Abstract
A new automated real-time PCR assay for the detection of rifampicin (RIF) and isoniazid (INH) resistance in Mycobacterium tuberculosis (MTB) was evaluated. A total of 163 clinical samples (128 pulmonary and 35 extra-pulmonary) were processed using four PCR assay kits: Abbott RealTime MTB RIF/INH, Genotype MTBDRplus, Xpert/MTB RIF, and Anyplex MTB/MDR. The results of phenotypic drug-susceptibility testing using BACTECMGIT 960 were used as reference. The sensitivity and specificity of the new Abbott RealTime MTB RIF/INH assay in comparison with phenotypic testing was 96.3% (95%CI 87.32%–100%) for RIF and 100% (95%CI 99.3%–100%) for INH; the sensitivity was 78.8% (95%CI 66.8%–90.9%) and the specificity was 100% (95%CI 98.9%–100%). The Abbott RealTime MTB RIF/INH test could be a valid method for detecting the most common mutations in strains resistant to RIF and INH.
Keywords: Abbott RealTime MTB RIF/INH Resistance assay, M. tuberculosis, mutations
Introduction
Drug-resistant tuberculosis (TB) continues to threaten global TB control and remains a major public health concern in many countries. Globally, an estimated 3.3% of new cases and 20% of previously treated cases have multidrug-resistant TB (MDR-TB). The World Health Organization’s (WHO’s) End TB Strategy calls for the early diagnosis of TB and universal drug-susceptibility testing (DST), highlighting the critical role of laboratories in the post-2015 era in rapidly and accurately detecting TB and drug resistance.1,2 Surveillance of drug resistance in TB over the past 2 decades has informed and guided the response to the epidemic. Molecular tests for detecting drug resistance to rifampicin (RIF) alone or in combination with resistance to isoniazid (INH) have been recommended for use by WHO.3
The latest WHO data offer an estimated 10.4 million new (incident) TB cases worldwide, of which 1.2 million (11%) were people living with HIV, 480,000 new cases of MDR-TB, and ~190,000 deaths from MDR-TB. On average, an estimated 9.7% of people with MDR-TB have extensive drug resistant-TB. It is estimated that up to 50 million people may be infected with drug-resistant TB.2
The WHO European region is the area most affected by MDR-TB in the entire world. Of the ten countries in the world with the highest burden of MDR-TB, nine are in the European region, with ~190,000 deaths from MDR-TB per year. In 2015, estimated percentages of MDR among new and previously treated TB cases in this region were 16% and 48%, respectively.4
There was a US$2 billion funding shortfall for the implementation of TB treatment in 2016 and over US$1 billion for TB research. In industrialized countries, TB treatment costs approximately US$250,000 for patients with drug-resistant TB. MDR-TB is currently responsible for 150,000 deaths per year. WHO reports that only 7% of MDR-TB cases are diagnosed.1,2
Until recently, systems for detecting resistance using solid media were slow and laborious. The development of a modern liquid-medium, non-radiometric system (Bactec™ MGIT™ 960 System; BD, Franklin Lakes, NJ, USA) has done much to speed up the detection process, allowing the testing of any patient requiring it.5
Recent innovations in molecular tests are facilitating the shift from periodic surveys to routine surveillance. Rapid molecular tests such as Genotype MTBDRplus (Hain Life-science, Nehren, Germany),6 Xpert/MTB RIF assay (Cepheid AB, Bromma, Sweden),7 and Anyplex MTB/MDR (Seegene Technologies, Concord, CA, USA)8 provide results much faster than conventional methods, do not require sophisticated laboratory infrastructure, and decrease cost.
WHO indicates the approval of use of these molecular tests for rapid detection of MDR-TB should be decided by the ministries of health of each country in the context of national plans for the proper management of patients with MDR-TB. These tests must be adequately validated for use in sputum smear positive for Mycobacterium tuberculosis (MTB). The use of commercial methods, experienced centers, and adequate funding are recommended. Therefore, phenotypic tests are still necessary.
The Abbott RealTime MTB RIF/INH Resistance assay (Abbott Laboratories, Abbott Park, IL, USA) is a new companion assay to the Abbott RealTime MTB,9 for the qualitative detection of RIF and INH resistance in MTB-positive respiratory samples via a fully automated process.
In this study, we have evaluated the new automated Abbott RealTime MTB RIF/INH Resistance assay using pulmonary and extra-pulmonary clinical samples, in a comparison with three other marketed PCR test kits, using a standard culture phenotypic system as the reference method.
Materials and methods
A total of 163 frozen decontaminated clinical samples (128 pulmonary and 35 extra-pulmonary) were processed (Figure 1). The sources of the samples were: 103 sputum, 25 bronchial aspirate, 21 biopsy, one fistula of chest wall, one cervical fistula, three gastric aspirate, and nine exudates. Under the directive of the Official Gazette of the Government of Spain, Real Decreto 1716/2011, no ethics approval is required for the use of anonymized clinical samples which constitute customary professional diagnostic procedures.
Figure 1.
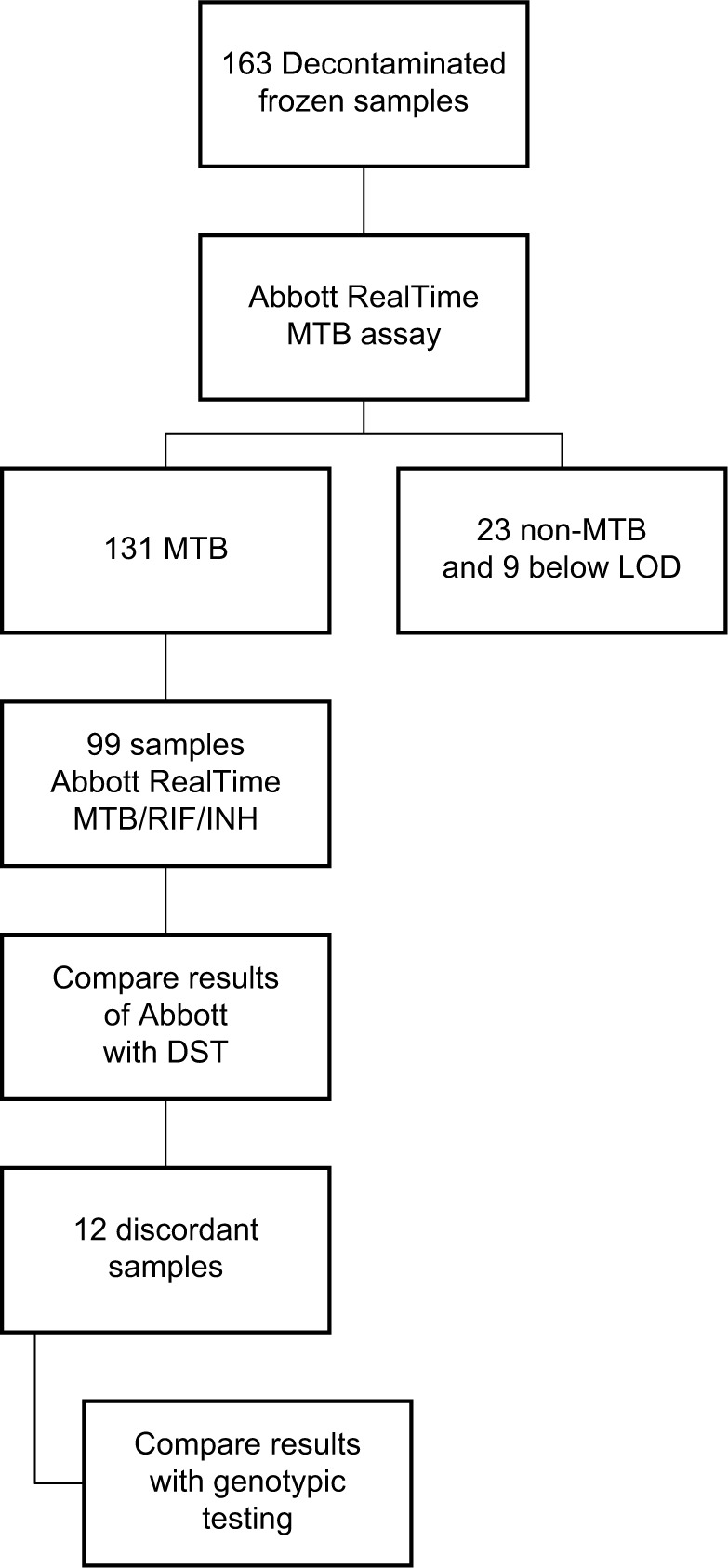
Flow diagram for Abbott RealTime MTB RIF/INH evaluation.
Abbreviations: MTB, Mycobacterium tuberculosis; RIF, rifampicin; INH, isoniazid; LOD, level of detection; DST, drug-susceptibility testing.
All samples were processed with the new automated Abbott RealTime MTB RIF/INH Resistance assay. To inactivate MTB, inactivation reagent was added to the samples (in a 1:3 ratio) and they were incubated for 1 hour. DNA extraction, amplification, and detection were performed automatically with the instrument Abbott m2000rt, which had been set up with the appropriate reagents according to manufacturer’s instructions. After inactivation, samples were inoculated on a 24-well plate – 22 samples and two controls, positive and negative. The extraction and preparation of DNA was performed with the lysing reagent and magnetic microparticles. When identification results were “MTB detected”, the detection of resistance was considered. Then, the amplification mixture was prepared and added to the Abbott system, and the amplification was made. Targets were rpoB (RIF resistance), katG and inhA promoter regions (resistance to INH). The detection was determined by the joint interpretation of 12 signals from different probes: eight wild-type rpoB probes and probes at katG e inhA.10
In this study, 131 cases of MTB were identified with the RT MTB, and the resistance to RIF and INH was determined in 99 samples, with Abbott RealTime MTB INH/RIF (RT MTB INH/RIF) (23 samples were identified as non-TB mycobacteria and nine samples with low level of detection [LOD] were not considered in this study). MTB resistance to RIF was detected with eight rpoB wild-type probes and eight mutation probes; MTB resistance to INH was detected with two wild-type probes and two mutation probes, one wild-type probe with mutation probes in inhA and one wild-probe with mutation in katG. All samples had been previously identified with Genotype CM/AS (Hain Lifescience, Nehren, Germany). The results previously obtained of phenotypic DST using BACTECMGIT 960 were used for reference. When there were discrepancies between results from the molecular test Abbott RealTime MTB INH/RIF (RT MTB INH/RIF) and phenotypic DST, the following tests were performed, according to the manufacturer’s instructions, to confirm the results: Genotype MTBDR plus, Xpert/MTB RIF assay, and Anyplex MTB/MDR.
Statistical analysis
Statistical analyses were carried out using EpiData version 3.1 software (EpiData Association, Copenhagen, Denmark) to calculate sensitivity, specificity, negative and positive predictive values, and kappa index of concordance between techniques. We determined 95% CIs.
Results
Of the 163 frozen clinical samples processed with the Abbott RealTime MTB, in nine (seven pulmonary and two extra-pulmonary) samples, extracted DNA was detected as “low”. One hundred and thirty-one samples were identified as MTB and 23 samples were identified as non-MTB. All samples were processed with the Genotype Mycobacteria CM/AS (131 MTB and 23 different non-TB mycobacteria).
Of the 131 MTB samples, 99 were tested for resistance, using RealTime MTB INH/RIF (RT MTB INH/RIF). The results were compared with the phenotypic test (Table 1). Of the 72 samples that were phenotypically susceptible to RIF, no rpoB mutation was detected by Abbott RealTime MTB RIF/INH, and of the 27 samples that were resistant to RIF, Abbott RealTime MTB INH/RIF detected resistance mutations in 26 of them. Only in one phenotypically RIF resistant sample was the mutation not detected. With the Genotype MTBDRplus, Xpert/MTB RIF assay, and Anyplex MTB/MDR, the mutation was detected. For the RIF resistance mutation, the Abbott RealTime MTB RIF/INH test demonstrated 96.3% (95%CI 87.32%–100%) sensitivity and 100% (95%CI 99.3%–100%) specificity. The kappa coefficient between the Abbott Real-Time MTB RIF/INH Resistance assay and the indirect DST results for RIF was 0.97 (95%CI 0.92–1), classified according to the Landis and Koch scale as “almost perfect agreement”. Of the 47 samples that were phenotypically susceptible to INH, no mutations in InhA or Kat G were detected. Of the 52 phenotypically INH resistant samples, eleven showed no mutations with any of the molecular methods tested. For INH, the Abbott RealTime MTB RIF/INH test displayed 78.8% (95%CI 66.8%–90.9%) sensitivity and 100% (95%CI 98.9%–100%) specificity. The kappa coefficient between the Abbott RealTime MTB RIF/INH Resistance assay and the indirect DST results was 0.78 (95%CI 0.66–0.90), “substantial agreement” according to the Landis and Koch scale.
Table 1.
Comparison of the Abbott RealTime MTB RIF/INH Resistance assay to the indirect DST results for RIF and INH (sample results [n =99])
| Indirect DST | |||||
|---|---|---|---|---|---|
| Rifampicin | Isoniazid | ||||
| Resistant | Susceptible | Resistant | Susceptible | ||
| Abbott RealTime MTB RIF/INH | Resistant | 26 (96.3%) | 0 (0%) | 41 (78.8%) | 0 (0%) |
| Susceptible | 1 (3.7%) | 72 (100%) | 11 (21.2%) | 47 (100%) | |
| Sensitivity: 96.3% (95%CI 87.3%–100%) Specificity: 100% (95%CI 99.3%–100%) PPV: 100% (95%CI 99.1%–100%) vs 100% (95%CI 98.3%–100%) NPV: 98.6% (95%CI 95.3%–100%) |
Sensitivity: 78.8% (95%CI 66.8%–90.9%) Specificity: 100% (95%CI 98.9%–100%) PPV: 100% (95%CI 98.8–100%) NPV: 81.0% (95%CI 70.1%–92.0%) |
||||
Abbreviations: MTB, Mycobacterium tuberculosis; RIF, rifampicin; INH, isoniazid; DST, drug-susceptibility testing; PPV, positive predictive value; NPV, negative predictive value.
Twelve samples with discrepancies were compared with other genotypic tests (Table 2). In the phenotypically RIF resistant sample, the mutation was not detected with Abbott RealTime MTB INH/RIF (RT MTB INH/RIF); the other genotypic test detected this mutation (H526D).
Table 2.
Comparison of 12 discordant results between Abbott RealTime MTB RIF/INH Resistance assay and DST with other genotypic tests
| Indirect DST | |||||
|---|---|---|---|---|---|
| Rifampicin | Isoniazid | ||||
| Resistant | Susceptible | Resistant | Susceptible | ||
| Abbott RealTime MTB RIF/INH | Resistant | 0 | 0 | 0 | 0 |
| Susceptible | 1 | 0 | 11 | 0 | |
| Genotype MTBDRplus | Resistant | 1 | 0 | 0 | 0 |
| Susceptible | 0 | 0 | 11 | 0 | |
| Anyplex MTB/MDR | Resistant | 1 | 0 | 0 | 0 |
| Susceptible | 0 | 0 | 11 | ||
| Xpert/MTB RIF | Resistant | 1 | 0 | ND | ND |
| Susceptible | 0 | 0 | ND | ND | |
Abbreviations: MTB, Mycobacterium tuberculosis; RIF, rifampicin; INH, isoniazid; DST, drug-susceptibility testing; ND, non-determined.
For INH, eleven samples were phenotypically resistant; no mutations were detected with any of the molecular methods tested.
Discussion
The Abbott RealTime MTB RIF/INH is a novel, fully automated real-time PCR system for diagnosis of MTB and resistance to RIF and INH in clinical respiratory samples.
The aim of this study was to evaluate this method using pulmonary and extra-pulmonary samples compared with three other commercial PCR test kits. The results obtained using Abbott RealTime MTB RIF/INH corresponded with those obtained using the three molecular techniques and with phenotypic DST, except in 12 samples. Only one of these samples was discordant for RIF, which is possibly due to the use of frozen samples,11 and the eleven remaining samples were discordant for INH, due to other genes involved in resistance to this drug. The sensitivity for RIF was 96.3%, and the sensitivity for INH was 78.8%.
The sensitivity and specificity results of Abbott RealTime MTB RIF/INH have been published in recent papers. Kostera et al10 performed a study to evaluate this test in pulmonary samples in comparison with phenotypic results, demonstrating a sensitivity to RIF of 94.8%. There were five samples phenotypically resistant which were not detected by Abbott RealTime MTB RIF/INH. When sequencing was performed, four of them were found to be wild type, and mutation was detected in one sample. The specificity for RIF was 100%. Comparing these results with Genexpert, a sensitivity of 95.8% and a specificity of 100% were detected. For INH, the sensitivity was 88.3% and the specificity was 93.3%. Eleven samples were resistant according to DST and were not detected by Abbott RealTime MTB RIF/INH. The sequencing results showed that, in only one sample, the Abbott RealTime MTB RIF/INH assay did not detect the mutation (S315T1). Thus, Abbott RealTime MTB RIF/INH results presented statistically equivalent sensitivity and specificity as compared to Genexpert and Genotype MTBDRplus.10
A study conducted by Hoffman-Thiel et al11 evaluated Abbott RealTime MTB RIF/INH in respiratory and extra-pulmonary samples. Comparing the results with phenotypic testing (DST) and Genotype MTBDRplus, a concordance of 100% was obtained for RIF, and Abbott RealTime MTB RIF/INH failed in only one sample. A mutation to INH in katG (S315t), which was detected by Genotype MTBDRplus and phenotypic testing, showed high sensitivity with paucibacillary samples.11
A recent study was carried out in respiratory samples with Abbott RealTime MTB RIF/INH in China, comparing the resistance results with the phenotypic test MGIT 960 SIRE and Sanger sequencing. For RIF, 100% concordance was obtained using genotypic testing, and 78.2% using phenotypic testing.12
TB and HIV coinfection is a problem in some countries such as South Africa, where there is a high incidence. An Abbott RealTime MTB RIF/INH study has been carried out in Johannesburg and results have been compared with Genexpert MTB/RF. Of the 206 individuals studied, 73% were HIV positive and better results were obtained compared to Genexpert. The most important disadvantage in the study was the high percentage (33%) of samples with a low LOD.13
Like the previously published studies, our research had its limitations. Due to the use of frozen samples from our collection to carry out our study, results could have been affected. In this case, Abbott RealTime MTB RIF/INH did not detect the resistance to RIF in a sample, while the other methods did detect it. This may be because they were carried out with samples that were not fresh; however, there was only one discordant sample.
The main advantage of this method is that the whole sample preparation, identification, and mutation detection process is totally automatic and allows automatic processing of a large number of samples. Its sensitivity and specificity is comparable to other tests.
Conclusion
The Abbott RealTime MTB RIF/INH test could be a valid method for detecting the most common mutations in strains resistant to RIF and INH. Although the test is designed for respiratory samples, based on our results with positive extra-pulmonary clinical samples, it seemed to have correctly detected the mutations of resistance, although further research is required to confirm these findings. Furthermore, the whole process is automated; it is no more expensive than any of the other methods and has an advantage over Genexpert – the detection of INH.
Acknowledgments
This work was supported by the Spanish Ministry of Health and Consumer Affairs, Institute de Health Carlos III-ERDF, Spanish Network for Research into Infectious Diseases (REIPI RD06/0008).
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.World Health Organization . Guidelines for surveillance of drug resistance in tuberculosis. Geneva, Switzerland: WHO; 2016. 2016. [Accessed November 21, 2017]. Available from: http://apps.who.int/iris/bitstream/10665/174897/1/9789241549134_eng.pdf?ua=1. [Google Scholar]
- 2.World Health Organization . Global tuberculosis report 2016. Geneva, Switzerland: WHO; 2016. [Accessed November 21, 2017]. Available from: http://apps.who.int/medi-cinedocs/documents/s23098en/s23098en.pdf. [Google Scholar]
- 3.World Health Organization, 2016 . The use of molecular line probe assays for the detection of resistance to isoniazid and Rifampicin: Policy update. Geneva, Switzerland: WHO; 2016. [Accessed November 21,2017]. Available from: http://apps.who.int/iris/bitstream/10665/250586/1/9789241511261-eng.pdf?ua=1. [Google Scholar]
- 4.European Center for Disease Prevention and Control/WHO Regional Office for Europe . Tuberculosis surveillance and monitoring in Europe, 2017. ECDC; 2017. [Accessed November 21, 2017]. Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/ecdc-tuberculosis-surveillance-monitoring-Europe-2017.pdf. [Google Scholar]
- 5.Rusch-Gerdes S, Pfyffer GE, Casal M, Chadwick M, Siddiqi S. Multicenter laboratory validation of the BACTEC MGIT 960 technique for testing susceptibilities of Mycobacterium tuberculosis to classical second-line drugs and newer antimicrobials. J Clin Microbiology. 2006;44(3):688–692. doi: 10.1128/JCM.44.3.688-692.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Causse M, Ruiz P, Gutierrez JB, Zerolo J, Casal M. Evaluation of new GenoType (R) MTBDRplus for detection of resistance in cultures and direct specimens of Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2008;12(12):1456–1460. [PubMed] [Google Scholar]
- 7.Causse M, Ruiz P, Gutierrez-Aroca JB, Casal M. Comparison of two molecular methods for rapid diagnosis of extrapulmonary tuberculosis. J Clin Microbiol. 2011;49(8):3065–3067. doi: 10.1128/JCM.00491-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Causse M, Ruiz P, Gutierrez JB, Vaquero M, Casal M. New Anyplex (TM) II MTB/MDR/XDR kit for detection of resistance mutations in M. tuberculosis cultures. Int J Tuberc Lung Dis. 2015;19(12):1542–1546. doi: 10.5588/ijtld.15.0235. [DOI] [PubMed] [Google Scholar]
- 9.Tang N, Frank A, Pahalawatta V, et al. Analitycal and clinical performance of Abbott RealTime MTB, an assay for detection of Mycobacterium tuberculosis in pulmonary specimens. Tuberculosis (Edinb) 2015;95(5):613–619. doi: 10.1016/j.tube.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Kostera J, Leckie G, Tang N, Lampinen J, Szostak M, Abravaya K, Wang H. Analytical and clinical performance characteristics of the Abbott RealTime MTB RIF/INH Resistance, an assay for the detection of Rifampicin and Isoniazid resistant Mycobacterium tuberculosis in pulmonary specimens. Tuberculosis (Edinb) 2016;101:137–143. doi: 10.1016/j.tube.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann-Thiel S, Molodtsov N, Antonenka U, Hoffmann H. Evaluation of the Abbott RealTime MTB and RealTime MTB INH/RIF assays for direct detection of Mycobacterium tuberculosis complex and resistance markers in respiratory and extra-pulmonary specimens. J Clin Microbiol. 2016;54(12):3022–3027. doi: 10.1128/JCM.01144-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam KK, Leung KS, To SW, et al. Direct detection of Mycobacterium tuberculosis and drug resistance in respiratory specimen using Abbott Real Time MTB detection and RIF/INH resistance assay. Diagn Microbiol Infect Dis. 2017;89(2):118–124. doi: 10.1016/j.diagmicrobio.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Scott L, David A, Noblr L, et al. Performance of the Abbott Real Time MTB and MTB RIF/INH assays in a setting of high tuberculosis and HIV Coinfection in South Africa. J Clin Microbiol. 2017;55(8):2491–2501. doi: 10.1128/JCM.00289-17. [DOI] [PMC free article] [PubMed] [Google Scholar]


