SUMMARY
Immune response (Ir) genes, originally proposed by Baruj Benacerraf to explain differential antigen-specific responses in animal models, have become synonymous with the major histocompatibility complex (MHC). We discovered a non-MHC-linked Ir gene in a T cell receptor (TCR) locus that was required for CD8+ T cell responses to the Plasmodium berghei GAP5040–48 epitope in mice expressing the MHC class I allele H-2Db. GAP5040–48-specific CD8+ T cell responses emerged from a very large pool of naive Vβ8.1+ precursors, which dictated susceptibility to cerebral malaria and conferred protection against recombinant Listeria monocytogenes infection. Structural analysis of a prototypical Vβ8.1+ TCR-H-2Db-GAP5040–48 ternary complex revealed that germline-encoded complementarity-determining region 1β residues present exclusively in the Vβ8.1 segment mediated essential interactions with the GAP5040–48 peptide. Collectively, these findings demonstrated that Vβ8.1 functioned as an Ir gene that was indispensable for immune reactivity against the malaria GAP5040–48 epitope.
Keywords: Immune response genes, TCR bias, naive CD8+ T cells, malaria, experimental cerebral malaria
INTRODUCTION
Genetic regulation of the immune system is a central determinant of health and disease. Based on the observation that random-bred guinea pigs challenged with simple antigens segregate into responder and non-responder groups, Benacerraf and colleagues proposed the notion of autosomal dominant immune response (Ir) genes (Benacerraf and Germain, 1978). Ir genes were first identified as major histocompatibility complex (MHC)-linked (McDevitt and Chinitz, 1969), and subsequently found to encode specific allotypes of antigen-presenting molecules, such as MHC class II (MHC II) (Benacerraf, 1974; Benacerraf and McDevitt, 1972). Although unidentified at the time, a determinative role was also suggested for the putative T cell receptor (TCR) (Benacerraf and Germain, 1978). MHC II-mediated antigen presentation is required to activate T helper cells and generate antibody responses to T-dependent antigens (Owens and Zeine, 1989). In addition, the host must assemble TCRs capable of engaging specific MHC-peptide (MHCp) complexes with sufficient avidity to trigger immune reactivity (Davis et al., 2003; van der Merwe and Dushek, 2011). Numerous studies of inbred animals have linked the absence of specific immune responses with a lack of appropriate MHC alleles (Marshak et al., 1977; Zinkernagel, 1978). In contrast, despite major advances in our understanding of antigen recognition over the last four decades, it remains unclear if germline-encoded segments of the TCR can function as Ir genes.
CD8+ T cells recognize MHC I-restricted peptides via heterodimeric TCRs (Davis and Bjorkman, 1988; Townsend et al., 1985). A vast number of different TCRs can be generated from a limited number of germline-encoded segments through the process of V(D)J gene recombination with junctional diversification and subsequent random pairing of the somatically rearranged TCRα and TCRβ chains (Cabaniols et al., 2001; Chothia et al., 1988; Davis and Bjorkman, 1988; Rossjohn et al., 2015; Turner et al., 2006). As a consequence, each individual harbors an extensive repertoire of naive CD8+ T cells, which ensures broad recognition of a large number of foreign antigens presented by MHC I (Goldrath and Bevan, 1999). To meet the diversity criterion within space limitations, however, only a few naive precursors are specific for any given epitope (Blattman et al., 2002; Obar et al., 2008), and robust antigen-driven proliferation is required to establish effector and memory CD8+ T cell populations (Busch et al., 1998; Goldrath and Bevan, 1999). It is estimated that most naive antigen-specific repertoires in mice do not contain more than 10–300 CD8+ T cells (Obar et al., 2008). Larger pre-immune repertoires comprising 1,000–1,500 naive CD8+ T cells have been reported for the murine cytomegalovirus (MCMV) M45985–993 and vaccinia virus (VacV) B8R20–27 epitopes (Jenkins and Moon, 2012), but it remains unclear if these specific precursor pools define the upper limits of antigen reactivity in the post-thymic landscape of clonotypically distributed TCRs.
Clonal selection ensures the recruitment of biologically and structurally optimal immune receptors from the naive repertoire (Malherbe et al., 2004; Price et al., 2005), frequently leading to biased TCR usage among memory CD8+ T cell populations (Miles et al., 2011; Turner et al., 2006). In extreme cases, non-peptidic antigens restricted by non-classical MHC molecules elicit innate-like responses dominated by semi-invariant TCRs (Bendelac et al., 1997; Godfrey et al., 2015; Van Rhijn et al., 2015). Here, we found that a similar phenomenon can regulate conventional CD8+ T cell immunity. We demonstrated that an epitope from the Plasmodium berghei (P. berghei) ANKA glideosome-associated protein (GAP5040–48), a key pathogenic target in experimental cerebral malaria (ECM), was associated with the largest naive CD8+ T cell repertoire yet described in laboratory mice (Jenkins and Moon, 2012). We also revealed the molecular properties of these GAP5040–48-specific TCRs, demonstrating exclusive use of the Vβ8.1 segment underpinned by direct interactions with the malarial peptide. Mice lacking the Vβ8.1 gene did not respond to the GAP5040–48 epitope after P. berghei infection and did not develop ECM. Moreover, the very large pool of naive precursors conferred enhanced control of primary infection with recombinant Listeria monocytogenes (L. monocytogenes) expressing the GAP5040–48 epitope. Collectively, these findings extend the notion of Ir genes to incorporate germline-encoded components of antigen-specific TCRs.
RESULTS
GAP5040–48-specific CD8+ T cells exhibit an extreme TCR bias
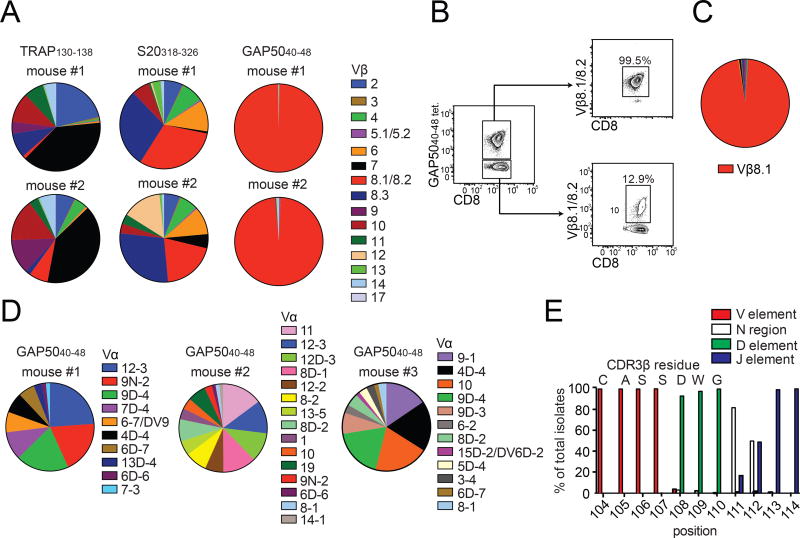
ECM in susceptible C57Bl/6 (B6) mice infected with P. berghei ANKA (Engwerda et al., 2005) is a valuable model of severe malarial disease (Brewster et al., 1990). It is established that the development of ECM is critically dependent on pathogenic CD8+ T cells (Amani et al., 2000; Haque et al., 2011; Yanez et al., 1996) expressing Vβ8+ TCRs (Boubou et al., 1999; Mariotti-Ferrandiz et al., 2016), especially those specific for the H-2Db-restricted GAP5040–48 epitope (Howland et al., 2013). However, it is not known why GAP5040–48-specific CD8+ T cells are pathogenic in ECM. To address this issue, we set out to generate TCR retrogenic mice harboring monoclonal or oligoclonal CD8+ T cell populations specific for individual epitopes derived from P. berghei, namely thrombospondin-related adhesion protein (TRAP)130–138, sporozoite-specific protein 20 (S20)318–326 and GAP5040–48 (Holst et al., 2006; Hafalla et al., 2013; Howland et al., 2013). As a first step, we immunized three separate groups of B6 mice with peptide-pulsed dendritic cells (DCs) followed 7 days later by recombinant L. monocytogenes expressing the same epitope (LM-GAP5040–48). This accelerated prime-boost approach (Badovinac et al., 2005) elicited large CD8+ T cell responses specific for TRAP130–138, S20318–326 and GAP5040–48 (Fig. S1A). We then used the corresponding MHC I tetramers and a panel of anti-mouse Vβ antibodies to profile the constituent malaria-specific TCRs.
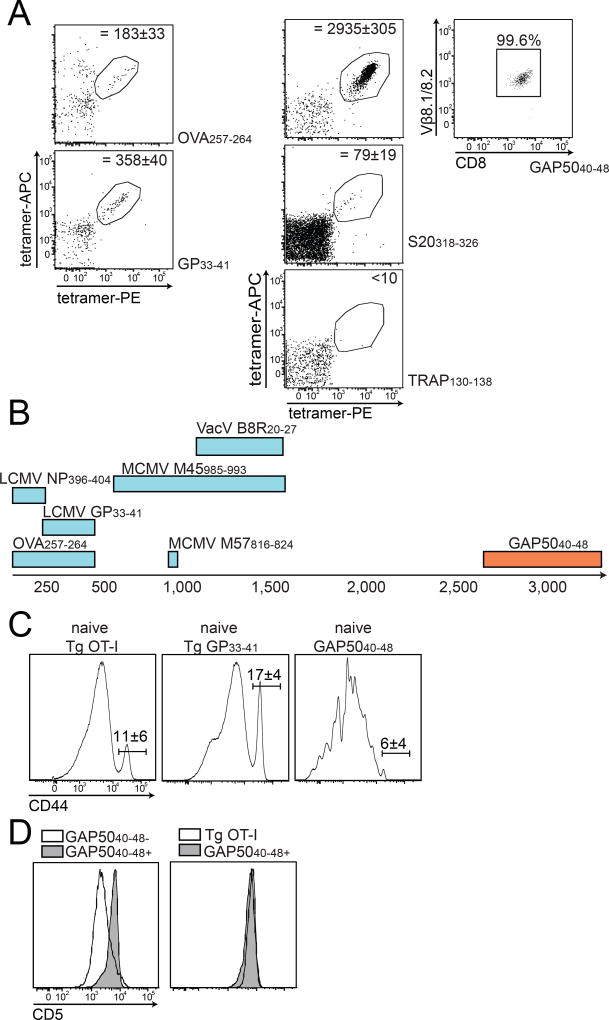
CD8+ T cells specific for TRAP130–138 and S20318–326 expressed several different TCR Vβ segments with distinct preferences (Fig. 1A and S1B). For example, almost 75% of the S20318–326-specific repertoire was focused on Vβ8.1/8.2 and Vβ8.3, while the TRAP130–138-specific repertoire was dominated by Vβ2 and Vβ7. In contrast, >99% of GAP5040–48-specific CD8+ T cells expressed Vβ8.1/8.2, which cannot be discriminated by antibody staining (Fig. 1A and S1B). These segments comprised <15% of the corresponding GAP5040–48 tetramer− repertoires in the same mice (Fig. 1B). Moreover, an identical TCR bias was observed after inoculation of sporozoites by mosquito bite or immunization with radiation-attenuated sporozoites or DC-GAP5040–48 alone (data not shown), and similar results were obtained with other strains of mice expressing the MHC I allele H-2Db, including CB6F1 and BALB.b (Fig. S1C). The GAP5040–48 epitope therefore mobilized an almost exclusive repertoire of TCR Vβ-defined memory CD8+ T cells.
Figure 1. GAP5040–48-specific CD8+ T cells express biased TCRs.
(A, B) Mice were primed with peptide-coated DCs and boosted on day 7 with recombinant L. monocytogenes expressing the same epitope. (A) The TCR Vβ repertoire was assessed using splenocytes isolated from immunized mice by staining with the indicated tetramers and Vβ-specific antibodies. Results are shown for two mice for each antigen specificity. Data represent four independent experiments (n = 5 mice/group). Cumulative data are shown in Supplementary Figure 1B. (B) Extreme focusing of the GAP5040–48-specific repertoire does not reflect the overall frequency of Vβ8.1/8.2+ clonotypes in the CD8+ T cell compartment. Data represent three independent experiments (n = 4–5 mice/group). (C, D) Confirmation of TCR bias at the transcriptional level. Sequences were derived from GAP5040–48-specific CD8+ T cells isolated from the spleens of mice previously immunized with DC-LM-GAP5040–48 (total = 316 molecular clones). (C) Population-level analysis revealed an extreme bias towards Vβ8.1. (D) TCR Vα sequence distributions from three different mice are depicted. No obvious bias was apparent in the Vα repertoire. (E) Origin of CDR3β amino acids expressed relative to the total number of molecular Vβ8.1+ clones (n = 5 mice). Vβ: red; Dβ: green; Jβ: purple; N segments: white. (See also Figures S1–S3 and Table S1).
To gain a deeper understanding of this extreme bias, we sorted GAP5040–48-specific CD8+ T cells directly ex vivo from DC-LM-GAP5040–48-immunized B6 mice and performed an unbiased molecular analysis of all expressed TRA and TRB gene rearrangements (Quigley et al., 2011). Sequence analysis showed that >98% of transcripts encoded Vβ8.1 (Fig. 1C and Table S1). In contrast, a variety of TRAV gene segments were detected, indicating promiscuous pairing of diverse TCRα chains with a constrained repertoire of Vβ8.1+ TCRβ chains (Fig. 1D). Almost 40% of retrogenic T cells generated with a GAP5040–48-specific TCRβ chain bound the GAP5040–48 tetramer in naive mice, while only ~1% of retrogenic T cells generated with a TRAP130–138-specific TCRβ chain bound the TRAP130–138 tetramer (Fig. S2). These data suggested that the Vβ8.1 segment was a major recognition element for GAP5040–48-specific CD8+ T cells.
Analysis of the third TCRβ chain complementarity-determining region (CDR3β) revealed additional features of the GAP5040–48-specific repertoire (Fig. S3A–C, Table S1). In particular, we identified a common CDR3β length (~60% of translated sequences incorporated 14 amino acids) (Fig. S3A), an almost uniform bias towards TRBD2 gene usage within a single reading frame (Fig. S3B) and a strong preference for TRBJ gene segments from the Jβ2 cluster (Fig. S3C). Moreover, the Vβ-Dβ junction lacked N additions, and a germline-encoded CDR3β motif (104CASSDWG110) was present in >95% of sequences (Table S1 and Fig. 1E). These data demonstrated that a highly convergent pattern of gene rearrangements “licensed” the Vβ8.1-driven immune response to the GAP5040–48 epitope (Quigley et al., 2010).
The C-terminal region of the GAP5040–48 peptide is a “hotspot” for TCR recognition
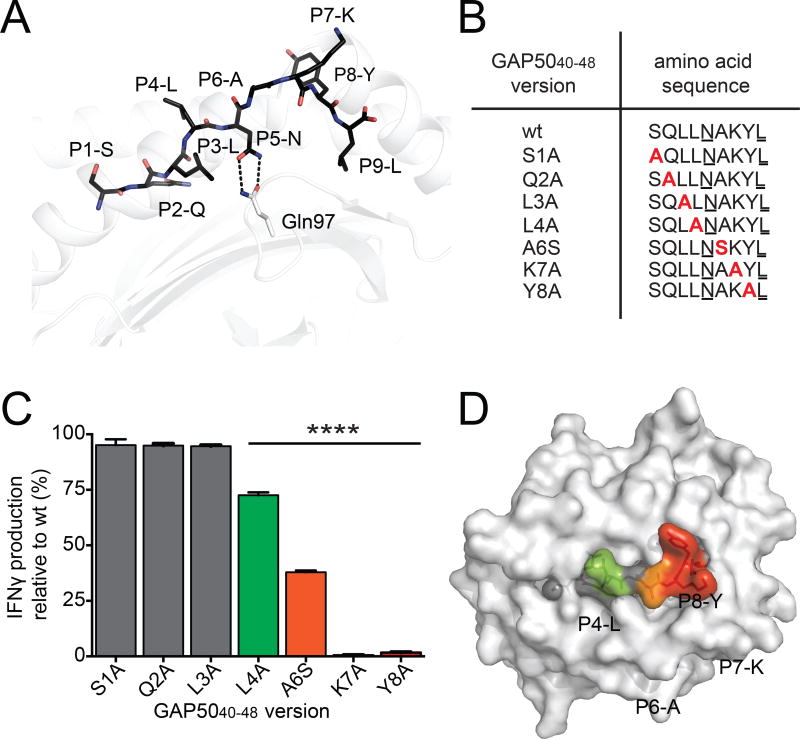
To understand the molecular basis of this extreme TCR bias, we first determined the structure of the H-2Db-GAP5040–48 complex at a resolution of 2.2 Å (Table S2). The bound peptide adopted a canonical extended conformation (Young et al., 1994), with four solvent-exposed amino acid residues (P4-L, where L is leucine; P6-A, where A is alanine; P7-K, where K is lysine; and P8-Y, where Y is tyrosine) representing potential contacts for the TCR (Fig. 2A).
Figure 2. TCR interactions focus on the C-terminal region of the GAP5040–48 peptide.
(A) The structure of the GAP5040–48 peptide in complex with H-2Db. The GAP5040–48 peptide is represented as black sticks in the antigen-binding cleft of H-2Db (white cartoon). The P2-Q and P9-L anchor residues are buried within the antigen-binding cleft, and P5-N acts as a secondary anchor residue forming hydrogen bonds (black dashes) with Gln97 (white stick) at the base of the antigen-binding cleft of H-2Db (white cartoon). (B) A panel of peptide mutants was generated by replacing amino acids with Ala or Ser at selected positions (red). Anchor residues are underlined. (C) Splenocytes isolated from GAP5040–48-immunized mice were restimulated for 5 h in vitro using wt or mutant peptides. The production of IFNγ induced by stimulation with the different mutant peptides is expressed relative to the production of IFNγ induced by stimulation with the wt peptide. Data represent two independent experiments performed in triplicate. Bars represent mean ± SD. Significance was assessed using a one-way ANOVA (****p < 0.0001). (D) The effect of peptide substitutions on recognition of the H-2Db-GAP5040–48 complex by specific CD8+ T cell clones. The surface of H-2Db is shown in white. Peptide residues that were not mutated or did not affect IFNγ production are shown in gray. Green represents up to a 25% decrease in T cell activation; orange represents up to a 70% decrease in T cell activation; red represents a total abrogation of T cell activation. (See also Figure S4 and Table S2).
To determine which GAP5040–48 residues were important for TCR recognition, we replaced individual amino acids with alanine (Ala), except for the wildtype (wt) Ala residue at position 6 (P6), which was replaced with serine (Ser) (Fig. 2B). We did not mutate the critical H-2Db anchor residues at P5 or P9 (Valkenburg et al., 2013). The wt and mutant peptides were then used at saturating concentrations in intracellular cytokine staining assays to determine the impact of each residue on the functional reactivity of GAP5040–48-specific CD8+ T cells (Fig. 2C and S4). Interferon-γ (IFNγ) production relative to the wt peptide was unaffected by the substitutions at P1, P2 and P3 (Fig. 2C). In contrast, the mutations at P4-L and P6-A diminished IFNγ production, while the mutations at P7-K and P8-Y abolished IFNγ production (Fig. 2C). The C-terminal P7-K and P8-Y residues were therefore crucial for GAP5040–48-specific CD8+ T cell activation and represented “hotspots” for TCR engagement (Fig. 2D).
GAP5040–48 peptide “hotspots” interact exclusively with the TCRβ chain
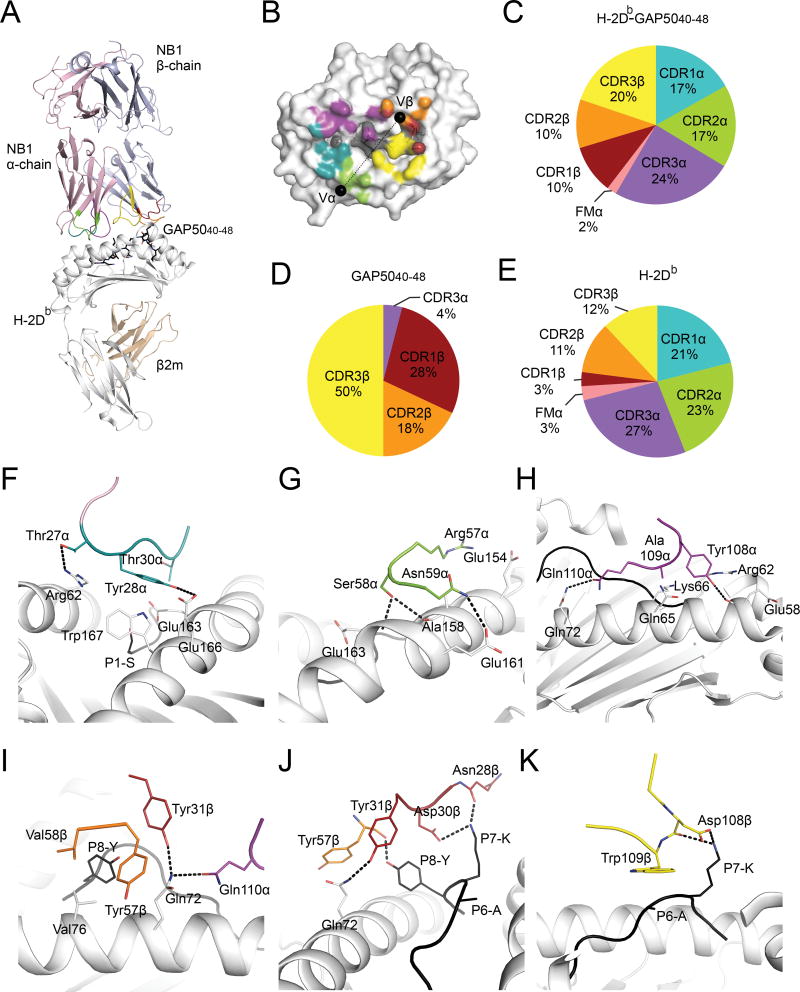
Next, we determined the structure of a prototypical Vβ8.1+ TCR, derived from the GAP5040–48-specific CD8+ T cell clone NB1 (Table S3), in complex with H-2Db-GAP5040–48 (Fig. 3A, Table S2). The NB1 TCR docked centrally atop the H-2Db cleft, with an angle of 43° and a buried surface area (BSA) of app roximately 2,200 Å2, values that fall within the range previously determined for TCR-MHCp complexes (Rossjohn et al., 2015) (Fig. 3B). All six CDR loops were involved to a varying extent in the interaction with H-2Db-GAP5040–48 (Fig. 3C). Namely, the CDR3α and CDR3β loops contributed 24% and 20% of the BSA, respectively, while the CDR1α and CDR2α loops each contributed ~17% of the BSA (Fig. 3C). Somewhat unexpectedly given the extreme TCR Vβ bias, the germline-encoded CDR1β and CDR2β loops each contributed only ~10% of the BSA. Thus, the cumulative β chain contribution to the overall BSA was substantially smaller (39%) relative to the cumulative α chain contribution (61%) (Fig. 3C). However, >95% of TCR-GAP5040–48 peptide interactions were mediated by the TCRβ chain (Fig. 3D, Table S4), while the TCRα chain primarily contacted the MHC molecule (74% of the BSA) (Fig. 3E, Table S4).
Figure 3. GAP5040–48-specific TCR bias is peptide-driven.
(A) The crystal structure of the NB1 TCR (α chain: pink cartoon; β chain: purple cartoon) bound to the GAP5040–48 peptide (black sticks) presented by H-2Db (heavy chain: white cartoon; β2m, wheat cartoon). (B) The CDR loop contribution to the BSA is represented as teal (CDR1α), green (CDR2α), purple (CDR3α), red (CDR1β), orange (CDR2β) or yellow (CDR3β). Black spheres represent the center of mass for the Vα and Vβ. (C–E) The contribution (%) of the CDR loops to the interaction with H-2Db-GAP5040–48 (C), GAP5040–48 (D) and H-2Db (E). (F–K) Contact residues between H-2Db-GAP5040–48 and CDR1α (F), CDR2α (G), CDR3α (H), CDR1β (I, J), CDR2β (J) and CDR3β (K). The CDR color scheme is maintained though panels F–K. Residues that make contacts with the GAP5040–48 peptide or H-2Db (white) are depicted as black and white sticks, respectively. The H-2Db molecule is represented as white cartoon, and hydrogen bonds are shown as black dashes. (See also Figure S5 and Tables S2–S5).
The CDR1α loop stretched between the peptide-binding α-helices of H-2Db, with tyrosine (Tyr) 28α playing a principal role by contacting glutamic acid (Glu)163, Glu166 and tryptophan (Trp)167 at the N-terminal end of the cleft (Fig. 3F, Table S4). The CDR2α loop sat above the α2-helix, with arginine (Arg)57α lying flat above residues Glu154 and Ala158 (Fig. 3G). The CDR3α loop contacted a large stretch of the α1-helix (spanning residues 58–72), with hydrophobic and hydrogen bond contacts exclusively mediated by germline-encoded residues from the Jα segment (108YAQ110, where Q is glutamine) (Fig. 3H). The NB1 TCRα chain engaged the H-2Db molecule with a large footprint over the N-terminal region of the antigen-binding cleft (Fig. 3B). Mutational analyses of H-2Db binding performed using three distinct GAP5040–48-specific CD8+ T cell clones further indicated that different Vβ8.1+ TCRs used an identical MHC I docking strategy (Fig. S5, Table S5).
Interactions between the NB1 TCRα chain and the GAP5040–48 peptide were limited to two van der Waals interactions between the CDR3α loop and P4-L (Table S4). In contrast, the NB1 TCR Vβ8.1 region dominated contacts with the bound epitope (44 of 46 contacts) (Table S4). Moreover, all three CDRβ loops contacted the C-terminus of the peptide (P6–8) (Fig. 3I–K), encompassing the previously identified “hotspots” for CD8+ T cell recognition (Fig. 2D). The CDR1β loop interacted with both P7-K and P8-Y via its 28N-DY31 motif (where N is asparagine and D is aspartic acid). More specifically, P7-K and P8-Y formed a notch in which aspartic acid (Asp)30β was inserted as a peg, forming a salt bridge with P7-K (Fig. 3J). In addition, asparagine (Asn)28β hydrogen bonded via its main chain with P7-K, while the aromatic group of Tyr31β sat above P8-Y (Fig. 3J). Contacts were also established between P8-Y and the CDR2β loop via Tyr57. Accordingly, P8-Y was closely sequestered upon TCR binding (Fig. 3J). The conserved CDR3β motif 108DW109 (where W is tryptophan), which was most frequently germline-encoded within the D gene (Fig. 1E), interacted with P6-A and P7-K (Fig. 3K). The large Trp109β side chain lay flat on the top of P6-A, acting as a lid covering the central part of the epitope, while Asp108β and P7-K formed a salt bridge (Fig. 3K).
TCRβ chain interactions with the H-2Db molecule were modest in comparison to the TCRα chain (Table S4). The CDR1β loop contacted only glutamine (Gln)72 via Tyr31β (Fig. 3I, J). The side chains of valine (Val)58β from the CDR2β loop abutted Val76 from H-2Db, establishing an interaction network extended by Tyr57β-mediated contacts with Gln72, Arg75 and Val76. The conserved CDR3β 108DW109 motif from the TRBD2 gene segment formed interactions with the hinge region of the α2-helix, whereby Trp109β nestled between histidine (His)155 and Ser150. Thus, germline-encoded TCRβ chain residues played a minor role in contacting the H-2Db molecule.
The observation that the prototypical NB1 TCR interacted with GAP5040–48 extensively via its β chain suggested that the peptide itself drove the extreme bias towards Vβ8.1. This notion contrasts with prevailing dogma, which asserts that germline bias arises as a consequence of allotype-specific MHC reactivity (Garcia et al., 2009).
CDR1 residues unique to Vβ8.1 are critical for GAP5040–48 peptide recognition
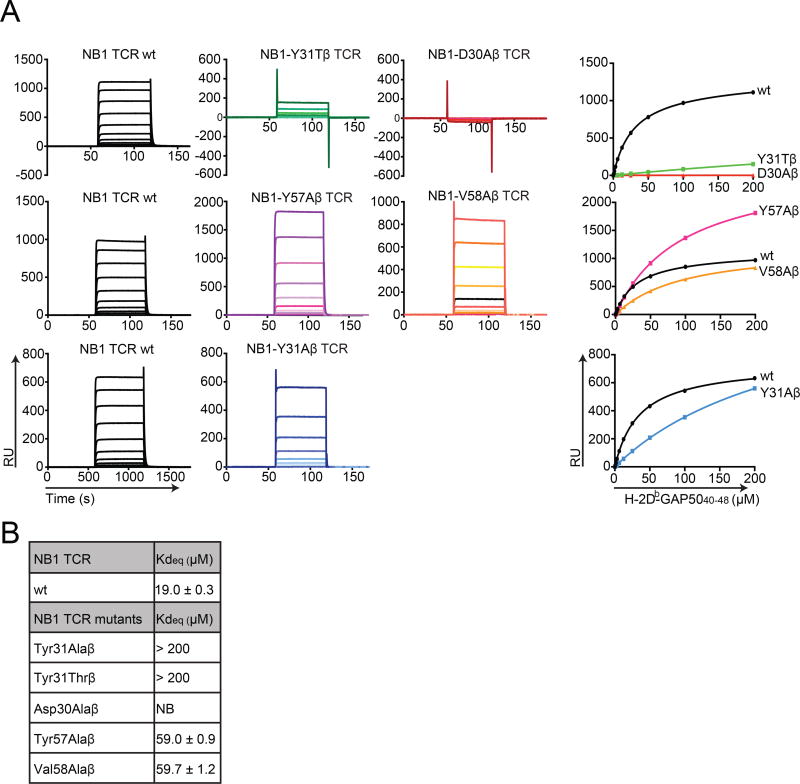
These structural insights allowed us to identify candidate factors underlying the exclusive recruitment of Vβ8.1+ TCR clonotypes into the immune repertoire. To determine the precise amino acid sequences involved in this process, we initially focused on the germline-encoded CDR1β and CDR2β loops. Sequence alignments revealed that the CDR1β loops encoded by six TRBV genes shared a common Asp30β, while the Asp30β-Tyr31β motif was unique to Vβ8.1. In the other five TRBV genes, Tyr31β was replaced by a smaller threonine (Thr)/Ser residue (Table S6A). Three TRBV genes shared a common Val58β residue in the encoded CDR2β loop, but only Vβ8.1 incorporated the additional Tyr57β required to sequester P8-Y (Table S6B).
Although the conserved CDR3β region interacted with both the peptide and H-2Db, we wanted to determine if the germline-encoded Vβ8.1 residues were essential for TCR-mediated recognition of H-2Db-GAP5040–48. We therefore conducted affinity measurement studies using targeted mutants of the NB1 TCR. Ala mutagenesis was performed on residues Asp30β and Tyr31β in the CDR1β loop and residues Tyr57β and Val58β in the CDR2β loop (Fig. 4A and B). In addition, Tyr31β was mutated to Thr to mimic the CDR1β loop encoded by the five other TRBV genes that share Asp30β (Fig. 4A and B). Affinity values for each TCR mutant, measured by surface plasmon resonance (SPR), were compared with the affinity of the wt TCR. The results showed that mutations in the CDR2β loop exerted a moderate effect, decreasing binding affinity by 3-fold compared with the NB1 TCR (Fig. 4A and B). In contrast, mutation of Tyr31β to either Ala or Thr in the CDR1β loop substantially reduced binding to H-2Db-GAP5040–48, resulting in Kdeq values >200 µM (Fig. 4A and B). As shown in Fig. 3I and Table S4, Tyr31β contacted both H-2Db and P8-Y. Additionally, mutation of Asp30β to Ala abolished binding to H-2Db-GAP5040–48 (Fig. 4A and B). The Asp30β residue contacted the peptide alone via a salt bridge with P7-K, one of the two defined “hotspot” residues for TCR recognition (Fig. 3J and Table S4).
Figure 4. Binding affinities of NB1 TCR mutants for H-2Db-GAP5040–48.
(A) Surface plasmon resonance (SPR) analysis of NB1 wt (black) and the indicated mutant (colored) TCRs across a range of H-2Db-GAP5040–48 concentrations up to a maximum of 200 µM. Representative SPR binding curves are shown for the NB1 wt TCR (black) and NB1 mutant TCRs with the following β chain substitutions: Y31A (green), D30A (red), Y57A (pink), V58A (orange) and Y31A (blue). Each NB1 mutant TCR was tested in parallel with the NB1 wt TCR. (B) Summary table representing equilibrium binding affinities of NB1 TCR mutants for H-2Db-GAP5040–48. NB: no binding; RU: response units. Data represent two independent experiments performed in duplicate (mean ± SEM). (See also Tables S6A and S6B).
The unique germline-encoded CDR1β 30DY31 motif within the Vβ8.1 segment therefore underpinned both the extreme TCR bias and the residue-specific patterns of epitope recognition that characterized the CD8+ T cell response to H-2Db-GAP5040–48.
The naive GAP5040–48-specific CD8+ T cell repertoire is extraordinarily large
Extreme TCR focusing described in the literature appears to result primarily from the selection of high-affinity clonotypes during repeated or persistent infection (Busch and Pamer, 1999; Malherbe et al., 2004; Price et al., 2005; Savage et al., 1999), but recombinatorial bias and structural constraints dictate the available pre-immune repertoire (Miles et al., 2011; Neller et al., 2015; Turner et al., 2006). We therefore adapted a tetramer-based enrichment protocol (Moon et al., 2007) to enumerate the naive repertoires specific for the P. berghei epitopes TRAP130–138, S20318–326 and GAP5040–48. Parallel experiments were performed with tetramers representing epitopes derived from ovalbumin (H-2Kb-OVA257–264) and lymphocytic choriomeningitis virus glycoprotein (H-2Db-LCMV GP33–41) to allow comparison with previously reported evaluations of the naive antigen-specific T cell repertoire (Jenkins and Moon, 2012).
Precursor numbers specific for OVA257–264 and GP33–41 were consistent with prior reports (183 ± 33 and 358 ± 40 cells, respectively, Fig. 5A) (Jenkins and Moon, 2012). The naive S20318–326-specific repertoire was relatively small (~79 ± 19 cells), while fewer than 10 cells were counted in the TRAP130–138-specific repertoire (Fig. 5A). In contrast, as described recently (Gordon et al., 2015), the GAP5040–48-specific naive repertoire was extraordinarily large (2,935 ± 305 cells, Fig. 5A). The upper limit for a naive antigen-specific CD8+ T cell repertoire in mice was previously established at 1,200–1,500 cells for the MCMV M45985–993 and VacV B8R20–27 epitopes (Jenkins and Moon, 2012). The GAP5040–48-specific repertoire therefore constituted the largest naive antigen-specific CD8+ T cell pool yet described (Fig. 5B). Moreover, >99% of naive GAP5040–48-specific CD8+ T cells expressed Vβ8.1 (Fig. 5A). These findings indicated that the extreme TCR Vβ bias observed in the memory CD8+ T cell pool did not evolve as a consequence of repertoire focusing in response to antigenic challenge, but instead reflected intrinsic recognition of the GAP5040–48 epitope characterized by absolute dependence on the interaction with Vβ8.1.
Figure 5. The naive GAP5040–48-specific CD8+ T cell repertoire is extremely large.
(A) CD8+ T cells specific for OVA257–264, GP33–41, GAP5040–48, S20318–326 and TRAP130–138 were enriched from the spleens and macroscopic lymph nodes of naive C57Bl/6 mice and used to calculate the final numbers of tetramer+ cells indicated in each plot. Target cells were identified as CD8+ CD90.2+ CD11b− CD11c− B220− tetramer(APC)+ tetramer(PE)+. Depicted numbers of naive precursors were calculated as mean ± SD from 11 mice for the GAP5040–48-specific repertoire. Right panel: Vβ8.1/8.2 expression on naive GAP5040–48-specific CD8+ T cells. (B) Sizes of the naive repertoires specific for various CD8+ T cell epitopes. The number of naive GAP5040–48-specific precursors is represented as a range from 2,700–3,250 cells (total n = 11 mice). (C, D) Phenotypic characterization of naive GAP5040–48-specific CD8+ T cells. (C) Expression of CD44 on naive GAP5040–48-specific CD8+ T cells was compared with CD8+ T cells obtained from naive Tg OT-I and P14 mice. (D) Expression of CD5 on naive GAP5040–48-specific CD8+ T cells was compared with total naive, non-GAP5040–48-specific CD8+ T cells (left panel) and with naive Tg OT-I cells (right panel). Data represent two independent experiments (n = 4 mice/group). Bars depict mean ± SD.
Phenotypically, the GAP5040–48-specific precursors resembled classical naive CD8+ T cells, comprised of a large CD44lo population (>90%) and a small CD44hi “virtual memory” population (<10%) akin to OT-I and P14 TCR-Tg cells from naive mice (Fig. 5C) (Akue et al., 2012). Expression of the CD5 surface protein, which acts as a surrogate marker for the strength of TCR activation induced by self-derived MHCp complexes during thymic selection and frequently correlates with T cell avidity for antigen (Fulton et al., 2015; Mandl et al., 2013), was uniformly higher on naive GAP5040–48-specific CD8+ T cells compared with the majority of non-GAP5040–48-specific naive CD8+ T cells in the same host and mirrored levels expressed by OT-I cells, which express high-affinity TCRs (Fig. 5D) (Kedl et al., 2000). These data suggested that the extremely large pool of Vβ8.1+ precursors specific for the GAP5040–48 epitope arose as a consequence of strong thymic selection.
Absence of Vβ8.1 compromises the GAP5040–48-specific CD8+ T cell response
The results of our structural and mutagenesis studies indicated that immune responses to GAP5040–48 required the presence of Vβ8.1. To test this hypothesis, we studied the generation of GAP5040–48-specific CD8+ T cell responses in C57/L (H-2b) mice, which lack Vβ8 (Behlke et al., 1986). For this purpose, C57/L mice and control B6 mice were exposed to infection with 103 P. berghei ANKA sporozoites, a challenge that induces ECM in susceptible mouse strains.
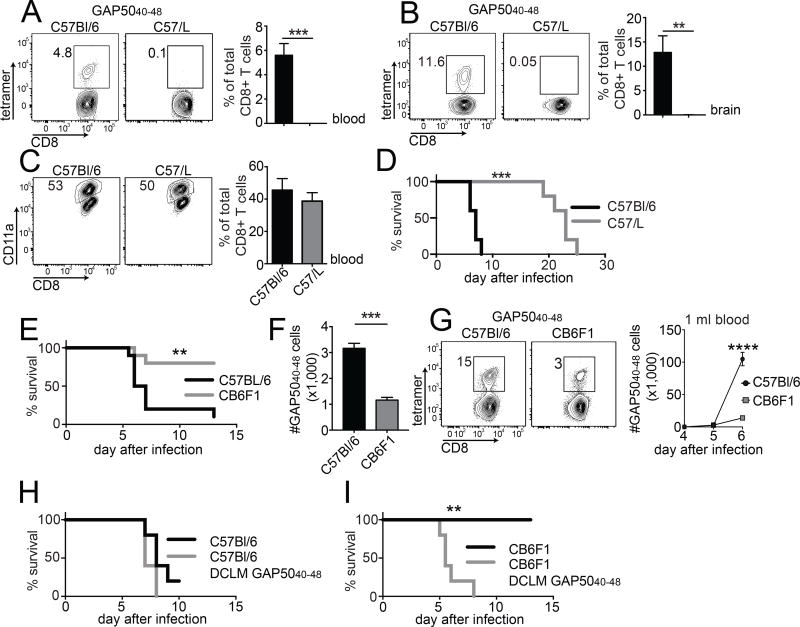
In contrast to B6 mice, which generated GAP5040–48-specific responses comprising 4–5% of the total CD8+ T cell pool in peripheral blood and >10% of the total CD8+ T cell pool in the brain by day 7 post-infection, the GAP5040–48-specific response in C57/L mice was undetectable in both compartments (Fig. 6A and B). This finding strongly suggested that the GAP5040–48-specific CD8+ T cell response was critically dependent on Vβ8.1. In addition, B6 × C57/L F1 mice mounted substantial Vβ8.1+ GAP5040–48-specific CD8+ T cell responses after infection with LM-GAP5040–48 or P. berghei ANKA (data not shown), excluding negative selection of GAP5040–48-specific naive precursors in C57/L mice. However, C57/L mice lack other Vβ genes (Vβ5, Vβ8, Vβ9, Vβ11 and Vβ12) (Behlke et al., 1986), which could potentially compromise the ability to mount alternative responses to GAP5040–48. Countering this argument, the magnitude of the total P. berghei-specific CD8+ T cell response, as detected by the frequency of CD8+ T cells expressing surrogate activation markers (CD11ahi CD8lo) (Rai et al., 2009) at day 7 post-infection, was essentially identical in B6 and C57/L mice (Fig. 6C). Moreover, C57/L mice did not develop ECM, instead they survived beyond day 9 post-infection and succumbed to high-level parasitemia after ~3 weeks (Fig. 6D). Thus, the Vβ8.1 gene was necessary for the generation of detectable GAP5040–48-specific CD8+ T cell responses and the development of ECM.
Figure 6. Vβ8.1 is required for the generation of GAP5040–48-specific CD8+ T cells and the development of ECM.
(A, B) C57Bl/6 and C57/L mice were infected with 1,000 P. berghei ANKA sporozoites. H-2Db-GAP5040–48 tetramer+ cells were quantified in the blood (A) and brains (B) at day 7 post-infection (representative plots: left; summary graph: right). (C) Magnitude of the activated CD8+ T cell response at day 7 post-infection, expressed as % CD11ahi CD8lo of total CD8+ T cells (representative plots: left; summary graph: right). Naive control values were subtracted from individual values. Data represent two independent experiments (n = 3 mice/group). Bars depict mean ± SD. Significance was assessed using an unpaired, two-tailed t test (**p < 0.01, ***p < 0.001). (D) Survival curves for C57Bl/6 mice and C57/L mice after infection with P. berghei ANKA. Data represent three independent experiments (n = 5 mice/group). Significance was assessed using the Mantel-Cox log rank test (***p = 0.001). (E) C57Bl/6 and CB6F1 mice were infected with 106 P. berghei-parasitized red blood cells (pRBCs). Mice were monitored for the development of ECM symptoms and scored for survival over a period of two weeks. Data represent three independent experiments (n = 5 mice/group). Significance was assessed using the Mantel-Cox log rank test (**p = 0.0025). (F) GAP5040–48-specific CD8+ T cells were tetramer-enriched from naive C57Bl/6 and CB6F1 mice and quantified to estimate repertoire size. Cumulative results are shown from two independent experiments (total n = 4 mice/group). Bars depict mean ± SEM. Significance was assessed using an unpaired, two-tailed t test (****p <0.0001). (G) C57Bl/6 and CB6F1 mice were infected with 106 pRBCs. The GAP5040–48-specific CD8+ T cell response was followed in the blood of infected mice using H-2Db-GAP5040–48 tetramers. All C57Bl/6 mice succumbed to ECM by days 6 and 7, while all CB6F1 mice survived. Data represent two independent experiments (n = 5 mice/group). Bars depict mean ± SD. Significance was assessed using an unpaired, two-tailed t test (****p < 0.0001). (H, I) C57Bl/6 and CB6F1 mice were injected IV with 5 × 105 GAP5040–48 peptide-coated DCs and then infected 7 days later with recombinant attenuated LM-GAP5040–48. At a memory time point after LM infection, DC-LM-GAP5040–48-immunized or non-immunized mice were infected with 106 pRBCs. Survival curves are shown for C57Bl/6 mice (H) and CB6F1 mice (I). Data represent two independent experiments (n = 5 mice/group). Significance was assessed using the Mantel-Cox log rank test (p = 0.0993 in panel D; **p = 0.0018 in panel E).
GAP5040–48-specific CD8+ T cells are pathogenic in ECM
A recent publication suggested that GAP5040–48-specific CD8+ T cells play a key role in the pathogenesis of ECM in susceptible B6 mice (Howland et al., 2013). Mechanistically, accumulation of GAP5040–48-specific CD8+ T cells in the brain could lead to eventual rupture of the blood-brain-barrier, which triggers well recognized neurological symptoms leading to rapid death (7–9 days post-infection) (Howland et al., 2013). In contrast to susceptible B6 mice, which all succumbed to ECM by day 7 post-infection with P. berghei ANKA-parasitized red blood cells (pRBCs), only 20% of CB6F1 mice succumbed to ECM (Fig. 6E). We therefore considered the possibility that a strain-specific difference in the size of the naive GAP5040–48-specific CD8+ T cell repertoire might underlie these differential outcomes. Tetramer pull-down of naive GAP5040–48-specific CD8+ T cells revealed that CB6F1 mice harbored ~3-fold fewer precursors than B6 mice (Fig. 6F). Moreover, this smaller naive pool resulted in a smaller GAP5040–48-specific CD8+ T cell response in CB6F1 mice compared with B6 mice at day 6 post-infection with P. berghei ANKA pRBCs (Fig. 6G). These results suggested that a GAP5040–48-specific CD8+ T cell response threshold might be required for the induction of ECM. To confirm a threshold-dependent pathogenic role for GAP5040–48-specific CD8+ T cells in ECM, we generated a large GAP5040–48-specific CD8+ T cell response in CB6F1 or B6 mice prior to infection with P. berghei ANKA. We observed no meaningful difference in the onset of ECM symptoms or the mortality rate between previously immunized and non-immunized B6 mice, demonstrating that sufficient numbers of GAP5040–48-specific effector CD8+ T cells can be generated from the large pool of naive precursors in this susceptible strain (Fig. 6H). In contrast, prior immunization against GAP5040–48 was required to render all CB6F1 mice susceptible to ECM after P. berghei ANKA infection (Fig. 6I). Collectively, these findings demonstrated that a numerical threshold delimited the pathogenicity of GAP5040–48-specific CD8+ T cells in the etiology of ECM.
GAP5040–48-specific CD8+ T cells protect against LM-GAP5040–48
Although pathogenic in the context of ECM, we hypothesized that the rapid generation of a substantial GAP5040–48-specific response from the large number of naive precursors may protect against infection controlled primarily by CD8+ T cells. To address this possibility, we determined if B6 mice could better control primary infection with virulent LM-GAP5040–48 compared with virulent wt LM.
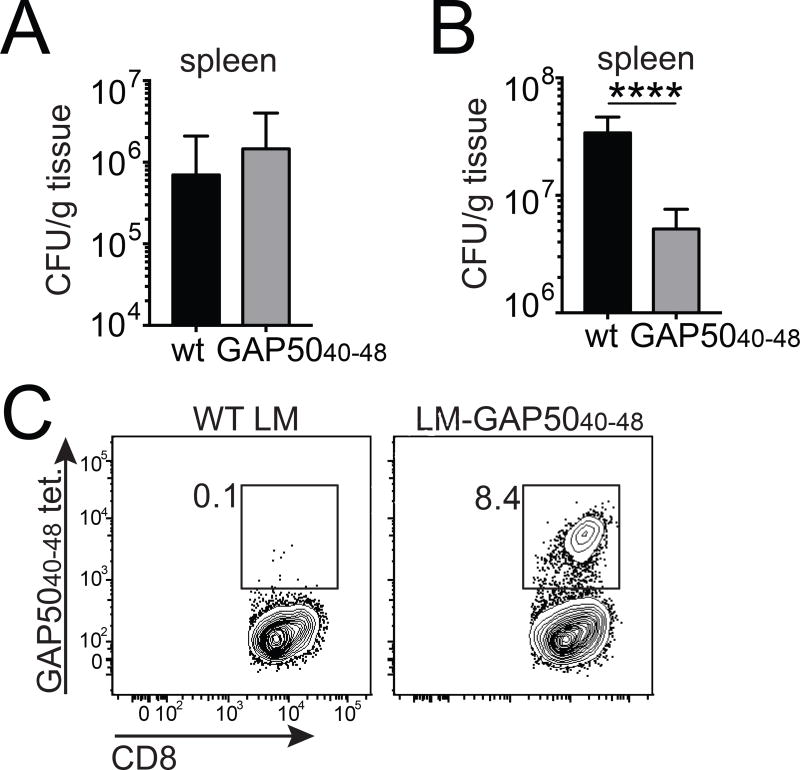
To ensure comparability, we first established that the two different strains of LM exhibited similar virulence in the absence of a GAP5040–48-specific CD8+ T cell response. For this purpose, we infected BALB/c mice (H-2d), which were non-responsive to the H-2Db-restricted GAP5040–48 epitope (Fig. 7A). Bacterial burden assessed in the spleens of infected mice 5 days after infection with 5 × 103 colony-forming units (CFU) of each strain was similar between the two groups, suggesting equivalent virulence in vivo (Fig. 7B). In contrast, we measured ~10-fold fewer bacteria in the spleens of LM-GAP5040–48-infected B6 mice compared with wt LM-infected B6 mice (Fig. 7C–D). This superior control of bacterial infection in LM-GAP5040–48-infected B6 mice was accompanied by a large GAP5040–48 specific response, comprising >8% of the total CD8+ T cell pool in the spleen (Fig. 7E). The expansion of GAP5040–48-specific effector cells from a large pool of naive precursors therefore conferred protection in the context of an infection controlled by CD8+ T cells.
Figure 7. Large numbers of GAP5040–48-specific CD8+ T cells control primary L. monocytogenes infection.
(A) BALB/c mice were infected IV with 5×103 CFU of wt LM or recombinant LM-GAP5040–48. Bacterial burden was measured in the spleen on day 5. Data represent two independent experiments (n = 4 mice/group). Bars depict mean ± SD. (B) C57Bl/6 mice were infected IV with 104 CFU of wt LM or recombinant LM-GAP5040–48. Bacterial burden was measured in the spleen on day 5. Cumulative results are shown from three independent experiments (total n = 14–15 mice/group). Bars depict mean ± SEM. Significance was assessed using an unpaired, two-tailed t test (****p<0.0001). (C) Representative flow cytometry plots showing H-2Db-GAP5040–48 tetramer staining of splenocytes isolated from infected C57Bl/6 mice at day 5 post-infection.
DISCUSSION
Conceptual frameworks developed in the 1970s, prior to a detailed understanding of the molecular interactions that govern T cell antigen recognition, established the notion of genetically controlled immune responsiveness. Overwhelming experimental data have since accumulated to validate one original proposition that MHC-linked Ir genes dictate the presentation of specific antigens in immunogenic form (Benacerraf and Germain, 1978). There is also some evidence to support the idea that germline-encoded components of the TCR interact preferentially with defined MHC allotypes (Garcia et al., 2009; Scott-Browne et al., 2009), building on the earlier theoretical work of Niels Jerne (Jerne, 1971). However, it has not been shown previously that heritable elements of an antigen receptor can license immune reactivity, a scenario postulated almost four decades ago as an alternative model to explain the Ir gene phenomenon (Benacerraf and Germain, 1978). In this study, we demonstrated that CD8+ T cell responses specific for the murine malaria epitope GAP5040–48 were entirely dependent on amino acid residues unique to the TCR Vβ8.1 segment, thereby providing insights into the genetic mechanisms that control adaptive immunity.
As a consequence of structural constraints focused primarily on the CDR1β loop, both naive and antigen-expanded H-2Db-restricted GAP5040–48-specific CD8+ T cells almost invariably express Vβ8.1+ TCRs. Similarly extreme biases are typically associated with innate-like responses to non-peptidic antigens presented by non-classical MHC molecules, although immune reactivity among these unconventional T cell subsets is generally endowed by a conserved TCRα chain (Van Rhijn et al., 2015). Narrow TCR repertoires specific for peptide epitopes have previously been linked with low numbers of naive precursors (Moon et al., 2007) and clonal selection during the genesis of memory populations, especially in the presence of constant or repetitive antigen stimulation (Busch and Pamer, 1999; Price et al., 2005; Savage et al., 1999). Paradoxically, the pre-immune GAP5040–48-specific repertoire was the largest yet described in mice, numbering ~3,000 cells in a naive B6 host. Moreover, this substantial precursor pool did not seem to arise as a consequence of expansion into the virtual memory subset (Haluszczak et al., 2009), as >90% of naive GAP5040–48-specific CD8+ T cells expressed low levels of CD44 (Akue et al., 2012). Although the self-derived peptides associated with positive selection of these antigen-specific precursors remain unknown, the uniformly high expression of CD5 by naive GAP5040–48-specific CD8+ T cells suggested the existence of strong TCR-mediated interactions in the thymus (Mandl et al., 2013). It is also notable that Vβ8 gene segments are highly represented in laboratory mouse strains (Wilson et al., 2001). High numbers of naive GAP5040–48-specific CD8+ T cells therefore most likely emerged as a consequence of both permissive thymic selection and the frequent generation of Vβ8.1+ transcripts during the process of somatic recombination. In addition, the presence of a highly conserved Vβ-Dβ junction lacking N additions suggested that convergent gene rearrangements, which occur more commonly on a probabilistic basis (Quigley et al., 2010), effectively licensed the extreme penetrance of the Vβ8.1 gene-encoded phenotype.
So what are the immunological consequences of such a large pool of antigen-specific precursors in the naive CD8+ T cell compartment? Earlier work indicated that CD8+ T cells expressing Vβ8.1/Vβ8.2+ TCRs are highly prevalent during malaria infection and contribute to the pathogenesis of ECM (Mariotti-Ferrandiz et al., 2016). In addition, it had been shown previously that high-dose tolerization with the GAP5040–48 peptide abrogates disease susceptibility in B6 mice (Howland et al., 2013). These observations can be explained by our finding that an extremely large pool of Vβ8.1+ antigen-specific naive precursors (~3,000 cells) underpinned the immunodominant CD8+ T cell response to GAP5040–48 and the susceptibility of B6 mice to ECM. In contrast, CB6F1 mice harbored a diminished pool of antigen-specific naïve precursors (~1,000 cells), leading to smaller GAP5040–48-specific CD8+ T cell responses and relative resistance to ECM. This causal association was confirmed by the observation that increasing the number of GAP5040–48-specific CD8+ T cells in CB6F1 mice prior to infection rendered them susceptible to ECM. It remains unclear if similar germline biases can dictate immune responses to malarial antigens in humans, but our data nonetheless provide proof-of-concept that genetic associations with disease outcome can be extended to loci encoding components of the TCR. Moreover, the peak incidence of severe anemia during malaria infection occurs in children under 2 years of age, while cerebral malaria occurs most commonly in children aged 3–5 years who have prior exposure to infection (Struik and Riley, 2004). These epidemiological patterns are consistent with the possibility that larger pre-infection repertoires of malarial antigen-specific T cells predispose to the development of cerebral malaria.
GAP5040–48-specific memory CD8+ T cells are not protective during the liver-stage of murine malaria (Doll et al., 2016; Horne-Debets et al., 2016; van der Heyde et al., 1993; Vinetz et al., 1990), and the capacity of CD8+ T cells to protect during blood-stage malaria remains controversial (Horne-Debets et al., 2016; Imai et al., 2010; van der Heyde et al., 1993; Vinetz et al., 1990). Thus, the pathogenic effects of GAP5040–48-specific CD8+ T cells in ECM most likely resulted from a large population of precursors, which expanded rapidly after infection to generate substantial numbers of effector cells, leading to sustained immune activation without elimination of the pathogen. In contrast, rapid expansion from the same population of precursors enhanced immune control of primary infection with L. monocytogenes expressing the GAP5040–48 epitope. An unusually large naive CD8+ T cell pool can therefore be pathogenic or protective, depending on the nature of the infectious challenge.
In conclusion, our data provide direct evidence that a germline-encoded TCR segment can determine immune responsiveness to an exogenous peptide antigen, thereby extending the concept of Ir genes beyond the MHC. The unique features associated with this phenomenon may allow novel interventions to improve vaccine efficacy and limit immune pathology in humans, pending further studies to identify similar genetic associations between disease outcome and heritable components of the TCR.
METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John T. Harty (john-harty@uiowa.edu).
Experimental Model and Subject Details
C57Bl/6 (H-2b), BALB/c (H-2d) and CB6F1 (H-2b×d) mice were purchased from the National Cancer Institute (Frederick, MD). BALB.b (H-2b) and C57/L (H-2b) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All mice were housed with appropriate biosafety containment at the University of Iowa Animal Care Unit. The animals were treated and handled in accordance with guidelines established by the Institutional Animal Care and Use Committee. All experiments were performed using female mice, aged 6–8 weeks. P. berghei ANKA clone 234-parasitized Anopheles stephensi mosquitos were produced in house or obtained from New York University. For ECM infection, mice were injected intravenously (IV) with 103 viable sporozoites or intraperitoneally (IP) with 106 pRBCs. pRBCs were obtained from mice infected with a frozen stock 4–5 days previously. Numbers of transferred pRBCs were estimated by counting infected cells in Giemsa-stained blood smears. For virulent LM infection (wt or LM-GAP5040–48), BALB/c mice were infected IV with 5×103 CFU, and C57Bl/6 mice were infected IV with 104 CFU. Bacterial burden in the spleen was measured according to a previously published protocol (Badovinac and Harty, 2000).
Method Details
DC-LM immunizations
For the purpose of TCR clonotyping, mice were immunized using a DC-LM prime-boost regimen (Badovinac et al., 2005). FMS-like tyrosine kinase-3 ligand (Flt-3L)-induced splenic DCs were LPS-matured, isolated after collagenase/DNAse digestion (Pham et al., 2009), and incubated with the following peptides for 2 h at a final concentration of 2 µM: TRAP130–138 (SALLNUDNL), S20318–326 (VNYSFLYLF) or GAP5040–48 (SQLLNAKYL). CD11c+ cells were then enriched using anti-CD11c MicroBeads (Miltenyi Biotec). A total of 5×105 purified DCs in saline were used to prime each mouse IV. After 7 days, mice were boosted IV with 107 CFU of recombinant attenuated LM-TRAP130–138, LM-S20318–326 or LM-GAP5040–48. All recombinant attenuated LM strains were both actA- and inlB-deficient. The number of injected bacteria was verified by plating on tryptic soy agar supplemented with 50 µg/ml streptomycin.
TCR clonotyping
C57Bl/6 mice were immunized against GAP5040–48 using the DC-LM prime-boost regimen. After 5 weeks, splenocytes were isolated and stained with LIVE/DEAD Fixable Aqua and anti-CD16/CD32 (FCR4G8; Thermo Fisher Scientific). Antigen-specific cells were labeled using a BV421-conjugated version of the GAP5040–48 tetramer. Lineage markers were identified using anti-CD3-Cy5PE (17A2; BioLegend), anti-CD4-Alexa Fluor 700 (RM4–5; BD Biosciences) and anti-CD8-BV711 (53-6.7; BioLegend). Viable CD3+ CD4− CD8+ GAP5040–48 tetramer+ cells were then sorted at >98% purity using a custom-built 20-parameter FACSAria II (BD Biosciences). TCR clonotyping was performed from 5,000 sorted cells per mouse using a template-switch anchored RT-PCR (Quigley et al., 2011). Amplicons were subcloned, sampled, sequenced and analyzed as described previously (Price et al., 2005). In all cases, TCR nomenclature was translated from the IMGT database via web-based alignment of molecular transcripts (http://www.imgt.org).
Tetramer-based CD8+ T cell enrichment
The size of the naive antigen-specific CD8+ T cell repertoire was quantified using a modified version of a previously published protocol (Moon et al., 2007). Spleen, popliteal, inguinal, brachial, axillary and cervical lymph nodes were isolated from naive mice and processed to a single cell suspension. Prepared cells were then labeled with an equimolar mixture of PE- and APC-conjugated tetramers representing each defined epitope. After incubation for 1 h at 4°C, cells wer e surface stained for CD8, CD90.2 and exclusion markers (B220, CD11b and CD11c). Labeled cells were subsequently captured and enumerated using anti-PE and anti-APC magnetic beads with an AutoMACS separator (Miltenyi Biotec).
Peptide mutants
Mutant GAP5040–48 peptides were generated by replacing individual amino acids with Ala or Ser (for P6-Ala), barring the anchor residues at P5 and P9 (BioSynthesis Inc.). Splenocytes were isolated from DC-LM-GAP5040–48-immunized mice during the memory phase (>40 days post-immunization) and restimulated for 5 h in the presence of brefeldin A with either wt or mutant peptides at a final concentration of 500 nM. All restimulations were performed in triplicate. IFNγ-producing CD8+ T cells were identified using a standard intracellular cytokine staining protocol (Doll et al., 2016). Data are expressed relative to the percentage of CD8+ T cells producing IFNγ in response to the wt peptide.
Generation of TCRβ retrogenic mice
TCRβ retrogenic mice were generated on a C57Bl/6 background using retrovirus-mediated stem cell gene transfer (Bettini et al., 2013; Holst et al., 2006). The relevant antigen-specific TCRβ sequences were obtained from CD8+ T cell clones derived from mice immunized with GAP5040–48 (Vβ8.1+) or TRAP130–138 (Vβ9+).
Isolation of mononuclear cells from the brains of P. berghei ANKA-infected mice
Mononuclear cells were isolated from brain tissue using a previously published protocol with minor modifications (Zhao et al., 2009). Brains harvested after intravascular exclusion were processed and digested with collagenase D (1 mg/ml; Roche) and DNase (0.1 mg/ml; Sigma-Aldrich) for 30 min at 37°C. Dissociated brain tissue was then passed through a 70 µm nylon mesh cell strainer, spun down, resuspended and centrifuged for 20 min over 37% Percoll. Mononuclear cells were collected from the pellet, and RBCs were lysed using 1X VitaLyse (CMDG).
Protein expression, purification and crystallization
The α and β chains of the TCR clones NB1, KL1 and KL4 were expressed separately as inclusion bodies and refolded with engineered disulfide linkages in the constant domains as described previously (Day et al., 2011). Ala substitutions in the H-2Db and NB1 TCR sequences were introduced using site-directed mutagenesis. Soluble wt and mutant H-2Db heterodimers containing the GAP5040–48 peptide were prepared as described previously (Day et al., 2011). Crystals of H-2Db-GAP5040–48 (5 mg/ml) or the NB1 TCR in complex with H-2Db-GAP5040–48 (6 mg/ml) in 10 mM Tris-HCl (pH 8) and 150 mM NaCl were grown by the hanging-drop, vapor-diffusion method at 20°C with a protein/reservoir drop ratio of 1:1. Crystals of free H-2Db-GAP5040–48 were obtained in 0.1 M Tris-HCl (pH 8.5), 0.2 M lithium sulfate and 25–30% PEG8000. Crystals of the NB1 TCR in complex with H-2Db-GAP5040–48 were obtained in 18% PEG3350, 2% ethylene glycol, 0.2 M CaCl2 and 0.1 M HEPES (pH 7.2).
Data collection and structure determination
Crystals were soaked in a cryoprotectant solution containing mother liquor with the PEG8000 concentration increased to 30% (v/v) and then flash frozen in liquid nitrogen. Data were collected on the MX2 beamline at the Australian Synchrotron using the ADSC-Quantum 315r CCD detectors (at 100K). Data were processed with XDS software (Kabsch, 2010) and scaled using SCALA software (Evans, 2006) from the CCP4 suite (Collaborative Computational Project, 1994). The NB1 TCR structure was determined by molecular replacement using the Phaser program (Read, 2001) with the LC13 TCR as the search model (Protein Data Bank accession number, 1KGC) (Kjer-Nielsen et al., 2002), and the H-2Db structure was determined by molecular replacement using the Phaser program (Read, 2001) with H-2Db as the search model (Protein Data Bank accession number, 4L8D) (Valkenburg et al., 2013). Manual model building was conducted using Coot software (Emsley and Cowtan, 2004), followed by maximum-likelihood refinement with Buster. The NB1 TCR was numbered according to the IMGT unique numbering system (Lefranc, 2003). The final models were validated using the Protein Data Bank validation web site, and the final refinement statistics are summarized in Table S2. All molecular graphics representations were created using PyMol (DeLano, 2002).
Surface plasmon resonance
Surface plasmon resonance experiments were conducted at 25°C on a BIAcore 3000 instrument using 10 mM Tris-HCl (pH 8) and 150 mM NaCl supplemented with 1% BSA and 0.005% surfactant P20. The TCRs (wt or mutant) were immobilized on research-grade CM5 chips via standard amine coupling. All experiments were carried out at least twice in duplicate as described previously (Gras et al., 2009) across a H-2Db-GAP5040–48 (wt or mutant) concentration range of 0.78–200 µM. Data were analyzed using BIAevaluation version 3.1 with the 1:1 Langmuir binding model.
Quantification and Statistical Analysis
Statistical differences between two study groups were evaluated using an unpaired, two-tailed t test. Statistical differences between more than two study groups were evaluated using a one-way ANOVA with Tukey’s multiple comparison post-hoc test. Bar graphs display mean ± SD for representative experiments and mean ± SEM for combined experiments. Statistical significance was assigned as *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. Statistical analyses were performed using Prism 7 software (GraphPad).
Data and Software Availability
Structural coordinates were submitted to the Protein Data Bank with accession codes 5WLI for H-2Db-GAP5040–48 and 5WLG for the NB1 TCR in complex with H-2Db-GAP5040–48.
Supplementary Material
Highlights.
P. berghei GAP5040–48-specific naive and memory CD8+ T cells uniformly express Vβ8.1
The GAP5040–48-specific CD8+ T cell response is entirely dependent on Vβ8.1
Large naïve repertoire of GAP5040–48-specific CD8+ T cells is pathogenic in malaria
The Vβ8.1 gene-encoded CDR1β loop interacts directly with GAP5040–48
Acknowledgments
We thank Marc Jenkins for helpful discussions, Vladimir Badovinac and Stanley Perlman for constructive comments on the manuscript, Lisa Hancox and Steven Moieffer for excellent technical support, and staff at the New York University Insectary Core and the University of Iowa Flow Cytometry Core. This work was supported by grants from the National Institutes of Health (NIH) to JTH (AI42767, AI85515, AI95178 and AI100527), the Australian Research Council (ARC) and the Australian National Health and Medical Research Council (NHMRC) to JR, and the Wellcome Trust to DAP (100326/Z/12/Z). NVBB was supported by NIH grant T32 AI007511, and SLU was supported by NIH grant T32 AI007343. SG is a Monash Senior Research Fellow, DAP is a Wellcome Trust Senior Investigator, and JR is an ARC Australian Laureate Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
NVBB, SG, KL, TMJ, LP, SLU, CF, DAP, JR and JTH designed the study. NVBB, SG, KL, TMJ, LP, SLU, KLM and CF conducted experiments. NVBB, SG, KL, TMJ, LP, SLU, KLM, CF, DAP, JR and JTH analyzed data and interpreted results. NVBB and SG drafted the manuscript. NVBB, SG, KL, TMJ, LP, SLU, CF, DAP, JR and JTH edited the manuscript.
References
- Akue AD, Lee JY, Jameson SC. Derivation and maintenance of virtual memory CD8 T cells. J Immunol. 2012;188:2516–2523. doi: 10.4049/jimmunol.1102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amani V, Vigario AM, Belnoue E, Marussig M, Fonseca L, Mazier D, Renia L. Involvement of IFN-gamma receptor-medicated signaling in pathology and anti-malarial immunity induced by Plasmodium berghei infection. Eur J Immunol. 2000;30:1646–1655. doi: 10.1002/1521-4141(200006)30:6<1646::AID-IMMU1646>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Harty JT. Adaptive immunity and enhanced CD8+ T cell response to Listeria monocytogenes in the absence of perforin and IFN-gamma. J Immunol. 2000;164:6444–6452. doi: 10.4049/jimmunol.164.12.6444. [DOI] [PubMed] [Google Scholar]
- Badovinac VP, Messingham KA, Jabbari A, Haring JS, Harty JT. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat Med. 2005;11:748–756. doi: 10.1038/nm1257. [DOI] [PubMed] [Google Scholar]
- Behlke MA, Chou HS, Huppi K, Loh DY. Murine T-cell receptor mutants with deletions of beta-chain variable region genes. PNAS. 1986;83:767–771. doi: 10.1073/pnas.83.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benacerraf B. Editorial: Immune response genes. Scand J Immunol. 1974;3:381–386. doi: 10.1111/j.1365-3083.1974.tb01269.x. [DOI] [PubMed] [Google Scholar]
- Benacerraf B, Germain RN. The immune response genes of the major histocompatibility complex. Immunol Rev. 1978;38:70–119. doi: 10.1111/j.1600-065x.1978.tb00385.x. [DOI] [PubMed] [Google Scholar]
- Benacerraf B, McDevitt HO. Histocompatibility-linked immune response genes. Science. 1972;175:273–279. doi: 10.1126/science.175.4019.273. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity, and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- Bettini ML, Bettini M, Nakayama M, Guy CS, Vignali DA. Generation of T cell receptor-retrogenic mice: improved retroviral-mediated stem cell gene transfer. Nat Protoc. 2013;8:1837–1840. doi: 10.1038/nprot.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattman JN, Antia R, Sourdive DJ, Wang X, Kaech SM, Murali-Krishna K, Altman JD, Ahmed R. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J Exp Med. 2002;195:657–664. doi: 10.1084/jem.20001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubou MI, Collette A, Voegtle D, Mazier D, Cazenave PA, Pied S. T cell response in malaria pathogenesis: selective increase in T cells carrying the TCR V(beta)8 during experimental cerebral malaria. Int Immunol. 1999;11:1553–1562. doi: 10.1093/intimm/11.9.1553. [DOI] [PubMed] [Google Scholar]
- Brewster DR, Kwiatkowski D, White NJ. Neurological sequelae of cerebral malaria in children. Lancet. 1990;336:1039–1043. doi: 10.1016/0140-6736(90)92498-7. [DOI] [PubMed] [Google Scholar]
- Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J Exp Med. 1999;189:701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch DH, Pilip I, Pamer EG. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J Exp Med. 1998;188:61–70. doi: 10.1084/jem.188.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabaniols JP, Fazilleau N, Casrouge A, Kourilsky P, Kanellopoulos JM. Most alpha/beta T cell receptor diversity is due to terminal deoxynucleotidyl transferase. J Exp Med. 2001;194:1385–1390. doi: 10.1084/jem.194.9.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Boswell DR, Lesk AM. The outline structure of the T-cell alpha beta receptor. EMBO J. 1988;7:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dai S, Huseby ES, Rubtsova K, Scott-Browne J, Crawford F, Macdonald WA, Marrack P, Kappler JW. Crossreactive T cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Davis MM, Krogsgaard M, Huppa JB, Sumen C, Purbhoo MA, Irvine DJ, Wu LC, Ehrlich L. Dynamics of cell surface molecules during T cell recognition. Annu Rev Biochem. 2003;72:717–742. doi: 10.1146/annurev.biochem.72.121801.161625. [DOI] [PubMed] [Google Scholar]
- Day EB, Guillonneau C, Gras S, La Gruta NL, Vignali DA, Doherty PC, Purcell AW, Rossjohn J, Turner SJ. Structural basis for enabling T-cell receptor diversity within biased virus-specific CD8+ T-cell responses. PNAS. 2011;108:9536–9541. doi: 10.1073/pnas.1106851108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org.
- Doll KL, Pewe LL, Kurup SP, Harty JT. Discriminating Protective from Nonprotective Plasmodium-Specific CD8+ T Cell Responses. J Immunol. 2016;196:4253–4262. doi: 10.4049/jimmunol.1600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Engwerda C, Belnoue E, Gruner AC, Renia L. Experimental models of cerebral malaria. Curr Top Microbiol Immunol. 2005;297:103–143. [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Fulton RB, Hamilton SE, Xing Y, Best JA, Goldrath AW, Hogquist KA, Jameson SC. The TCR's sensitivity to self peptide-MHC dictates the ability of naive CD8(+) T cells to respond to foreign antigens. Nat Immunol. 2015;16:107–117. doi: 10.1038/ni.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. 2015;16:1114–1123. doi: 10.1038/ni.3298. [DOI] [PubMed] [Google Scholar]
- Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- Gordon EB, Hart GT, Tran TM, Waisberg M, Akkaya M, Kim AS, Hamilton SE, Pena M, Yazew T, Qi CF, et al. Targeting glutamine metabolism rescues mice from late-stage cerebral malaria. PNAS. 2015;112:13075–13080. doi: 10.1073/pnas.1516544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras S, Burrows SR, Kjer-Nielsen L, Clements CS, Liu YC, Sullivan LC, Bell MJ, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The shaping of T cell receptor recognition by self-tolerance. Immunity. 2009;30:193–203. doi: 10.1016/j.immuni.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Hafalla JC, Bauza K, Friesen J, Gonzalez-Aseguinolaza G, Hill AV, Matuschewski K. Identification of targets of CD8(+) T cell responses to malaria liver stages by genome-wide epitope profiling. PLoS Pathog. 2013;9:e1003303. doi: 10.1371/journal.ppat.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluszczak C, Akue AD, Hamilton SE, Johnson LD, Pujanauski L, Teodorovic L, Jameson SC, Kedl RM. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A, Best SE, Unosson K, Amante FH, de Labastida F, Anstey NM, Karupiah G, Smyth MJ, Heath WR, Engwerda CR. Granzyme B expression by CD8+ T cells is required for the development of experimental cerebral malaria. J Immunol. 2011;186:6148–6156. doi: 10.4049/jimmunol.1003955. [DOI] [PubMed] [Google Scholar]
- Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, Vignali DA. Generation of T-cell receptor retrogenic mice. Nat Protoc. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- Horne-Debets JM, Karunarathne DS, Faleiro RJ, Poh CM, Renia L, Wykes MN. Mice lacking Programmed cell death-1 show a role for CD8(+) T cells in long-term immunity against blood-stage malaria. Sci Rep. 2016;6:26210. doi: 10.1038/srep26210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howland SW, Poh CM, Gun SY, Claser C, Malleret B, Shastri N, Ginhoux F, Grotenbreg GM, Renia L. Brain microvessel cross-presentation is a hallmark of experimental cerebral malaria. EMBO Mol Med. 2013;5:916–931. doi: 10.1002/emmm.201202273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Shen J, Chou B, Duan X, Tu L, Tetsutani K, Moriya C, Ishida H, Hamano S, Shimokawa C, et al. Involvement of CD8+ T cells in protective immunity against murine blood-stage infection with Plasmodium yoelii 17XL strain. Eur J Immunol. 2010;40:1053–1061. doi: 10.1002/eji.200939525. [DOI] [PubMed] [Google Scholar]
- Jenkins MK, Moon JJ. The role of naive T cell precursor frequency and recruitment in dictating immune response magnitude. J Immunol. 2012;188:4135–4140. doi: 10.4049/jimmunol.1102661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerne NK. The somatic generation of immune recognition. Eur J Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, Marrack P. T cells compete for access to antigen-bearing antigen-presenting cells. J Exp Med. 2000;192:1105–1113. doi: 10.1084/jem.192.8.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Clements CS, Brooks AG, Purcell AW, McCluskey J, Rossjohn J. The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 2002;10:1521–1532. doi: 10.1016/s0969-2126(02)00878-x. [DOI] [PubMed] [Google Scholar]
- Lefranc MP. IMGT databases, web resources and tools for immunoglobulin and T cell receptor sequence analysis, http://imgt.cines.fr. Leukemia. 2003;17:260–266. doi: 10.1038/sj.leu.2402637. [DOI] [PubMed] [Google Scholar]
- Mach N, Gillessen S, Wilson SB, Sheehan C, Mihm M, Dranoff G. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 2000;60:3239–3246. [PubMed] [Google Scholar]
- Malherbe L, Hausl C, Teyton L, McHeyzer-Williams MG. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity. 2004;21:669–679. doi: 10.1016/j.immuni.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity. 2013;38:263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti-Ferrandiz E, Pham HP, Dulauroy S, Gorgette O, Klatzmann D, Cazenave PA, Pied S, Six A. A TCRbeta Repertoire Signature Can Predict Experimental Cerebral Malaria. PLoS One. 2016;11:e0147871. doi: 10.1371/journal.pone.0147871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak A, Doherty PC, Wilson DB. The control of specificity of cytotoxic T lymphocytes by the major histocompatibility complex (AG-B) in rats and identification of a new alloantigen system showing no AG-B restriction. J Exp Med. 1977;146:1773–1790. doi: 10.1084/jem.146.6.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt HO, Chinitz A. Genetic control of the antibody response: relationship between immune response and histocompatibility (H-2) type. Science. 1969;163:1207–1208. doi: 10.1126/science.163.3872.1207. [DOI] [PubMed] [Google Scholar]
- Miles JJ, Douek DC, Price DA. Bias in the alphabeta T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol Cell Biol. 2011;89:375–387. doi: 10.1038/icb.2010.139. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neller MA, Ladell K, McLaren JE, Matthews KK, Gostick E, Pentier JM, Dolton G, Schauenburg AJ, Koning D, Fontaine Costa AI, et al. Naive CD8(+) T-cell precursors display structured TCR repertoires and composite antigen-driven selection dynamics. Immunol Cell Biol. 2015;93:625–633. doi: 10.1038/icb.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens T, Zeine R. The cell biology of T-dependent B cell activation. Biochem Cell Biol. 1989;67:481–489. doi: 10.1139/o89-078. [DOI] [PubMed] [Google Scholar]
- Pham NL, Badovinac VP, Harty JT. A default pathway of memory CD8 T cell differentiation after dendritic cell immunization is deflected by encounter with inflammatory cytokines during antigen-driven proliferation. J Immunol. 2009;183:2337–2348. doi: 10.4049/jimmunol.0901203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley MF, Almeida JR, Price DA, Douek DC. Unbiased molecular analysis of T cell receptor expression using template-switch anchored RT-PCR. Curr Protoc Immunol. 2011;Chapter 10(Unit10):33. doi: 10.1002/0471142735.im1033s94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley MF, Greenaway HY, Venturi V, Lindsay R, Quinn KM, Seder RA, Douek DC, Davenport MP, Price DA. Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. PNAS. 2010;107:19414–19419. doi: 10.1073/pnas.1010586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai D, Pham NL, Harty JT, Badovinac VP. Tracking the total CD8 T cell response to infection reveals substantial discordance in magnitude and kinetics between inbred and outbred hosts. J Immunol. 2009;183:7672–7681. doi: 10.4049/jimmunol.0902874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struik SS, Riley EM. Does malaria suffer from lack of memory? Immunol Rev. 2004;201:268–290. doi: 10.1111/j.0105-2896.2004.00181.x. [DOI] [PubMed] [Google Scholar]
- Townsend AR, Gotch FM, Davey J. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell. 1985;42:457–467. doi: 10.1016/0092-8674(85)90103-5. [DOI] [PubMed] [Google Scholar]
- Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- Valkenburg SA, Quinones-Parra S, Gras S, Komadina N, McVernon J, Wang Z, Halim H, Iannello P, Cole C, Laurie K, et al. Acute emergence and reversion of influenza A virus quasispecies within CD8+ T cell antigenic peptides. Nat Commun. 2013;4:2663. doi: 10.1038/ncomms3663. [DOI] [PubMed] [Google Scholar]
- van der Heyde HC, Manning DD, Roopenian DC, Weidanz WP. Resolution of blood-stage malarial infections in CD8+ cell-deficient beta 2-m0/0 mice. J Immunol. 1993;151:3187–3191. [PubMed] [Google Scholar]
- van der Merwe PA, Dushek O. Mechanisms for T cell receptor triggering. Nat Rev Immunol. 2011;11:47–55. doi: 10.1038/nri2887. [DOI] [PubMed] [Google Scholar]
- Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15:643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinetz JM, Kumar S, Good MF, Fowlkes BJ, Berzofsky JA, Miller LH. Adoptive transfer of CD8+ T cells from immune animals does not transfer immunity to blood stage Plasmodium yoelii malaria. J Immunol. 1990;144:1069–1074. [PubMed] [Google Scholar]
- Wilson A, Marechal C, MacDonald HR. Biased V beta usage in immature thymocytes is independent of DJ beta proximity and pT alpha pairing. J Immunol. 2001;166:51–57. doi: 10.4049/jimmunol.166.1.51. [DOI] [PubMed] [Google Scholar]
- Yanez DM, Manning DD, Cooley AJ, Weidanz WP, van der Heyde HC. Participation of lymphocyte subpopulations in the pathogenesis of experimental murine cerebral malaria. J Immunol. 1996;157:1620–1624. [PubMed] [Google Scholar]
- Young AC, Zhang W, Sacchettini JC, Nathenson SG. The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection. Cell. 1994;76:39–50. doi: 10.1016/0092-8674(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhao J, Perlman S. De novo recruitment of antigen-experienced and naive T cells contributes to the long-term maintenance of antiviral T cell populations in the persistently infected central nervous system. J Immunol. 2009;183:5163–5170. doi: 10.4049/jimmunol.0902164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinkernagel RM. Thymus and lymphohemopoietic cells: their role in T cell maturation in selection of T cells' H-2-restriction-specificity and in H-2 linked Ir gene control. Immunol Rev. 1978;42:224–270. doi: 10.1111/j.1600-065x.1978.tb00264.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.