Abstract
Tetraspanin CD82, also known as KAI1, was revealed as an attractive prognostic tumor biomarker in recent studies. However, some results of these studies remained debatable and inconclusive. Therefore, we conducted a meta-analysis to clarify the precise predictive value of CD82 in various neoplasms. Qualified studies were identified up to April 27, 2017, by searching PubMed, EMBASE, and the Web of Science. In total, 29 eligible studies were ultimately enrolled in this meta-analysis. Pooled hazard ratios (HRs) with 95% CIs of overall survival and disease/recurrence/progression-free survival were calculated to evaluate the correct prognostic role of CD82. Statistical analysis demonstrated that high expression of CD82 was significantly associated with enhanced overall survival (HR =0.56, 95% CI: 0.47–0.67) and disease/recurrence/progression-free survival (HR =0.42, 95% CI: 0.30–0.59) in cancer patients. Furthermore, we also conducted the subgroup analysis and the results revealed that CD82 was associated with favorable outcomes in cancer patients. Taken together, CD82 could be a promising biomarker for predicting the prognosis of patients with malignant neoplasms, and the biological functions of CD82 are of great research value of the subject.
Keywords: CD82, KAI1, prognosis, meta-analysis
Introduction
Tetraspanins are a family of 34 proteins, which are involved in diverse functions such as B- and T-cell activation, platelet aggregation, migration, proliferation, morphogenesis, and tumor cell progression.1 The key feature of tetraspanins is their potential to associate with each other and with a multitude of molecules from other protein families.2,3 CD82, also known as KAI1, belongs to tetraspanin family associated not only with extensive physiological processes, but also in pathological situations such as cancer invasion and metastasis,4,5 and its differential expressions are found in various normal and malignant tissues,6–8 which indicate that CD82 may play a pivotal role in cancer growth, progression, motility, invasion, and metastasis.9
A number of studies have demonstrated that decreased CD82 expression in tumor tissues was associated with unfavorable survival in cancer patients.10–15 Whereas in some individual studies focused on gastric carcinoma,16 osteosarcoma,17 colorectal carcinoma,18 and clear cell renal cell carcinoma,19 increased expression of CD82 might predict diverse, even opposing outcome. The discrepancies between these studies highlight the importance of evaluating the prognostic significance of CD82 in human malignant neoplasms. Therefore, we conducted this systematic review using meta-analysis to shed light on the relationship between CD82 expression and the prognosis of patients with carcinoma.
Methods
Search strategy
We searched online databases, including PubMed, EMBASE, and Web of Science, to identify relevant literature published until April 27, 2017. For the literature retrieval, combinations of the keywords were used as follows: (“cancer” or “carcinoma” or “neoplasm” or “tumor” or “tumour”) and (“KAI1” or “CD82”) and (“prognostic” or “prognosis” or “survival” or “outcome” or “recurrence” or “relapse”). The following criteria should be considered to select the published studies: 1) human, English publications; 2) a relationship of CD82 expression with cancer prognosis. In addition, we searched for studies published in Chinese to comprehensively understand the role of miRNA-205 in cancer. In order to supplement our literature search, the bibliographies in these studies were also carefully scanned.
Quality assessment
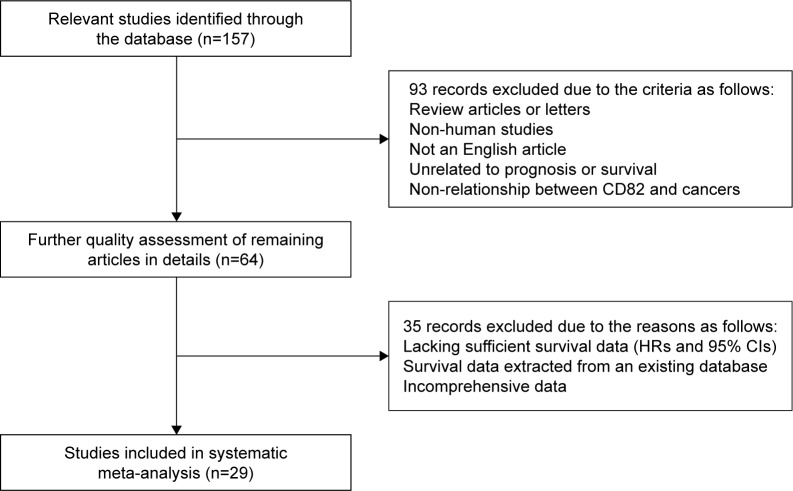
To evaluate the retrieved studies, they should include clear definitions of the following: 1) the study population and country; 2) the study design; 3) assay method to determine CD82 expression: immunohistochemistry (IHC), quantitative reverse transcription-polymerase chain reaction (qRT-PCR), or Western-blot; 4) the prognosis or survival assessment; 5) the detected tumor and pathology information; 6) the cutoff point of CD82; and 7) the follow-up duration (Table 1). Sensitivity analyses and published bias were performed to promote the quality of this meta-analysis. A flow diagram of the study selection process is presented in Figure 1.
Table 1.
Main characteristics of studies included in the meta-analysis
| First author, publication year | Case nationality | Dominant ethnicity | Median or mean age | Study design | Malignant disease | Main type of pathology | Detected sample | Assay method | Survival analysis | Source of HR | Maximum months of follow-up |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zhu, 201710 | China | Asian | 59.4 | R | Colorectal carcinoma | AdenoCa | Tissue | IHC | OS | Reported | 96 |
| Lu, 201612 | China | Asian | 57.7 | R | Gastric carcinoma | AdenoCa | Tissue | IHC | OS | Reported | 95 |
| Singh, 201611 | India | Asian | 49 | R | Breast cancer | AdenoCa | Tissue | qRT-PCR | OS | SC | 36 |
| Guo, 201516,a | China | Asian | 48 | R | Gastric carcinoma | AdenoCa | Tissue | IHC | OS | SC | 60 |
| Guo, 201516,b | China | Asian | 48 | R | Gastric carcinoma | AdenoCa | Tissue | qRT-PCR | OS | SC | 60 |
| Wu, 201532 | China | Asian | 62.1 | R | Colorectal carcinoma | AdenoCa | Tissue | IHC | OS | Reported | 108 |
| Han, 201536 | China | Asian | 45.6 | R | Breast cancer | AdenoCa | Tissue | IHC | OS | Reported | NM |
| Yu, 201413 | China | Asian | 62.1 | R | LSCC | SqCa | Tissue | IHC | OS/DFS | Reported | NM |
| Kwon, 201419 | Korea | Asian | 55.8 | R | CCRCC | AdenoCa | Tissue | IHC | OS | Reported | 223 |
| Tang, 201414,c | Canada | Caucasian | 60 | R | Melanoma | Melanoma | Tissue | IHC | OS | Reported | 120 |
| Tang, 201414,d | Canada | Caucasian | 60 | R | Melanoma | Melanoma | Tissue | IHC | OS | Reported | 60 |
| Zhang, 201337 | China | Asian | 58.7 | R | LSCC | SqCa | Tissue | IHC | OS | SC | NM |
| Zhang, 201338 | China | Asian | 59.6 | R | LSCC | SqCa | Tissue | WB | OS | SC | 84 |
| Wu, 201215 | China | Asian | 58.6 | R | NSCLC | AdenoCa | Tissue | IHC | OS | Reported | 88 |
| Knoener, 201233 | Germany | Caucasian | NM | R | Gastric carcinoma | AdenoCa | Tissue | IHC | OS | SC | 163 |
| Guo, 200939 | China | Asian | 60 | R | Hepatocellular carcinoma | AdenoCa | Tissue | IHC | OS | SC | 24 |
| Protzel, 200834 | Germany | Caucasian | 66.5 | R | Penile carcinoma | SqCa | Tissue | IHC | OS | SC | 125 |
| Miyazaki, 200540 | Japan | Asian | 61 | R | ESCC | SqCa | Tissue | IHC | OS | SC | 60 |
| Leavey, 200517 | America | Caucasian | 11 | R | Osteosarcoma | Sarcoma | Tissue | IHC | OS/PFS | SC | NM |
| Farhadieh, 200441 | Australia | Caucasian | 65 | R | OSCC | SqCa | Tissue | IHC | OS/DFS | Reported | 258 |
| Goncharuk, 200442 | America | Caucasian | 65 | R | NSCLC | AdenoCa | Tissue | IHC | OS | SC | NM |
| Su, 200450 | Japan | Asian | 64 | R | Bladder cancer | TCC | Tissue | IHC | RFS | Reported | 78 |
| Hashida, 200318 | Japan | Asian | 62.8 | R | Colorectal carcinoma | AdenoCa | Tissue | IHC | OS/DFS | Reported | 85.9 |
| Imai, 200243 | Japan | Asian | 62.6 | R | OSCC | SqCa | Tissue | qRT-PCR | OS | SC | 60 |
| Schindl, 200144 | Austria | Caucasian | 56.8 | R | Epithelial ovarian cancer | AdenoCa | Tissue | IHC | OS/DFS | Reported | 130 |
| Miyazaki, 200045 | Japan | Asian | 61.8 | R | ESCC | SqCa | Tissue | IHC | OS | SC | 195.2 |
| Yang, 200046 | America | Caucasian | NM | R | Breast cancer | AdenoCa | Tissue | IHC | OS | SC | NM |
| Sho, 199847 | Japan | Asian | 66 | R | Pancreatic cancer | AdenoCa | Tissue | IHC | OS | SC | 62 |
| Huang, 199848 | Japan | Asian | 50 | R | Breast cancer | AdenoCa | Tissue | IHC | OS/DFS | Reported | NM |
| Higashiyama, 199735 | Japan | Asian | 63.7 | R | NSCLC | AdenoCa | Tissue | IHC | OS | Reported | 61.3 |
| Adachi, 199649 | Japan | Asian | 60 | R | NSCLC | AdenoCa | Tissue | qRT-PCR | OS | Reported | 58 |
Notes:
Data extracted from one study due to different assay methods (IHC and qRT-PCR).
Data extracted from one study due to different follow-up times (120 and 60 months). Study design is described as retrospective.
Abbreviations: AdenoCa, adenocarcinoma; CCRCC, clear cell renal cell carcinoma; DFS, disease-free survival; ESCC, esophageal squamous cell carcinoma; HR, hazard ratio; IHC, immunohistochemistry; LSCC, laryngeal squamous cell carcinoma; NM, not mentioned; NSCLC, non-small-cell lung cancer; OS, overall survival; OSCC, oral squamous cell carcinoma; PFS, progression-free survival; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; R, retrospective; RFS, recurrence-free survival; SC, survival curve; SqCa, squamous carcinoma; TCC, transitional cell carcinoma; WB, Western-blot.
Figure 1.
Flow diagram of the study selection process.
Abbreviation: HRs, hazard ratios.
Data extraction
All data from eligible studies were extracted independently; ambiguous data were reviewed in detail. Parameters of these literatures were extracted from each single paper, including the first author’s surname, publication year, patients’ median or mean age, nationality, dominant ethnicity, number of patients, investigating method, cutoff value, follow-up time, and hazard ratios (HRs) for prognostic outcomes (overall survival [OS] and disease/recurrence/progression-free survival [DFS/RFS/PFS]) along with their 95% CI and p-values. If only Kaplan–Meier curves were available, data were extracted from graphical survival curves to extrapolate HRs with 95% CIs using previously described methods.20–22 All of the aforementioned data are comprehensively detailed in Tables 1 and 2.
Table 2.
HRs and 95% CIs of patient survival or cancer progression relating to CD82 expression in eligible studies
| First author, publication year | Assay method | Cutoff point | Case number
|
OS
|
DFS/RFS/PFS
|
|||
|---|---|---|---|---|---|---|---|---|
| High expression | Low expression | HR (95% CI) (U/M) | p-value | HR (95% CI) (U/M) | p-value | |||
| Zhu, 201710 | IHC | IRS scores ≥3 (range of 0–12) | 65 | 139 | 0.302 (0.184–0.497)M | <0.001 | NM | NM |
| Lu, 201612 | IHC | IRS scores ≥3 (range of 0–12) | 134 | 191 | 0.648 (0.492–0.854)M | 0.002 | NM | NM |
| Singh, 201611 | qRT-PCR | NM | 46 | 29 | 0.66 (0.26–1.68)Ue | 0.047 | NM | NM |
| Guo, 201516,a | IHC | >10% of tumor cells stained | 28 | 100 | 1.07 (0.22–5.24)Ue | <0.05 | NM | NM |
| Guo, 201516,b | qRT-PCR | NM | 40 | 88 | 0.62 (0.19–1.98)Ue | <0.05 | NM | NM |
| Wu, 201532 | IHC | IRS scores ≥3 (range of 0–12) | 56 | 118 | 0.430 (0.269–0.687)M | <0.001 | NM | NM |
| Han, 201536 | IHC | IRS scores ≥3 (range of 0–12) | 137 | 188 | 0.617 (0.462–0.823)M | 0.001 | NM | NM |
| Yu, 201413 | IHC | IRS scores ≥3 (range of 0–12) | 34 | 49 | 0.226 (0.094–0.545)M | 0.001 | 0.278 (0.119–0.647)M | 0.003 |
| Kwon, 201419 | IHC | .0% of tumor cells stained | 98 | 546 | 1.513 (1.057–2.165)M | 0.024 | f | f |
| Tang, 201414,c | IHC | IRS scores ≥8 (range of 0–12) | 114 | 303 | 0.42 (0.29–0.63)M | 0.0000147 | NM | NM |
| Tang, 201414,d | IHC | IRS scores ≥8 (range of 0–12) | 114 | 303 | 0.59 (0.36–0.96)M | 0.029 | NM | NM |
| Zhang, 201337 | IHC | Accumulated points ≥80 | 52 | 48 | 0.54 (0.27–1.05)Ue | NM | NM | NM |
| Zhang, 201338 | WB | NM | 43 | 43 | 0.36 (0.17–0.77)Ue | NM | NM | NM |
| Wu, 201215 | IHC | IRS scores >1 (range of 0–12) | 19 | 31 | 0.039 (0.007–0.236)M | 0.00 | NM | NM |
| Knoener, 201233 | IHC | IRS scores ≥3 (range of 0–12) | 168 | 103 | 0.77 (0.55–1.07)Ue | 0.2305 | NM | NM |
| Guo, 200939 | IHC | >10% of tumor cells stained | 21 | 59 | 0.81 (0.28–2.32)Ue | 0.022 | NM | NM |
| Protzel, 200834 | IHC | >50% of tumor cells stained | 17 | 13 | 0.34 (0.02–4.90)Ue | 0.0042 | NM | NM |
| Miyazaki, 200540 | IHC | Ki index ≥39 | 48 | 43 | 0.48 (0.22–1.03)Ue | 0.0023 | NM | NM |
| Leavey, 200517 | IHC | NM | 16 | 31 | 1.14 (0.27–4.81)Ue | 0.28 | 0.99 (0.35–2.85)Ue | 0.24 |
| Farhadieh, 200441 | IHC | >10% of tumor cells stained | 15 | 42 | 0.52 (0.27–1.01)M | 0.053 | 0.4 (0.2–0.8)M | 0.009 |
| Goncharuk, 200442 | IHC | Median | 77 | 27 | 0.47 (0.23–0.97)Ue | 0.034 | NM | NM |
| Su, 200450 | IHC | >50% of tumor cells stained | 54 | 33 | NM | NM | 0.255 (0.111–0.588)M | 0.0013 |
| Hashida, 200318 | IHC | Scores ≥120 (range of 0–300) | 63 | 83 | 0.903 (0.297–2.747)M | 0.858 | 1.054 (0.431–2.577)M | 0.909 |
| Imai, 200243 | qRT-PCR | NM | 25 | 18 | 0.51 (0.06–4.38)Ue | NM | NM | NM |
| Schindl, 200144 | IHC | Scores ≥4 (range of 3–7) | 48 | 59 | 0.45 (0.22–0.92)M | 0.0282 | 0.3 (0.09–0.93)M | 0.0372 |
| Miyazaki, 200045 | IHC | >10% of tumor cells stained | 36 | 19 | 0.41 (0.17–1.00)Ue | 0.024 | NM | NM |
| Yang, 200046 | IHC | >5% of tumor cells stained | 36 | 36 | 0.72 (0.20–2.61)Ue | NM | NM | NM |
| Sho, 199847 | IHC | >50% of tumor cells stained | 15 | 25 | 1.16 (0.29–4.64)Ue | 0.018 | NM | NM |
| Huang, 199848 | IHC | HSCORE >50 | 44 | 65 | 0.442 (0.109–1.789)M | 0.2528 | 0.360 (0.149–0.870)M | 0.0234 |
| Higashiyama, 199735 | IHC | >50% of tumor cells stained | 65 | 135 | 0.700 (0.502–0.976)M | 0.037 | f | f |
| Adachi, 199649 | qRT-PCR | Gene conservation rate value >1.2 | 35 | 116 | 0.416 (0.175–0.986)M | 0.046 | NM | NM |
Notes: The source of HR and 95% CI was extracted from survival curves or article reports.
Data extracted from one study due to different assay method (IHC and qRT-PCR).
Data extracted from one study due to different follow-up time (120 and 60 months).
HR calculated from survival curves.
Excluded due to its heterogeneity and publication bias.
Abbreviations: IHC, immunohistochemistry; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; WB, western-blot; NM, not mentioned; IRS, immunoreactivity score; HSCORE, the intensity and respective percentage cells that stain at each intensity were multiplied to reach a HSCORE that ranged from 0 to 300; OS, overall survival; DFS, disease-free survival; RFS, recurrence-free survival; PFS, progression-free survival; SC, survival curve; HR, hazard ratio; CI, confidence interval; U, univariate analysis; M, multivariate analysis.
Statistical analysis
We quantified the effect of heterogeneity via I2=100%× (Q - df)/Q. A random-effects model (DerSimonian–Laird method) was applied if p<0.10 or I2>50%; otherwise, a fixed-effects model (Mantel–Haenszel method) was used instead.23 In addition, we classified the enrolled studies into subgroups to reduce the influence of heterogeneity. The publication bias was evaluated by the Begg’s funnel plot and Egger linear regression test with a funnel plot.24 Sensitivity analysis was also tested. All p-values were calculated using a two-sided test, and p<0.05 was considered statistically significant. Statistical analyses were performed via the Stata 12.0 (StataCorp, College Station, TX, USA) and Microsoft Excel (V.2007, Microsoft Corporation, Redmond, WA, USA).
Results
Summary of enrolled studies
A total of 157 studies were retrieved from PubMed, EMBASE, and Web of Science. After initial scanning of titles and abstracts, 93 studies were excluded because they were review articles/letters or non-English publications, were not associated with CD82 or prognosis/survival. The remaining 64 records were downloaded as full text and accessed very carefully. Among them, 35 potentially suitable studies were excluded because they lacked sufficient survival data (HRs and 95% CIs), did not report comprehensive data, or extracted their survival data from an existing database. Ultimately, 29 studies were considered eligible for our meta-analysis. The inclusion and exclusion reasons for candidate studies are presented in detail in Figure 1.
The main features of the 29 enrolled studies are systematically summarized in Tables 1 and 2. Twenty-two studies reported patient OS, one focused on RFS, and six investigated OS as well as DFS or PFS. Eight of the studies focused on Caucasian populations, which mainly came from European countries, and 21 focused on Asian populations, of which 10 were from China, 9 from Japan, 1 from India, and 1 from Korea. As for cancer type, malignant neoplasms assessed in these studies included colorectal carcinoma, gastric carcinoma, breast cancer, laryngeal squamous cell carcinoma (LSCC), esophageal squamous cell carcinoma (ESCC), and oral squamous cell carcinoma (OSCC). Among the 29 studies, the pathological types of adenocarcinoma (AdenoCa), squamous carcinoma (SqCa), transitional cell carcinoma (TCC), sarcoma, and melanoma were covered. All of these studies were retrospective in design and determined CD82 expression using tissue samples. IHC and qRT-PCR were used in the majority of all eligible studies to detect CD82 expression, and Western-blot analysis was conducted in one study. When we analyzed the HR and 95% CI in each study, we extracted two sets of data from the same article due to different assay methods or different follow-up times in two studies (Guo et al16 and Tang et al14). The source of HR and 95% CI was extracted from survival curves or article reports.
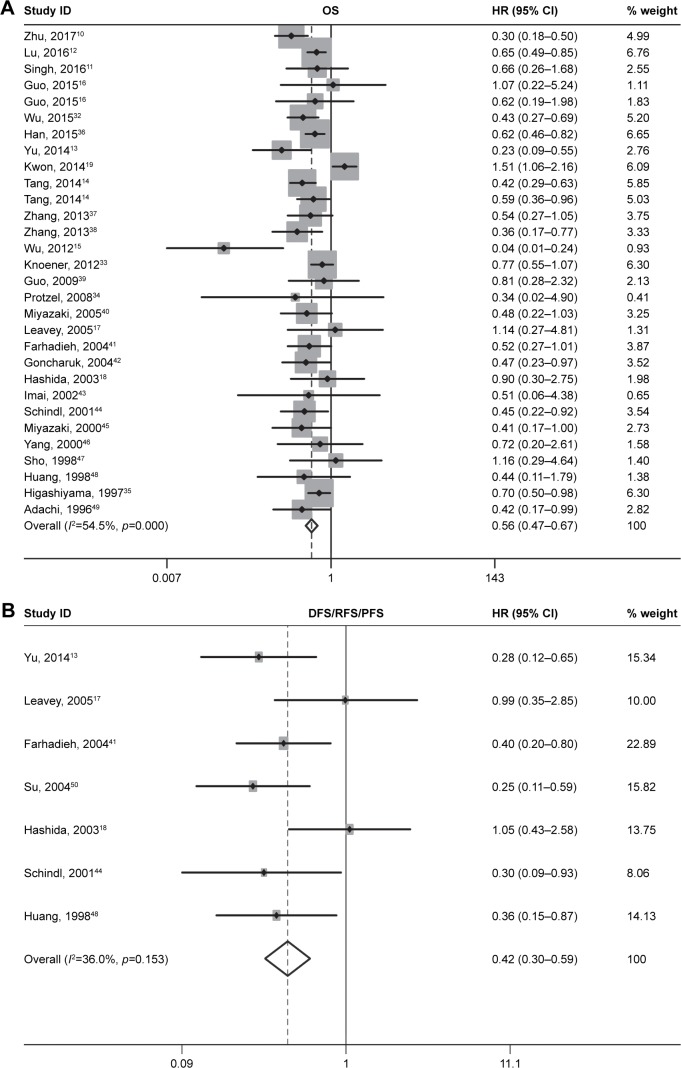
OS associated with CD82 expression
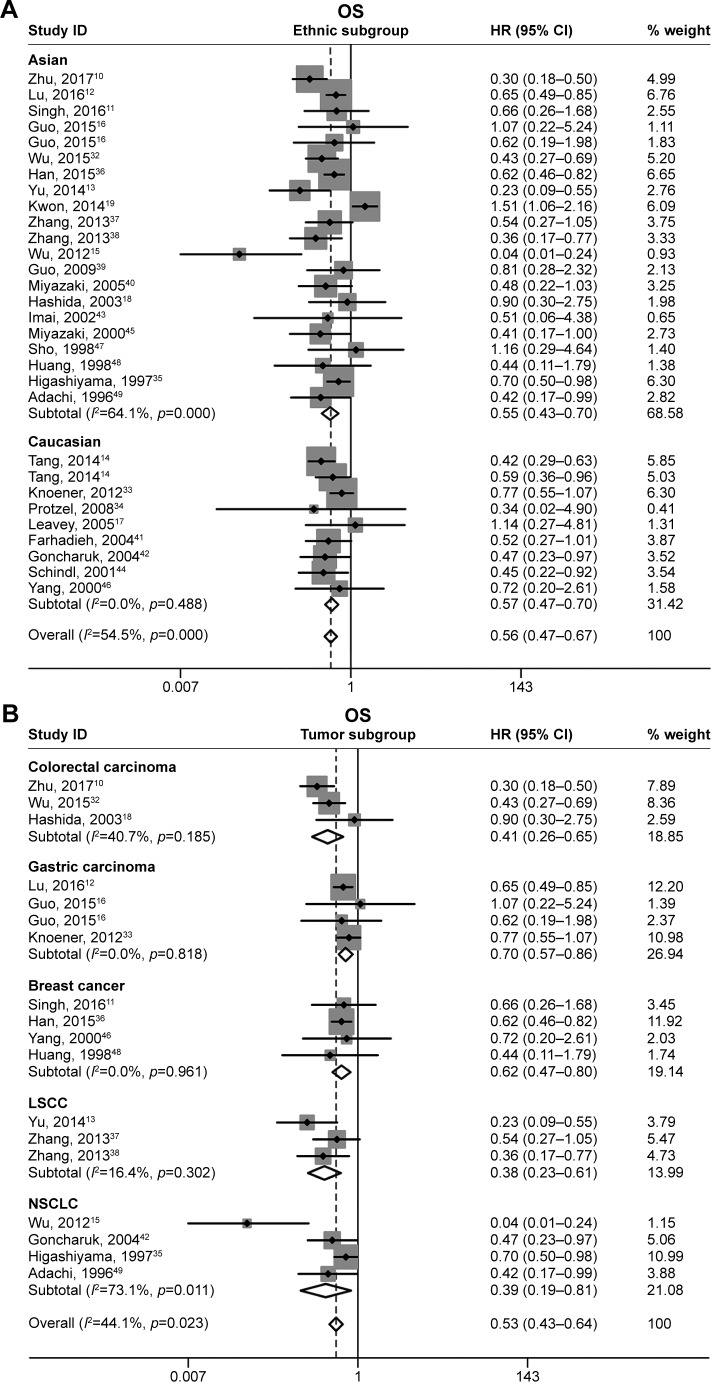
A total of 28 original studies were included to analyze the OS, with a random-effects model on account of the moderate heterogeneity (p=0.000, I2=54.5%). The results indicated that CD82-positive expression was significantly associated with favorable OS in cancer patients (pooled HR =0.56, 95% CI: 0.47–0.67, p<0.05; Figure 2A). Subgroup study was then performed; increased CD82 expression was significantly associated with enhanced OS in the Asian patients (pooled HR =0.55, 95% CI: 0.43–0.70) as well in Caucasian (pooled HR =0.57, 95% CI: 0.47–0.70; Figure 3A). In tumor subgroup analysis, we found high expression of CD82 correlated with longer OS in colorectal carcinoma, gastric carcinoma, breast cancer, LSCC, and non-small-cell lung cancer (NSCLC) (Figure 3B). Due to insufficient studies, correlation between CD82 and OS in other tumor types have not been further analyzed. Stratification analyses for other subgroups are presented in detail in Figure 3C and D.
Figure 2.
Forest plots of combined analyses associated with CD82 expression.
Notes: (A) OS and (B) DFS/RFS/PFS. Weights are from random effects analysis.
Abbreviations: DFS, disease-free survival; HR, hazard ratio; ID, identifier; OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival.
Figure 3.
Forest plots of stratified analysis of the OS.
Notes: (A) Stratified by ethnic subgroup, (B) stratified by tumor subgroup, (C) stratified by pathological subgroup, and (D) stratified by assay method subgroup. Weights are from random effects analysis.
Abbreviations: AdenoCa, adenocarcinoma; HR, hazard ratio; ID, identifier; IHC, immunohistochemistry; LSCC, laryngeal squamous cell carcinoma; NSCLC, non-small-cell lung cancer; OS, overall survival; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SqCa, squamous carcinoma.
DFS/RFS/PFS associated with CD82 expression
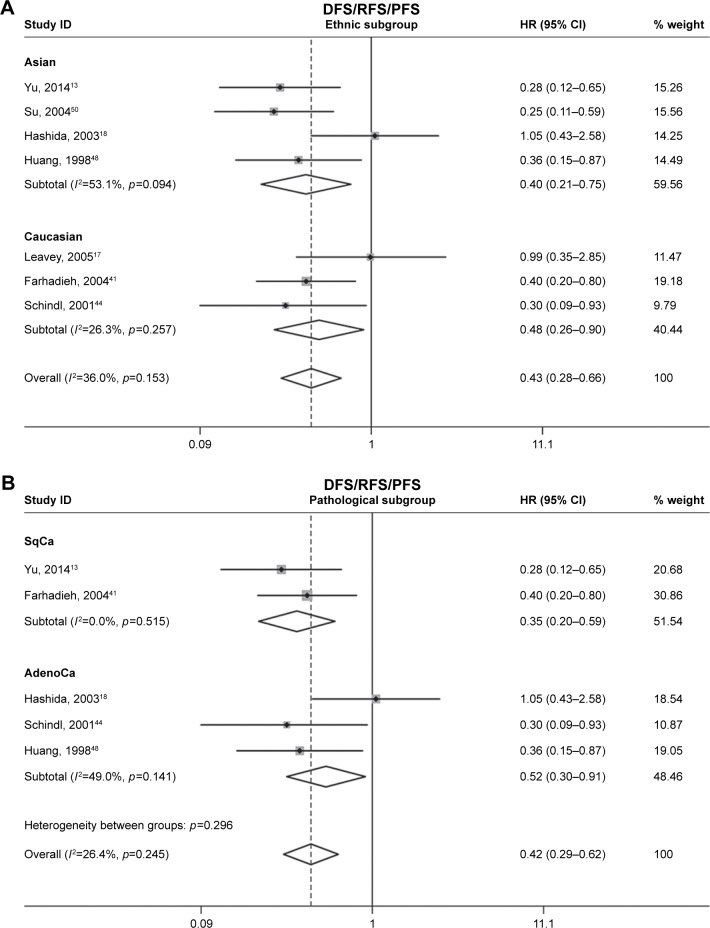
Seven of the studies analyzed DFS/RFS/PFS. The heterogeneity between these studies was low (p=0.153, I2=36.0%); thus, a fixed-effects model was applied to calculate a pooled HR of 0.42 (95% CI: 0.30–0.59). This result demonstrated that CD82 overexpression predicted low risk of cancer progression (Figure 2B). Ethnic and pathological subgroup analysis for DFS/RFS/PFS also demonstrated the protective effect of CD82 and that it may play a key role of progression in cancer patients (Figure 4).
Figure 4.
Forest plots of stratified analysis of the DFS/RFS/PFS.
Notes: (A) Stratified by ethnic subgroup and (B) stratified by pathological subgroup. Weights are from random effects analysis.
Abbreviations: AdenoCa, adenocarcinoma; DFS, disease-free survival; HR, hazard ratio; ID, identifier; OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival; SqCa, squamous carcinoma.
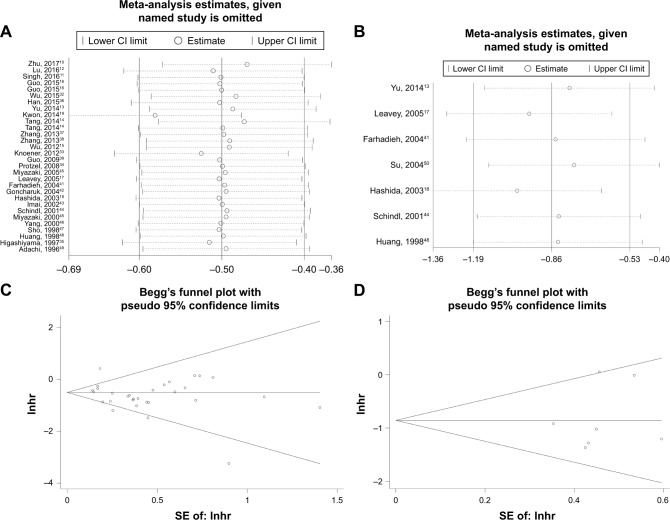
Sensitivity analyses
In order to determine the robustness of the above results and evaluate the stability of results, a sensitivity analysis was performed by Stata12.0 software. Individual data that could affect the final conclusions have been deleted in advance (see footnote “f” in Table 2). The analyzed result from a fixed model suggested that our results are comparatively credible and stable (Figure 5A and B).
Figure 5.
Sensitivity analysis under specific model and Begg’s funnel plots of publication bias test.
Notes: (A) Effect of individual studies on the combined HR for OS, (B) effect of individual studies on the combined HR for DFS/RFS/PFS, (C) Begg’s funnel plots of OS, and (D) Begg’s funnel plots of DFS/RFS/PFS.
Abbreviations: DFS, disease-free survival; HR, hazard ratio; OS, overall survival; PFS, progression-free survival; RFS, recurrence-free survival.
Publication bias
Begg’s funnel and the Egger’s test were used to evaluate the possible publication bias in this meta-analysis. The funnel plots of the publication bias are presented in Figure 5C and D. p-values calculated from Egger’s test with higher detection effectiveness were 0.135 for OS, 0.610 for DFS/RFS/PFS, respectively, indicating no significant publication bias in the meta-analysis.
Discussion
In recent years, elaborate efforts have been invested to detect promising biomarkers for patients with multiple carcinomas. Tetraspanins are a family of integral membrane proteins with four transmembrane helices, a small extracellular loop, and a large extracellular loop. Recent studies have revealed the importance of tetraspanins in solid tumors and hematologic malignancies, and the expression of tetraspanins is associated with the tumor biologic characteristics.4 Some members of this family are known as metastasis suppressor genes, while others are supposed to promote tumor progression.1
CD82 is considered to be important in tetraspanin family given its differential expression between cancer and normal tissues. CD82 is downregulated in many types of cancers and loss of CD82, both protein and mRNA, is strongly correlated with poor prognosis in many malignancies.9,25 Current understanding of CD82 function indicates that it is likely to be involved in detachment, motility/invasion, and cell survival, which are associated with various adhesion receptors (eg, integrins), receptor tyrosine kinases (eg, epithelial growth factor receptor [EGFR] and c-Met), and other signaling pathway molecules.4,9,26 CD82 can interact with other tetraspanin proteins (eg, CD151 and CD81), integrins (eg, α3β1, α4β1, and α5β1), and chemokines to regulate the migration, adhesion, and signaling of cells.9,27,28 Integrins are not the only molecules that CD82 regulate, and studies have revealed that tetraspanins play a critical role in regulating receptor tyrosine kinase signaling in immune cells.29,30 Odintsova et al reported that CD82 was a regulator of epithelial growth factor (EGF)-induced signaling and showed that the association of EGFR with the tetraspanin is critical in EGFR desensitization.31 The diverse biological activities and molecular mechanisms of CD82 may partially contribute to the tumor prognosis in cancer patients.
In this meta-analysis, we first evaluated the correlation of CD82 with prognostic outcomes (OS, DFS/RFS/PFS) of patients with various cancers systemically. We also performed subgroup, sensitivity, and heterogeneity analyses to explore the effects of dominant characteristics from available studies. Many studies have reported that decreased expression of CD82 often predicts unfavorable outcome in cancers and the level of CD82 was negatively correlated with invasion of depth, vessel invasion, lymph node metastasis, distant metastasis, and TNM stages.10,16,32–34 However, there are also some individual studies that came to a diverse, even opposing, conclusion in our included literature. Our prognosis analysis revealed the pooled HR of 0.56 (OS) and 0.42 (DFS/RFS/PFS), demonstrating that increased CD82 expression could be a favorable prognostic factor of various tumors. Similarly, in subgroup analysis based on the characteristics of the individual studies, we observed the statistically significant outcomes when data were stratified according to ethnicity, pathology, assay method, and tumor types. Although AdenoCa and squamous cell carcinoma have oncologically different characteristics, according to our study, high expression of CD82 correlated with longer OS or DFS/RFS/PFS in both types. Due to few relevant studies and small sample size, results of some tumors and pathologic types were not presented in the forest plot when we conducted the subgroup analysis. This means the conclusion should be considered with caution when we come across these neoplasms, including hepatocellular carcinoma, clear cell renal cell carcinoma, melanoma, osteosarcoma, and pancreatic cancer. Therefore, more existing investigations toward these tumors are needed for further evidence.
After extracting the data from studies, sensitivity analysis was conducted to check which individual data could affect the final conclusions. When we analyzed the stability of results, we found that if the data toward DFS/RFS/PFS from Higashiyama et al35 and Kwon et al19 were included in our review, they might have significant impact on the pooled significance. Therefore, we deleted these two sets of data in advance to ensure the stability of our analysis. The final results were assured that exclusion of any individual study alters little change of the pooled significance. We attributed this to the different ethnicity, tumor type, and pathological type, as well as the different source of HR. No obvious publication bias was detected in this meta-analysis, indicating our analysis was stable.
Despite the meta-analysis was performed with rigorous statistics, our conclusion still has several limitations for the following reasons. First, heterogeneity existed in the OS analyses and it was likely due to the different characteristics of the patients, such as age, ethnicity, tumor type, and pathological type, as well as the different source of HR. Second, most studies established their own varied expression cutoff, and a standard cutoff value was hard to define so that the pooled outcome may be different with the actual value. This may cause a bias in the results of the effectiveness of CD82 as a prognostic factor. Third, no independent investigation on Negroid was included in this meta-analysis, which might undermine the comprehensiveness to some extent. Fourth, the validity of results might be impaired due to the lack of prospective studies. Taking these limitations into consideration, our results should be interpreted rigorously, and more well-designed studies are needed to verify the function of CD82 in various carcinomas.
Conclusion
In summary, the significant relationship between high CD82 expression and favorable prognostic in various neoplasms was clearly revealed in this meta-analysis. The results indicated that CD82 could be a promising biomarker for predicting the prognosis of patients with malignant neoplasms, and the biological functions of CD82 are of great research value of the subject.
Acknowledgments
This study was funded by National Natural Science Foundation of China (grant numbers 81270685 and 81402321), and Project of Nanjing Science and Technology Committee (201605001).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Zoller M. Gastrointestinal tumors: metastasis and tetraspanins. Z Gastroenterol. 2006;44(7):573–586. doi: 10.1055/s-2006-926795. [DOI] [PubMed] [Google Scholar]
- 2.Yunta M, Lazo PA. Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell Signal. 2003;15(6):559–564. doi: 10.1016/s0898-6568(02)00147-x. [DOI] [PubMed] [Google Scholar]
- 3.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 4.Yang YG, Sari IN, Zia MF, Lee SR, Song SJ, Kwon HY. Tetraspanins: spanning from solid tumors to hematologic malignancies. Exp Hematol. 2016;44(5):322–328. doi: 10.1016/j.exphem.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Richardson MM, Jennings LK, Zhang XA. Tetraspanins and tumor progression. Clin Exp Metastasis. 2011;28(3):261–270. doi: 10.1007/s10585-010-9365-5. [DOI] [PubMed] [Google Scholar]
- 6.You J, Madigan MC, Rowe A, Sajinovic M, Russell PJ, Jackson P. An inverse relationship between KAI1 expression, invasive ability, and MMP-2 expression and activity in bladder cancer cell lines. Urol Oncol. 2012;30(4):502–508. doi: 10.1016/j.urolonc.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Iiizumi-Gairani M, Okuda H, et al. KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NFkappaB complex in human prostate cancer. J Biol Chem. 2011;286(21):18949–18959. doi: 10.1074/jbc.M111.232637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christgen M, Christgen H, Heil C, et al. Expression of KAI1/CD82 in distant metastases from estrogen receptor-negative breast cancer. Cancer Sci. 2009;100(9):1767–1771. doi: 10.1111/j.1349-7006.2009.01231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranti CK. Controlling cell surface dynamics and signaling: how CD82/KAI1 suppresses metastasis. Cell Signal. 2009;21(2):196–211. doi: 10.1016/j.cellsig.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Zhu B, Zhou L, Yu L, et al. Evaluation of the correlation of vasculogenic mimicry, ALDH1, KAI1 and microvessel density in the prediction of metastasis and prognosis in colorectal carcinoma. BMC Surg. 2017;17(1):47. doi: 10.1186/s12893-017-0246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh R, Bhatt ML, Singh SP, et al. Expression levels of Tetraspanin KAI1/CD82 in breast cancers in North Indian females. Asian Pac J Cancer Prev. 2016;17(7):3431–3436. [PubMed] [Google Scholar]
- 12.Lu G, Zhou L, Zhang X, et al. The expression of metastasis-associated in colon cancer-1 and KAI1 in gastric adenocarcinoma and their clinical significance. World J Surg Oncol. 2016;14(1):276. doi: 10.1186/s12957-016-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu L, Zhou L, Wu S, et al. Clinicopathological significance of cancer stem cells marked by CD133 and KAI1/CD82 expression in laryngeal squamous cell carcinoma. World J Surg Oncol. 2014;12:118. doi: 10.1186/1477-7819-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang Y, Cheng Y, Martinka M, Ong CJ, Li G. Prognostic significance of KAI1/CD82 in human melanoma and its role in cell migration and invasion through the regulation of ING4. Carcinogenesis. 2014;35(1):86–95. doi: 10.1093/carcin/bgt346. [DOI] [PubMed] [Google Scholar]
- 15.Shiwu WU, Lan Y, Wenqing S, Lei Z, Yisheng T. Expression and clinical significance of CD82/KAI1 and E-cadherin in non-small cell lung cancer. Arch Iran Med. 2012;15(11):707–712. [PubMed] [Google Scholar]
- 16.Guo J, Fan KX, Xie LI, et al. Effect and prognostic significance of the KAI1 gene in human gastric carcinoma. Oncol Lett. 2015;10(4):2035–2042. doi: 10.3892/ol.2015.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leavey PJ, Timmons C, Frawley W, Lombardi D, Ashfaq R. KAI-1 expression in pediatric high-grade osteosarcoma. Pediatr Dev Pathol. 2006;9(3):219–224. doi: 10.2350/11-05-0137.1. [DOI] [PubMed] [Google Scholar]
- 18.Hashida H, Takabayashi A, Tokuhara T, et al. Clinical significance of transmembrane 4 superfamily in colon cancer. Br J Cancer. 2003;89(1):158–167. doi: 10.1038/sj.bjc.6601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon HJ, Min SY, Park MJ, et al. Expression of CD9 and CD82 in clear cell renal cell carcinoma and its clinical significance. Pathol Res Pract. 2014;210(5):285–290. doi: 10.1016/j.prp.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta-analysis with time-to-event outcomes. Stat Med. 2002;21(22):3337–3351. doi: 10.1002/sim.1303. [DOI] [PubMed] [Google Scholar]
- 21.DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. 2015;45(Pt A):139–145. doi: 10.1016/j.cct.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 24.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng J, Huang C, Wren JD, et al. Tetraspanin CD82: a suppressor of solid tumors and a modulator of membrane heterogeneity. Cancer Metastasis Rev. 2015;34(4):619–633. doi: 10.1007/s10555-015-9585-x. [DOI] [PubMed] [Google Scholar]
- 26.Romanska HM, Berditchevski F. Tetraspanins in human epithelial malignancies. J Pathol. 2011;223(1):4–14. doi: 10.1002/path.2779. [DOI] [PubMed] [Google Scholar]
- 27.Malik FA, Sanders AJ, Jiang WG. KAI-1/CD82, the molecule and clinical implication in cancer and cancer metastasis. Histol Histopathol. 2009;24(4):519–530. doi: 10.14670/HH-24.519. [DOI] [PubMed] [Google Scholar]
- 28.Lee J, Byun HJ, Lee MS, et al. The metastasis suppressor CD82/KAI1 inhibits fibronectin adhesion-induced epithelial-to-mesenchymal transition in prostate cancer cells by repressing the associated integrin signaling. Oncotarget. 2017;8(1):1641–1654. doi: 10.18632/oncotarget.13767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaguillaumie A, Harriague J, Kohanna S, et al. Tetraspanin CD82 controls the association of cholesterol-dependent microdomains with the actin cytoskeleton in T lymphocytes: relevance to co-stimulation. J Cell Sci. 2004;117(Pt 22):5269–5282. doi: 10.1242/jcs.01380. [DOI] [PubMed] [Google Scholar]
- 30.Odintsova E, Voortman J, Gilbert E, Berditchevski F. Tetraspanin CD82 regulates compartmentalisation and ligand-induced dimerization of EGFR. J Cell Sci. 2003;116(Pt 22):4557–4566. doi: 10.1242/jcs.00793. [DOI] [PubMed] [Google Scholar]
- 31.Odintsova E, Sugiura T, Berditchevski F. Attenuation of EGF receptor signaling by a metastasis suppressor, the tetraspanin CD82/KAI-1. Curr Biol. 2000;10(16):1009–1012. doi: 10.1016/s0960-9822(00)00652-7. [DOI] [PubMed] [Google Scholar]
- 32.Wu Q, Yang Y, Wu S, et al. Evaluation of the correlation of KAI1/CD82, CD44, MMP7 and beta-catenin in the prediction of prognosis and metastasis in colorectal carcinoma. Diagn Pathol. 2015;10:176. doi: 10.1186/s13000-015-0411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pal’Tseva EM, Samofalova O, Gorbacheva I, Tsar’Kov PV. Expression of some biomarkers in primary colon adenocarcinomas and their lymph node metastases. Arkh Patol. 2012;74(4):12–18. [PubMed] [Google Scholar]
- 34.Protzel C, Kakies C, Kleist B, Poetsch M, Giebel J. Down-regulation of the metastasis suppressor protein KAI1/CD82 correlates with occurrence of metastasis, prognosis and presence of HPV DNA in human penile squamous cell carcinoma. Virchows Arch. 2008;452(4):369–375. doi: 10.1007/s00428-008-0590-0. [DOI] [PubMed] [Google Scholar]
- 35.Higashiyama M, Kodama K, Yokouchi H, et al. KAI1/CD82 expression in nonsmall cell lung carcinoma is a novel, favorable prognostic factor: an immunohistochemical analysis. Cancer-Am Cancer Soc. 1998;83(3):466–474. doi: 10.1002/(sici)1097-0142(19980801)83:3<466::aid-cncr15>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 36.Han Z, Chen Z, Zheng R, Cheng Z, Gong X, Wang D. Clinicopathological significance of CD133 and CD44 expression in infiltrating ductal carcinoma and their relationship to angiogenesis. World J Surg Oncol. 2015;13:56. doi: 10.1186/s12957-015-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang BH, Liu W, Li L, et al. KAI1/CD82 and MRP1/CD9 serve as markers of infiltration, metastasis, and prognosis in laryngeal squamous cell carcinomas. Asian Pac J Cancer Prev. 2013;14(6):3521–3526. doi: 10.7314/apjcp.2013.14.6.3521. [DOI] [PubMed] [Google Scholar]
- 38.Zhang B, Liu W, Li L, et al. KAI1/CD82 and cyclin D1 as biomarkers of invasion, metastasis and prognosis of laryngeal squamous cell carcinoma. Int J Clin Exp Pathol. 2013;6(6):1060–1067. [PMC free article] [PubMed] [Google Scholar]
- 39.Guo C, Liu QG, Zhang L, Song T, Yang X. Expression and clinical significance of p53, JunB and KAI1/CD82 in human hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2009;8(4):389–396. [PubMed] [Google Scholar]
- 40.Miyazaki T, Kato H, Kimura H, et al. Evaluation of tumor malignancy in esophageal squamous cell carcinoma using different characteristic factors. Anticancer Res. 2005;25(6B):4005–4011. [PubMed] [Google Scholar]
- 41.Farhadieh RD, Smee R, Ow K, et al. Down-regulation of KAI1/CD82 protein expression in oral cancer correlates with reduced disease free survival and overall patient survival. Cancer Lett. 2004;213(1):91–98. doi: 10.1016/j.canlet.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Goncharuk VN, Del-Rosario A, Kren L, et al. Co-downregulation of PTEN, KAI-1, and nm23-H1 tumor/metastasis suppressor proteins in non-small cell lung cancer. Ann Diagn Pathol. 2004;8(1):6–16. doi: 10.1016/j.anndiagpath.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Imai Y, Sasaki T, Shinagawa Y, Akimoto K, Fujibayashi T. Expression of metastasis suppressor gene (KAI1/CD82) in oral squamous cell carcinoma and its clinico-pathological significance. Oral Oncol. 2002;38(6):557–561. doi: 10.1016/s1368-8375(01)00120-8. [DOI] [PubMed] [Google Scholar]
- 44.Schindl M, Birner P, Breitenecker G, Oberhuber G. Downregulation of KAI1 metastasis suppressor protein is associated with a dismal prognosis in epithelial ovarian cancer. Gynecol Oncol. 2001;83(2):244–248. doi: 10.1006/gyno.2001.6366. [DOI] [PubMed] [Google Scholar]
- 45.Miyazaki T, Kato H, Shitara Y, et al. Mutation and expression of the metastasis suppressor gene KAI1 in esophageal squamous cell carcinoma. Cancer-Am Cancer Soc. 2000;89(5):955–962. doi: 10.1002/1097-0142(20000901)89:5<955::aid-cncr3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, Wei L, Tang C, Slack R, Montgomery E, Lippman M. KAI1 protein is down-regulated during the progression of human breast cancer. Clin Cancer Res. 2000;6(9):3424–3429. [PubMed] [Google Scholar]
- 47.Sho M, Adachi M, Taki T, et al. Transmembrane 4 superfamily as a prognostic factor in pancreatic cancer. Int J Cancer. 1998;79(5):509–516. doi: 10.1002/(sici)1097-0215(19981023)79:5<509::aid-ijc11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 48.Huang CI, Kohno N, Ogawa E, Adachi M, Taki T, Miyake M. Correlation of reduction in MRP-1/CD9 and KAI1/CD82 expression with recurrences in breast cancer patients. Am J Pathol. 1998;153(3):973–983. doi: 10.1016/s0002-9440(10)65639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adachi M, Taki T, Ieki Y, Huang CL, Higashiyama M, Miyake M. Correlation of KAI1/CD82 gene expression with good prognosis in patients with non-small cell lung cancer. Cancer Res. 1996;56(8):1751–1755. [PubMed] [Google Scholar]
- 50.Su JS, Arima K, Hasegawa M, et al. Decreased expression of KAI1 metastasis suppressor gene is a recurrence predictor in primary pTa and pT1 urothelial bladder carcinoma. Int J Urol. 2004;11(2):74–82. doi: 10.1111/j.1442-2042.2004.00752.x. [DOI] [PubMed] [Google Scholar]