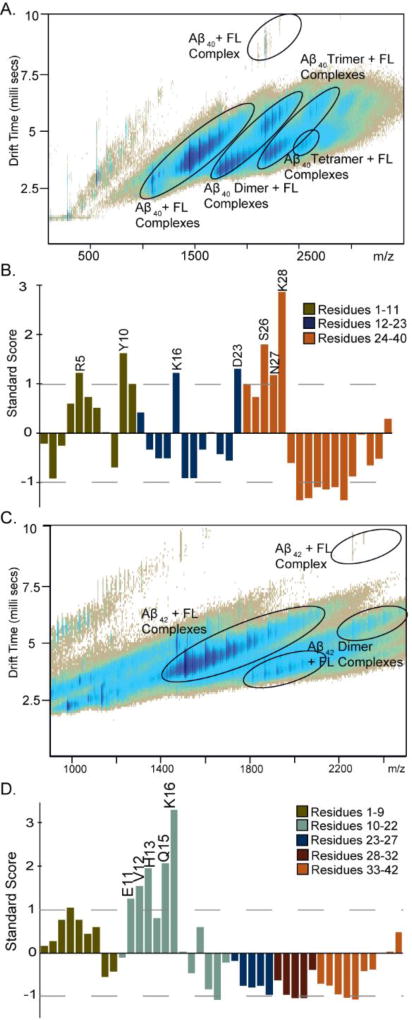
As our data strongly indicates that it is primarily the hydrophobic C-terminus of leucine enkephalin that is responsible for its binding to Aβ, we moved to focus our experimental efforts on the dipeptide FL. IM-MS data acquired from solutions continuing 20 µM Aβ1–40 and FL ranging from 10 – 80 µM, reveals the formation of Aβ1–40:FL complexes having stoichiometries of 1:1 – 1:4, 2:1 – 2:5, 3:1 – 3:4, and 4:1 (Figure 2A). Detailed arrival time distributions can be found in the supplemental material. Companion MD simulations of Aβ1–40:FL 1:1 complex yields evidence of significant interactions between the dipeptide and R5, Y10, K16, D23, S26, N27, and K28 within the Aβ sequence (Figure 2B). This result largely agrees with our analysis of Aβ1–40: leucine enkephalin models discussed previously (Figure 1C), which predicted R5, Y10, K16, D23, and K28 as likely FL binding sites. While Aβ1–40 is found in greater quantities in vivo, Aβ1–42 is theorized to be the more pathogenic variant and aggregates more rapidly, making it more difficult to study in vitro. IM-MS data shown in Figure 2C indicates that Aβ1–42 also binds FL, exhibiting KD and complex stoichiometry values similar to that of Aβ1–40. Low energy models generated for Aβ1–42:FL 1:1 complex exhibit a strong preference toward binding within the hydrophobic core of Aβ1–42, specifically with residues E11, V12, H13, Q15, and K16 (Figure 2D), an observation that is generally retained from our Aβ1–40 MD and IM-MS data as well.
Figure 2.
Interaction between Aβ with the dipeptide FL. A) IM-MS data shows the presence of Aβ1–40 in complex with FL as a monomer, dimer, trimer, and tetramer at 20 µM Aβ: 60 µM FL. Multiple copies of FL bind to each oligomer under these concentrations, while at lower FL concentrations (10 and 20 µM) only 1:1 complexes are observed. B and D) Plots of the standard score (Z-score) for Aβ residues within 4Å of the bound FL. Larger values denote contacts of greater significance on the standard deviation (σ) scale. Negative values denote contacts of reduced significance. C) IM-MS data shows the presence of Aβ1–42 in complex with FL as both monomer and dimer, with up to 4 copies of FL binding to each oligomer at 20 µM Aβ: 60 µM FL. As with Aβ1–40, with lower concentrations of FL, fewer FL copies are bound.
In addition, the measured KD values for Aβ1–40 binding with leucine enkephalin (supplemental) and Aβ1–40 binding with FL are within an order of magnitude, a difference we consider to be negligible due to the variability of our KD measurements. This observation further confirms the importance of phenylalanine and leucine in Aβ binding. If other residues were also critical for leucine enkephalin binding, we would expect that the additional inter-molecular interactions would produce an aggregate KD value for FL binding significantly greater than the ~60 µM observed here.
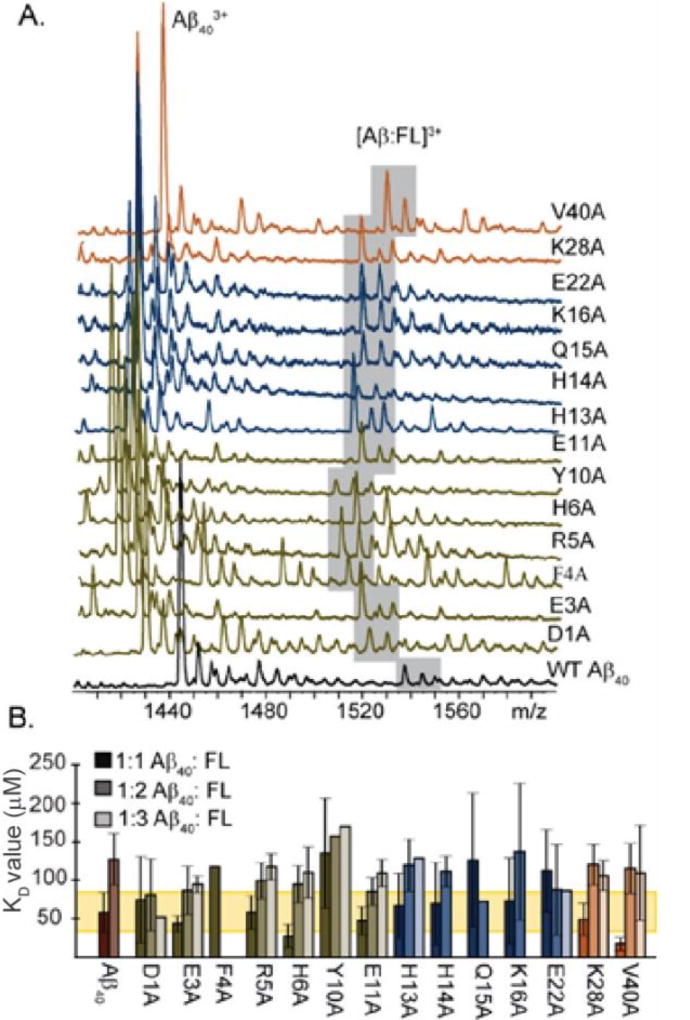
To more-clearly elucidate the binding location of FL within the Aβ1–40 sequence, KD values were extracted from the MS data acquired for all alanine substituted Aβ variants listed above, in addition to control variants: D1A, E3A, F4A, H6A, H13A, and V40A (Figure 3B). All control residues represent regions of the Aβ sequence that IM-MS filtered computational models did not highlight as critical for leucine enkephalin binding. Unavoidable signal variations in our data resulting from the complex mixture of ions analyzed in our experiments bring about relatively large standard deviations for some of our KD measurements. Here, we conservatively assign significance to values that differ by greater than one standard deviation from the mean.
Figure 3.
Site directed amino acid substitution effects on FL biding. A) Shown is a representative spectra of 20 µM Aβ1–40 variant incubated with 80 µM FL for 1 hour on ice. The grey boxed region indicates the 1:1 complex detected at a 3+ charge state. Signal for the 1:1 complexes of Y10A: FL and Q15A: FL show decreases in intensity when compared to the WT interaction. B) KD values calculated for each amino acid substitution. Error bars indicate 1 standard deviation (σ). The yellow bar is ±1 (σ) of the 1:1 complex. Of the 1:1 interactions observed, only Y10A, Q15A, and E22A are above 1 σ different than the WT. All 1:2 interactions fell within standard deviation of the WT 1:2 interaction and a WT 1:3 interaction could not be measured in triplicate due to signal-to-noise constraints.
Of the fourteen alanine variants studied, only three produced KD values significantly greater than those recorded for the WT Aβ1–40:FL 1:1 complex (KD = 58.1 ± 25.9 µM). These variants include Y10A, Q15A, and E22A (KD = 135 ± 71.3 µM, 126 ± 86.6 µM, 112 ± 53 µM respectively). Neighboring variants E11A, H14A and K16A do not deviate significantly from WT. In fact, the mutations R5A, K16A, and K28A all fall within error of the WT value, demonstrating the lysine and arginine residues of Aβ have no impact on FL binding. Of the control cases, D1A, E3A and H13A behaved similarly to the WT as expected. The 1:1 complex of F4A:FL was only observed twice in seven separate samples, having a measured KD value of 117 µM (data was insufficient to project experimental error). In contrast to these results, two variants, H6A and V40A, produced significantly reduced KD values relative to WT, 27.5 ± 14.8 µM and 17.2 ± 8.5 µM respectively. (Figure 3B). Taken together, these data indicate that residues Y10, Q15, and E22 likely provide key molecular contacts that stabilize the Aβ1–40:FL 1:1 complex, while H6 and V40 provide a destabilizing or screening effect that, when removed, improves the strength of the resulting complex significantly.
Multiple FL binding events to monomeric Aβ (1:2 and 1:3 Aβ:FL) yielded weaker KD values, generally, than those associated with the 1:1 interactions observed for all variants where 1:2 or 1:3 complexes were detected. Additionally, all KD measurements for 1:2 complexes were within error of each other, providing no clear evidence of a preferred FL binding site in the higher-order complexes observed here. KD values for recorded for binding between alanine variants and leucine enkephalin, as well as the values acquired for higher order Aβ: leucine enkephalin complexes, are available in Supporting Information.
In order to validate that the structures of the variant forms of Aβ1–40 are not significantly altered from that of the WT sequence, CD spectroscopy data was acquired for all alanine variants and WT Aβ peptides (Supporting Information). While Y10A and Q15A peptides have CD signatures similar to that of Aβ1–40, the CD spectrum acquired for E22A exhibits a pronounced β-sheet structure similar to that of Aβ1–42. The known familial mutation E22G (Arctic mutation) of Aβ1–40 has shown an increased propensity for aggregation and behavior mimicking Aβ1–42.21 The alanine variant E22A differs from this mutation by only a methyl group and was suspected to mimic E22G structurally, making the CD results unsurprising. As such, CD data supports an analysis of our IM-MS KD data interpreted on the basis of only the altered molecular contacts within the FL:Aβ binding surface for Y10A and Q15A, but strongly suggests that E22A KD data is a product of a significantly altered peptide conformation.
Using all the data described above, we filtered the output of MD simulations to produce a range of models that describe the molecular details of the Aβ1–40:FL 1:1 complex observed in our experiments. Our workflow begins by filtering all models that have CCS values outside of our experimental error (652.6 ± 22.1 Å2 for the Aβ1–40:FL complex). Next, we remove all models that lack clear interactions (contact within 4 Å) between FL and Y10 and Q15, which our experimental data suggest are key elements for the efficient formation of a stable Aβ1–40:FL complex.
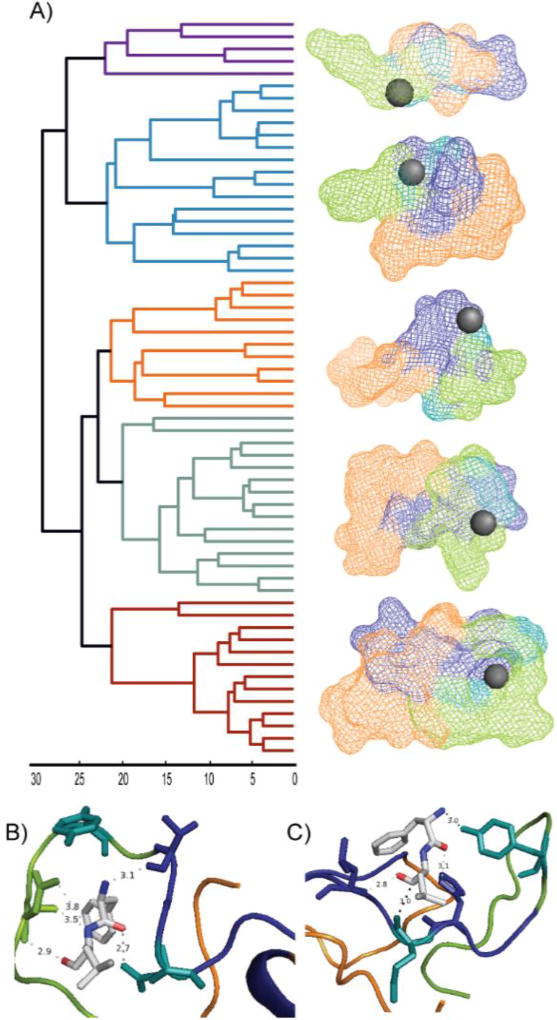
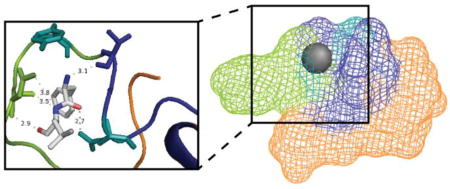
Hierarchical clustering of this filtered model pool yields five structural families. Figure 4A shows the output of our clustering algorithm, along with low-resolution averaged peptide structures that depict an averaged location for FL binding on the Aβ surface. In most cases, FL is found between the N-terminus and the hydrophobic core while the c-terminus is further away. A detailed, manual analysis of these five structural families reveals common themes in the molecular contacts created between FL and the Q15 residue within the Aβ sequence. Two examples of these details are shown in Figure 4B and 4C, where we highlight bonding of FL and Aβ residue Q15.
Figure 4.
Hierarchical clustering analysis and model visualization of Aβ1–40:FL complexes. A) Clustering hierarchy, and the averaged family structure for each cluster. Each leaf of the dendrogram represents an individual Aβ1–40:FL complex and the height of the branches represents the average linkage distance as measured by pairwise RMSD (Å). B and C) High resolution representative models of Aβ:FL complex. Q15 and Y10 (teal) primarily interact with the leucine of FL. N-terminus (green), hydrophobic core (blue), C-terminus (orange), and FL (grey, oxygen in red, nitrogen in blue).
An interaction distance of 2.8 Å between the carbonyl oxygen of L within FL, and an ε nitrogen (atom name NE2 in pdb format) of Q15 is observed in the lowest energy structure, matching our experimental restraints (data not shown). The ligand configuration is stabilized by R5, H6, and D7 with hydrogen bonding distances of 3.1 Å and 3.2 Å between R5 and D7 with the carbonyl oxygen of F, and a 3.8 Å interaction distance between the F nitrogen and H6. In this particular model, Y10 is too far away from the carbonyl oxygen on L to hydrogen bond. In another low energy structure, FL is wrapped tightly within the Aβ structure (Figure 4B). Here, hydrogen bonding occurs between the NE2 position of Q15 and the carbonyl oxygen on F (2.7 Å). While there are no contacts less than 4 Å between FL and Y10 in this model, the ring structure surrounds the FL pocket. The interaction is stabilized through an interaction having a 3.1 Å distance between carboxylic acid oxygen on V12 and the backbone nitrogen on F, in addition to 3 apparent hydrogen bonds between D7 and L.
Despite the Y10 not being close enough for hydrogen bonding in the two models discussed thus far, another low energy structure from a separate cluster does demonstrate such interactions (Figure 4C). Here, Y10 displays evidence of a hydrogen bond between the backbone nitrogen of F (distance of 3.0 Å). In addition, Q15 is within 3.0 Å of the oxygen on Q15 and the alpha carbon of L. The interaction is further stabilized through hydrogen bonding between an oxygen of L and N of L17 (2.8 Å), as well as a δ nitrogen (atom name ND1 in pdb format) of H14 and the oxygen of F (3.1 Å). We note that all FL hydrogen bonding captured within our models occur along the dipeptide backbone. As such hydrogen bonds are relatively weak interactions, we believe our MD data only captures a subset of all potential interactions. Side-chain specific interactions are thus also possible, and perhaps likely, in these complexes, but remain undocumented in the three lowest energy models detailed here.
Discussed thus far, all KD and computational analysis has focused on the analysis of FL binding to monomeric Aβ. IM-MS data circled in Figure 2A however, clearly shows the interaction of FL with small soluble oligomers of Aβ1–40. We calculated the percentage of oligomers found in both unbound and bound states. Of the total monomeric population detected, 34 ± 12 % is found bound to at least 1 FL. In contrast, 69 ± 4.5% of the dimer population is bound to FL and 50 ± 20% of the trimers are bound. This suggests preference for binding to the dimer and trimer populations over the monomer in WT Aβ1–40, despite the monomer being the dominant species in our samples at time of measurement. Binding with the tetramer was not detected in enough replicates to calculate a precise value for the average bound fraction.
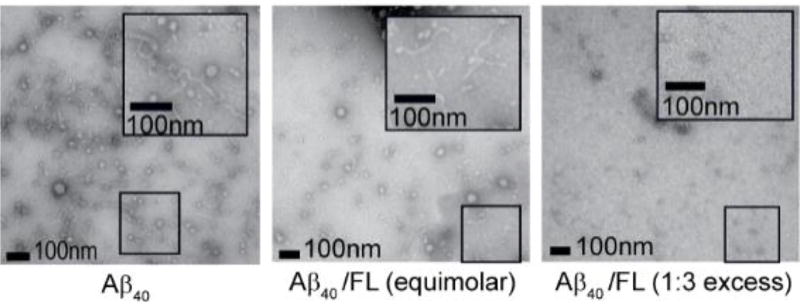
In order to evaluate the ability of FL to alter Aβ fibril formation, TEM images were acquired for Aβ1–40 samples dosed with varying amounts of FL. The features observed in the TEM data shown in Figure 5 have a marked dependence upon FL concentration, with long mature fibril structures observed for Aβ samples where no FL is added, transitioning to truncated fibril structures in samples where equimolar FL is added, and finally a complete abrogation of Aβ fibril formation when excess FL is present. Truncated fibrils observed by TEM upon equimolar FL addition to Aβ samples are less dense when compared to control samples, further indicating that mature fibril formation is inhibited under these conditions. The weakly-stained forms observed under conditions of FL excess appear universally amorphous, indicating that while Aβ aggregation still occurs, it likely does so through an altered route that does not share a morphology connected with the production of toxic species, which have been observed in previous studies and similarly interpreted.22
Figure 5.
TEM images of Aβ1–40 incubated with FL (as labeled), displaying reduced fibril formation upon FL addition, in a dose-dependent fashion.
Conclusions
Through a comprehensive set of IM-MS experiments and MD simulations for an exhaustive series of amino acid variants, we have identified the two C-terminal residues of leucine enkephalin, FL, as critical for its previously-reported interaction with Aβ. A subsequent IM-MS analysis of Aβ 1:1 complex formed with the isolated FL dipeptide from leucine enkephalin revealed both equivalent binding strengths to the full length construct. Through detailed IM-MS and CD spectroscopy covering 14 alanine variants of Aβ1–40, carefully selected through an analysis of MD simulation output, we have narrowed the potential sites of FL contact on the Aβ1–40 surface to the Y10 and Q15 residues. Models extensively filtered through a range of structural biology data indicate that FL primarily interacts with Q15 through hydrogen bonding to the FL backbone, and that the dipeptide is positioned between the N-terminus and the hydro-phobic core in a manner similar to previous Aβ binders of therapeutic note.5, 11, 20 With regard to small soluble oligomers we note improved attachment over the monomers. Critically, TEM data shows that FL inhibits Aβ1–40 fibril formation, which further points to its potential as a foundation for future AD therapeutics. Additional computational modeling and further experiments are required to give details concerning the binding of FL to Aβ at the dimer level and above, although we speculate it to access a similar mode as found for the monomer. To move forward, we look towards the chemical modification FL in the creation of a therapeutic library for IM-MS screening. Modifications should reduce potential cellular toxicity and increase binding affinity. Additionally, amidated and acetylated FL variants are of interest, as such modifications in the Aβ1–42 C-terminal fragment VVIA have been shown to decrease the toxicity of resultant Aβ oligomers.23 It is worth noting that the binding location of FL is somewhat rare among previously described peptidic Aβ binders, with most accessing the C-terminus and hydrophobic core rather than the N-terminal region of the peptide23 It is likely that further IM-MS screens, in combination with FL optimization, will lead to a more detailed mechanism of Aβ fibril formation inhibition in general.
Methods
Reagents were purchased from commercial sources and used without further purification unless otherwise specified. Aβ1–40 was purchased from Anaspec. Alanine mutations were custom synthesized and received at a purity of at least 95% (GenScript). Aβ variants (1 mg) were dissolved in 200 µL 1% (v/v) ammonium hydroxide and diluted with pH 6.9 100 mM ammonium acetate to a total volume of 1500 µL. Peptides were then injected into a Slide-A-Lyzer Dialysis Cassette (2K MWCO, 3 mL volume). 100mM ammonium acetate was added to fill volume of cassette capacity, and dialysis was performed over a 24 h period. Samples were then removed from the cassette, flash frozen with liquid nitrogen and lyophilized overnight. Peptides were reconstituted using 200 µL 1% (v/v) ammonium hydroxide and diluted with pH 6.9 100 mM ammonium acetate to a total volume of 1500 µL. Peptide concentration for the stock solution was calculated from absorbance at 280nm (ε = 1490 M−1 cm-1). Peptides were aliquoted and stored at −80 °C. Leucine enkephalin acetate hydrate (YGGFL) and prepared in pH 6.9 100mM ammonium acetate. FL was synthesized by AnaSpec, and prepared in dimethylsulfoxide.
IM-MS
Mass spectra were collected on a quadrupole-ion mobility-time-of-flight (TOF) mass spectrometer (Synapt G1 HDMS, Waters) with a nano-electrospray ionization (nESI) source. Protein ions were generated using a nESI source and optimized to allow transmission of noncovalent protein complexes using electrospray capillaries prepared as described previously.23 Protein complex ions were generated using an aliquot of the sample (ca. 7 µL) sprayed from the nESI emitter at a capillary voltage of 1.2kV. The source was operated in positive ion mode with the sample cone at 35 V and extraction cone at 1.0V. The bias voltage was 21 V, with backing pressure at 2.32 mbar and TOF pressure at 1.81 × 10−6 mbar. The travelling-wave IM separator was operated at a pressure of approximately 3.0 mbar of nitrogen. Mass spectra were calibrated externally using a solution of cesium iodide (100 mg mL−1) and analyzed using MassLynx 4.1 and Driftscope 2.0 software. CCS (Ω) measurements were externally calibrated using a database of known values in helium, using values for peptides and proteins that bracket the likely CCS and ion mobility values of the unknown ions.24, 25 Samples were prepared by mixing stock solutions (as prepared above) of neuropeptide and Aβ1–40 variants at pH 6.9, to generate a final Aβ1–40 concentration of 20 µM, with 2.88% v/v DMSO present in all samples. Samples were incubated on ice for 1 hour prior to IM-MS analysis.
Docking Studies
Flexible ligand docking studies were preformed using AutoDock Vina26. FL was created using PyMOL and studies were conducted against the Aβ1–40 monomer (aqueous solution NMR structure PDB 2LFM)27 and Aβ1–42 (aqueous solution NMR structure PDB 1Z0Q).28 Twenty docking simulations with Aβ1–40 and thirty docking simulations with Aβ1–42, one with each conformation within the ensemble uploaded to the PDB, were performed. The structures for FL and Aβ were prepared for use with Auto Dock Vina using AutoDock Tools.26 All hydrogens were added to Aβ and torsions were kept as the default selected in AutoDock Tools. The exhaustiveness for the docking studies was set at 8, resulting in 9 output models for each Aβ conformation. Docked models of FL were visualized with Aβ using PyMOL.
MD Simulations
Simulations were started using the minimized Aβ solution NMR structures (PDB 2LFM or PDB 1Z0Q).27, 28 Charges were applied to R5, K16, and K28 to mimic the 3+ charge state observed in the gas phase. The simulations were performed using periodic boundary conditions in a dodecahedron with the minimum distance between the simulated molecules and the box wall being 1.0 nm. GROMACS ligand topology was prepared using the GlycoBioChem PRODRG2 server.29 The MD simulations were carried out using the GROMACS software package30 and GROMOS96 force field.31 To constrain the bond length in the Aβ and FL, the LINCS algorithm was used, allowing an integration time step of 2 fs. Long-range electrostatic interactions were treated with the particle mesh Ewald method. Temperature was maintained using the method of Berendsen et al.32 The FL and the Aβ were separately coupled to external temperature bath with a temperature-coupling constant of 0.1 ps.
The system was energy-minimized by steepest decent for 500 steps. After equilibration, simulated annealing was performed for the Aβ: FL complex in the gas-phase. The three most basic Aβ side chains (R5, K16, and K28) were charged. The system was heated from 300 K to 500 K over 100 ps, then cooled down to 300 K over the next 100 ps. The cycle was repeated over 20 ns in order to allow for escape from local minima and enhance equilibration. For the Aβ1–40:FL complex, 20 independent simulated annealing runs, each running for a total of 20 ns, were performed from the lowest energy complexes generated by AutoDock Vina. For the Aβ1–42:FL complex, 30 independent simulated annealing runs, each running for a total of 20 ns, were performed from the lowest energy complexes generated by AutoDock Vina. From the MD trajectory generated, 100 structures were sampled at 300 K and the CCS was calculated with Mobcal using the trajectory method algorithm.33, 34 Models of the Aβ:FL complexes were visualized in PyMOL. In total 2000 structures were generated for Aβ1–40 and 3000 structures for Aβ1–42. Of the structures which were within ± 3% of the experimentally determined CCS, the 200 structures with lowest energy were analyzed to determine the Aβ residues within 4 Å of FL. Standardized values (Z-scores) were calculated for each residue of Aβ1–40 and Aβ1–42 and plotted in standard deviation (σ) space in order to determine the relative likelihood of FL binding within a given region of Aβ.
Hierarchical Clustering Analysis
The CCS-filtered structures of Aβ1–40:FL, generated from MD simulations in the previous step were subjected to another filter for agreement with site-directed amino acid substation experiments (FL was within 4 Å of Aβ residue Y10 and Q15). This reduced the set of possible structures from 963 to 60. We then characterized the structural families present in the dataset using hierarchical clustering. Aβ candidate structures were temporarily stripped of their ligands while being aligned using the Kabsch algorithm,35 and pairwise RMSD values were calculated for each structure in the dataset. These pairwise RMSD values were then used as a distance matrix for hierarchical clustering using the average linkage clustering module from Scipy.36 Intra and inter-cluster RMSD values were analyzed and it was found that the dataset could reasonably be broken into 5 clusters, allowing for facile interpretation of the dataset. The results of the clustering are represented as a dendrogram where each leaf represents an individual structure, and the height of the branches represents the distance between structures or clusters in RMSD space. For each structural cluster, an average structure was calculated to represent the structural family, and all ligands were then superimposed on the representative structure, revealing strong structure-specific ligand clustering, in addition to an averaged ligand location. Structures were visualized in PyMOL.
KD Measurements by MS
KD values for peptide:Aβ complexes were calculated using the relative intensity of each species from the mass spectra, as described previously.37 In our previous work,20 we modified this method to accommodate multiple ligand binding events with the following assumptions: (1) the spray and detection efficiency of all species are similar, (2) the ligand concentration is sufficiently high so that [L]eq remains constant and (3) the ligand binds to the complex one at a time in a stepwise fashion. Here we present further modification to better represent multiple ligand binding stoichiometry.
For the equilibrium binding of A (unbound protein) and B (a ligand/binding partner):
| (1) |
| (2) |
Where Rx is an equilibrium quotient between the bound form of the protein (Cx, having × ligands attached) when interacting with ligand B, and is unbound for (A).
| (3) |
Eqn (3) above defines Rx for all bound forms of the protein (Ci), where y is the number of species A in the complex and:
| (4) |
Allows for the determination of KD for any given step in the sequential equilibrium described in eqn (1), where C0 = A. Standard deviation values (σ) for the KD measurements reported here are shown from at least three replicate measurements.
TEM
Negatively stained specimens for transmission electron microscopy were prepared by applying 5 µL of peptide solution to hydrophilic 400 mesh carbon-coated formvar support films mounted on copper grids (Ted Pella, Inc.). The samples were allowed to adhere for 3 min, rinsed twice with distilled water, and stained for 60–90 s with 1% uranyl acetate (Ted Pella, Inc.). All samples were imaged at an accelerating voltage of 60 kV in a CM-100 electron microscope (Philips, Inc.). Samples were prepared in 100 mM ammonium acetate, with 2.88% DMSO at a concentration of 25 µM Aβ and allowed to incubate at room temperature for 24 h.
Supplementary Material
Acknowledgments
Funding Sources
This research was supported jointly by the UM protein folding disease (PFD) initiative, part of the University of Michigan Health System (UMHS) Strategic Research Initiative and Fast Forward programs, and National Institutes of Health (National Institute of General Medical Sciences, GM095832).
The authors wish to thank M. Ivanova (UM) for the collection of TEM images. Additionally J. Clark (UM) for his assistance and advice in determining modes of binding. R. Kerr (UM) for his discussion of Aβ variants and preparation ideas. K. Korshaven and M. Wolf (UM) for assistance in troubleshooting CD.
ABBREVIATIONS
- IM-MS
Ion mobility-mass spectrometry
- AD
Alzheimer’s Disease
- TEM
Transmission Electron Microscopy
- CD
Circular Dichroism
- KD
Dissociation Constant
- LE
Leucine Enkephalin
- Aβ
Amyloid β peptide
Footnotes
ASSOCIATED CONTENT
Supporting Information. Detailed annotations of 3D IM-MS data, 2D MS data, expected and M/Z for each complex, CD data, as well as details of leucine enkephalin interactions with Aβ variants can be found in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
Experiments were conceived of by MTS-H and BTR. IM-MS Aβ experiments, docking studies, MD simulations, detailed modeling analysis, KD measurements, and TEM analysis were performed by MTS-H. JDE was responsible for development of the hierarchical clustering method. The manuscript was written by MTS-H and BTR. All authors have given approval to the final version of the manuscript.
References
- 1.Young LM, Saunders JC, Mahood RA, Revill CH, Foster RJ, Ashcroft A, Radford S. ESI-IMS–MS: A method for rapid analysis of protein aggregation and its inhibition by small molecules. Methods. 2015 doi: 10.1016/j.ymeth.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young LM, Saunders JC, Mahood RA, Revill CH, Foster RJ, Tu L-H, Raleigh DP, Radford SE, Ashcroft AE. Screening and classifying small-molecule inhibitors of amyloid formation using ion mobility spectrometry–mass spectrometry. Nature chemistry. 2015;7:73–81. doi: 10.1038/nchem.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woods LA, Platt GW, Hellewell AL, Hewitt EW, Homans SW, Ashcroft AE, Radford SE. Ligand binding to distinct states diverts aggregation of an amyloid-forming protein. Nature chemical biology. 2011;7:730–739. doi: 10.1038/nchembio.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young LM, Cao P, Raleigh DP, Ashcroft AE, Radford SE. Ion mobility spectrometry–mass spectrometry defines the oligomeric intermediates in amylin amyloid formation and the mode of action of inhibitors. Journal of the American Chemical Society. 2013;136:660–670. doi: 10.1021/ja406831n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyung S-J, DeToma AS, Brender JR, Lee S, Vivekanandan S, Kochi A, Choi J-S, Ramamoorthy A, Ruotolo BT, Lim MH. Insights into antiamyloidogenic properties of the green tea extract (−)-epigallocatechin-3-gallate toward metal-associated amyloid-β species. Proceedings of the National Academy of Sciences. 2013;110:3743–3748. doi: 10.1073/pnas.1220326110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck MW, Oh SB, Kerr RA, Lee HJ, Kim SH, Kim S, Jang M, Ruotolo BT, Lee J-Y, Lim MH. A rationally designed small molecule for identifying an in vivo link between metal–amyloid-β complexes and the pathogenesis of Alzheimer's disease. Chemical Science. 2015;6:1879–1886. doi: 10.1039/c4sc03239j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pithadia AS, Kochi A, Soper MT, Beck MW, Liu Y, Lee S, DeToma AS, Ruotolo BT, Lim MH. Reactivity of diphenylpropynone derivatives toward metal-associated amyloid-β species. Inorganic chemistry. 2012;51:12959–12967. doi: 10.1021/ic302084g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleiholder C, Do TD, Wu C, Economou NJ, Bernstein SS, Buratto SK, Shea J-E, Bowers MT. Ion Mobility Spectrometry Reveals the Mechanism of Amyloid Formation of Aβ (25–35) and Its Modulation by Inhibitors at the Molecular Level: Epigallocatechin Gallate and Scyllo-inositol. Journal of the American Chemical Society. 2013;135:16926–16937. doi: 10.1021/ja406197f. [DOI] [PubMed] [Google Scholar]
- 9.Gessel MM, Wu C, Li H, Bitan G, Shea J-E, Bowers MT. Aβ (39–42) modulates Aβ oligomerization but not fibril formation. Biochemistry. 2011;51:108–117. doi: 10.1021/bi201520b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Gessel MM, Wisniewski ML, Viswanathan K, Wright DL, Bahr BA, Bowers MT. Z-Phe-Ala-diazomethylketone (PADK) disrupts and remodels early oligomer states of the Alzheimer disease Aβ42 protein. Journal of Biological Chemistry. 2012;287:6084–6088. doi: 10.1074/jbc.C111.328575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkitadze MD, Bitan G, Teplow DB. Paradigm shifts in Alzheimer's disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. Journal of neuroscience research. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 12.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 13.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. Journal of Biological Chemistry. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 14.Ono K, Condron MM, Teplow DB. Structure–neurotoxicity relationships of amyloid β-protein oligomers. Proceedings of the National Academy of Sciences. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nature chemistry. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitan G, Fradinger EA, Spring SM, Teplow DB. Neurotoxic protein oligomers—what you see is not always what you get. Amyloid. 2005;12:88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- 17.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to clusters and biomolecules. Journal of Mass Spectrometry. 1997;32:577–592. [Google Scholar]
- 18.Bohrer BC, Merenbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Biomolecule analysis by ion mobility spectrometry. Annual review of analytical chemistry (Palo Alto, Calif.) 2008;1:293. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benesch JL, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chemical reviews. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- 20.Soper MT, DeToma AS, Hyung S-J, Lim MH, Ruotolo BT. Amyloid-β–neuropeptide interactions assessed by ion mobility-mass spectrometry. Physical Chemistry Chemical Physics. 2013;15:8952–8961. doi: 10.1039/c3cp50721a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paivio A, Jarvet J, Graslund A, Lannfelt L, Westlind-Danielsson A. Unique physicochemical profile of β-amyloid peptide variant Aβ1–40E22G protofibrils: Conceivable neuropathogen in arctic mutant carriers. Journal of molecular biology. 2004;1:145–159. doi: 10.1016/j.jmb.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 22.Walsh D, Klyubin I, Fadeeva J, Rowan M, Selkoe D. Amyloid-beta oligomers: their production, toxicity and therapeutic inhibition. Biochemical Society Transactions. 2002;30:552–557. doi: 10.1042/bst0300552. [DOI] [PubMed] [Google Scholar]
- 23.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature protocols. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 24.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Ion Mobility–Mass Spectrometry Reveals Long-Lived, Unfolded Intermediates in the Dissociation of Protein Complexes. Angewandte Chemie International Edition. 2007;46:8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 25.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Analytical chemistry. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 26.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. A partially folded structure of amyloid-beta (1–40) in an aqueous environment. Biochemical and biophysical research communications. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomaselli S, Esposito V, Vangone P, van Nuland NA, Bonvin AM, Guerrini R, Tancredi T, Temussi PA, Picone D. The α-to-β Conformational Transition of Alzheimer's Aβ-(1–42) Peptide in Aqueous Media is Reversible: A Step by Step Conformational Analysis Suggests the Location of β Conformation Seeding. ChemBioChem. 2006;7:257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 29.SchuÈttelkopf AW, Van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallographica Section D: Biological Crystallography. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 30.Lindahl E, Hess B, Van Der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. Journal of Molecular Modeling. 2001;7:306–317. [Google Scholar]
- 31.van Gunsteren WF, Billeter S, Eising A, Hünenberger PH, Krüger P, Mark AE, Scott W, Tironi IG. Biomolecular simulation: The {GROMOS96} manual and user guide 1996 [Google Scholar]
- 32.Berendsen HJ, Postma JPM, van Gunsteren WF, DiNola A, Haak J. Molecular dynamics with coupling to an external bath. The Journal of chemical physics. 1984;81:3684–3690. [Google Scholar]
- 33.Mesleh M, Hunter J, Shvartsburg A, Schatz G, Jarrold M. Structural information from ion mobility measurements: effects of the long-range potential. The Journal of Physical Chemistry. 1996;100:16082–16086. [Google Scholar]
- 34.Shvartsburg AA, Jarrold MF. An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chemical Physics Letters. 1996;261:86–91. [Google Scholar]
- 35.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallographica Section A: Crystal Physics, Diffraction, Theoretical and General Crystallography. 1976;32:922–923. [Google Scholar]
- 36.Jones E, Oliphant T, Peterson P. {SciPy}: Open source scientific tools for {Python} 2014 [Google Scholar]
- 37.Wang W, Kitova EN, Klassen JS. Influence of solution and gas phase processes on protein-carbohydrate binding affinities determined by nanoelectrospray Fourier transform ion cyclotron resonance mass spectrometry. Analytical chemistry. 2003;75:4945–4955. doi: 10.1021/ac034300l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.