Abstract
Self-assembly is a fundamental concept and a powerful approach in molecular science. However, creating functional materials with the desired properties through self-assembly remains challenging. In this work, through a combination of experimental and computational approaches, we report on the self-assembly of small amphiphilic dendrons into nanosized supramolecular dendrimer micelles with a degree of structural definition similar to traditional covalent high-generation dendrimers. We demonstrate that, with the optimal balance of hydrophobicity and hydrophilicity, one of the self-assembled nanomicellar systems, totally devoid of toxic side effects, is able to deliver small interfering RNA (siRNA) and achieve effective gene silencing both in cells - including the highly refractory human hematopoietic CD34+ stem cells - and in vivo, thus paving the way for future biomedical implementation. This work presents a case study of the concept of generating functional supramolecular dendrimers via self-assembly. The ability of carefully designed and gauged building blocks to assemble into supramolecular structures opens new perspectives on the design of self-assembling nanosystems for complex and functional applications.
Keywords: Amphiphilic dendrimers, self-assembly, nonviral vectors, siRNA delivery, supramolecular micelles, multiscale molecular modeling
Table of contents
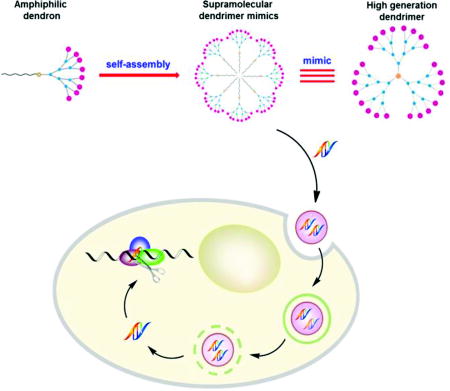
Supramolecular dendrimers created from small amphiphilic dendrons are able to mimic covalently constructed high-generation dendrimers for siRNA delivery. An optimal balance between the hydrophobic alkyl chain length and the hydrophilic dendritic portion is crucial for self-assembly of these amphiphilic dendrons into micellar nanostructures, and critically impacts the effectiveness of siRNA delivery and functional gene silencing.
1. Introduction
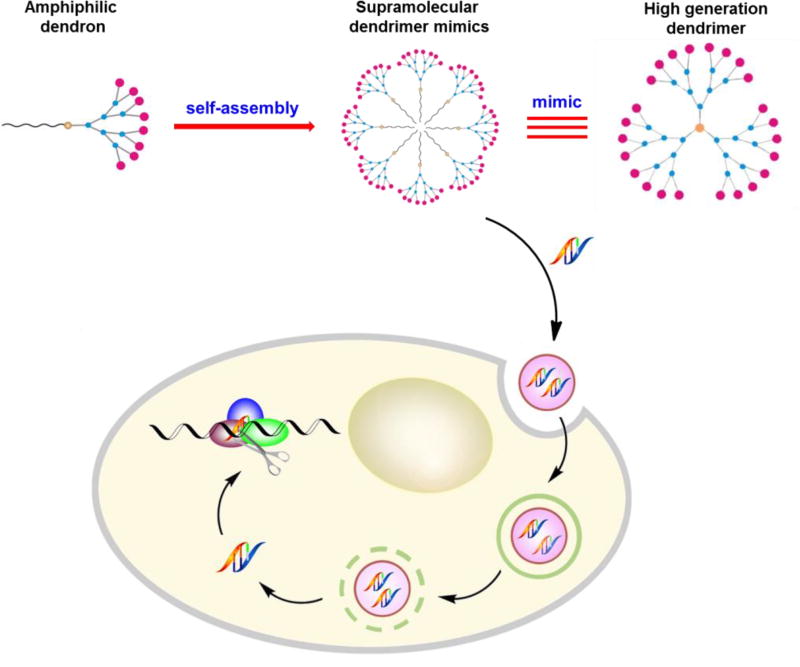
Self-assembly is a fundamental concept in modern molecular science for the creation of new materials.[1–4] It relies on the cumulative effects of multiple non-covalent interactions to assemble molecular building blocks into supramolecular entities in a reversible, controllable, and specific way, yet with relatively little synthetic effort. Of particular value is the ability of self-assembled structures to behave as more than the sum of their individual parts, and exhibit completely new properties.[5] As an example, we present here our investigation into the self-organization of small amphiphilic dendrons into spherical, nanosized supramolecular micellar dendrimers, the underlying mechanism for their functional delivery of small interfering RNA (siRNA), and the resulting gene silencing efficiency (Figure 1).
Figure 1.
Schematic presentation of self-assembly of amphiphilic dendrons into supramolecular dendrimers to resemble covalent high-generation dendrimers for functional siRNA delivery and gene silencing.
Small interfering RNA molecules (siRNAs) are able to specifically and efficiently turn off gene expression, offering an enormous opportunity for exploitation in biomedical applications such as gene-based disease treatment.[6–8] However, the main challenge facing siRNA therapeutics is their safe and efficient delivery.[9, 10] Indeed, siRNAs are too negatively charged to spontaneously cross biomembranes; at the same time, they are vulnerable to nuclease attack within biofluids. In principle, an ideal siRNA delivery vector should assemble the nucleic acid molecule within a compact complex, protect it from degradation, promote its cellular uptake, allow it to escape from endolysosomal compartments, and finally release it into the cytoplasm where the RNAi machinery is located.[9, 10] The most common non-viral vectors currently employed in siRNA delivery can be classified into two major categories: lipids and polymers.[11–13] Within the latter group, cationic dendrimers are emerging as promising nanovectors by virtue of their well-defined structure, intriguing features of multivalency, and high cargo payload confined within a nanosized volume.[14, 15]
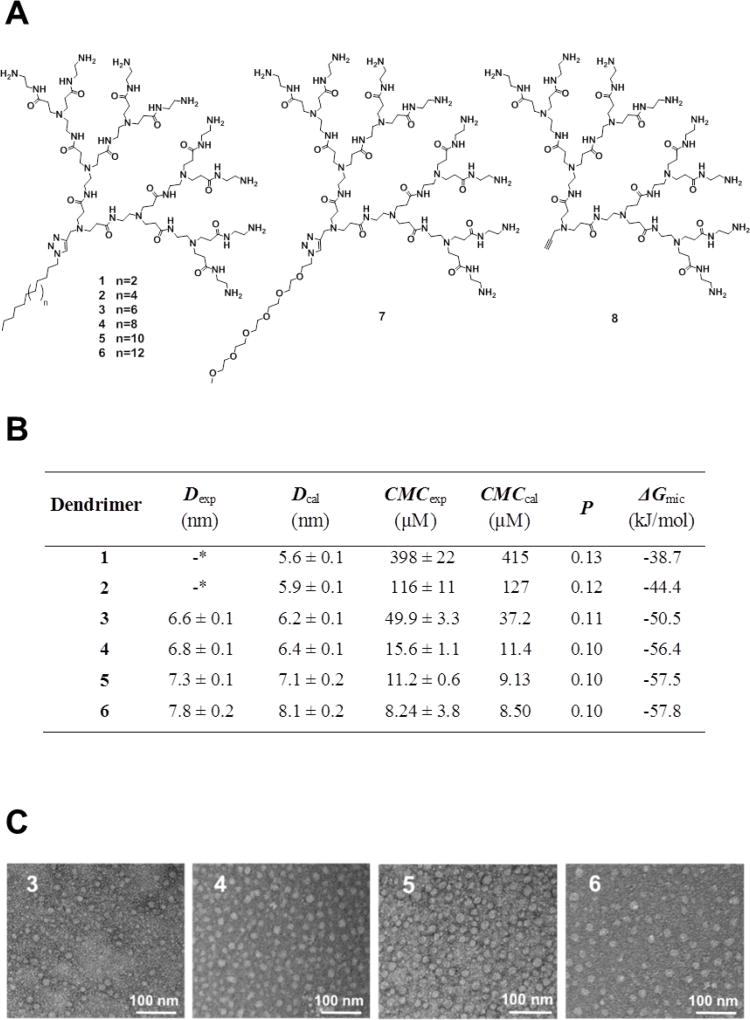
In the past decade, our group has been actively engaged in developing structurally flexible high-generation poly(amidoamine) (PAMAM) dendrimers[16–18] as potent and effective nanovectors for siRNA delivery in vitro and in vivo in various disease models,[19–22] with one of them being scheduled for clinical trials. More recently, we have shifted our interest towards amphiphilic molecules bearing dendritic portions as polar heads,[23–25] which are able to self-assemble into nanosized supramolecular structures of various size and shape.[3],[24],[26] In this work, drawing on our considerable interest in the field, and with the aim of obtaining more insightful understanding of self-assembling dendrimer nanovectors for siRNA delivery, we designed, synthesized and characterized a series of dendrons featuring hydrophobic tails of different lengths (1–6) (Figure 2A). Additionally, we conceived dendrons 7 and 8 as non-amphiphilic references with 7 bearing a hydrophilic pentaethylene glycol (PEG) chain and 8 being a small hydrophilic poly(amidoamine) dendron without any long hydrophobic chain. We systematically investigated these dendrons for their ability to self-assemble, their siRNA binding activity and their siRNA delivery capacity using both experimental approaches and computer modeling. Our results demonstrate that the optimal balance of the hydrophobic and hydrophilic components critically controls their self-organization, which in turn significantly affects their performance in siRNA delivery. Indeed, these supramolecular dendrimers show a degree of structural definition similar to that of traditional covalent high-generation dendrimers (Figure 1), and one of them possesses excellent siRNA delivery and gene silencing activity even in the highly refractory human hematopoietic CD34+ stem cells. Our study opens new perspectives on the design of self-assembling nanosystems for complex and functional applications.
Figure 2.
(A) Dendrons 1–8 investigated in this study; (B) The experimental/predicted values of self-assembled micellar supramolecular dendrimer diameter (Dexp, Dcal) and critical micelle concentration (CMCexp, CMCcal), the calculated packing parameter (P) and free energy of micellization (ΔGmic) for dendrons 1–6. (C) TEM images of the self-assembled supramolecular micellar structures formed by 3, 4, 5 and 6 in buffer at pH 7.4.
2. Results and Discussion
2.1. Self-assembly of amphiphilic dendrons into micellar supramolecular structures
The amphiphilic dendrons 1–7 studied in this work were synthesized via “click” chemistry[23] (Scheme S1). Dynamic light scattering (DLS) analysis revealed that solutions containing 3–6 predominantly formed nanoparticles around 7.0 nm in size (Figure 2B and Figure S1) while no stable nanostructures were observed for 1, 2, 7, and 8. The apparent self-assembly of 3–6 unveiled by DLS data was confirmed by transmission electron microscopic (TEM) imaging, which clearly revealed the formation of small spherical particles (Figure 2C) with sizes in the range of typical micellar nano-objects. Moreover, an increase in the alkyl chain length from C16 to C22 (3 to 6) was accompanied by an increase in the size of the micelles (Dexp) from 6.6 to 7.8 nm, consistent with the values (Dcal) predicted by related molecular simulations[27–30] (Figure 2B and Table S1). Of note, the dimensions of these nanomicelles are similar to those of structurally flexible high-generation dendrimers,[17] indicating that these self-assembled nanostructures have a degree of structural definition similar to traditional covalent high-generation dendrimers, i.e., they can be considered as supramolecular dendrimer mimics (Figure 1).
We further determined the values of the critical micelle concentrations (CMCexp) of each nanovector using the hydrophobic fluorescent probe pyrene.[31] As shown in Figure 2B, increasing the alkyl chain length (1 to 6) resulted in a drop of CMCexp values from 398 to 8.6 µM, confirming the expected inverse relationship between the length of the hydrophobic portion and CMCexp, i.e., dendrons bearing longer hydrophobic chains self-assemble and pack more efficiently than those featuring shorter aliphatic tails. The relatively large values of CMCexp for 1 and 2 provide an explanation for the absence of micelle formation observed during DLS analysis, whereas for 7 and 8, which are devoid of amphiphilic character, no CMCexp could be determined.
We next inspected the self-assembly of these dendrons using computational methods. For 1–6, the estimated value of their critical packing parameter P, an effective ratio of the relative sizes of the polar and apolar domains of an amphiphilic compound, was well below 0.33 (Figure 2B), implying that these molecules have a tendency to self-assemble into spherical micelles.[32, 33] This was in line with the results obtained with TEM imaging (Figure 2C) and mesoscopic molecular simulation (Figure S2). In addition, the associated values of the micellization free energy (ΔGmic) were largely negative, indicating that micelle formation was a spontaneous and highly favorable process for the amphiphilic molecules 1–6. However, ΔGmic decreased substantially (i.e., became more negative and hence more favorable) with increasing hydrophobic chain length from 1 to 6 (Figure 2B). Accordingly, the predicted CMC values (CMCcal) followed the expected decreasing trend as the hydrophobic character of the dendrimers increased, in agreement with the experimental data. Since the hydrophilic dendron architecture was the same in all dendrimers, the main differential contribution to ΔGmic, and hence to CMCcal, is related to the length of the hydrophobic component (see SI for more details). Altogether, this combined experimental/theoretical approach allowed us to assess the impact of the proper balance between hydrophobic and hydrophilic components of amphiphilic dendrons on the chemico-physical characteristics of the resulting multivalent self-assembled micelles.
2.2. Formation of siRNA/dendrimer nanoparticles protects siRNA from degradation
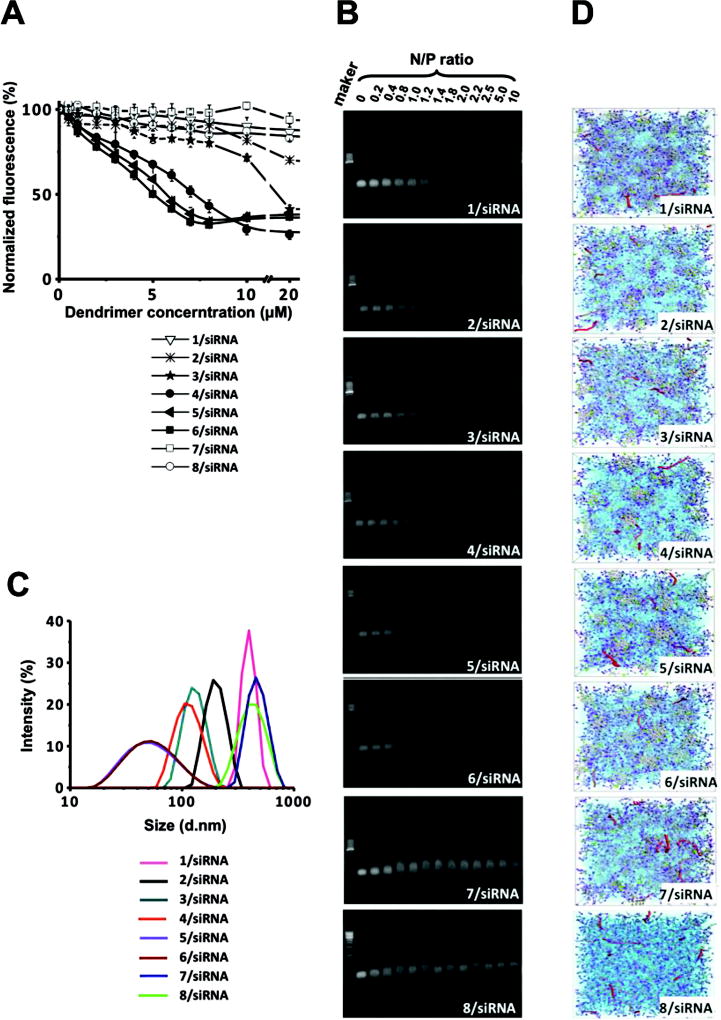
We next examined the ability of these supramolecular dendrimers to bind siRNA, a prerequisite for their possible application in siRNA delivery. The binding of these dendrimers to siRNA was assessed using a fluorescent ethidium bromide (EB) displacement assay, which is based on competition between EB and the dendrimer in binding siRNA. The value of C50, or the concentration of dendron at which the fluorescent intensity of intercalated EB is reduced by 50%, denotes the siRNA binding efficiency. The no-tail dendron 8, the PEG chain dendron 7, and dendron 1 bearing the shortest alkyl chain were all unable to effectively displace EB from siRNA (Figure 3A), presumably due to their inability to self-assemble under the assay conditions. For the molecular series 2–6, the C50 value decreased with increasing alkyl chain length, dendron 6 being the most efficient in siRNA binding (Figure 3A and Table S2). Notably, these data were consistent with the results of gel shift assays (Figure 3B), which confirmed the correlation between alkyl chain length and siRNA binding strength: the dendron bearing the longest alkyl chain was the most effective siRNA binder. In other words, these results showed that the dendrons endowed with the best self-assembly properties are the most effective in achieving siRNA binding. This was further confirmed by DLS investigation of the siRNA/dendrimer complexes (Figure 3C). Dendrons 1, 2, 7 and 8 formed very big aggregates with siRNA, which are far too large in dimension for eventual delivery.[34, 35] On the other hand, dendrons 3–6, which bear C16–C22 alkyl chains, were able to form siRNA nanoparticles around 100 nm in size, perfectly matching the size range required for effective delivery. The formation of compact and uniform 4/siRNA nanoparticles was further affirmed by additional TEM imaging (Figure S3)
Figure 3.
siRNA/dendrimer binding complexes revealed by (A) ethidium bromide assay; (B) gel retardation, (C) dynamic light scattering and (D) mesoscale modeling. Color legend in D: siRNA, red sticks; purple and lilac, hydrophilic dendron beads; light grey, hydrophobic tail beads; green, linker beads. Water and counterions are portrayed as a light blue field for clarity.
Next, mesoscale molecular simulations were performed to investigate the interactions between these dendrimers and the siRNA molecules. The morphology of the self-assembled supramolecular dendrimers and siRNAs is presented in Figure 3D. As can be seen, at the same N/P ratio for 1 through to 6, the siRNA molecules are more uniformly distributed and better encased within the micellar network, indicative of efficient protection from the surrounding environment. In contrast, for the non-self-assembling systems like 7 and 8, the siRNA molecules are less well protected from possible degradation by external agents (e.g., RNase).
2.3. Best performance of Dendron 4 for functional delivery of siRNA and the underlying rationale
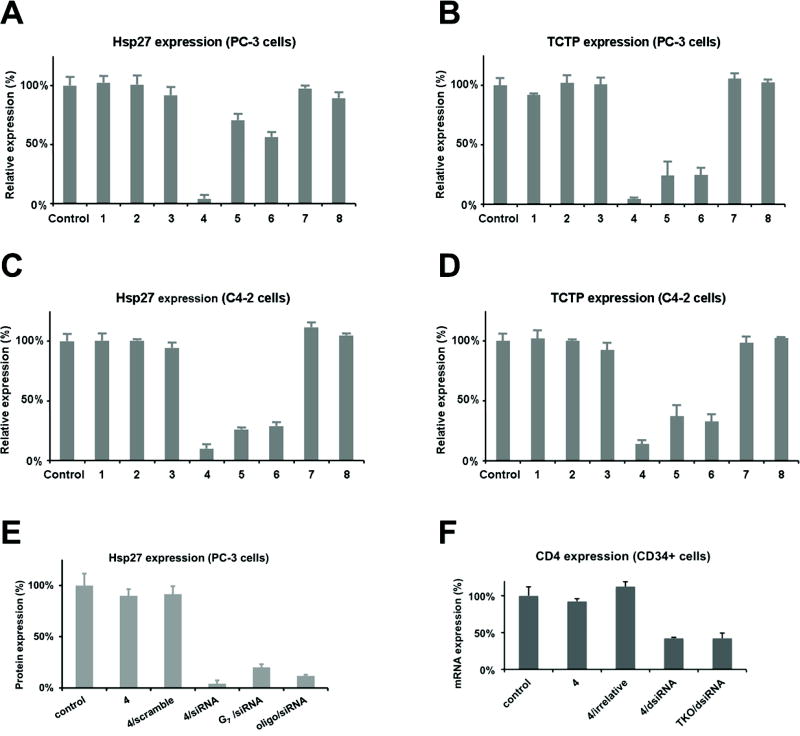
To investigate the siRNA delivery capacity of the supramolecular dendrimers formed by the amphiphilic dendrons 1–6, siRNA molecules targeting either heat shock protein 27 (Hsp 27)[36] or translationally controlled tumor protein (TCTP)[37] were tested in human castration-resistant prostate cancer PC-3 and C4-2 cell lines (Figure 4 and Figure S4). Concomitantly, the related cellular toxicity was also assessed (Figure S5). As shown in Figure 4A–D, 4 clearly displayed the best siRNA delivery capacity, leading to the most potent gene silencing. The gene silencing activity of 4 was better than that exhibited by the commercial reagent Oligofectamine and by the structurally flexible generation 7 dendrimer G7 previously developed by us (Figure 4E).[38] In addition, we showed that this potent gene silencing was specific, as the scramble siRNA delivered by 4 did not produce any noticeable effect (Figure 4E). More importantly, 4 mediated effective siRNA delivery and specific gene silencing even in human hematopoietic CD34+ stem cells (Figure 4F and Figure S4C) - an extremely challenging cell type for nucleic acid delivery - without any evident cytotoxicity (Figure S5E and Figure S5F).
Figure 4.
Dendrimer-mediated siRNA delivery and gene silencing of heat shock protein 27 (Hsp27) and translationally controlled tumor protein (TCTP) in human prostate cancer PC-3 (A, B) and C4-2 (C, D) cells. Hsp27 and TCTP protein levels were quantified by western blotting 72 h after treatment with the corresponding siRNA/dendrimer complexes at an N/P ratio of 10. (E) Hsp27 expression in human prostate cancer PC-3 cells treated with 50 nM Hsp27 siRNA and 4 at an N/P ratio of 10 compared with Hsp27 expression in cells treated with the high-generation dendrimer G7. (F) CD4 expression in human hematopoietic CD34+ stem cells treated with 50 nM CD4 dsiRNA and 4 at an N/P ratio of 5. The commercial transfection reagents Oligofectamine (oligo) and TransIT-TKO (TKO) was used as controls.
It is evident that the supramolecular dendrimers formed from dendrons containing longer hydrophobic tails (5 and 6) were less effective, whereas those bearing shorter aliphatic (1–3), hydrophilic (PEG) (7) or no chains (8) failed to elicit any obvious gene silencing (Figure 4A–D). These results can be sensibly rationalized in light of the previous discussion: because of the intrinsic incapacity of 7 and 8 to self-assemble, or the inability of 1–3 to generate stable micelles at the concentrations employed in the transfection experiments, none of these compounds was able to generate robust supramolecular dendrimers to mimic high-generation dendrimers for effective siRNA delivery. In other words, the positively charged dendron structure alone is not sufficient to efficiently bind, protect and deliver the negatively charged siRNA.
In order to understand why 4 was better than 5 and 6 in siRNA delivery and gene silencing, we generated 4/siRNA, 5/siRNA and 6/siRNA complexes using fluorescent labelled siRNA and examined their cellular uptake by means of flow cytometry. Our results show that the cellular uptake was similar for all three complexes (Figure S6). This finding, together with the observation that the 4/siRNA complex delivered siRNA more effectively than the 5/siRNA and 6/siRNA complexes, led us to speculate that the siRNA release process might be more efficient for 4/siRNA than for 5/siRNA and 6/siRNA because the siRNA complexes formed with the supramolecular dendrimers generated by 5 or 6 were too stable to disassemble. This stability would impair siRNA release within cells, ultimately decreasing the efficiency of gene silencing by 5/siRNA and 6/siRNA.
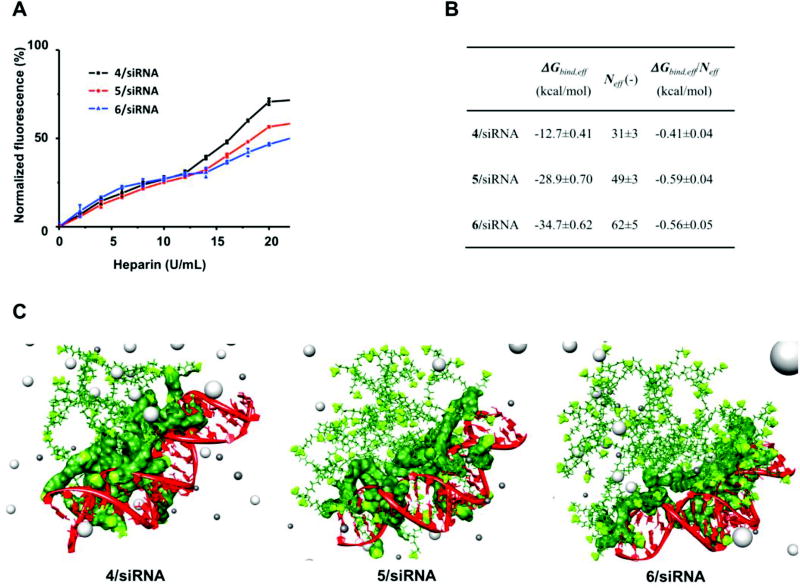
To verify this hypothesis, we examined siRNA dissociation from the corresponding siRNA/dendrimer complexes using the EB assay in the presence of heparin, a highly negatively charged polysaccharide that competes with siRNA for dendrimer binding. As illustrated in Figure 5A, when the heparin concentration was increased, the siRNA molecules were more effectively displaced from their complex with 4 than from their complexes with 5 and 6.
Figure 5.
(A) siRNA release from complexes with 4–6 was assessed using heparin-coupled ethidium bromide fluorescent assays. (B) siRNA/dendrimer binding data as derived from molecular dynamics simulations for 4–6: free energy of effective binding (ΔGbind,eff), number of effective charges (Neff) and effective-charge-normalized free energy of binding (ΔGbind,eff/Neff) for siRNA/dendrimer complexes. (C) Atomistic MD simulations of amphiphilic dendron self-assembly in the presence of siRNA. In all panels, the dendrons 4–6 are shown as forest green, with the terminal charged amine groups highlighted as chartreuse green sticks-and-balls. The siRNA is represented as a red ribbon and some representative Cl− and Na+ ions and counterions are portrayed as light and dark gray spheres, respectively. Water is omitted in all panels for clarity. The binding interface region between each micelle and siRNA is also shown in each complex.
To further corroborate this finding, we performed a quantitative characterization of siRNA/dendrimer interactions by atomistic molecular dynamics (MD) simulations[27–30] starting from the morphological information gathered by mesoscopic modeling. As presented in Figure 5B, the supramolecular dendrimer formed by 4 possessed a total surface charge of +64. 31 of these positive charges (48%) were effectively engaged in complexing siRNA (Figure 5C), resulting in a charge-normalized effective free energy of binding (ΔGbind.eff/Neff) value of −0.41 kcal/mol (Figure 5B). In contrast, the self-assembled dendrimers generated by 5 and 6 were not only able to exploit 49/80 (61%) and 62/102 (61%) positive charges, respectively, to constantly bind siRNA (see Figure 5C), but they also did it more effectively, as demonstrated by their more favorable ΔGbind.eff/Neff values of −0.59 and −0.56 kcal/mol (Figure 5B). These higher values of ΔGbind.eff/Neff clearly characterize 5 and 6 as the strongest siRNA binders of our series, in agreement with the experimental evidence presented above. Collectively, these results demonstrate that under our experimental conditions the self-assembled nanomicelles formed by 4 are endowed with the best siRNA releasing ability alongside optimal siRNA binding strength. These properties allowing the nanomicelles to carry the siRNA cargo along its journey to the cell and efficiently release the loaded siRNA upon reaching its final destination. The sensible molecular rationale for this is that an optimal balance is achieved between the hydrophobic chain length and the hydrophilic dendritic entity in 4, providing this amphiphile with the ideal properties to efficiently self-assemble into micellar supramolecular nanostructures. Such nanovectors easily form complexes with siRNA before being disassembled during endosome release, ultimately resulting in the best siRNA delivery and the most potent gene silencing out of all the dendrons presented in this work.
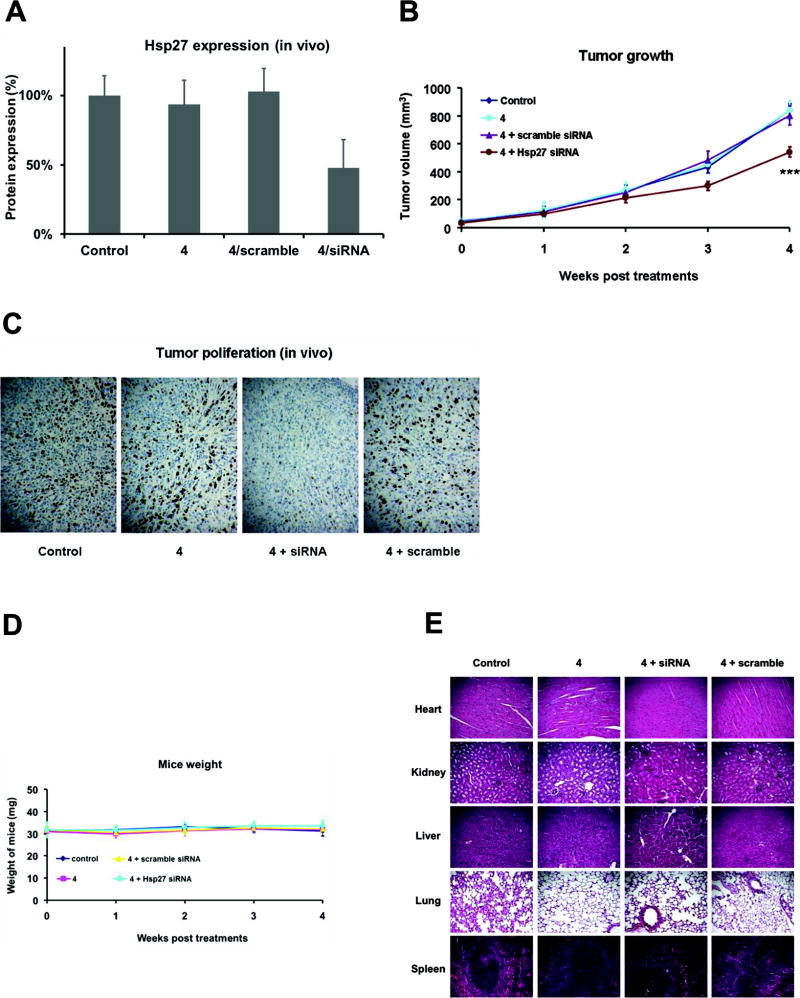
Finally, we carried out in vivo evaluation of 4-mediated systemic delivery of an siRNA targeting Hsp27 in a prostate cancer xenografted nude mice model. We observed significant gene silencing (Figure 6A and Figure S7) and effective inhibition of tumor growth 4 weeks post-treatment (Figure 6B). Additional immunohistochemistry with Ki67 staining also confirmed the inhibition of tumor proliferation (Figure 6C). The observed successful in vivo activity can be ascribed to the powerful capacity of 4 to deliver siRNA alongside the ability of the nanosized 4/siRNA complexes to home in on the tumor via the Enhanced Permeation and Retention (EPR) effect.[39,40]
Figure 6.
Effective 4-mediated systemic siRNA delivery and gene silencing in mice. Nude mice bearing prostate cancer PC-3 tumors of 30–50 mm3 were randomly selected for treatment with Hsp27 siRNA/4 and with PBS, 4 alone, and scrambled siRNA/4 as controls (3 mg/kg siRNA and 4 at an N/P ratio of 5). Treatments were administered by intraperitoneal injection during a period of 4 weeks with an injection frequency of twice per week (A–D). Effective gene silencing of Hsp27 was measured by western blotting and quantified by Image J software (A). (B) Inhibition of tumor growth and (C) the antiproliferation activity in tumors assessed by measuring tumor size and by Ki-67 immunohistochemical staining, respectively. (D) The body weight of the mice was recorded during the whole treatment period. (E) In vivo toxicity assessment of 4/siRNA by sectioning and HES staining of the major mouse organs including heart, kidney, liver, lung, and spleen.
We further evaluated the toxicity of 4 by examination of inflammatory cytokines (Figure S8), blood biochemistry, liver and kidney functions (Figure S9), and pathological changes in major organs (Figure S10). Neither 4 nor its siRNA complex showed any evident toxicity. No significant animal weight loss (Figure 6D) or discernible toxicity to different organs was observed in the xenograft mice during the entire treatment (Figure 6E). Collectively, these results support the utilization of the amphiphilic compound 4 as an siRNA vector for future therapeutic implementation.
3. Conclusions
Molecular engineering of functional material through self-assembly is an extremely interesting concept, but still faces great challenges. Indeed, self-assembly creates non-covalent supramolecular constructions through weak interactions, which are often critically impacted by factors such as molecular composition, physical and chemical properties, and geometrical features of the building blocks. Therefore, understanding the relationship between these features and the underlying supramolecular organization mechanism is of paramount importance in establishing functional materials through self-assembly.
RNA interference (RNAi) is a robust method for post-transcriptional silencing of genes using small, double-stranded RNA (siRNA) with sequence homology-driven specificity, providing a simple, fast and cost-effective alternative to existing gene targeting approaches. However, living organisms are well protected by intra- and extracellular barriers against invasion of foreign genetic material. Therefore, siRNA delivery systems have to be carefully designed to overcome these barriers. From this perspective, we designed, synthesized and characterized a series of amphiphilic molecules based on a hydrophilic PAMAM dendron head and a hydrophobic portion consisting of a linear hydrocarbon chain of variable length.
Interestingly, these amphiphilic dendrons self-assembled and generated supramolecular nanostructures which resemble and mimic the structurally well-defined covalent high-generation dendrimers. By using a powerful combination of experimental and theoretical approaches we gained unique and detailed insights into the ability of the amphiphilic dendrons to self-assemble into supramolecular dendrimers for functional siRNA delivery. Our investigation revealed that an optimal balance between the hydrophobic chain length and the hydrophilic dendritic portion of the amphiphilic dendrimers is instrumental in their self-assembly and their siRNA delivery activity. This work presents a case study of the concept of generating functional supramolecular dendrimers via self-assembly. The ability of carefully designed and gauged building blocks to spontaneously assemble into complex supramolecular structures underpins developments in a wide range of technologies, from materials science to biology and medicine, with huge potential for implementation in future applications.
4. Materials and methods
Synthesis and characterization of amphiphilic dendrimers
The detailed synthetic procedure is described in the Supporting Information.
In vitro transfection
Prostate cancer PC-3 and C4-2 cells
One day before transfection, 1.5×105 PC-3 or C4-2 cells were seeded in 6 cm dishes in 4 mL of fresh complete medium containing 10% FBS. Before transfection, a solution of the siRNA/amphiphilic dendrimer complex was prepared accordingly. The desired amount of siRNA and amphiphilic dendrimer was diluted in 200 µL of Opti-MEM transfection medium. The solutions were mixed with a vortex for 10 s, and then left for 10 min at room temperature. The amphiphilic dendrimer was added to the siRNA solution, homogenized for 10 s with a vortex and left for 30 min at room temperature. Then 1.6 mL serum-free medium was added into the complex solution and the final volume brought to 2 mL. Before addition of the transfection complexes, the complete medium with serum was removed and cells were washed once with PBS. Then, 2 mL of the complex solution was added and incubated at 37 °C in the absence of 10% FBS. After 8 h of incubation, the transfection mixture was replaced with complete medium containing 10% FBS, and maintained under normal growth conditions for a further incubation period of 72 h. The commercial transfection reagent Oligofectamine (Thermo Fisher Scientific) was used as a control according to the manufacturer’s instructions.
Human hematopoietic CD34+ stem cells
4×105 CD34+ cells per well were seeded in 24-well tissue culture plates in 300 µL fresh complete medium. Before transfection, complexes of dsiRNA/dendrimer reagents were prepared. The desired amount of dsiRNA and dendrimer reagent was diluted in 50 µL of serum-free medium (Opti-MED). The dendrimer solution was mixed gently and incubated for 10 min at room temperature. After the 10-minute incubation, the diluted dsiRNA and the dendrimer reagent were mixed gently and incubated for 30 minutes at room temperature. Then 100 µL of dsiRNA/dendrimer complex was added to each well containing cells and medium and mixed gently by rocking the plate back and forth. The cells were incubated at 37°C in a CO2 incubator for 24–48 h for further assays. The commercial transfection reagent Trans-TKO (Mirus) was used as a control according to the manufacturer’s instructions.
In vivo gene silencing in tumor-xenografted nude mice
Institutional guidelines for the proper and humane use of animals in research were followed. Approximately 10 × 106 PC-3 cells in 0.1 mL of DMEM (Thermo Fisher Scientific) which was supplemented with 10% FBS were inoculated subcutaneously into the flank region of 5-week-old male xenograft nude mice (Charles River Laboratories, Larbresles, France) via a 27-gauge needle. As previously described, when PC-3 tumors reached 30–50 mm3 (usually 2 to 3 weeks after injection), mice were randomly selected for treatment with Hsp27 siRNA/amphiphilic dendrimer 4, scrambled siRNA/amphiphilic dendrimer 4, amphiphilic dendrimer 4, or PBS buffer alone. Each experimental group consisted of 6–7 mice. After randomization, PBS buffer, 3 mg/kg Hsp27 siRNA, amphiphilic dendrimer 4 at an N/P ratio (= [total terminal amines in the dendrimer 4]/[phosphates in the siRNA]) = 5, Hsp27 siRNA/amphiphilic dendrimer 4 or scrambled siRNA/amphiphilic dendrimer 4 complexes, were injected intraperitoneally twice per week for 4 weeks. The size of the tumors and the weight of mice were recorded once per week. After 4 weeks of treatments, mice were sacrificed. Tumors were removed from the animals and divided into two parts. One part of each tumor was frozen in liquid nitrogen and then stored at −80 °C for RNA and protein extraction. The in vivo expression of Hsp27 protein was measured by western blot as described before. The other part of each tumor was fixed in 4% paraformaldehyde for immunohistochemistry. The heart, kidney, liver, lung and spleen were removed from the animals and fixed in 4% paraformaldehyde for hematoxylin and eosin (HES) staining.
Computational methods
Atomistic and mesoscopic simulations were performed in the framework of a multiscale modeling strategy developed by our group.[17],[26],[28],[30],[41] This approach is based on the systematic elimination of computationally expensive degrees of freedom while retaining implicitly their influence on the remaining degrees freedom in the mesoscopic model, thus allowing us to reproduce more realistic systems and to significantly extend the accessible time scales. Atomistic molecular dynamics calculation were carried out using the AMBER 14[42] suite of programs and mesoscale simulations were performed in the framework of the Dissipative Particle Dynamics theory[43]. For further details, refer to the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by Association pour la Recherche sur les Tumeurs de la Prostate (LP, XL), Ligue Nationale Contre le Cancer (LP), Association Française contre les Myopathies (XL), CNRS, INSERM, PACA Canceropôle, INCa, China Scholarship Council (CC, CL, YW, JT), Bourse d’Excellence Eiffel (YT), the international ERA-Net EURONANOMED European Research project “Target4Cancer”. The financial support from ESTECO s.r.l. (Grant DDOS) and from University of Trieste (FRA project “Combined Computational/Experimental Evidences for Multivalent Amphiphilic Nanoscale Carriers”) is gratefully acknowledged. Access to CINECA supercomputing facility was granted through the sponsored HPC Italian Supercomputing Resource Allocation (ISCRA) projects MONALISA and SIMBIOSY. We are grateful to Dr. Claudia Andrieu for her help with the ethidium bromide assay and Ms. Virginie Baylot for TCTP siRNA. CC, PP and XL contributed equally to this work.
Footnotes
Supporting Information is available from the Wiley Online Library.
Authors’ contributions
LP conceived the project. CC, PP, XL, SP, LP designed experiments; CC and TY synthesized the dendrimers; PR and JJR contributed the biological models; CC, CL, YW, JT and SG characterized the dendrimers and the related nanoparticles. XL performed cell uptake, in vitro and in vivo experiments on siRNA delivery in prostate cancer models; JZ carried out experiments with CD34+ stem cells and cell uptake assay, and QC in vivo toxicity assessment; PP, EL, VDL, MF and SP performed computer modeling. CC, PP, XL, QC, SG, FQ, PR, ZL, SP, LP analyzed data; CC, XL, SP, LP wrote the manuscript. All authors contributed to and proof-read the manuscript.
References
- 1.Lehn J-M. Science. 2002;295 doi: 10.1126/science.1071063. [DOI] [PubMed] [Google Scholar]
- 2.Webber MJ, Appel EA, Meijer EW, Langer R. Nat. Mater. 2015;15:13. doi: 10.1038/nmat4474. [DOI] [PubMed] [Google Scholar]
- 3.Percec V, Wilson DA, Leowanawat P, Wilson CJ, Hughes AD, Kaucher MS, Hammer DA, Levine DH, Kim AJ, Bates FS, Davis KP, Lodge TP, Klein ML, DeVane RH, Aqad E, Rosen BM, Argintaru AO, Sienkowska MJ, Rissanen K, Nummelin S, Ropponen J. Science. 2010;328:1009. doi: 10.1126/science.1185547. [DOI] [PubMed] [Google Scholar]
- 4.Aida T, Meijer EW, Stupp SI. Science. 2012;335:813. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitty A. Nat. Chem. Biol. 2008;4:435. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- 6.Haussecker D, Kay MA. Science. 2015;347:1069. doi: 10.1126/science.1252967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crunkhorn S. Nat. Rev. Drug Discov. 2013;12:178. doi: 10.1038/nrd3962. [DOI] [PubMed] [Google Scholar]
- 8.Castanotto D, Rossi JJ. Nature. 2009;457:426. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanasty R, Dorkin JR, Vegas A, Anderson D. Nat. Mater. 2013;12:967. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 10.Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discov. 2009;8:129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Huang L. Mol. Ther. 2010;18:669. doi: 10.1038/mt.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyata K, Nishiyama N, Kataoka K. Chem. Soc. Rev. 2012;41:2562. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- 13.Wagner E. Acc. Chem. Res. 2011;45:1005. doi: 10.1021/ar2002232. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Rocchi P, Peng L. New J. Chem. 2012;36:256. [Google Scholar]
- 15.Sheikhi Mehrabadi F, Fischer W, Haag R. Curr. Opin. Solid State Mater. Sci. 2012;16:310. [Google Scholar]
- 16.Zhou J, Wu J, Hafdi N, Behr J-P, Erbacher P, Peng L. Chem. Commun. 2006:2362. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 17.Liu X, Wu J, Yammine M, Zhou J, Posocco P, Viel S, Liu C, Ziarelli F, Fermeglia M, Pricl S, Victorero G, Nguyen C, Erbacher P, Behr J-P, Peng L. Bioconjugate Chem. 2011;22:2461. doi: 10.1021/bc200275g. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Liu C, Catapano CV, Peng L, Zhou J, Rocchi P. Biotechnol. Adv. 2014;32:844. doi: 10.1016/j.biotechadv.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Neff CP, Liu X, Zhang J, Li H, Smith DD, Swiderski P, Aboellail T, Huang Y, Du Q, Liang Z, Peng L, Akkina R, Rossi JJ. Mol. Ther. 2011;19:2228. doi: 10.1038/mt.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reebye V, Sætrom P, Mintz PJ, Huang K-W, Swiderski P, Peng L, Liu C, Liu X, Lindkær-Jensen S, Zacharoulis D, Kostomitsopoulos N, Kasahara N, Nicholls JP, Jiao LR, Pai M, Spalding DR, Mizandari M, Chikovani T, Emara MM, Haoudi A, Tomalia DA, Rossi JJ, Habib NA. Hepatology. 2014;59:216. doi: 10.1002/hep.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kala S, Mak ASC, Liu X, Posocco P, Pricl S, Peng L, Wong AST. J. Med. Chem. 2014;57:2634. doi: 10.1021/jm401907z. [DOI] [PubMed] [Google Scholar]
- 22.Cui Q, Yang S, Ye P, Tian E, Sun G, Zhou J, Sun G, Liu X, Chen C, Murai K, Zhao C, Azizian KT, Yang L, Warden C, Wu X, D'Apuzzo M, Brown C, Badie B, Peng L, Riggs AD, Rossi JJ, Shi Y. Nat. Commun. 2016;7 doi: 10.38/ncomms10637. [DOI] [Google Scholar]
- 23.Yu T, Liu X, Bolcato-Bellemin A-L, Wang Y, Liu C, Erbacher P, Qu F, Rocchi P, Behr J-P, Peng L. Angew. Chem. Int. Ed. Engl. 2012;51:8478. doi: 10.1002/anie.201203920. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Zhou J, Yu T, Chen C, Cheng Q, Sengupta K, Huang Y, Li H, Liu C, Wang Y, Posocco P, Wang M, Cui Q, Giorgio S, Fermeglia M, Qu F, Pricl S, Shi Y, Liang Z, Rocchi P, Rossi JJ, Peng L. Angew Chem. Int. Ed. Engl. 2014;53:11822. doi: 10.1002/anie.201406764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Liu C, Zhou J, Chen C, Qu F, Rossi JJ, Rocchi P, Peng L. Nanoscale. 2015;7:3867. doi: 10.1039/c4nr04759a. [DOI] [PubMed] [Google Scholar]
- 26.Wei T, Chen C, Liu J, Liu C, Posocco P, Liu X, Cheng Q, Huo S, Liang Z, Fermeglia M, Pricl S, Liang X-J, Rocchi P, Peng L. Proc. Natl. Acad. Sci. U. S. A. 2015;112:2978. doi: 10.1073/pnas.1418494112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posocco P, Laurini E, Dal Col V, Marson D, Karatasos K, Fermeglia M, Pricl S. Curr. Med. Chem. 2012;19:5062. doi: 10.2174/0929867311209025062. [DOI] [PubMed] [Google Scholar]
- 28.Barnard A, Posocco P, Pricl S, Calderon M, Haag R, Hwang ME, Shum VWT, Pack DW, Smith DK. J. Am. Chem. Soc. 2011;133:20288. doi: 10.1021/ja2070736. [DOI] [PubMed] [Google Scholar]
- 29.Jones SP, Gabrielson NP, Wong C-H, Chow H-F, Pack DW, Posocco P, Fermeglia M, Pricl S, Smith DK. Mol. Pharm. 2011;8:416. doi: 10.1021/mp100260c. [DOI] [PubMed] [Google Scholar]
- 30.Posocco P, Pricl S, Jones S, Barnard A, Smith DK. Chem. Sci. 2010;1:393. [Google Scholar]
- 31.Mazor S, Vanounou S, Fishov I. Chem. Phys. Lipids. 2012;165:125. doi: 10.1016/j.chemphyslip.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 32.Israelachvili JN, Mitchell DJ, Ninham BW. BBA - Biomembranes. 1977;470:185. doi: 10.1016/0005-2736(77)90099-2. [DOI] [PubMed] [Google Scholar]
- 33.Nagarajan R. Langmuir. 2001;18:31. [Google Scholar]
- 34.Wheeler JJ, Palmer L, Ossanlou M, MacLachlan I, Graham RW, Zhang YP, Hope MJ, Scherrer P, Cullis PR. Gene Ther. 1999;6:271. doi: 10.1038/sj.gt.3300821. [DOI] [PubMed] [Google Scholar]
- 35.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Nat. Rev. Genet. 2014;15:541. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 36.Acunzo J, Katsogiannou M, Rocchi P. Int. J. Biochem. Cell. 2012;44:1622. doi: 10.1016/j.biocel.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Acunzo J, Baylot V, So A, Rocchi P. Cancer Treat. Rev. 2014;40:760. doi: 10.1016/j.ctrv.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 38.Liu XX, Rocchi P, Qu FQ, Zheng SQ, Liang ZC, Gleave M, Iovanna J, Peng L. Chem. Med. Chem. 2009;4:1302. doi: 10.1002/cmdc.200900076. [DOI] [PubMed] [Google Scholar]
- 39.Matsumura Y, Maeda H. Cancer Res. 1986;46:6387. [PubMed] [Google Scholar]
- 40.Chauhan VP, Jain RK. Nat. Mater. 2013;12:958. doi: 10.1038/nmat3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posocco P, Liu X, Laurini E, Marson D, Chen C, Liu C, Fermeglia M, Rocchi P, Pricl S, Peng L. Mol. Pharm. 2013;10:3262. doi: 10.1021/mp400329g. [DOI] [PubMed] [Google Scholar]
- 42.Case DA, Berryman JT, Betz RM, Cerutti DS, Cheatham ITE, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Luchko T, Luo R, Madej B, Merz KM, Monard G, Needham P, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Salomon-Ferrer R, Simmerling CL, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, York DM, Kollman PA. AMBER. University of California; San Francisco, CA, USA: 2015. [Google Scholar]
- 43.Groot RD, Warren PB. J. Chem. Phy. 1997;107:4423. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.