Abstract
A deeper understanding of the metastatic process is required for the development of new therapies that improve patient survival. Metastatic tumor cell growth and survival in distant organs is facilitated by the formation of a pre-metastatic niche composed of hematopoietic cells, stromal cells, and extracellular matrix (ECM). Perivascular cells, including vascular smooth muscle cells (vSMCs) and pericytes, are involved in new vessel formation and in promoting stem cell maintenance and proliferation. Given the well-described plasticity of perivascular cells, we hypothesize that perivascular cells similarly regulate tumor cell fate at metastatic sites. Using perivascular cell-specific and pericyte-specific lineage-tracing models, we trace the fate of perivascular cells in the pre-metastatic and metastatic microenvironments. We show that perivascular cells lose the expression of traditional vSMC/pericyte markers in response to tumor-secreted factors and exhibit increased proliferation, migration, and ECM synthesis. Increased expression of the pluripotency gene Klf4 in these phenotypically-switched perivascular cells promotes a less differentiated state characterized by enhanced ECM production that establishes a pro-metastatic fibronectin-rich environment. Genetic inactivation of Klf4 in perivascular cells decreases pre-metastatic niche formation and metastasis. Our data reveal a previously unidentified role for perivascular cells in pre-metastatic niche formation and uncover novel strategies for limiting metastasis.
Microenvironmental signals arising early in pre-metastatic sites are among the key determinants of successful metastatic colonization. Previously, we defined activated stromal cells, altered extracellular matrix (ECM), and recruited bone marrow-derived cells (BMDCs) as components of a tumor-conducive microenvironment at distant sites in response to factors released by the primary tumor, termed the pre-metastatic niche1. Expansion of PDGFRα+ stromal cells and an associated localized increase in fibronectin supports the recruitment of hematopoietic cells to the pre-metastatic niche1. These recruited hematopoietic cells develop into myeloid cells at pre-metastatic sites and exhibit immunosuppressive features that support metastatic tumor cell colonization and proliferation2–5. While there is an increased understanding of the role of myeloid cells in the pre-metastatic environment and tumor metastases, less is known about the contribution of stromal cells to pre-metastatic niche formation and their functional role in metastatic outgrowth.
Perivascular cells, including vascular smooth muscle cells (vSMCs) and pericytes, support vascular stability through close contact and signaling crosstalk with the endothelium, and their contractile role in regulating blood vessel tone, diameter, and permeability6–9. Growing evidence suggests that perivascular cells are also the key stromal component of stem cell niches in which they regulate stem cell maintenance and proliferation, and as such are critical to tissue regeneration and organ homoeostasis10,11. Perivascular cells are traditionally identified by a combination of contractile genes such as Myh11, Acta2, and Tagln (vSMCs), and cell surface marker proteins such as NG2, PDGFRB, and RGS5 (pericytes)12–14, with extensive overlap in marker expression observed in vSMC and pericyte populations15. Perivascular cells also exhibit remarkable plasticity in the settings of inflammation and vascular disease7, where they lose expression of contractile genes such as Myh11, Acta2 and Tagln, and acquire a proliferative, migratory, and ECM synthesizing/modulating phenotype6–8,14,16. This process has been termed phenotypic switching. Recent studies have demonstrated that perivascular cells can acquire macrophage and mesenchymal stem cell (MSC) phenotypes in atherosclerosis, dependent on expression of Klf4 and Oct4, where they play a critical role in pathogenesis17,18. As a result of phenotypic switching, perivascular cells can be overlooked or misidentified due to lack of classic markers17,19. Although there is evidence for perivascular phenotypic switching in inflammation and vascular disease, the role of perivascular cells in pre-metastatic niche formation and metastatic progression remains under appreciated.
Here we describe perivascular cell phenotypic switching in pre-metastatic lung, coincident with KLF4 and PDGFRα expression, in response to a growing, localized primary tumor or to tumor secreted factors. Tumor-induced perivascular cell phenotypic switching initiates pre-metastatic niche formation at least in part by contributing to fibronectin protein production that promotes tumor cell metastatic behavior. Disruption of perivascular cell-specific Klf4 expression inhibits perivascular phenotypic switching and decreases metastasis. Our results reveal a novel role for perivascular cells in pre-metastatic niche formation and identify KLF4 as a critical inducer of perivascular cell phenotypic switching. By identifying perivascular cell plasticity in the pre-metastatic niche, we uncover a new opportunity to redirect stromal involvement in this setting and limit metastatic progression.
Results
Lineage-traced perivascular cells demonstrate that phenotypic switching occurs in pre-metastatic sites
Perivascular cell phenotypic switching is characterized by loss of marker gene expression such as Myh11 and Acta2, and an increase in proliferation, migration, and ECM production7. Due to this reduced perivascular marker gene expression and the potential misidentification of these cells in their activated state, in vivo studies that carefully trace and investigate the function of phenotypically switched perivascular cells are required. To determine whether perivascular cells undergo phenotypic switching in pre-metastatic tissue, we used the recently described Myh11-ERT-creT2 ROSA-STOP-flox-eYFP lineage-tracing mice, wherein the perivascular-specific gene Myh11 promoter drives an inducible cre-recombinase (designated as “Myh11 lineage-tracing mice”) (Supplementary Fig. 1a)17,19. In adult Myh11 lineage-tracing mice, tamoxifen induces stable expression of eYFP in vSMCs and pericytes, and enables the detection of cells expressing the Myh11 gene only at the time of tamoxifen administration, including pre-existing SMCs/pericytes and their progeny, even when this perivascular marker expression is subsequently lost17,19. Importantly, we found that nearly all MYH11+ cells in the lungs of healthy Myh11 lineage-tracing mice treated with tamoxifen were eYFP+ and co-expressed MYH11 (Supplementary Fig. 1b). YFP+ cells were also ACTA2+, a known marker of perivascular cells and myofibroblasts (Supplementary Fig. 1c).
To interrogate the role of perivascular cells during metastatic development, we orthotopically injected metastatic melanoma B16-F10 or metastatic rhabdomyosarcoma M3-9M tumors into syngeneic Myh11 lineage-tracing mice and analyzed pre-metastatic lung at multiple time points for evidence of perivascular phenotypic switching in eYFP-expressing cells that have lost expression of perivascular markers MYH11 and ACTA2 (Supplementary Fig. 1d-e). We found that there is an increase in the total number of eYFP+ cells in the lungs of B16-F10 (Fig 1a-c, e, Supplementary Fig 2a) and M3-9M (Fig 1 h-j) tumor-bearing mice from day 10 to day 18 post-tumor injection. By careful confocal imaging across multiple pre-metastatic time-points, we demonstrated the loss of perivascular markers MYH11 and ACTA2, the displacement of perivascular cells away from blood vessels by day 3, the appearance of YFP+ cells away from blood vessels by day 5, and increase in eYFP+ MYH11- ACTA2- cells found in clusters within the parenchyma separated from the vasculature of pre-metastatic lung by day 10 post-tumor injection (Fig 1e, Supplementary Fig 2a). The total number of eYFP+ cells and the proportion of phenotypically modulated eYFP+ cells that were MYH11- ACTA2- was increased in pre-metastatic lung (Fig 1c, f, j, k). This expanded population of eYFP+ cells may be due to vSMC recruitment to these pre-metastatic sites, proliferation in situ, or a combination of both. Consistent with the increase in eYFP+ cell number identified in stained tissue sections, we determined that many eYFP+ MYH11- ACTA2- cells expressed the proliferation marker Ki67 (Supplementary Fig 2b). More eYFP+ cells were found at farther distances from blood vessels in the lungs of both B16-F10 (Fig 1d) and M3-9M (Fig 1l) tumor-bearing mice compared to control mice. These findings suggest that perivascular cells in pre-metastatic lung exhibited enhanced proliferation and migration either from existing lung vasculature or other unappreciated sites, which are hallmarks of perivascular phenotypic switching. This increase in phenotypically modulated perivascular cells was specific to pre-metastatic lung tissue in that a similar expansion of perivascular cells was not seen in other tissues of tumor-bearing mice such as the liver (Supplementary Fig. 3), to which the B16-F10 and M3-9M models rarely metastasize, implying that this process is associated with site-specificity of metastasis.
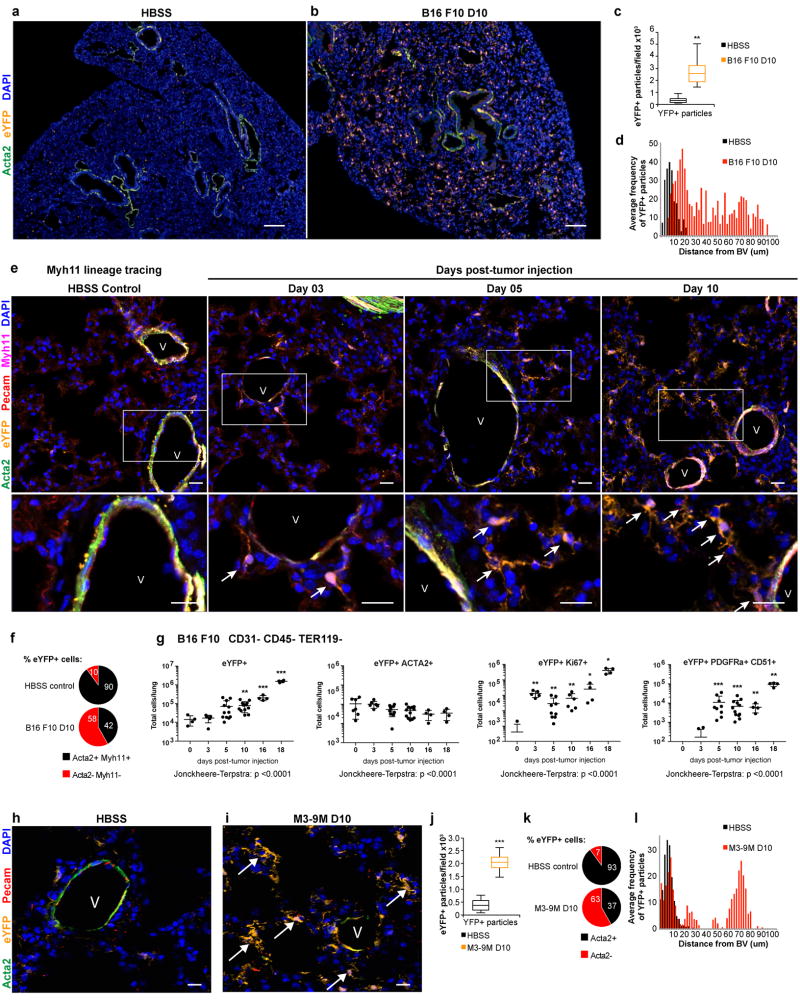
Figure 1. Perivascular phenotypic switching is activated in pre-metastatic lung.
(a-b) The pre-metastatic lungs of B16-F10 melanoma tumor-bearing Myh11 lineage-tracing mice (b), at day 10 post-tumor injection, exhibit an expansion of eYFP-expressing lineage-traced perivascular cells (eYFP, in orange) that can be found throughout the parenchyma, in contrast to the lungs of non-tumor bearing mice (a) where eYFP-expressing lineage-traced cells are found only around blood vessels and surrounding airways, and express the perivascular marker smooth muscle alpha-actin (ACTA2, in green). DAPI nuclear stain is shown in blue. Bars = 50 µm.
(c) Computational analysis quantifying DAPI+ eYFP+ particles in immunofluorescent images from lungs of B16-F10 tumor-bearing Myh11 lineage-tracing mice at day 10 post tumor injection and litter-mate HBSS-injected Myh11 lineage-tracing mice, that demonstrate an increase in the number of eYFP+ DAPI+ particles per 20× field in tumor bearing compared to HBSS treated mice. n = 8 mice per treatment group. **P < 0.001 , Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(d) Computational analysis quantifying the proximity of DAPI+ eYFP+ particles to the nearest blood vessel (BV) in immunofluorescent images from lungs of B16-F10 tumor-bearing Myh11 lineage-tracing mice at day 10 post tumor injection compared to litter-mate HBSS-injected control Myh11 lineage-tracing mice, that demonstrates an increase in the average frequency of DAPI+ eYFP+ particles at longer distances from the nearest blood vessel in pre-metastatic lungs, compared to lungs from litter-mate HBSS-injected controls. n = 8 mice per treatment group. ***P < 0.0001, ANOVA.
(e) A time-course of pre-metastatic lungs from B16-F10 melanoma tumor-bearing Myh11 lineage-tracing mice, from days 3, 5, and 10 post-tumor injection, compared to non-tumor-bearing HBSS-injected control Myh11-lineage-tracing mice. eYFP+ (orange), MYH11+ (magenta) and ACTA2+ (green) cells around blood vessels in HBSS Myh11 lineage-tracing mice. eYFP+ (orange) MYH11+ (magenta) cells that are not ACTA2+ (green) near PECAM+ (red) blood vessels (V) were observed at day 3, while the appearance of eYFP+ MYH11- cells farther away from blood vessels (V) increased over time (days 5–10), and the accumulation of eYFP+ MYH11- cells at even farther distances from blood vessels was marked by day 10. Arrows designate phenotypically switched eYFP+ perivascular cells. Insets are shown below each time-point. Bar = 20 µm.
(f) Quantification from confocal images of the percent of lineage traced eYFP+ cells that were also MYH11+ and ACTA2+ per field, comparing the lungs of Myh11 lineage-tracing HBSS control mice (top) to those of pre-metastatic B16-F10 day 10 tumor-bearing mice (bottom). Mean depicted for n = 8 mice per treatment group; SD = 3.447 for HBSS; SD = 4.281 for B16-F10 day 10.
(g) Flow cytometric analysis of Myh11 lineage-tracing lungs over a time-course of eYFP+ cells in B16-F10 pre-metastatic lungs compared to lungs from litter-mate HBSS-injected controls (day 0). Left depicts total eYFP+ cells which are increased at days 10, 16, and 18 compared to day 0. Left-middle depicts eYFP+ ACTA2+ cells which decrease across time-points. Right-middle depicts eYFP+ Ki67+ cells which are significantly increased at all tumor-bearing time-points compared to day 0. Right depicts eYFP+ PDGFRa+ CD51+ cells which increase on day 5, 10, 16 and 18, compared to day 0. Sample sizes (and p-value vs day 0 by ANOVA) for far left and far right panels are as follows: day 0 n = 4; day 3 n = 5; day 5 n = 12; day 10 n = 12; day 16 n = 4; day 18 n = 4. Sample sizes for middle left panel is as follows: day 0 n = 7; day 3 n = 5; day 5 n = 9; day 10 n = 12; day 16 n = 4; day 18 n = 4. Sample sizes for middle right panel is as follows: day 0 n = 7; day 3 n = 5; day 5 n = 9; day 10 n = 6; day 16 n = 4; day 18 n = 4. *P < 0.05, **P < 0.001, ***P < 0.0001, and ****P < 0.00001, ANOVA. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
(h-i) Perivascular phenotypic switching is observed in an additional pre-metastatic model, the M3-9M metastatic rhabdomyosarcoma model. Compared to the lungs of non-tumor-bearing Myh11 lineage-tracing mice (h), pre-metastatic tumor-bearing Myh11 lineage-tracing mice (i) at day 10 post-tumor injection exhibit an increase in the number of eYFP-expressing lineage-traced cells (orange), designated by arrows, that are no longer found around blood vessel (V) components such as endothelial cells labeled by PECAM1 (red) and vascular smooth muscle cells labeled by ACTA2 (green). Bar = 20 µm.
(j) Computational analysis quantifying DAPI+ eYFP+ particles in immunofluorescent images from pre-metastatic lungs of M3-9M rhabdomyosarcoma tumor-bearing Myh11 lineage-tracing mice at day 10 post-tumor-injection, that demonstrate an increase in the number of eYFP+ DAPI+ particles per 4×3 tiled fields compared to HBSS treated Myh11 lineage-tracing mice. n = 7 HBSS; n = 8 M3-9M. ***P < 0.0001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(k) Quantification from confocal images of the percent of lineage traced eYFP+ cells that were ACTA2+ per field, in the lungs of Myh11 lineage-tracing HBSS control mice (top) compared to those in the lungs of pre-metastatic M3-9M rhabdomyosarcoma day 10 tumor-bearing mice (bottom). Mean depicted for n = 8 mice per treatment group; SD = 2.325 for HBSS; SD = 1.613 for M3-9M day 10.
(l) Computational analysis quantifying the proximity of DAPI+ eYFP+ particles to the nearest blood vessel (BV) in immunofluorescent images from lungs of M3-9M tumor-bearing mice at day 10 post-tumor injected compared to litter-mate HBSS-injected controls in Myh11 lineage-tracing mice, that demonstrates an increase in the average frequency of DAPI+ eYFP+ particles at longer distances from the nearest blood vessel in pre-metastatic lungs, compared to lungs from litter-mate HBSS-injected controls. n = 8 mice per treatment group.
Using flow cytometric analysis, we confirmed and quantified the expansion of eYFP+ cells in pre-metastatic lungs of B16-F10 tumor-bearing mice over a range of time-points (Fig 1g, Supplementary Fig. 4a, b). Pathology review, IVIS imaging, and flow cytometry were utilized to assay for dual luciferase+ mCherry+ tumor cells and confirm that days 5 through 16 post-tumor injection were below the level of detection for metastasis (Supplementary Fig 1f, g). A significant 4.39-fold increase in total eYFP+ cell number was observed in the pre-metastatic lungs of B16-F10 tumor-bearing Myh11 lineage-tracing mice at day 10 post-tumor injection and persisting to later time-points, compared to HBSS-injected non-tumor-bearing Myh11 lineage-tracing littermate control mice (Fig. 1g). A concurrent increase in eYFP+ Ki67+ cells was observed at all pre-metastatic time-points, including at day 3 post-tumor injection prior to the increase in eYFP+ cells at day 10 post-tumor injection. Ki67 marks cells in all stages of the cell cycle leading to cell division, thus this early time-point may represent the early transition from a quiescent to cycling eYFP+ cell population20. Together these data indicate that eYFP+ cells proliferate in pre-metastatic lung (Fig 1g, Supplementary Fig 4d). Similarly, a 3.2-fold increase in eYFP+ cells was observed in the pre-metastatic lungs of M3-9M tumor-bearing Myh11 lineage-tracing mice compared to HBSS-injected non-tumor-bearing Myh11 lineage-tracing littermate control mice (Supplementary Fig. 4c). A decrease in total eYFP+ ACTA2+ cells over time was observed in pre-metastatic lung of B16-F10 tumor-bearing Myh11 lineage-tracing mice (Fig 1g). These data were consistent with our analysis of immunofluorescently stained tissue sections of the similar but more specific eYFP+ ACTA2+ Myh11+ cell population, highlighting the complimentary nature of analysis by confocal imaging and flow cytometry to examine perivascular cells in pre-metastatic lung (Fig 1e, f, j, k). Together these data are indicative of marker down-regulation that is characteristic of perivascular phenotypic switching.
To further characterize perivascular lineage-tracing eYFP+ cells in our pre-metastatic models, we evaluated pericyte markers in eYFP+ cells in the Myh11 lineage-tracing model. We discovered many eYFP+ pericytes in the microcirculation of the heart, skeletal muscle and lung (Supplementary Fig. 5). A subset of eYFP+ cells within the microvasculature of non-tumor-bearing lungs of Myh11 lineage-tracing mice expressed the pericyte markers PDGFRβ and NG2 (Supplementary Fig. 5c). We identified eYFP+ cells dissociated from the microvasculature within pre-metastatic lungs of B16-F10 tumor-bearing Myh11 lineage-tracing mice (Supplementary Fig. 5c). Of note, Myh11 lineage traced eYFP+ cells in the pre-metastatic lungs of mice with B16-F10 tumors did not express macrophage markers F4/80 or LGALS3 as reported in atheromas17 (Supplementary Figure 4e). While PDGFRα+ cells are found in normal healthy lung, this total population was increased only at later pre-metastatic time-points (day 18) (Supplementary Fig 6a, c). By contrast, eYFP+ PDGFRα+ cells dramatically increased as early as day 10 post-tumor injection (Supplementary Fig. 6a), and we found that eYFP+ PDGFRα+ CD51+ cells increased throughout the time course in pre-metastatic lungs (Fig 1g). This was consistent with previous findings that identified the expansion of a PDGFRα+ fibroblast-like population in the pre-metastatic niche1. Together, these data demonstrate that pre-metastatic lungs contain an expanded population of eYFP+ perivascular cells that are not in close association with blood vessels and may contribute to PDGFRα+ activated stromal cell population previously described in the pre-metastatic niche.
NG2+ pericytes contribute to the activated perivascular population in the pre-metastatic niche
In addition to the stable eYFP labeling of vSMCs in the Myh11 lineage-tracing model, eYFP+ pericytes were observed in many tissues, including the microvasculature of the heart and spinotrapezius muscle (Supplementary Fig 5a-b). Further, eYFP+ cells in the lungs of non-tumor-bearing Myh11 lineage-tracing mice expressed the pericyte markers NG2 and PDGFRβ (Fig 2a, Supplementary Fig 5c), and the total number of eYFP+ NG2+ cells increases in pre-metastatic lungs over time (Fig 2b). Together, these data suggested that pericytes could substantially contribute to the expansion of Myh11 lineage-tracing eYFP+ cells observed in pre-metastatic lung. Given that NG2+ perivascular cells are a known key component of stem cell niches, particularly in the bone marrow where they support hematopoietic stem cell maintenance11 and fetal liver where they induce hematopoietic stem cell proliferation10, we sought to determine the contribution of the NG2-expressing pericyte population to the perivascular Myh11 eYFP+ population in pre-metastatic tissue.
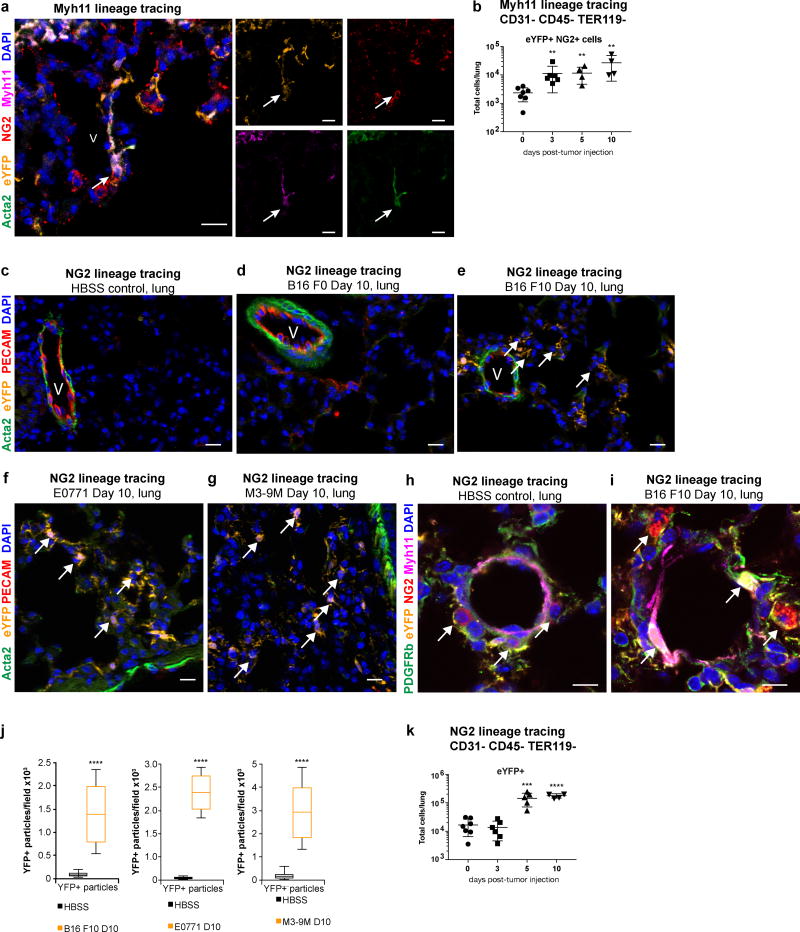
Figure 2. NG2-expressing pericytes are activated in pre-metastatic lung.
(a) eYFP+ cells (orange) in the lungs of Myh11 lineage-tracing mice express the pericyte marker NG2 (red) around blood vessels (V), and express the perivascular markers ACTA2 (green) and Myh11 (magenta). DAPI nuclear stain is shown in blue. 20× magnification confocal images with single channel fluorescence images to the right. Arrow depicts eYFP+ NG2+ MYH11+ ACTA2+ cell. Bar = 20 µm
(b) Flow cytometric analysis of Myh11 lineage-tracing lungs over a time-course of eYFP+ NG2+ cells in B16-F10 pre-metastatic lungs compared to lungs from litter-mate HBSS-injected controls (day 0), demonstrating a significant increase in eYFP+ NG2+ cells in pre-metastatic lungs of Myh11 lineage-tracing mice over time. n = 6 day 0; n = 6 day 3; n = 5 day 5; n = 5 day 10. **P < 0.001, ANOVA. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
(c-g) The pre-metastatic lungs of B16-F10 melanoma tumor-bearing NG2 lineage-tracing mice (e), at day 10 post-tumor injection, exhibit an expansion of eYFP-expressing lineage-traced pericytes (eYFP, orange) that can be found throughout the parenchyma away from the vasculature, in contrast to the lungs of litter-mate HBSS-injected controls (c), where eYFP-expressing lineage-traced cells are rare, found only around blood vessels (V) stained by PECAM (red), and express the perivascular marker smooth muscle alpha-actin (ACTA2, in green). The NG2 lineage-tracing mice injected with the lowly metastatic B16-F0 melanoma cell line does not exhibit appreciable eYFP+ cell expansion into the lung parenchyma (d). The pre-metastatic lungs of E0771 tumor-bearing mice (f) and M3-9M tumor-bearing mice (g) exhibit an expansion of eYFP+ cells. DAPI nuclear stain is shown in blue. Bar = 20 µm.
(h-i) Confocal imaging showing NG2+ (red) PDGFRb+ (green) MYH11+ (magenta) eYFP+ perivascular cells (orange) no longer in close association with the blood vessel in the lungs of B16-F10 melanoma tumor-bearing NG2 lineage-tracing mice (j) at day 10 post-tumor injection (arrows), a pre-metastatic time-point, in contrast to NG2+ PDGFRb+ MYH11+ eYFP-expressing cells, which are found around blood vessels in HBSS-injected litter-mate controls. DAPI nuclear stain is shown in blue. Bar = 10 µm.
(j) Computational analysis quantifying DAPI+ eYFP+ particles in immunofluorescent images from lungs of three pre-metastatic models at day 10 post-tumor-injected NG2 lineage-tracing mice, that demonstrate an increase in the number of eYFP+ DAPI+ particles per 4×3 tiled image field compared to HBSS-injected NG2 lineage-tracing mice. Left depicts analysis of pre-metastatic lungs from B16 F10 metastatic melanoma tumor-bearing mice compared to litter-mate HBSS-injected controls (n = 8 mice per treatment group). Middle depicts analysis of pre-metastatic lungs from E0771 metastatic breast carcinoma tumor-bearing mice compared to litter-mate HBSS-injected controls (HBSS n = 9, E0771 n = 8). Right depicts analysis of pre-metastatic lungs from M3-9M metastatic rhabdomyosarcoma tumor-bearing mice compared to non-tumor-bearing litter-mate HBSS-injected controls (HBSS n = 6; M3-9M n = 7. ****P < 0.00001, Student’s t-test. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
(k) Flow cytometric analysis of NG2 lineage-tracing mice at days 3, 5 and 10 post-tumor injection with the metastatic B16 F10 cell line demonstrates an increase in the total number of eYFP+ pericyte-derived cells found in pre-metastatic lungs compared to non-tumor-bearing control HBSS injected mice. day 0 n = 6; day 3 n = 5; day 5 n = 5; day 10 n = 6. ***P < 0.0001, ****P < 0.00001, ANOVA. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
To elucidate whether pericytes in particular participate in pre-metastatic niche formation, we generated conditional NG2-ERT-creT2 ROSA-STOP-flox-eYFP lineage-tracing mice (designated as “NG2 lineage-tracing mice”) (Supplementary Fig. 7a)21. Upon tamoxifen injection in adult mice, NG2-expressing pericytes are stably labeled by eYFP in many tissues (Supplementary Fig 7b, c). Using this NG2 lineage-tracing model, we observed an expansion of NG2 lineage-tracing eYFP+ cells in the lungs of B16-F10, M3-9M, and E0771 tumor-bearing mice that was comparable to the expanded perivascular population identified in the tumor bearing Myh11 lineage-tracing mice, suggesting that NG2+ pericytes represent an expanded perivascular population in the pre-metastatic setting (Fig. 2c-j, Supplementary Fig 7b). We confirmed and quantified this expansion by immunofluorescent image analysis and flow cytometric analysis (Fig. 2j-k, Supplementary Fig. 7d). eYFP+ cells were only rarely identified in the lungs of NG2 lineage-tracing mice that received non-metastatic melanoma B16-F0 tumors, suggesting that the increase in NG2 lineage-tracing eYFP+ cells correlated with metastatic potential (Fig. 2d). Together, the data reveal that perivascular cells, including NG2-expressing pericytes, exhibit expansion in the parenchyma of pre-metastatic lungs in a number of murine models of metastasis.
Tumor-derived factors induce KLF4 expression in perivascular cells of the pre-metastatic niche
Perivascular cell phenotypic switching is dependent on the expression of KLF4, Krüppel-like factor 4, in several disease models7,8,14,16,17,22 (Fig. 3a). Direct KLF4 binding onto the promoters of vSMC marker genes results in down-regulation of marker gene expression and a transition from a contractile SMC phenotype to a plastic, reactive state17,22,23. We observed an increase in the percentage of eYFP+ cells that were KLF4+ in the pre-metastatic lungs of B16-F10 tumor-bearing Myh11 lineage-tracing mice at day 10 post tumor injection compared to HBSS-injected Myh11 lineage-tracing littermate control mice (Fig 3b-e). As such, we sought to determine whether tumor-derived factors might induce KLF4 expression in perivascular cells that, in turn, mediates perivascular cell plasticity and phenotypic switching.
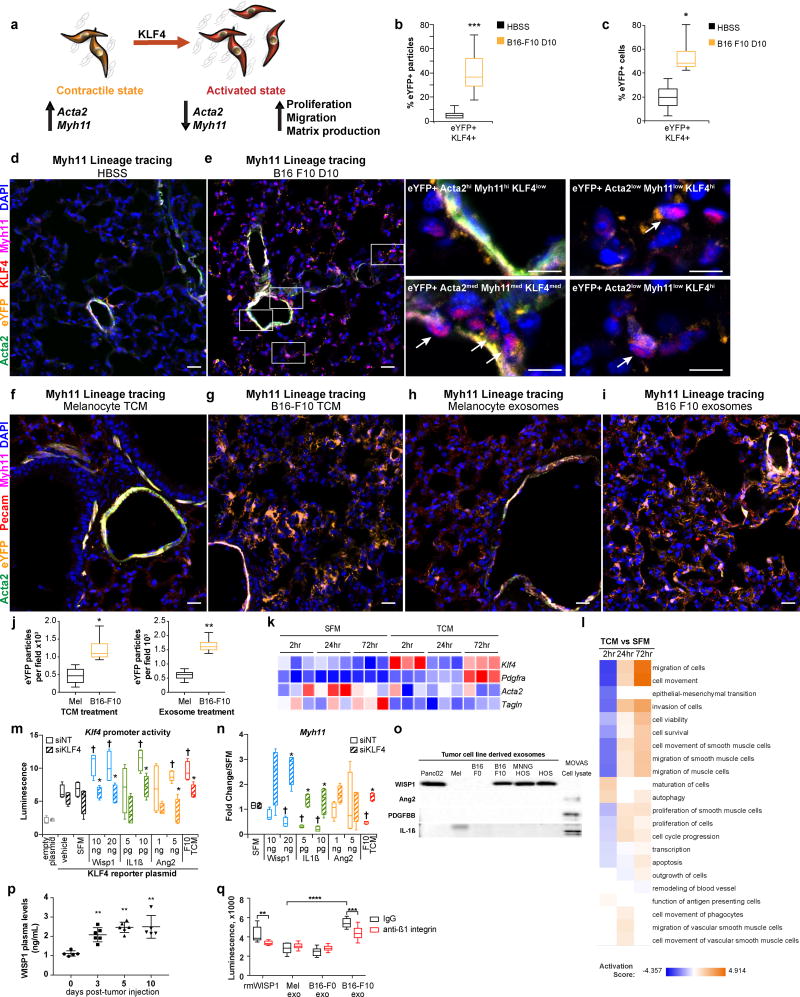
Figure 3. Metastatic tumor-derived factors promote activation of KLF4-dependent perivascular cell phenotypic switching.
(a) A schematic describing the hallmarks of perivascular cell phenotypic switching. Smooth muscle cells and pericytes in their contractile state are found surrounding blood vessels, where they express marker genes such as ACTA2 and MYH11. In response to inflammatory microenvironmental cues, perivascular cells express KLF4 and enter a synthetic state where they down-regulate expression of marker genes, and increase proliferation, migration, and ECM production.
(b) Computational analysis quantifying the percentage of DAPI+ eYFP+ particles that were KLF4+ in immunofluorescent images from pre-metastatic lungs at day 10 post-B16 F10 tumor-injection in Myh11 lineage-tracing mice, that demonstrate an increase in the number of eYFP+ DAPI+ particles that were KLF4+ compared to HBSS-injected control Myh11 lineage-tracing mice. n = 8 per treatment group. ***P < 0.0001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(c) Flow cytometric analysis of Myh11 lineage-tracing lungs from B16 F10 tumor injected mice at day 10 post-tumor injection compared to lungs from litter-mate HBSS-injected Myh11 lineage-tracing controls demonstrates a significant increase in the proportion of eYFP+ cells that are also KLF4+ found in pre-metastatic lungs. n = 9 mice in each group. *P < 0.05, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(d-e) In contrast to HBSS-injected control Myh11 lineage-tracing mice (d), the pre-metastatic lungs of B16-F10 melanoma tumor-bearing Myh11 lineage-tracing mice (e), at day 10 post-tumor injection, contain a subset of eYFP+ lineage-traced perivascular cells (eYFP, in orange) that express the pluripotency marker KLF4 (red), are no longer restricted to the perivascular location, and begin to lose expression of the perivascular markers ACTA2 (green) and Myh11 (magenta). Zoomed insets of eYFP+ populations in pre-metastatic lung shown at right. These eYFP+ ACTA2low Myh11low KLF4hi perivascular cells denoted by arrows represent a hallmark of phenotypic switching. Background subtracted MFI was used to designate low-med-hi designations: ACTA2 low <= 100, 100 < med < 1000, 1000 <= hi; MYH11 low <= 200, 200 < med < 1000, 1000 <= hi; KLF4 low <= 250, 250 < med < 800, 1000 <= hi. DAPI nuclear stain is shown in blue. Bars = 20 µm, inset bars = 10 µm.
(f-g) The lungs of non-tumor-bearing Myh11 lineage-tracing mice exposed to metastatic B16-F10 tumor conditioned media (TCM) (g) exhibit an expansion of eYFP+ lineage-traced perivascular cells (eYFP, in orange), similar to that found in pre-metastatic lungs, in contrast to the lungs of non-tumor bearing mice exposed to non-malignant melanocyte TCM (f) where eYFP+ lineage-traced cells are found around PECAM+ blood vessels (red) and surrounding airways, and express the perivascular markers ACTA2 (green) and Myh11 (magenta). DAPI nuclear stain is shown in blue. Myh11 lineage-tracing mice were i.p. injected with TCM from the metastatic B16 F10 or non-tumor melanoma cell lines once daily 6 times over 14 days. Bars = 20 µm.
(h-i) The lungs of non-tumor-bearing Myh11 lineage-tracing mice exposed to metastatic B16-F10 tumor-derived exosomes (i) exhibit an expansion of eYFP+ lineage-traced perivascular cells (eYFP, in orange), similar to that found in pre-metastatic lungs, in contrast to the lungs of non-tumor bearing mice exposed to non-malignant melanocyte exosomes (h) where eYFP+ lineage-traced cells are found only around PECAM+ blood vessels (red) and surrounding airways, and express the perivascular markers ACTA2 (green) and MYH11 (magenta). DAPI nuclear stain is shown in blue. Myh11 lineage-tracing mice were i.p. injected with exosomes isolated from the conditioned media of the metastatic B16 F10 and non-tumor melanoma cell lines once daily 6 times over 14 days. Bars = 20 µm.
(j) Computational analysis quantifying DAPI+ eYFP+ particles in immunofluorescent images from lungs of Myh11 lineage-tracing mice that were i.p. injected with either TCM (left panel) or exosomes (right panel) from non-tumor melanocyte or metastatic tumor B16 F10 cell lines, that demonstrate an increase in the number of eYFP+ DAPI+ particles per 20× field in B16 F10 TCM or exosome treated mice compared to melanocyte TCM or exosome treated mice.
Each measurement represents five stained fields per mouse. n = 3 mice per treatment group. *P < 0.05, **P < 0.001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(k) Heatmap of RNA-Seq analysis of vSMCs cultured in vitro demonstrating up-regulated gene expression of the activation markers Klf4 and Pdgfra and down-regulation of perivascular markers Acta2, Myh11 and Tagln in response to metastatic B16-F10 conditioned media (F10 TCM), as compared to serum free media (SFM) cultured cells, where red indicates higher expression and blue indicates lower expression.
(l) Ingenuity Pathway Analysis of highly enriched pathways (positive activation score) in the gene expression data collected from vSMCs cultured in vitro with B16-F10 TCM compared to SFM, demonstrating activation of proliferation and migration pathways (orange) over 72 hours.
(m) vSMCs cultured in vitro were transfected with a Klf4 promoter reporter plasmid that upon activation of the Klf4 promoter reporter induces expression of secretable Gaussia luciferase. Transfected vSMCs were then subjected to siRNA targeting with either Klf4-targeting (siKLF4) or a non-targeting control sequence (siNT). 24 hours after transfection, media was replaced with serum-free media containing one of the following recombinant proteins: WISP1, Ang2, or IL1ß. 16 hours after treatment with recombinant proteins, secreted luciferase levels were quantified, and demonstrated that multiple recombinant proteins activate Klf4 promoter activity in vSMCs in vitro. *P < 0.05 indicates comparison between siNT and siKLF4, Student’s t-test; †P < 0.05 indicates comparison between SFM and cytokine treatment, Student’s t-test. n = 3 independent experiments. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(n) vSMCs cultured in vitro were subjected to siRNA targeting with either Klf4-targeting (siKLF4) or a non-targeting control sequence (siNT). 24 hours after transfection, media was replaced with serum-free media containing one of the following recombinant proteins: WISP1, ANG2, or IL1ß. 24 hours after treatment with recombinant proteins, gene expression was profiled by qRT-PCR to demonstrate that vSMC cultured with any of the three recombinant proteins decreases expression of perivascular marker gene Myh11 in a KLF4-dependent manner. Gene expression expressed as fold change over serum free media control for either siNT or siKLF4 treatment. *P < 0.05 indicates comparison between siNT and siKLF4, Student’s t-test; †P < 0.05 indicates comparison between SFM and cytokine treatment, Student’s t-test. n = 3 independent experiments. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(o) Western blotting of tumor cell line derived exosomes demonstrating that of the three candidate cytokines, only WISP1 detected in exosomes. Significantly, WISP1 was only found in the exosomes of metastatic tumor cell lines, including Panc02 pancreatic adenocarcinoma, B16-F10 melanoma, MMNG HOS osteosarcoma, and HOS osteosarcoma. WISP1 was not found in the non-tumor control melanoma cell line (Mel), nor non-metastatic tumor cell line B16-F0 exosomes. vSMC cell lysate (MOVAS) used as positive control for cytokine staining (far right).
(p) WISP1 levels increase in the plasma of pre-metastatic tumor-bearing mice over time, as measured by ELISA. Sample sizes are as follows: day 0 n = 5; day 3 n = 6; day 5 n = 6; day 10 n = 5. **P < 0.001, ANOVA. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
(q) Metastatic B16-F10 exosomes or recombinant WISP1 (rmWISP1) containing serum-free media that was pre-treated with neutralizing anti-ß1 integrin antibodies (red) prior to applying onto vSMCs in vitro exhibit a decreased ability to promote vSMC proliferation compared to B16-F10 exosomes or rmWISP1 pre-treated with IgG control antibodies (black). Control melanocyte exosomes or non-metastatic B16-F0 tumor exosomes do not induce vSMC proliferation, and show no change with ß1 integrin inhibition. Proliferation measured using CellTiter Glo. n = 10 technical replicates. **P < 0.001, ***P < 0.0001, ****P < 0.00001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
Previous investigations have demonstrated that in vivo administration of tumor-derived factors such as tumor-conditioned media (TCM) or tumor-derived exosomes potentiates pre-metastatic niche formation1,24–26. To determine whether perivascular cell activation in the pre-metastatic niche is initiated by tumor-derived factors, we intraperitoneally injected non-tumor-bearing Myh11-lineage-tracing mice to tumor-derived factors such as TCM or exosomes and assayed for phenotypic switching by analysis of Myh11 lineage-tracing eYFP+ cells that no longer express MYH11. eYFP+ MYH11- ACTA2- cells were increased in the lungs of Myh11 lineage-tracing mice exposed to tumor-derived factors that were no longer in close association with blood vessels, nor could be identified by perivascular marker expression, as was seen in tumor-bearing mice at pre-metastatic time points and indicative of their activated state (Fig 3f-j). Pre-conditioning of mice with B16-F10 exosomes or TCM followed by tail-vein injection of B16-F10 melanoma cells resulted in increased metastasis, indicating that pre-treatment with tumor-derived factors is sufficient for inducing perivascular cell plasticity and establishing a pre-metastatic niche that enhances tumor cell colonization and/or proliferation (Supplementary Fig 8a). These studies indicate that tumor-derived factors contribute to perivascular cell phenotypic switching and pre-metastatic niche formation.
The activated state of perivascular cells is characterized by a decrease in marker gene expression and an increase in proliferation, migration, and matrix production (Fig. 3a). To test whether tumor-derived factors induce KLF4-dependent perivascular phenotypic switching, we sought to establish an in vitro model by culturing vSMCs with either B16-F10 metastatic melanoma TCM or exosomes purified from B16-F10 TCM. qRT-PCR profiling of vSMCs exposed to TCM over the course of 72 hours revealed an increase in Klf4 gene expression as early as 2 hours post-treatment with TCM to levels similar to those seen by vSMCs treated with the cytokine PDGF-BB (Supplementary Fig 8b), a known inducer of KLF4-dependent phenotypic switching23,27. We observed a subsequent decrease in expression of vSMC marker genes, including Acta2, Myh11, and Tagln (Supplementary Fig. 8b). Notably, TCM derived from the non-metastatic melanoma B16-F0 cell line do not induce these gene expression changes, indicating that tumor factor-induced vSMCs phenotypic switching may be specific to tumor metastatic potential (Supplementary Fig 8b).
We extended these gene expression data by performing RNA-seq on cultured vSMCs exposed to TCM over the course of 72 hours. We confirmed that Klf4 gene expression is elevated in vSMCs treated with B16-F10 TCM, and that the expression of vSMC marker genes Acta2 and Tagln were decreased compared to vSMCs treated with serum-free media (Fig 3k). Pathway analysis demonstrated that gene sets related to proliferation, migration, and invasion, all hallmarks of perivascular phenotypic switching, were activated over the course of 72 hours (Fig 3l). We confirmed an increase in proliferation by vSMCs cultured in B16-F10 TCM (Supplementary Fig. 8d), which was indicative of their activated, phenotypically-modulated state. Consistent with our finding that eYFP+ PDGFRα+ cells were increased in the pre-metastatic lungs of B16-F10 tumor-bearing mice (Fig 1g, Supplementary Fig 6b-c), vSMCs that were cultured in vitro with TCM upregulated this activated stromal marker (Fig. 3k, Supplementary Fig 8b-c), suggesting that phenotypically-modulated vSMCs may represent a PDGFRα+ cell population that was previously reported in pre-metastatic tissue in response to tumor-derived factors1.
TCM from the metastatic B16-F10 cell line, but not from the non-metastatic B16-F0, is sufficient to induce vSMC phenotypic switching in vitro (Supplementary Fig 8b-c), indicating that factors secreted specifically by tumor cells with metastatic potential contribute to the activated vSMC phenotype. As such, we sought to determine which cytokines were elevated in the conditioned media from the metastatic B16-F10 cell line when compared to the non-metastatic B16-F0 cell line using a cytokine array (Supplemental Table 1). Of the 8 factors found to be elevated in this assay, WISP1, ANG2 and IL-1ß all have known roles in vascular biology and inflammation, and in some reports can induce KLF4 expression28–33. Using a Klf4 promoter reporter that results in the secretion of luciferase upon Klf4 promoter activity, combined with qRT-PCR, we determined that all three cytokines were capable of inducing Klf4 expression in vSMCs to levels comparable to that induced by B16-F10 TCM, and siKlf4 knock-down significantly inhibited this activation (Fig 3m). WISP1 and IL1ß, but not ANG2, induced KLF4-dependent suppression of vSMC marker gene expression (Fig 3n). However, ANG2 neutralizing antibody pre-treatment of vSMCs exposed to B16-F10 conditioned media blocked TCM-induced vSMC proliferation (Extended Fig 8e), indicating that ANG2-induced effects on vSMC activation may work coordinately through KLF4 and other effector proteins.
In addition to B16-F10 TCM, B16-F10 tumor cell-derived exosomes were also capable of inducing perivascular phenotypic switching in vivo (Fig 3h-j), and vSMC phenotypic switching in vitro (Supplementary Fig 8c). Isolated melanocyte-, B16-F0- and B16-F10-derived exosomes were found to be of similar size distribution, at a peak of 133 nm, 85 nm and 99 nm, respectively (Supplementary Fig. 8g) and contained exosomal and extracellular vesicle markers (Supplementary Fig 8h). Fluorescently labelled B16-F0 and B16-F10 exosomes were found capable of entering vSMCs (Supplementary Fig 8i). We thus determined whether any of the three candidate cytokines were contained in metastatic tumor-derived exosomes. We found that only WISP-1, and not IL1ß nor Ang2, was found in metastatic tumor cell derived exosomes including in B16-F10 melanoma, MNNG HOS and HOS osteosarcoma, and Panc02 pancreatic cell lines. WISP-1 was not found in exosomes derived from the non-metastatic B16-F0 cell line nor a non-tumor melanocyte cell line, suggesting that the presence of exosomal WISP1 production may be specific to metastatic tumor cell lines that in combination with non-exosomal cytokines such as IL1ß and Ang2 may enhance tumor metastatic behavior (Fig 3o, Supplementary Fig 8f, Supplementary Fig 9a, d). Circulating WISP-1 was also found to be elevated in B16-F10 tumor-bearing mice at pre-metastatic time-points (Fig 3p) to levels equivalent to that found in B16-F10 exosomes generated in vitro (Supplementary Fig 8f) and administered in vivo to promote perivascular phenotypic switching (Fig 3h-j), whereby this cytokine may contribute to pre-metastatic niche formation (Supplementary Fig 8a). Neutralizing anti-β1 integrin antibodies that are known to interfere with WISP1 activity inhibited both B16-F10 exosome- and rWISP1-induced vSMC proliferation in vitro (Fig 3q). Together, the data suggest that multiple factors secreted by the metastatic melanoma B16-F10 cell line, including but not limited to WISP-1 containing-tumor-derived exosomes, may work in concert through the induction of KLF4 expression to contribute to vSMC phenotypic switching characterized by downregulation of vSMC marker gene expression and increased vSMC proliferation.
Tumor-derived factors induce perivascular phenotypic switching and enhance ECM that supports tumor metastatic behavior
Phenotypically-modulated perivascular cells increase production of ECM proteins, such as fibronectin and various collagens, and ECM-remodeling enzymes, such as matrix metalloproteinases in many disease settings14,34. ECM modulation can dictate tumor cell fate and metastasis in a number of models35–38. As has been demonstrated previously1, we observed an increase in fibronectin staining in the lungs of B16-F10 tumor-bearing Myh11 lineage-tracing mice at day 10 post-tumor injection as compared to HBSS-treated littermate control mice (Fig 4a-b). Elevated fibronectin deposition in pre-metastatic lung was found more closely associated with eYFP+ cells than with eYFP- cells within the same immunofluorescently stained tissue sections (Fig 4c). eYFP+ aortic vSMCs from Myh11 lineage-tracing mice that were isolated and cultured ex vivo in either B16-F10 TCM or tumor-derived exosomes also exhibited an increase in fibronectin production when compared to those cultured in defined serum-free media (Fig 4d-e). We next used an in vitro model to test whether the matrix produced by phenotypically-modulated vSMCs provides a pro-metastatic environment and enhances metastatic behavior in tumor cells. vSMC phenotypic switching was induced in vitro by treatment with either B16-F10 TCM or tumor-derived exosomes for 72 hours, after which vSMCs were lysed from culture plates to leave behind vSMC-derived ECM (Fig 4f-g). B16-F10 tumor cells seeded onto plates containing ECM produced by phenotypically-modulated vSMCs with enhanced fibronectin (Fig 4g) resulted in increased B16-F10 tumor cell adhesion and proliferation compared to those B16-F10 tumor cells which were seeded on plates containing ECM produced by non-phenotypically modulated vSMCs (Fig. 4h, j). B16-F10 tumor cells increased migration across a transwell membrane towards ECM components derived from phenotypically-switched vSMCs educated by B16-F10 exosomes, B16-F10 TCM, or rWISP1, compared to ECM components derived from vSMCs conditioned with B16-F0 exosomes or melanocyte exosomes (Fig 4i). Fibronectin production by vSMCs cultured in B16-F10 TCM was KLF4-dependent in that siRNA-mediated knock-down of Klf4 resulted in a reduction of B16-F10 TCM-induced fibronectin production (Fig 4k, Supplementary Fig 9b), suggesting that activated vSMCs contribute to a pro-metastatic environment, at least in part, through KLF4-dependent fibronectin production. Indeed, B16-F10 tumor cells that are cultured on the ECM derived from phenotypically-switched vSMCs demonstrate an enhanced ability to form tumorspheres (Supplementary Fig 8j), suggesting that phenotypically-modulated vSMC-derived ECM production in response to tumor-derived factors promotes a stem-like environment that promotes features of metastasis-initiating tumor cells39,40.
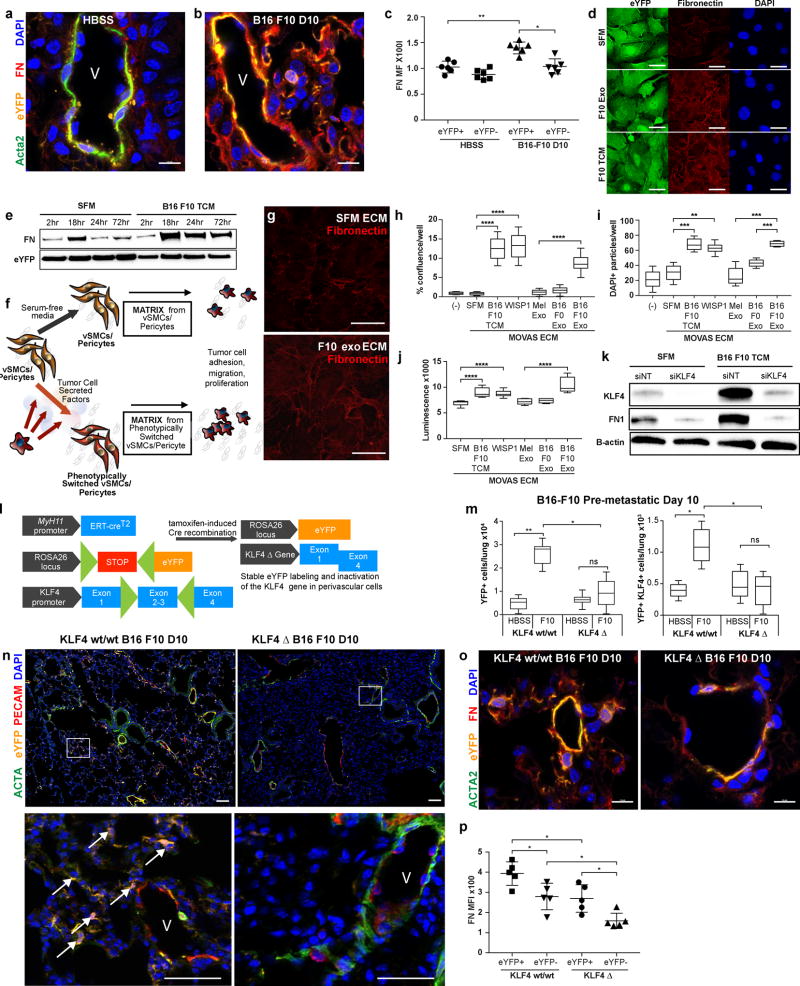
Figure 4. Activated smooth muscle cells secrete KLF4-dependent fibronectin-containing ECM that promotes tumor adhesion, migration and proliferation.
(a-b) An increase in fibronectin staining (red) is observed in Myh11 lineage-tracing pre-metastatic lungs (b) as compared to non-tumor-bearing HBSS injected litter-mate control mice (a). eYFP+ cells shown in orange and Acta2 staining shown in green. V denotes blood vessels. Bars = 10 µm.
(c) Background subtracted mean fluorescent intensity (MFI) of eYFP+ cells and eYFP- cells in HBSS control and B16-F10 pre-metastatic lung confocal images, as measured by two independent scorers. Each measurement represents five fields of view each from three stained tissue sections per mouse. n = 6 mice per treatment group. *P < 0.05, **P < 0.001, Student’s t-test. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
(d) eYFP+ lineage-tracing vSMCs cultured ex vivo in B16-F10 tumor-derived exosomes (F10 exo) or tumor conditioned media (F10 TCM) exhibit increased fibronectin staining (red). Bars = 10 µm.
(e) Western blotting of eYFP+ lineage-tracing vSMCs cultured ex vivo in B16-F10 tumor conditioned media (TCM) over the course of 72 hours exhibit an increase in fibronectin production, compared to eYFP+ cells cultured in defined serum-free media (SFM).
(f) Schematic describing vSMC-derived ECM experiments. vSMCs were cultured in vitro for 72 hours in either tumor-derived factors such as tumor conditioned media or tumor-derived exosomes, or serum free media as a control. The vSMCs were then lysed, resulting in culture dishes coated in vSMC-secreted ECM, and used for subsequent analysis of tumor cell behavior.
(g) The ECM of vSMCs cultured in B16 F10 exosomes, prepared as described in (f), exhibits an increase in fibronectin deposition as compared to ECM derived from vSMCs cultured in SFM. Bars = 10 µm.
(h) B16 F10 tumor cells were plated on ECM-coated plates prepared as described in (f), and then removed after 20 minutes and replaced with fresh media. The remaining tumor cells that were adhered to the plates were imaged via phase-contrast microscopy using an Incucyte Zoom, and percent confluence per well was measured using the associated software to demonstrate an increase in the number of tumor cells that adhered to the ECM of activated vSMCs cultured with B16 F10 TCM or B16 F10 exosomes (B16 F10 exo) compared to those that adhered to the ECM of vSMCs cultured with SFM or control melanocyte exosomes (Mel exo). Representative experiment of 2 independent experiments shown. ****P < 0.00001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(i) B16 F10 tumor cells were plated on transwell membranes placed on top of ECM-coated wells prepared as described in (f), and then fixed and stained for DAPI+ nuclei after 8 hours. The number of cells that migrated to the opposite side of the membrane were imaged via microscopy and quantified to demonstrate an increase in the number of tumor cells that migrated towards the ECM of activated vSMCs cultured with B16 F10 TCM or B16 F10 exosomes (B16 F10 exo) compared to those that migrated towards the ECM of vSMCs cultured with SFM or control melanocyte exosomes (Mel exo). Representative experiment of 2 independent experiments shown. **P < 0.001 ***P < 0.0001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(j) B16 F10 tumor cells were plated on ECM-coated plates prepared as described in (f), and then measured for proliferation using CellTiter Glo after 24 hours. B16-F10 tumor cells proliferated more when plated on the ECM of activated vSMCs cultured with B16 F10 TCM or B16 F10 exosomes (B16 F10 exo) compared to those tumor cells that were plated on the ECM of vSMCs cultured with SFM or control melanocyte exosomes (Mel exo). Representative experiment of 2 independent experiments shown. ****P < 0.00001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(k) vSMCs cultured in vitro were subjected to siRNA targeting with either KLF4-targeting (siKLF4) or a non-targeting control sequence (siNT). 24 hours after transfection, media was replaced with either serum-free media, or conditioned media of B16 F10 tumor cells (TCM). 48 hours after media treatment, protein was extracted and analyzed by western blot to demonstrate that tumor-derived factor induced expression of fibronectin (FN1) by vSMCs is KLF4-dependent.
(l) A schematic describing the Myh11 lineage-tracing KLF4 knock-out model. The Myh11 lineage-tracing model described in Fig 1a was crossed to the KLF4-flox mouse model such that cre-mediated recombination of the floxed codons of the KLF4 gene results in inactivated KLF4 alleles, in addition to stable eYFP labeling of mature vSMCs and pericytes. This resultant mouse model post-tamoxifen treatment is designated the KLF4 Δ mouse.
(m) Flow cytometric analysis of pre-metastatic lungs from KLF4 Δ tumor-bearing mice contain fewer total eYFP+ cells (left panel) and fewer eYFP+ KLF4+ cells (right panel) compared to tumor-bearing KLF4 wt/wt litter-mate controls at day 10 post-injection with metastatic melanoma B16-F10 by flow cytometry quantification. n = 5 per group. n.s. = not significant, *P < 0.05, **P < 0.001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(n) The pre-metastatic lungs of B16-F10 melanoma tumor-bearing Myh11 lineage-tracing KLF4 Δ mice, at day 10 post-tumor injection, demonstrate that the expansion of eYFP+ lineage-traced perivascular cells (eYFP, in orange) that is observed in KLF4 wild-type (wt/wt) pre-metastatic lung is reduced in the lungs of KLF4 Δ mice, where eYFP+ lineage-traced cells are found only around blood vessels and surrounding airways, and express the perivascular marker smooth muscle alpha-actin (ACTA2, in green). DAPI nuclear stain is shown in blue. Extended field of view tile images shown with white rectangles in each respective image denoting zoomed areas shown below to demonstrate that while KLF4 wt/wt Myh11 lineage-tracing mice exhibit many eYFP+ cells (arrows) that are no longer associated with blood vessels (V) labeled by PECAM1 for endothelial cells (red) and alpha smooth muscle actin (ACTA2) for vascular smooth muscle cells (green), few such eYFP+ cells can be found in the pre-metastatic lungs of KLF4 Δ mice at day 10 post-B16 F10 melanoma tumor injection. Bars = 50 µm, inset bars = 25 µm.
(o) The pre-metastatic lungs of B16 F10 melanoma tumor-bearing KLF4 Δ mice at 10 days post-tumor injection exhibit a decrease in fibronectin staining, as compared to lungs from tumor-bearing KLF4 wt/wt littermate controls at day 10 after B16 F10 melanoma injection. Bars = 10 µm.
(p) Background subtracted mean fluorescent intensity (MFI) of fibronectin staining in eYFP+ cells and eYFP- cells in confocal images of B16-F10 pre-metastatic lungs of KLF4 wild-type and KLF4Δ mice, as measured by two independent scorers. Each measurement represents five fields of view each from three stained tissue sections (15 total) per mouse. n = 5 mice per group. *P < 0.05, Student’s t-test. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
Klf4 knockout in perivascular cells limits phenotypic switching and pre-metastatic niche formation
To evaluate whether KLF4-dependent perivascular phenotypic switching is essential for pre-metastatic niche formation and metastasis, we crossed the perivascular cell-specific Myh11 lineage-tracing mice to Klf4 floxed (Klf4-flox) mice, effectively knocking out the Klf4 gene in all perivascular cells as previously described17. This Klf4-flox Myh11 lineage-tracing model enables simultaneous tamoxifen-inducible perivascular cell-specific Klf4 deletion (Klf4 Δ) and eYFP labeling prior to orthotopic tumor injection (Fig 4l). Despite the potential impact that perivascular phenotypic switching may have on primary tumor angiogenesis and tumor growth, we did not observe a significant difference in B16-F10 primary tumor diameters between Klf4 Δ and wild-type mice (Supplementary Fig.10a), nor involvement of eYFP+ perivascular cells in primary tumor architecture, with eYFP+ cells located only in normal adjacent tissue in full association with well-formed blood vessels (Supplementary Fig 10c). However, perivascular cell-specific Klf4 deletion resulted in a marked decrease in phenotypically-modulated eYFP+ ACTA2- cells in pre-metastatic lungs of B16-F10 tumor-bearing mice at day 10 post-tumor-injection when compared to the lungs of tumor-bearing Klf4 wild-type littermate control mice, as demonstrated by decreased numbers of phenotypically switched eYFP+ cells in the lung parenchyma by immunofluorescent staining and decreased numbers of eYFP+ cells and eYFP+ KLF4+ cells by flow cytometric analysis of whole lung (Fig. 4m-n). Consistent with a decrease in perivascular phenotypic switching with perivascular cell-specific Klf4 deletion, we observed decreased fibronectin in the lungs of B16-F10 tumor-bearing Klf4 Δ mice at day 10 post-tumor injection, compared to tumor-bearing Klf4 wild-type littermate control mice (Fig 4o-p), indicating that Klf4 expression in perivascular cells regulates fibronectin production during the process of pre-metastatic niche formation.
Perivascular cell-specific Klf4 deletion decreases early tumor cell colonization and metastatic burden
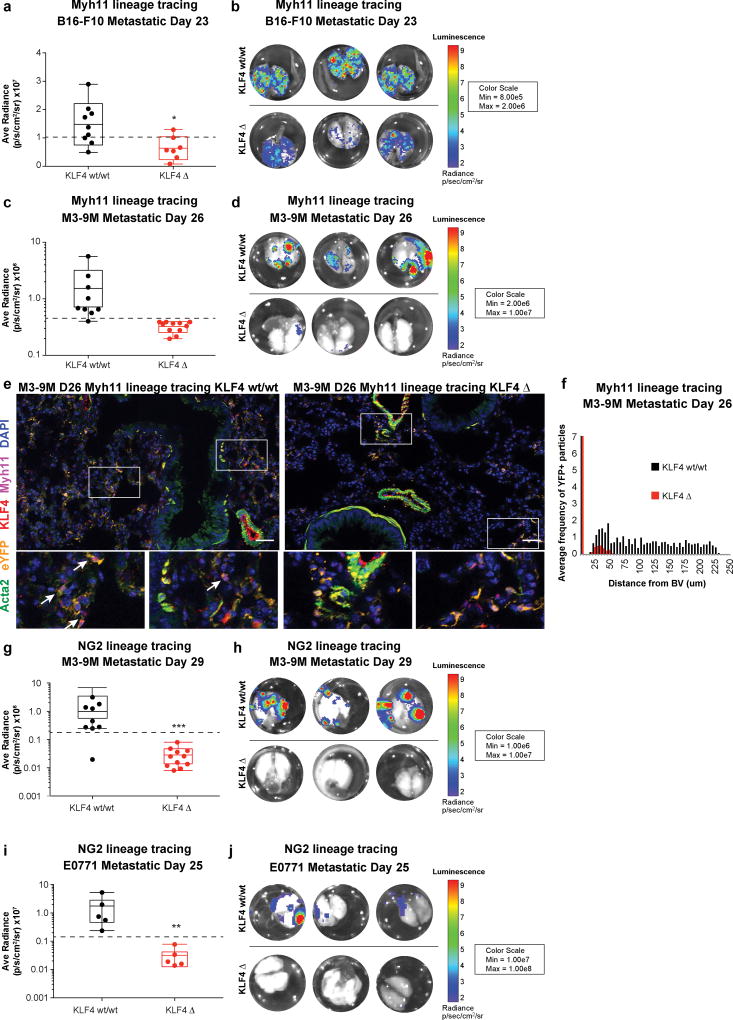
To determine the mechanistic role of KLF4-dependent perivascular mediated metastasis in early tumor cell seeding, we performed histological analysis of KLF4 wild-type and KLF4Δ lung sections at early metastatic time points and found decreased metastatic colonization (disseminated tumor cells and micrometastasis) in the lungs of KLF4Δ compared to KLF4 wild-type mice (Supplementary Fig 11a, b). To assess whether these changes in early tumor cell seeding impact metastatic burden, we stably transduced our B16-F10 metastatic melanoma, M3-9M metastatic rhabdomyosarcoma, and E0771 metastatic breast carcinoma cell lines with a mCherry-firefly luciferase fusion protein to detect the presence of tumor by immunofluorescence or bioluminescent signal. Analysis of late-stage metastatic lungs revealed a dramatic decrease in metastasis to the lungs of Myh11 lineage cell-specific Klf4 Δ mice in both the B16-F10 metastatic tumor model at day 23 post-tumor injection and M3-9M metastatic rhabdomyosarcoma model at day 26 post-tumor injection compared to Klf4 wild-type tumor-bearing mice (Figure 5a-d, Supplementary Fig. 11a, Supplementary Fig. 12a-b). No difference was seen in primary tumor growth in the B16-F10 or M3-9M models between Klf4 wild-type and Klf4 Δ mice by tumor volume measurements or bioluminescent signal (Supplementary Fig. 10a). While phenotypically modulated eYFP+ ACTA2- MYH11- cells were found in the late-stage metastatic lungs of both Klf4 Δ and wild-type mice, KLF4+ eYFP+ cells were minimally detected in Klf4 Δ lung sections in contrast to the abundant KLF4+ eYFP+ cells found in KLF4 wild-type lung sections, confirming perivascular cell-specific deletion of Klf4 in Klf4 Δ mice (Fig 5e). Further, there was a significant decrease in the average frequency of eYFP+ cells found at long distances from the closest PECAM+ blood vessels in the lungs of Klf4 Δ mice (Figure 5f).
Figure 5. Perivascular cell specific KLF4 deletion decreases metastasis.
(a) KLF4 Δ and KLF4 wt/wt Myh11 lineage-tracing mice were orthotopically injected with mCherry-luciferase expressing B16-F10 tumor cells, and lungs were measured for tumor-derived luciferase activity ex vivo on day 23 post-tumor injection. KLF4 Δ lung tissue at metastatic day 23 exhibit a decrease in ex vivo lung luminescence as compared to KLF4-wild-type littermate controls by student’s t test for average radiance per lung. Dotted line represents cut-off for low versus high luminescent signal used for analysis in (c). n = 9 KLF4 wt/wt; n = 7 KLF4 Δ. *ˆ < 0.05, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values not considered outliers.
(b) Representative images depicting bioluminescent signal from luciferase-expressing B16-F10 metastatic tumor nodules in the lungs of KLF4 Δ and wt/wt Myh11 lineage-tracing mice at day 23 post-tumor injection.
(c) KLF4 Δ and KLF4 wt/wt Myh11 lineage-tracing mice were orthotopically injected with mCherry-luciferase expressing M3-9M rhabdomyosarcoma tumor cells, and lungs were measured for tumor-derived luciferase activity ex vivo on day 26 post-tumor injection. KLF4 Δ lung tissue was analyzed at metastatic day 26 and exhibit a decrease in ex vivo lung luminescence as compared to KLF4 wt/wt littermate controls. Dotted line represents cut-off for absent/low versus high luminescent signal used for analysis in (f). n = 9 KLF4 wt/wt; n = 11 KLF4 Δ. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values not considered outliers.
(d) Representative images depicting bioluminescent signal from luciferase-expressing M3-9M metastatic tumor nodules in the lungs of KLF4 Δ and KLF4 wt/wt littermate control mice at day 23 post-tumor injection.
(e) The metastatic lungs of M3-9M rhabdomyosarcoma tumor-bearing Myh11 lineage-tracing KLF4 Δ mice (right), at day 26 post-tumor injection, demonstrate that the expansion of eYFP+ lineage-traced perivascular cells (eYFP, in orange) that is observed in KLF4 wild-type (wt/wt) (left) metastatic lung is reduced in the lungs of KLF4 Δ mice, where only a few eYFP+ lineage-traced cells are found away from blood vessels and surrounding airways, and express the perivascular markers ACTA2 (green) and MYH11 (magenta). DAPI nuclear stain is shown in blue. White rectangles encompass the areas depicted below, demonstrating that while many eYFP+ KLF4+ cells can be found in the metastatic lungs of KLF4 wild-type mice (denoted by arrows), none can be found in KLF4 Δ metastatic lungs, even when eYFP+ ACTA- MYH11- cells are observed. Bars = 50 um.
(f) Computational analysis quantifying the proximity of DAPI+ eYFP+ particles to the nearest blood vessel (BV) in immunofluorescent images from lungs at day 26 post-M3-9M injected KLF4 Δ mice, that demonstrates a decrease in the average frequency of DAPI+ eYFP+ particles at longer distances from the nearest blood vessel in KLF4 Δ lungs, compared to lungs from KLF4 wt/wt litter-mate M3-9M tumor bearing lungs. n = 9 KLF4 wt/wt; n = 11 KLF4 Δ. ****P < 0.00001, ANOVA.
(g) KLF4 Δ and KLF4 wt/wt NG2 lineage-tracing mice were orthotopically injected with mCherry-luciferase expressing M3-9M rhabdomyosarcoma tumor cells, and lungs were measured for tumor-derived luciferase activity ex vivo on day 29 post-tumor injection. KLF4 Δ lung tissue was analyzed at metastatic day 29 and exhibit a decrease in ex vivo lung luminescence as compared to KLF4 wt/wt littermate controls. Dotted line represents cut-off for absent/low versus high luminescent signal used for analysis in (k). n = 9 KLF4 wt/wt; n = 11 KLF4 Δ. ***P < 0.0001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values not considered outliers.
(h) Representative images depicting bioluminescent signal from luciferase-expressing M3-9M metastatic tumor nodules in the lungs of KLF4 Δ and KLF4 wt/wt NG2 lineage-tracing littermate control mice at day 29 post-tumor injection.
(i) KLF4 Δ and KLF4 wt/wt NG2 lineage-tracing mice were orthotopically injected with mCherry-luciferase expressing E0771 breast carcinoma tumor cells, and lungs were measured for tumor-derived luciferase activity ex vivo on day 25 post-tumor injection. KLF4 Δ lung tissue was analyzed at metastatic day 25 and exhibit a decrease in ex vivo lung luminescence as compared to KLF4 wt/wt littermate NG2 lineage-tracing controls. Dotted line represents cut-off for absent/low versus high luminescent signal used for analysis in (N). N=5 mice per group. **P < 0.001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values not considered outliers.
(j) Representative images depicting bioluminescent signal from luciferase-expressing E0771 metastatic tumor nodules in the lungs of KLF4 Δ and KLF4 wt/wt littermate NG2 lineage-tracing control mice at day 25 post-tumor injection.
Consistent with our findings in Myh11 lineage cell-specific KLF4 Δ mice, we found no significant difference in primary tumor growth (Supplementary Fig. 10a), but observed a dramatic decrease in metastasis to the lungs of NG2 lineage cell-specific Klf4 Δ mice in the M3-9M metastatic rhabdomyosarcoma model at day 29 (Fig 5 g-h, Supplementary Fig. 12c), and the E0771 metastatic breast carcinoma model at day 25 (Fig 5 i-j, Supplementary Fig. 12d). Together these data indicate that KLF4-dependent perivascular phenotypic switching is critical for metastatic progression in two genetic murine models of perivascular lineage-tracing and in three different orthotopic cancer models of spontaneous metastasis.
Inhibition of tumor cell binding to fibronectin recapitulates perivascular specific KLF4-dependent metastasis
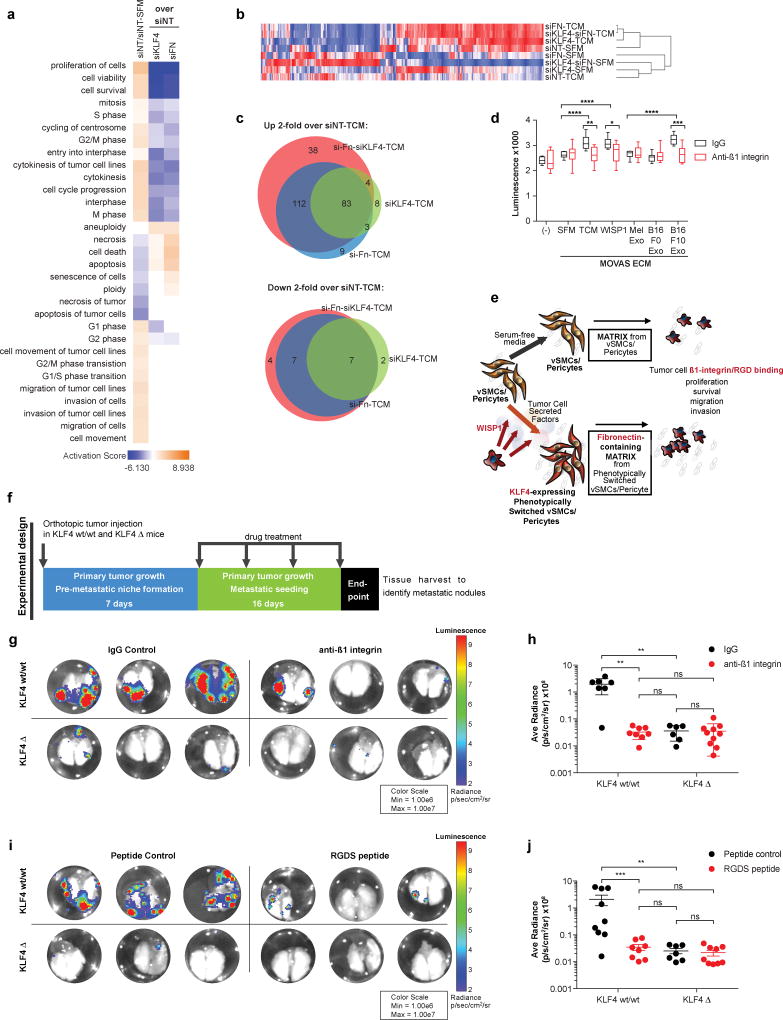
To assess the specific role of phenotypically-switched perivascular cell fibronectin deposition in promoting tumor metastasis, we utilized siRNA-mediated knock-down of Klf4, Fn1, or both genes together, in vSMCs cultured with B16-F10 TCM. Following vSMC lysis, we seeded B16-F10 tumor cells on the resultant vSMC-deposited ECM (Fig 4f) and performed RNA-seq to determine vSMC specific KLF4- and fibronectin-dependent changes in tumor cell metastatic behavior. Pathways related to tumor cell proliferation, viability, and survival were most activated in B16-F10 cells that were cultured on the ECM of vSMCs treated with TCM compared to those treated with SFM. These pathways were not activated when either Klf4 or Fn1 were knocked down (Fig 6a). Hierarchical clustering analysis demonstrated that siKlf4, siFn1, or siKlf4-siFn1 double knock-down resulted in B16-F10 cell gene expression profiles that clustered together, and clustered with B16-F10 cells that were cultured on the ECM of vSMCs treated with non-target siRNAs (siNT) and serum-free media (SFM) (Fig 6b). 83 genes in the siKlf4-TCM condition overlapped with the siFn-TCM and siKlf4-siFn-TCM conditions, and only 8 genes in the siKlf4-TCM condition did not overlap with either siFn-TCM or siKlf4-siFn-TCM conditions (Fig. 6c). These data suggest that the alteration of B16-F10 gene expression when cultured on vSMC-generated ECM is regulated by exposure to KLF4-dependent fibronectin deposition, which in turn results in activation of pathways associated with enhanced tumor cell survival, viability, and proliferation.
Figure 6. Inhibition of tumor cell binding to fibronectin recapitulates perivascular KLF4-dependent metastasis.
(a) Ingenuity Pathway Analysis of highly enriched pathways (positive activation score) in the gene expression data collected from B16-F10 tumor cells cultured in vitro on the ECM of vSMCs that were first subjected to siRNA-mediated knock-down of either Klf4, Fn1, or both Klf4 and Fn1 followed by activation in B16-F10 conditioned media (TCM) or SFM. Pathway analysis of B16-F10 tumor cells when cultured on the ECM of vSMCs treated with non-target siRNA (siNT), comparing B16-F10 TCM to SFM, demonstrate activation of cell survival, viability, and proliferation pathways (orange). These pathways are not activated in B16-F10 cells cultured on the ECM of siKlf4- or siFn1-treated vSMCs (blue).
(b) Hierarchical clustering of gene expression data collected from B16-F10 tumor cells cultured on the ECM of vSMCs as described in (a) demonstrates that the siFn-TCM, siKlf4-TCM, and siFn-siKlf4-TCM conditions cluster together, and cluster with the siNT-SFM condition, indicating that knockdown of either Klf4 or Fn in vSMCs results in the generation of ECM in response to TCM that is more similar to that produced by vSMCs cultured in SFM than when neither gene is knocked-down.
(c) Venn diagrams depicting the genes that were either 2-fold higher (top) or lower (bottom) in each knock-down condition when compared to siNT-TCM, demonstrating that the majority of the genes were commonly differentially regulated in the siKlf4, siFn, and siKlf4-siFn conditions.
(d) B16-F10 cells plated on the ECM of vSMCs that were first cultured in either TCM, recombinant WISP1, or B16-F10 exosomes (B16 F10 exo) exhibit increased proliferation when compared to the serum free media (SFM), melanocyte exosome (Mel exo), or B16-F0 exosome (B16-F0 exo) conditions. This proliferation is decreased with the treatment of B16-F10 tumor cells with neutralizing ß1 integrin antibodies to disrupt tumor cell binding to fibronectin. Representative experiment of 2 independent experiments shown. *P < 0.05, **P < 0.001, ***P < 0.0001, ****P < 0.00001, Student’s t-test. Center lines indicate median values, box edges indicate the 25th and 75th percentiles, and whiskers extend to the minimum and maximum values.
(e) Updated schematic summarizing in vitro findings that tumor-secreted factors, including WISP-1 containing exosomes, results in KLF4-dependent ECM that contains increased fibronectin, which in turn promotes ß1 integrin-dependent tumor cell proliferation.
(f) Schematic summarizing the experimental design for in vivo experiments shown in (G-J). KLF4 wild-type and KLF4 Δ mice are given orthotopic tumor injection, and allowed to establish pre-metastatic niche changes for 7 days post-tumor injection. For the 16 days following pre-metastatic niche formation, fibronectin binding was disrupted using either neutralizing ß1 integrin antibodies or RGDS peptide drugs, or corresponding control was administered. Metastasis was assessed in each mouse at the end of the drug treatment period.
(g) Representative images depicting bioluminescent signal from luciferase-expressing M3-9M metastatic tumor nodules in the lungs of KLF4 Δ and KLF4 wt/wt NG2-lineage-tracing littermate control mice treated with neutralizing ß1 integrin antibodies as described in (f) (right) compared to those treated with IgG control antibodies (left). Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
(h) Luminescent signal detected ex vivo in the lungs of KLF4 Δ and KLF4 wt/wt mice as described in (g), demonstrating a decrease in ex vivo lung luminescence in KLF4 wt/wt mice by anti-ß1 integrin treatment as compared to IgG control treatment, to levels similar to KLF4 Δ mice. n = 7 KLF4 wt/wt IgG; n = 8 KLF4 wt/wt anti-ß1 integrin; n = 6 KLF4 Δ IgG; n = 10 KLF4 Δ anti-ß1 integrin. n.s. = not significant, **P < 0.001, Student’s t-test. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
(i) Representative images depicting bioluminescent signal from luciferase-expressing M3-9M metastatic tumor nodules in the lungs of KLF4 Δ and KLF4 wt/wt NG2 lineage-tracing littermate control mice treated with RGDS peptides as described in (f) (right) compared to those treated with control peptides (left).
(j) Luminescent signal detected ex vivo in the lungs of KLF4 Δ and KLF4 wt/wt mice as described in (g), demonstrating a decrease in ex vivo lung luminescence in KLF4 wt/wt mice by RGDS peptide as compared to control peptide treatment, to levels similar to KLF4 Δ mice. n = 9 KLF4 wt/wt peptide control; n = 8 KLF4 wt/wt RGDS; n = 7 KLF4 Δ peptide control; n = 9 KLF4 Δ RGDS. n.s. = not significant, **P < 0.001, ***P < 0.0001, Student’s t-test. Center lines indicate median values and top and bottom lines indicate the 25th and 75th percentiles.
To confirm the role of activated vSMC-dependent fibronectin deposition on tumor cell proliferation, we used neutralizing ß1 integrin antibodies to disrupt B16-F10 tumor cell binding to fibronectin. We found that B16-F10 proliferation is significantly reduced with neutralizing β1-integrin antibody treatment whether the vSMCs were first activated using B16-F10 TCM, recombinant WISP1, or B16-F10 exosomes (Fig 6d). B16-F10 cells did not exhibit elevated proliferation when cultured on the ECM generated by vSMCs cultured with SFM, melanocyte exosomes, nor non-metastatic B16-F0 exosomes, and neutralizing ß1 integrin antibody treatment did not further reduce proliferation in these conditions (Fig 6d). Together, these data suggest that tumor cell secreted factors, including WISP1, induce KLF4 expression in perivascular cells that, in turn, result in enhanced deposition of fibronectin-containing ECM which promotes ß1 integrin-mediated tumor cell proliferation and survival (Fig 6e). To test this hypothesis further, we used our M3-9M metastatic rhabdomyosarcoma model in KLF4 wild-type and KLF4 Δ mice where, upon pre-metastatic niche formation, we treated mice with either neutralizing ß1 integrin antibodies or RGDS peptides to disrupt tumor cell-fibronectin binding (Fig 6f). Analysis of metastatic lungs revealed a dramatic decrease in metastasis with either anti-ß1 integrin (Fig 6g-h) or RGDS peptide treatment (Fig 6i-j) in KLF4 wild-type mice to levels comparable to KLF4 Δ mice treated with either anti-ß1 integrin antibodies, RGDS peptides, or their respective controls. Neither anti-ß1 integrin nor RGDS peptide treatment decreased primary tumor volume in either KLF4 wild-type or KLF4 Δ mice as compared to their respective controls (Supplementary Fig. 10b). Together these findings indicate that tumor cell ß1 integrin-mediated fibronectin binding contributes to the pro-metastatic functions of KLF4-dependent perivascular phenotypic switching.
These findings suggest that KLF4-dependent perivascular cell phenotypic transition is important for pre-metastatic niche formation, and that the lasting effects of KLF4 targeting can be observed in the late-stage metastatic microenvironment that ultimately dictates metastatic outcome. These results demonstrate the potential utility of perivascular plasticity as a novel target to inhibit metastasis. KLF4-targeting to prevent metastasis may be of greatest use in the adjuvant setting following primary tumor removal, or in combination with chemotherapy or other microenvironment modulating agents to decrease tumor metastatic-initiating potential.
Discussion
This study provides compelling evidence that perivascular cells in pre-metastatic sites undergo phenotypic switching whereby they activate KLF4, lose vSMC marker expression, are found in larger numbers away from the vasculature, and synthesize fibronectin-containing ECM that supports metastatic behavior in disseminated tumor cells. Genetic ablation of Klf4 in a perivascular cell-specific inducible model disrupts KLF4-dependent perivascular phenotypic switching, resultant perivascular cell-dependent fibronectin deposition, and decreases metastasis, highlighting the functional importance of these cells and their plasticity in pre-metastatic niche initiation. The use of lineage-tracing models enabled the identification of perivascular cells that would be otherwise misidentified as fibroblasts as a consequence of their activation state, given their common marker expression and production of ECM proteins41. Our results imply that although many of the key players in the primary tumor microenvironment are also found in the pre-metastatic microenvironment, stromal cells, including vSMCs/pericytes and fibroblasts, play distinct roles in these settings that are critical for tumor cell survival and proliferation.
Perivascular cells are well-recognized regulators of vascular permeability and provide repair mechanisms and vascular normalization signals to damaged blood vessels12,31,42–44. In primary tumor growth, pericytes are implicated in leaky and dysfunctional tumor vasculature12,45–47. This dual role of pericytes in tumor vasculature formation and vascular normalization is best exemplified by pericyte ablation studies, wherein specific depletion of PDGFRß- or NG2-expressing perivascular cells resulted in decreased primary tumor growth and enhanced metastasis31. Similarly, ablation of ACTA2-expressing cells enhances tumor progression48. Conversely, our findings provide direct evidence that targeting perivascular cell behavior, instead of entirely depleting pericytes, limits metastasis while effecting no change on primary tumor growth. Future studies are required to carefully delineate KLF4-dependent perivascular cell coverage around blood vessels in the context of pre-metastatic niche formation. Together these studies highlight the importance of modulating perivascular cell behavior and targeting the induction of key mediators of their phenotype, such as KLF4, to preserve their important role in vascular normalization.
In addition to their role in vascular stability and angiogenesis, NG2-expressing perivascular cells support stem cell niches in the bone marrow11 and fetal liver10 by mediating the activation and quiescence of hematopoietic stem cells. Recently, we and others have demonstrated that the bone marrow hematopoietic stem cell niche is altered during tumor development and results in enhanced mobilization of hematopoietic stem cells to pre-metastatic sites, where they enhance metastasis2. In this study, we demonstrate that phenotypically modulated perivascular cells play an early, key role in modulating the metastatic microenvironment to support stem-like features in tumor cells, such as enhanced tumorsphere formation, thus strengthening our understanding of the commonalities between the pre-metastatic niche and physiological stem cell niches49. Further studies of phenotypically activated NG2+ pericytes as the source of pluripotent mesenchymal cells that create perivascular niches11,50–52 in multiple organs as well as niches that support cancer initiating cells39,53,54 will need to be investigated.
Our results demonstrate a novel and previously unappreciated role for perivascular cell plasticity in pre-metastatic niche formation and identify KLF4 as a critical inducer of this plasticity. Although KLF4 has long been appreciated as an important mediator of perivascular plasticity in a number of disease models, including atherosclerosis and vascular injury8,16,17,22,23,27, as well as a context-dependent oncogene or tumor suppressor in a number of tumor types55–57, this is the first study to describe KLF4-mediated perivascular activation as essential to pre-metastatic niche formation and metastasis. Modulating perivascular cell phenotype and behavior introduces a new concept for therapeutic targeting of microenvironmental cells that focus on altering plasticity rather than lethality of this key cell population that also play crucial roles in tissue homeostasis, wound healing, and regeneration. This novel approach may lead to new therapeutic targets for tumor metastasis prevention, which are urgently needed in the clinic.
Online Methods
Detailed information on experimental design and reagents can be found in the Life Sciences Reporting Summary.
Mice
ROSA-STOP-flox-eYFP mice, MyH11-ERT-cre mice and NG2-ERT-cre mice were obtained from Jackson Laboratories. Only male mice inherit the MyH11-ERT-cre allele, and thus any experiments performed using this mouse model were done so using male mice that were ROSA-STOP-flox-eYFP +/+ and MyH11-ERT-cre +/−. The Klf4-flox mice were obtained through the Mutant Mouse Resource and Research Center (MMRRC), to whom they were donated by Klaus Kaestner. MyH11- ERT- cre mice were bred with the Klf4 floxed mice to obtain the Myh11-ERT-Cre Klf4 Δ mice. Genotyping was performed via PCR using previously published protocols. Cre activity was induced in mice aged 6–8 weeks via intraperitoneal injection (1 injection per day, 10 total injections, 10 mg ml−1 of tamoxifen in peanut oil). For drug treatment studies, mice were randomized within genotype grouping and drug treatment was performed blinded post-assignment. All experiments were approved by the NCI Animal Care and Use Committee and conducted under specific pathogen-free conditions at the NIH Animal Facility.
Cell lines
The B16-F0, B16-F10, E0771, and MOVAS cell lines were obtained from the ATCC. The M3-9M cell line was derived from the transgenic HGF/p53 knockout mice that develop spontaneous embryonal rhabdomyosarcoma tumors generated by Glenn Merlino and backcrossed to the Bl6 background by Crystal Mackall58. All cell lines were authenticated via microarray analysis and tested mycoplasma negative. All tumor cell lines were transduced with a lentivirus expressing a mCherry-firefly luciferase fusion protein and flow sorted for 98% mCherry expression to establish stable cell lines. eYFP+ SMCs were isolated from the aortas of MyH11 lineage-tracing mice post-tamoxifen injection, via flow sorting on eYFP+ expression after negative selection of CD45/CD31/TER119 by Milltenyi biotinylated beads and depletion columns. eYFP+ SMCs were kept in culture for up to 7 passages and then discarded.
B16-F0, B16-F10, MOVAS and eYFP+ SMC cell lines were grown in DMEM with 10% FBS and 1% penicillin/streptomycin/glutamine. The M3-9M cell line was grown in RPMI with 10% FBS and 1% penicillin/streptomycin/glutamine. Serum free media for eYFP+ SMCs is DMEM supplemented with 1% penicillin/streptomycin/glutamine, 0.2 mM ascorbic acid, 5 ug ml−1 transferrin, 2.8 ug ml−1 insulin and 6.25 ng ml−1 sodium-selenate. Serum-free media for all other cell lines are their growth media without FBS.
Orthotopic and tail-vein tumor models
Orthotopic tumor injections were administered with 5×105 tumor cells in single-cell suspension of 100 µL of Hank’s Balanced Salt Solution (HBSS) using a sterile 27.5-gauge needle. The B16-F0 and B16-F10 melanoma cell lines were administered intradermally in the flank. The M3-9M rhabdomyosarcoma cell line was administered in the gastrocnemius muscle of the right hind leg. The E0771 breast carcinoma cell line was administered into the mammary fat pad. Tail-vein tumor injections were administered with 1×105 tumor cells in single-cell suspension of 100 µL HBSS using a sterile 27.5-gauge needle into the lateral tail vein. Metastatic end-point was determined per experiment by maximum primary tumor burden and/or the appearance of moribund mice within all treatment groups as defined within protocols established by the NCI Animal Care and Use Committee.
Bioluminescent tumor cell tracking
For in vivo whole-animal imaging, anesthetized mice received 3 mg of D-luciferin (Gold Biotechnology) via intraperitoneal injection (i.p.). Luminescence readings were collected on a Xenogen IVIS 200, starting at 5 minutes post i.p. injection for 1 minute. For ex vivo tissue imaging, tissues were perfused with PBS, harvested, and incubated in 1 µg ml−1 D-luciferin in PBS. Luminescent readings were collected starting at 5 minutes later, for 1 minute. Analysis was conducted using the Xenogen Living Image software package.
Immunofluorescence
Tissues were first cardiac perfused with PBS followed by 4% PFA. Tissues were then harvested, fixed overnight in 4% PFA at 4°C, then transferred to 30% glucose and incubated at 4°C until tissues sink, or up to 3 days. Tissues were then embedded in OCT compound (Tissue-Tek) and cryosectioned for staining. Sections were air-dried for 2 hours, washed in PBS, permeabilized in 0.5% Triton-X, blocked in 10% fish skin gelatin/10% donkey serum, and then stained with the following antibodies: 4,6’-diamidino-2-phenylindole (DAPI) (Invitrogen) as a nuclear marker; goat anti-GFP (ab6673, Abcam) for lineage marker eYFP; mouse anti-ACTA2 (SMA) conjugated to FITC (1A4, Sigma); rat anti-Myh11 (KM3669, Kamiya); rabbit anti-PECAM1 (AB28364, Abcam); rabbit anti-KLF4 (HPA002926, Sigma); rabbit anti-fibronectin (ab23750, Abcam); rabbit anti-PDGFRa (APA5, Abcam); rabbit anti-PDGFRb (Y92, Abcam); rabbit anti-Ki67 (ab16667, Abcam); mouse anti-NG2 conjugated to biotin (ab5320b, Millipore); and chicken anti-mCherry (ab205402, Abcam). Donkey secondary antibodies were used where necessary (Jackson ImmunoResearch).
Cells were plated and treated on autoclaved glass coverslips placed in sterile 6-well plates. Upon conclusion of the experiment, cells were fixed in 4% PFA, permeabilized in 0.5% Triton-X, blocked in 10% donkey serum, and then stained as detailed above.
Images for phenotyping were acquired with a Zeiss Axio Observer Z1 inverted microscope fitted with an Apotome.2. Confocal fluorescence images were acquired with a Zeiss LSM880 laser scanning confocal microscope equipped with a 34-channel spectral detector and 20× plan-apochromat (air, N.A. 0.8) and 63× plan-apochromat (oil, N.A. 1.4) objective lens (Carl Zeiss Microscopy, LLC, Thornwood, NY). A five-channel imaging configuration was used to acquire images from multi-labeled fluorescent samples. The fluorophores used for imaging, with respective laser excitation and fluorescence emission detection windows, were DAPI (405 nm laser, 415–490 nm emission), FITC (488 nm laser, 490–535 nm emission), Cy3 (561 nm laser, 563–585 nm emission), AlexaFluor 594 (594 nm laser, 596–631 nm emission), and AlexaFluor 647 (633 nm laser, 655–742 nm emission). Confocal images were collected with a 1024×1024 pixel frame size, and with the 20× objective lens a 0.28 µm X-Y pixel size, 3.9 µm optical slice thickness, and 4× image frame averaging, and with the 63× objective lens 0.09 µm X-Y pixel size, 1.2 µm optical slice thickness, and 4× image frame averaging. Imaging parameters were kept constant for all images in an experiment. Isotype and single fluorophore labeled control samples were imaged using the same five channel imaging configuration so that the potential degree of fluorescence cross-talk between image channels could be compensated for during image analysis. For extended field of view tile imaging, image frames were collected with 5% overlap and stitched together into one large image file using Zeiss Zen blue (v. 2.3) image processing software. After background subtraction based on the isotype and single-label control samples, the brightness and contrast were adjusted for each fluorescence channel using linear histogram stretching and the adjustments were made using the same parameters for all images in a dataset. The resultant images were exported as Tiff files and composed into figures using Adobe Photoshop. Deconvolution and figure preparation was performed using the Zeiss Zen software package, and all particle analysis was conducted using the CellProfiler 2.1.1 software package (cellprofiler.org)59.
Image Analysis
eYFP+ particles were identified using CellProfiler 2.1.1 (cellprofiler.org)59, whereby Parent particles were identified based on DAPI positive nuclei, and eYFP+ Child particles were identified based on Parent particles surrounded by eYFP+ staining. The total number of eYFP+ Child particles was quantified in three 20× tiled images (4×4 average tile area) per mouse, and reported as average eYFP+ particles per tiled field. KLF4+ eYFP+ cells were identified in eYFP+ Child particles by KLF4 staining restricted to the nucleus of each eYFP+ particle. The average proximity of eYFP+ particles to blood vessels was computed by determining the radial distribution of PECAM to eYFP+ Child particles, reported in bins of 3 µm and 27 µm radial distances from the nucleus of each eYFP+ particle. Each eYFP+ particle was then analyzed to determine the closest bin containing PECAM signal. Each bin was summed per tiled image, and then reported as an average bin signal per mouse. Signal density of fibronectin staining was determined in ImageJ by recording the integrated density in at least 3 fields of each stained sample.
For image analysis, the original images were used for mean intensity measurements, and the background subtracted based on single-color and isotype control images. Briefly, individual regions of interest (ROIs) were drawn for different representative regions within the images to measure the mean intensity for each fluorescence channel. For the series of single label control samples, the mean intensity was also measured for each of non-labeled channels and the highest background value recorded was used to subtract from the measurements made from the corresponding image channel in the multi-label samples. 10–25 ROIs were drawn in each image centered around DAPI stained nuclei and 5 representative images per lung specimen were included for analysis per dataset. The background subtracted mean fluorescence intensities for YFP positive lineage traced cells and YFP negative cells were plotted using Prism. Each dataset was analyzed independently by two investigators.
Flow cytometry
Lungs were first perfused with PBS and then insufflated with 1 mg ml−1 Collagenase I, 10 µg mL-1 Dispase II and 20 µg mL−1 DNase I. Lungs were then harvested and placed in 10%FBS/5%BSA/PBS on ice. Lung tissue was finely minced, digested in collagenase I/Dispase II/DNase I at 37°C for 30 minutes with gentle agitation, and passed through a 70 µm nylon filter. Total cell counts per lung were determined using Trypan cell exclusion from single cell suspensions. Live cells were determined via exclusion of viability dye (eBioscience). eYFP+ cells were identified by their negative staining by antibodies to CD45 (30-F11, eBioscience), TER119 (TER119, eBioscience), and CD31 (390, Biolegend) in 0.5% BSA/PBS with 0.05% NaN3, and positive eYFP signal. Extracellular markers included CD51 (RMV-7, eBioscience), PDGFRα (APA5, eBioscience), NG2 (ab5320b, Millipore) Mac-2 (M3/38, eBioscience), CD11b (M1/70, eBioscience), and F4/80 (BM8, eBioscience). Where biotinylated antibodies were used, an APC-e780 streptavidin (eBioscience) staining step was added. For intracellular/nuclear staining, cell surface markers were first stained as stated above followed by fixation/permeabilization/staining for all other markers using a Foxp3/Transcription Factor Staining Buffer Set (eBioscience). Because fixation decreases eYFP emission, eYFP+ cells were identified using an anti-GFP antibody (5F12.4, eBioscience). Other intracellular/nuclear antibodies included Ki67 (SolA15, eBioscience), ACTA2 (1A4, eBioscience), and KLF4 (HPA002926, Sigma). Unconjugated KLF4 staining was followed by a PE-conjugated secondary (eBioscience). Flow cytometry data was collected on a BD LSRFortessa, and analyzed using the FlowJo software package (Tree Star). Cell populations were reported as the proportion of live single cells identified by flow, multiplied by the total number of cells isolated from each lung.
Tumor-conditioned media and exosome preparation
Tumor cells were plated at 1×105 cells ml−1 in T75 flasks and allowed to adhere overnight in growth medium. Growth medium was then removed, and the cells were washed in PBS and cultured in serum free media for 72 hours. This tumor-conditioned media was collected, filtered through a 0.22 µm filter, aliquoted and stored at −80°C for downstream experiments. Exosomes were isolated from tumor-conditioned media prior to storing at −80°C via ultracentrifugation at 25,000 rpm, 4°C, for 2 hours. Pelleted exosomes were re-suspended in 35 µL HBSS, quantified for protein content by BCA assay, and stored in 1 µg µL−1 aliquots at −80°C for downstream experiments. Aliquots were characterized using Nanosight to confirm uniform size distribution. For exosome labeling experiments, aliquots were thawed, diluted in 1 mL DilC, then mixed with 1 mL DilC containing 4 µL of PKH26 dye. After 5 minutes incubation at room temperature, 2 mL of 1% BSA was added for 1 minute to inhibit staining. Exosomes were then pelleted by ultracentrifugation and used directly in experiments. For in vivo pre-treatment experiments, 6 intraperitoneal injections were given over the course of 14 days of either 100 ul of tumor-conditioned media, or 10 ug of exosomes per injection.
Cytokine Array Analysis
Tumor-conditioned media from B16-F0 and B16-F10 was prepared as described above and then applied to RayBiotech C1000 antibody array as per manufacturer protocol. Membranes were analyzed by densitometry using the BioRad ChemiDoc imaging system.
Quantitative real-time PCR
Total RNA was isolated and precipitated using TRIzol followed by chloroform/ethanol washes. RNA concentration and quality was assessed using the Agilent 2100 Bioanalyzer. cDNA synthesis was performed using 1 µg RNA per sample and the Biorad iScript cDNA Synthesis Kit. qRT-PCR was performed using the Biorad SsoAdvanced master mix on a Biorad iCycler.
KLF4 knock-down and reporter plasmid
A KLF4 promoter-reporter plasmid that expresses the secretable Gaussia Luciferase was purchased from Genecopoeia, and transfected into plated MOVAS cells. 24 hours post-transfection, siRNAs targeting either KLF4 or non-target control were added to MOVAS cells according to manufacturer’s protocol (Dharmacon Accel siRNAs). 72 hours post-siRNA treatment, media was replaced with serum-free media containing recombinant proteins or tumor conditioned media. 16 hours after protein-containing treatment, media was removed and assayed for luciferase activity using the BioLux Gaussia Luciferase Assay Kit (New England BioLabs). Reporter plasmid activity is reported in luminescence as compared to empty plasmid control. Western Blots were also performed after TCM treatment of KLF4 knocked down vSMCs to probe for KLF4 (HPA002926, Sigma) and fibronectin (ab23750, Abcam), and were imaged using the BioRad ChemiDoc imaging system.
Western blotting
Exosomes or cell pellets were lysed in Cell Lysis Buffer (Cell Signaling) with protease and phosphatase inhibitor cocktail (Millipore). All lysates were cleared by centrifugation, and quantified by bicinchoninic acid (BCA) protein assay (ThermoFisher). 5 µg of protein from each sample were electrophoresed on 4–12% Bis-Tris Protein gels, and transferred to nitrocellulose membranes (ThermoFisher). Blots were probed with primary antibodies: anti-Angiopoietin-2 (2948, Cell Signaling), anti-WISP1 (ab178547, Abcam), anti-PDGF BB (ab23914, Abcam), anti-IL-1beta (ab200478, Abcam), anti-FN (1574-1, Epitomics), anti-KLF4 (HPA002926, Sigma), anti-GFP (ab6673, Abcam), anti-β-actin (8457s, Cell Signaling), anti-CD81 (Eat2 Thermo Scientific), anti-CD63 (ab193349, Abcam), anti-mouse CD9 (ab82390, Abcam), anti-human CD9 (ab92726, Abcam) , anti-Flotillin (18634, Cell Signaling), anti-Alix (2171, Cell Signaling), anti-Annexin V (8555, Cell Signaling), LAMP2 (ab203224, Abcam), anti-HSP70 (4876, Cell Signaling), anti-EpCAM (2626, Cell Signaling), and anti-GM130 (12480, Cell Signaling), then probed with the secondary antibody anti-rabbit IgG-HRP (7074s, Cell Signaling), or anti-mouse IgG-HRP (7076, Cell Signaling), or anti-Armenian hamster (PA1-32045, Thermo Scientific). HRP-based detection was performed using SuperSignal West Substrate (Thermo Scientific).
RNA sequencing
RNA sequencing was performed using the standard operating procedure of the CCR Genomics core facility. mRNA samples were prepared using Illumina TruSeq Stranded mRNA kit (Illumina). Samples were pooled and sequenced using the NextSeq500 platform in high output mode. Demultiplex was done allowing 1 mismatch in the barcodes. All reads for each sample were trimmed adapters and low quality bases using Trimmomatic software and aligned to the reference using STAR software. The data normalization and statistical analysis used to identify differentially expressed genes was performed using the Partek Flow installed in the NIH Helix cluster (https://partekflow.cit.nih.gov). The fastq was aligned to mm10 genome using STAR-2.4.1d aligner, the transcript abundance was estimated using the Partek E/M algorithm based on mm10-Ensembl Transcipts release 82 transcriptome model. Gene counts were normalized to total read count per sample and the differential gene expression (GSA) algorithm was used to detect differentially expressed genes. Pathway analysis was conducted using Ingenuity Pathway Analysis (Qiagen), using a 2-fold expression cut-off for all comparisons.
Wisp-1 ELISA
Exosomes as prepared above were assayed using the R&D Mouse/Rat WISP1/CCN4 Quantikine ELISA kit. Mouse plasma samples were obtained from terminal cardiac punctures and processed according to manufacturer’s recommendations.
In vitro SMC-derived matrix experiments
SMCs were plated at a density of 1×105 cells mL−1 and allowed to adhere to culture plates overnight. Cells were washed with PBS and media was replaced with either serum-free media, tumor-conditioned media, exosomes, or recombinant cytokines. 72 hours post-media treatment, SMCs were lysed in a solution of 1% SDS, 1% Triton-X-100, and 20 mM NH4OH for 5 minutes. The remaining matrix was washed in PBS 3 times and used immediately for downstream experiments.
Tumor cell adhesion was assessed by plating 1×104 tumor cells mL−1 onto SMC matrix, and then removing the tumor cells after 2 hours. Tumor cell confluence per well was determined using an IncuCyte Zoom (Essen BioScience) and accompanying software. Tumor proliferation was assessed using CellTiter Glo following the manufacturer’s provided protocol. Tumor migration was assessed by plating tumor cells on top of a 0.8 µm transwell membrane in serum-free media above an ECM-coated well. Percent migration was counted as the number of DAPI+ cells that were identified under the transwell membrane at 8 hours post-plating.
Tumorsphere formation was assessed by allowing tumor cells to proliferate on SMC-generated matrix for 72 hours, after which tumor cells were harvested, counted, and plated in ultra low-attachment plates (200 cells per well) in DMEM/F12 supplemented with B27 supplement, 20 ng mL−1 epidermal growth factor, 10 ng mL−1 basic fibroblast growth factor, 5 µg mL−1 insulin, and 0.4% FBS. Cells were cultured for 1 week, after which spheres were imaged using a phase-contrast microscope at 40× magnification. Tumorsphere were distinguished from cellular aggregates by their solid spherical shape with a diameter of at least 50 µm, and reported as the average number of tumorspheres per 40× field.
Statistical analysis
All analysis was performed using GraphPad Prism version 6.04, www.graphpad.com, with the exception of the Jonckheere-Terpstra test which was performed in R. Sample size was determined by running an a priori power analysis using G*Power with data collected from pilot experiments60. Data are expressed as mean +/− standard deviation. Unless otherwise stated, statistical differences were evaluated using an unpaired, nonparametric Student’s t-test. Data were evaluated for normality using the D’Agostino-Pearson omnibus normality test. Two-tailed Fisher’s exact test was used for categorical data. Jonckheere-Terpstra test was performed for data that represent independent values over time. Significant differences between experimental groups were represented as *P<0.05, **P<0.001, ***P<0.0001, and ****P<0.00001.
Supplementary Material
Acknowledgments
The authors would like to thank M. Kasai, J. Zhu, and K. Bhatt for their technical assistance.
Footnotes
Author Contributions
M.M. conducted most of the experiments, performed data analysis, generated most of the experimental mice, and was the primary writer of the manuscript. W.J. conducted in vivo metastasis studies, in vitro matrix experiments, generated RNA for sequencing, and analyzed exosomal content by western blot. M.E. conducted in vitro immunostaining and western blot experiments. J.K assisted with experimental mouse generation and conducted image analysis. D. B. and S.K. assisted with mouse experiments and generated/analyzed flow cytometry data. M.M.M. analyzed pathological tissue specimens. M.K. assisted with confocal image acquisition, analysis and figure preparation. H.L. and J.F.S. analyzed RNA sequencing data and assisted with figure preparation. O.A.C. and G.K.O. generated preliminary data, provided initial experimental mice, and provided advice throughout. R.N.K supervised the project, provided guidance on experimental design, data interpretation, and writing the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
Accession codes
All generated sequencing data are available at the Gene Expression Omnibus (GEO) with accession code GSE96972.
References
- 1.Kaplan RN, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giles AJ, et al. Activation of Hematopoietic Stem/Progenitor Cells Promotes Immunosuppression Within the Pre-metastatic Niche. Cancer Res. 2016;76:1335–1347. doi: 10.1158/0008-5472.CAN-15-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith HA, Kang Y. The metastasis-promoting roles of tumor-associated immune cells. J. Mol. Med. 2013;91:411–429. doi: 10.1007/s00109-013-1021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sceneay J, Parker BS, Smyth MJ, Möller A. Hypoxia-driven immunosuppression contributes to the pre-metastatic niche. Oncoimmunology. 2013;2:e22355. doi: 10.4161/onci.22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat. Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehling M, Mazzone M. Vessel Normalization in the Spot-LIGHT of Cancer Treatment. Trends Mol Med. 2016;22:85–87. doi: 10.1016/j.molmed.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc. Res. 2012;95:156–164. doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander MR, Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu. Rev. Physiol. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- 9.Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ. Res. 2005;97:512–523. doi: 10.1161/01.RES.0000182903.16652.d7. [DOI] [PubMed] [Google Scholar]
- 10.Khan JA, et al. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science. 2016;351:176–180. doi: 10.1126/science.aad0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunisaki Y, et al. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goel S, Wong AH-K, Jain RK. Vascular normalization as a therapeutic strategy for malignant and nonmalignant disease. Cold Spring Harb Perspect Med. 2012;2:a006486. doi: 10.1101/cshperspect.a006486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamzah J, et al. Vascular normalization in Rgs5-deficient tumours promotes immune destruction. Nature. 2008;453:410–414. doi: 10.1038/nature06868. [DOI] [PubMed] [Google Scholar]
- 14.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armulik A, Genové G, Betsholtz C. Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises. Dev. Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Salmon M, et al. KLF4 regulates abdominal aortic aneurysm morphology and deletion attenuates aneurysm formation. Circulation. 2013;128:S163–74. doi: 10.1161/CIRCULATIONAHA.112.000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shankman LS, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 2015;21:628–637. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherepanova OA, et al. Activation of the pluripotency factor OCT4 in smooth muscle cells is atheroprotective. Nat. Med. 2016;22:657–665. doi: 10.1038/nm.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Meth. 2013;10:171–177. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim KH, Sederstrom JM. Assaying Cell Cycle Status Using Flow Cytometry. Curr Protoc Mol Biol. 2015;111:28.6.1–11. doi: 10.1002/0471142727.mb2806s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22α promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circ. Res. 2012;111:685–696. doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida T, Gan Q, Owens GK. Kruppel-like factor 4, Elk-1, and histone deacetylases cooperatively suppress smooth muscle cell differentiation markers in response to oxidized phospholipids. American Journal of Physiology - Cell Physiology. 2008;295:C1175–82. doi: 10.1152/ajpcell.00288.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gajos-Michniewicz A, Duechler M, Czyz M. MiRNA in melanoma-derived exosomes. Cancer Lett. 2014;347:29–37. doi: 10.1016/j.canlet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Rafii S, Lyden D. S100 chemokines mediate bookmarking of premetastatic niches. Nat. Cell Biol. 2006;8:1321–1323. doi: 10.1038/ncb1206-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H1027–37. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M, Du Y, Xu Z, Jiang Y. Functional Effects of WNT1-Inducible Signaling Pathway Protein-1 on Bronchial Smooth Muscle Cell Migration and Proliferation in OVA-Induced Airway Remodeling. Inflammation. 2016;39:16–29. doi: 10.1007/s10753-015-0218-x. [DOI] [PubMed] [Google Scholar]
- 29.Reddy VS, Valente AJ, Delafontaine P, Chandrasekar B. Interleukin-18/WNT1-inducible signaling pathway protein-1 signaling mediates human saphenous vein smooth muscle cell proliferation. J. Cell. Physiol. 2011;226:3303–3315. doi: 10.1002/jcp.22676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le CTK, et al. Synergistic actions of blocking angiopoietin-2 and tumor necrosis factor-α in suppressing remodeling of blood vessels and lymphatics in airway inflammation. Am J. Pathol. 2015;185:2949–2968. doi: 10.1016/j.ajpath.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keskin D, et al. Targeting vascular pericytes in hypoxic tumors increases lung metastasis via angiopoietin-2. Cell Rep. 2015;10:1066–1081. doi: 10.1016/j.celrep.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexander MR, Murgai M, Moehle CW, Owens GK. Interleukin-1β modulates smooth muscle cell phenotype to a distinct inflammatory state relative to PDGF-DD via NF-κB-dependent mechanisms. Physiol. Genomics. 2012;44:417–429. doi: 10.1152/physiolgenomics.00160.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander MR, et al. Genetic inactivation of IL-1 signaling enhances atherosclerotic plaque instability and reduces outward vessel remodeling in advanced atherosclerosis in mice. J. Clin. Invest. 2012;122:70–79. doi: 10.1172/JCI43713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulauroy S, Di Carlo SE, Langa F, Eberl G, Peduto L. Lineage tracing and genetic ablation of ADAM12+ perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nat. Med. 2012;18:1262–1270. doi: 10.1038/nm.2848. [DOI] [PubMed] [Google Scholar]
- 35.Laklai H, et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bissell MJ, Radisky D. Putting tumours in context. Nat. Rev. Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessenbrock K, Plaks V, Werb Z. Matrix Metalloproteinases: Regulators of the Tumor Microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox TR, et al. LOX-Mediated Collagen Crosslinking Is Responsible for Fibrosis-Enhanced Metastasis. Cancer Res. 2013;73:1721–1732. doi: 10.1158/0008-5472.CAN-12-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.del Pozo Martin Y, et al. Mesenchymal Cancer Cell-Stroma Crosstalk Promotes Niche Activation, Epithelial Reversion, and Metastatic Colonization. Cell Rep. 2015;13:2456–2469. doi: 10.1016/j.celrep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oskarsson T, Batlle E, Massagué J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14:306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LeBleu VS, et al. Origin and function of myofibroblasts in kidney fibrosis. Nat. Med. 2013;19:1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baluk P, Falcón BL, Hashizume H, Sennino B, McDonald DM. Tumor Angiogenesis. Springer; Berlin Heidelberg: 2008. pp. 557–576. [DOI] [Google Scholar]
- 43.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 44.Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J. Clin. Invest. 2003;111:1287–1295. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morikawa S, et al. Abnormalities in pericytes on blood vessels and endothelial sprouts in tumors. Am. J. Pathol. 2002;160:985–1000. doi: 10.1016/S0002-9440(10)64920-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukumura D, Jain RK. Tumor microvasculature and microenvironment: targets for anti-angiogenesis and normalization. Microvasc. Res. 2007;74:72–84. doi: 10.1016/j.mvr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chambers AF, Groom AC, MacDonald IC. Metastasis: Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 48.Ozdemir BC, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murgai M, Giles A, Kaplan R. Physiological, Tumor, and Metastatic Niches: Opportunities and Challenges for Targeting the Tumor Microenvironment. Crit Rev Oncog. 2015;20:301–314. doi: 10.1615/critrevoncog.2015013668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanoun M, et al. Acute myelogenous leukemia-induced sympathetic neuropathy promotes malignancy in an altered hematopoietic stem cell niche. Cell Stem Cell. 2014;15:365–375. doi: 10.1016/j.stem.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat. Rev. Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kusumbe AP, et al. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature. 2016;532:380–384. doi: 10.1038/nature17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calabrese C, et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 54.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528:413–417. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu F, et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2005;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 57.Pandya AY, et al. Nuclear Localization of KLF4 Is Associated with an Aggressive Phenotype in Early-Stage Breast Cancer. Clin. Cancer Res. 2004;10:2709–2719. doi: 10.1158/1078-0432.ccr-03-0484. [DOI] [PubMed] [Google Scholar]
- 58.Meadors JL, et al. Murine rhabdomyosarcoma is immunogenic and responsive to T-cell-based immunotherapy. Pediatr Blood Cancer. 2011;57:921–929. doi: 10.1002/pbc.23048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology 2006 7:10. 2006;7:1. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faul F, Erdfelder E, Buchner A, Lang A-G. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.