Abstract
Carbapenem-resistant Enterobacteriaceae encountered in countries of the Arabian Peninsula usually produce OXA-48-like and New Delhi metallo-beta-lactamases (NDM) carbapenemases. However, a temporary increase in VIM-4-producing, clonally unrelated Enterobacteriaceae strains was described earlier in a Kuwaiti hospital. We investigated the genetic support of blaVIM-4 in six Klebsiella pneumoniae strains, one Escherichia coli, and one Enterobacter cloacae strain and compared it to that of VIM-4-producing isolates from other countries of the region. Five K. pneumoniae strains and the E. coli strain from Kuwait carried an ~165 kb IncA/C-type plasmid indistinguishable by restriction fragment length polymorphism. The complete sequence of one of them (pKKp4-VIM) was established. pKKp4-VIM exhibited extensive similarities to episomes pKP-Gr642 carrying blaVIM-19 encountered in Greece and to the partially sequenced pCC416 harboring blaVIM-4 detected in Italy. In other countries of the region, the only similar plasmid was the one detected in the isolate from the UAE. In all Kuwaiti strains, irrespective of the species and their VIM plasmids, the blaVIM-4 gene was located within the same integron structure (In416), different from those of other countries of the region. Our data show that the spread of this IncA/C plasmid and particularly that of the In416 integron caused a considerable, albeit temporary, increase in the rate of mostly clonally unrelated VIM-producing Enterobacteriaceae strains of multiple species. Monitoring of such events is of high importance as the interference with the spread of mobile genetic elements may represent a formidable challenge to infection control.
Keywords: Enterobacteriaceae, VIM carbapenemase, horizontal gene transfer, multidrug resistance, Middle East
Introduction
The emergence and spread of carbapenemase-producing Enterobacteriaceae (CPE) is a serious global threat that considerably limits therapeutic options available for life-threatening Gram-negative infections. Carbapenem-hydrolyzing enzymes have been described in the A, B and D classes of beta-lactamases.1 Group B enzymes, i.e., the metallo-beta-lactamases (MBLs), are especially worrisome, as recently introduced beta-lactamase inhibitors have no activity against them.2
Although it has been observed that countries of the Arabian Peninsula are burdened by CPE, there are as yet no systematic surveillance-based data regarding the magnitude of the problem. However, studies from the region have shown that locally class D OXA-48-like enzymes and New Delhi metallo-beta-lactamases (NDM) are the most common carbapenemases in Enterobacteriaceae with sporadic occurrence of KPC- and VIM-type enzymes.3–7 An exception to this trend was a temporarily increased prevalence of VIM-producing strains in Kuwait between 2009 and 2011. Early investigations in Kuwait showed that a few of these strains were clonally related only.8 In the current study, we investigate the role of mobile genetic elements in the increased number of VIM-positive isolates in Kuwait and compared the plasmids and integrons to other blaVIM-bearing mobile genetic elements identified in other countries of the region.
Materials and methods
Bacterial strains
Five Klebsiella pneumoniae (KKp1, KKp2, KKp4, KKp6 and KKp8), one Enterobacter cloacae (KEcl3) and one Escherichia coli (KEc7) were previously described as part of a VIM-producing Enterobacteriaceae outbreak in Kuwait.8 A further VIM producing K. pneumoniae (KW11) isolated in the same hospital during the same period was also included in the study. The characteristics of these isolates were compared to those of four VIM-producing E. cloacae (two [OM63 and OM69] from Oman, one [SA4/2] from the Kingdom of Saudi Arabia [KSA] and one [ABC104] from the UAE, respectively).4,6 All isolates were recovered from individual patients and were considered clinically relevant. The strains were stored at −80°C in Tryptic Soy Broth (Mast, Merseyside, UK) containing 20% glycerol.
Antibiotic susceptibility assays
Susceptibility to cefotaxime, ceftazidime, aztreonam, ertape-nem, meropenem, imipenem, ciprofloxacin, gentamicin, amikacin, trimethoprim/sulfamethoxazole, tetracycline, chloramphenicol and colistin (Col) was tested by broth microdilution, while susceptibility to fosfomycin (Fos) and tigecycline (Tig) was assessed by agar dilution.9 For the majority of antibiotics the Clinical and Laboratory Standards Institute (CLSI) clinical breakpoints were used for interpretation,9 with the exception of Col, Tig and Fos whereby the The European Committee on Antimicrobial Susceptibility Testing (EUCAST) criteria.10
Molecular typing
The XbaI-digested genomic DNA’s pulsed field gel electrophoresis (PFGE) pattern and the multi-locus sequence type of the isolates were established as described earlier.6,7 The Kuwaiti K. pneumoniae isolates were also compared by repetitive element sequence-based polymerase chain reaction (rep-PCR; DiversiLab; bioMerieux, Marcy l’Etoile France) using the Klebsiella kit, according to the manufacturer’s recommendation. Resistance genes (blaTEM, blaCTX-M, blaSHV, blaPER, blaAmpC, blaNDM, blaOXA-48-like, blaKPC, blaVIM, blaIMP, armA, rmtA, rmtB, rmtC, rmtD, rmtE, rmtF, qnrS, qepA, aac6-1b-cr, mcr-1, mcr-2) were detected as described.4,7,11,12 The specific alleles of beta-lactamase genes were determined by direct sequencing of the respective amplicons performed with the BigDye Cycle Terminator V.3.1 (Thermo Fisher Scientific, Waltham, MA, USA) using the 3130X Genetic Analyzer (Thermo Fisher Scientific).
Characterization of the genetic environment of blaVIM-4
The flanking region of the blaVIM-4 gene was determined by polymerase chain reaction (PCR) mapping and sequencing using primers designed (Table S1) according to the genetic surrounding of blaVIM published earlier (GenBank accession numbers AJ704863 and AY339625).13,14 Sequences were assembled with Clone Manager v9.0 (Sci-Ed Software, Cary, NC, USA) and annotated using Sequin (http://www.ncbi.nlm.nih.gov/Sequin) and submitted to GenBank.
Plasmid characterization
Plasmids were isolated and detected by the alkaline lysis method as described6 using E. coli 39R861 as plasmids molecular size standards.15 Mating out assays were performed with the clinical isolates using an azide-resistant derivative of rifampicin-resistant E. coli J53 (J53RAZ) as recipient. Transconjugants were selected on Tryptic Soy Agar containing 8 mg/L ceftazidime and 100 mg/L azide.7 If they were non-conjugative, heat shock transformation of the carbapenemase-bearing plasmids into E. coli DH5α was attempted.5 To prove the loclization of genes, electrophoretically separated plasmids isolated from the wild-type strains by the alkaline lysis method6 and those of the transconjugants or transformants were capillary transferred to Hybond N+ membranes that were subsequently hybridized with the appropriate digoxigenin-labeled (Roche Diagnostics GmbH, Mannheim, Germany) probes.6 The incompatibility (Inc) groups of the plasmids transferred were identified by PCR16,17 and confirmed by hybridization as mentioned earlier.7
Plasmids were purified from single plasmid containing E. coli K-12 derivatives using the Plasmid Maxiprep Kit (Qiagen NV, Venlo, the Netherlands). Restriction patterns of similarly sized plasmids belonging to the same Inc type were visually compared after digesting with HincII, HindIII and EcoRI restriction endonucleases. Furthermore, the complete sequence of pKKp4-VIM conjugally transferred from K. pneumoniae KKp4 into E. coli J53RAZ was established by next-generation sequencing on Illumina MiSeq platform (performed at the CCIB DNA Core Facility in Massachusetts General Hospital, Cambridge, MA, USA). The gaps between contigs assembled were closed by PCR and by direct sequencing of the amplicons. The complete plasmid sequence was assembled with Clone Manager v9.0, annotated using Sequin (http://www.ncbi.nlm.nih.gov/Sequin) and submitted to GenBank (MF582638).
Ethics approval
The ethics approval for the study was obtained from the Medical Ethics Committee of the Ministry of Health, Kuwait (288/MTT).
Results
Antibiotic susceptibility
The antibiotic susceptibility test results of the isolates are summarized in Table 1, while the respective minimal inhibitory concentration (MIC) values are provided in Table S1. All strains showed resistance to all beta-lactams tested. All Kuwaiti strains were multidrug resistant, with two being susceptible to Col only and one (K. pneumoniae KW11) being resistant to all antibiotics tested (Tables 1 and S2).
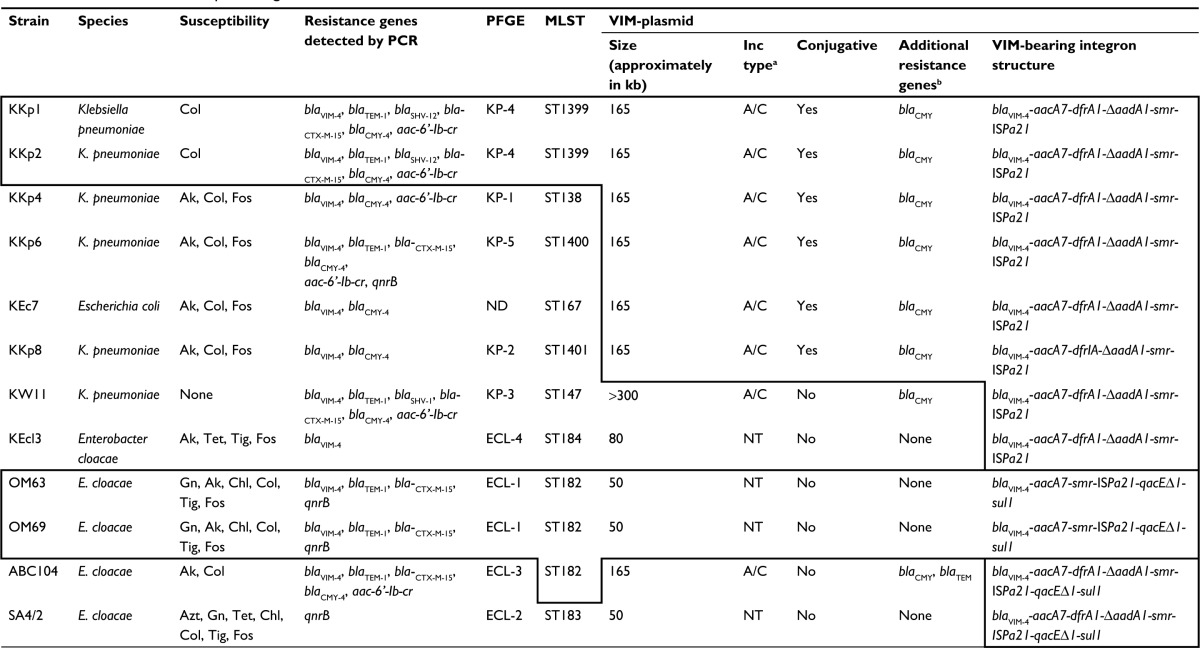
Table 1.
Characteristics of VIM-producing Enterobacteriaceae isolated in countries of the Arabian Peninsula
Notes: Features boxed by thick lines are identical.
As determined by PBRT.
As detected by hybridization.
Abbreviations: PCR, polymerase chain reaction; PFGE, pulsed field gel electrophoresis; Inc, incompatibility; Col, colistin; Ak, amikacin; Fos, fosfomycin; Tet, tetracycline; Tig, tigecycline; NT, non typable; Gn, gentamicin; Chl, chloramphenicol; Azt, aztreonam; PBRT, PCR-based replicon typing; MLST, multi-locus sequence typing; ND, not detected.
Molecular typing
All strains investigated carried a single carbapenemase, blaVIM-4. The molecular characteristics of the clinical isolates are summarized in Table 1. Confirming the data of the previous study, i.e., of the Kuwaiti strains, only K. pneumoniae KKp1 and KKp2 exhibited similar PFGE patterns (KP-4; Figure S1).8 With the exception of these two isolates, the sequence types (Table 1) and the rep-PCR patterns (Figure S2) of the other K. pneumoniae strains were all different. The sole E. cloacae (KEcl3) from Kuwait was different from the other four VIM-4-producing E. cloacae from the region both by PFGE and by multi-locus sequence typing (MLST) (Table 1 and Figure S1).
Comparison of the plasmids carrying blaVIM-4
Unlike in the previous study,8 we could conjugally transfer VIM-coding plasmids from six of the eight Kuwaiti strains as well as from the Saudi E. cloacae SA4/2. From the Omani E. cloacae isolates (OM63 and OM69), the VIM plasmids were transferred by transformation. From K. pneumoniae KW11 and E. cloacae KEcl3 and ABC104, neither conjugations nor transformations were successful.
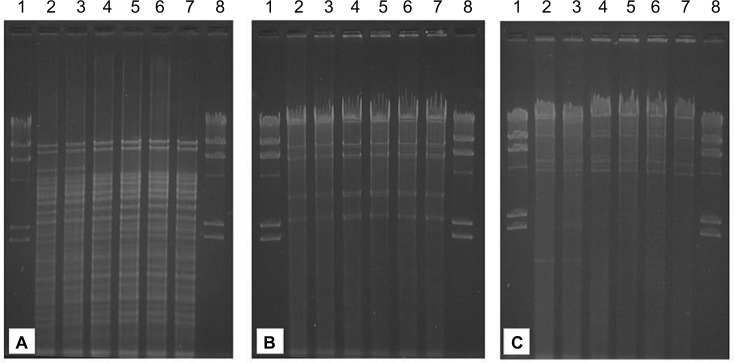
As confirmed by PCR and by Southern hybridization (Figure S3), in addition to the two clonally related K. pneumoniae (KKp1 and KKp2), three unrelated K. pneumoniae (KKp4, KKp6 and KKp8) isolates and the E. coli (KEc7) from Kuwait harbored blaVIM-4 on IncA/C Inc-type plasmids of ~165 kb. Beyond blaVIM-4, these plasmids also carried the blaCMY-4 gene (Table 1). The RFLP patterns of these plasmids were identical (Figure 1). E. cloacae ABC104 from the UAE (described earlier6) also carried blaVIM-4 and blaCMY-4 on similar-sized IncA/C Inc-type plasmid, but we were unable to compare the RFLP of this plasmid to the Kuwaiti ones as we could not generate a single VIM plasmid-containing derivative of this isolate. In K. pneumoniae KW11, the blaVIM-4 was located on a nontransferable IncA/C type, >300 kb plasmid, which also carried blaCMY-4. As shown in Table 1 and in Figure S3, the Kuwaiti, Saudi and Omani E. cloacae isolates all carried blaVIM-4 on smaller plasmids lacking blaCMY-4, which could not be identified by the PCR-based replicon typing (PBRT). In case of KKp1 and KKp2, the conjugal transfer of the blaVIM-bearing plasmids was accompanied by their fusion with IncN-type plasmids (Figure S4). No attempts were made, within the frames of the current study, to clarify the molecular details of this fusion.
Figure 1.
Restriction fragment length polymorphism of conjugative IncA/C plasmids of the Kuwaiti isolates.
Notes: (A) Digestion with HincII. (B) Digestion with EcoRI. (C) Digestion with HindIII. Lanes 1 and 8, lambda phage DNA digested with HindIII; Lane 2, pKKp1-VIM; Lane 3, pKKp2-VIM; Lane 4, pKKp4-VIM; Lane 5, pKKp6-VIM; Lane 6, pKEc7-VIM and Lane 7, pKKp8-VIM.
Complete sequence of pKKp4-VIM
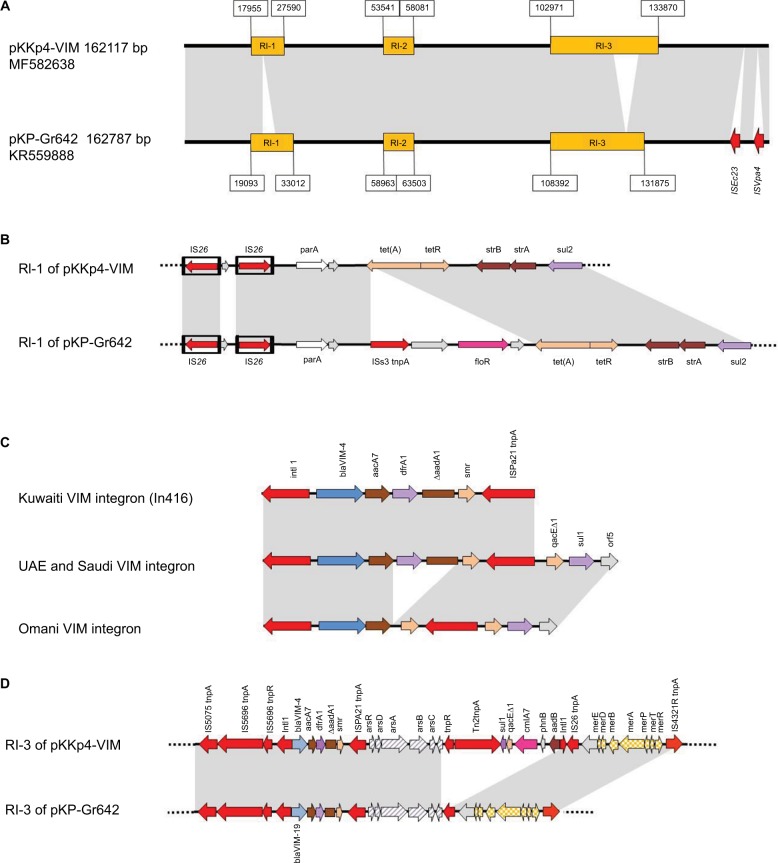
To obtain a more detailed picture on the conjugative IncA/C-type plasmids dominating the isolates from Kuwait, the entire sequence was determined from a transconjugant containing pKKp4-VIM derived from K. pneumoniae KKp4. The plasmid was a 162117 bp long, type 1 IncA/C2 plasmid with respective regions for replication, conjugative transfer and plasmid maintenance (GenBank Accession No. MF582638). It was highly similar to pKP-Gr642, a type 1 IncA/C2-type plasmid of a Greek clinical K. pneumoniae isolate carrying blaVIM-19 (Figure 2A)18. Apart from the plasmid backbone, pKKp4-VIM harbored three resistance islands: RI-1, RI-2 and RI-3 (Figure 2A). On RI-1 tet(A), strA, strB and sul2 genes are located (Figure 2B). The RI-2 consists of ISEcp1, blaCMY-4, blc and sugE genes. The third resistance island RI-3 contains an In416 with blaVIM-4, aacA7, dfrA1, DaadA1 and smr gene cassettes, a Tn8802 with arsenic resistance operon, an In-t4-like integron and a mercury resistance operon (Figure 2D).
Figure 2.
Structure of pKKp4-VIM.
Notes: (A) Comparison of the complete pKKp4-VIM to pKP-Gr642. (B) Comparison of RI-1 of pKKp4-VIM and pKP-Gr642. (C) Comparison of the three blaVIM-4-bearing integron variants. (D) Comparison of RI-3 of pKKp4-VIM to RI-3 of pKP-Gr642. Gray areas represent ≥95% similarity.
Genetic surrounding of blaVIM-4 in all isolates
PCR mapping and sequencing revealed that irrespective of the species or plasmid Inc type, the integron structure of all Kuwaiti isolates was identical to the one in pKKp4-VIM, i.e., blaVIM-4 was located on an In416 integron, which lacked the 3′ conserved sequences (CS). On the other hand, in E. cloacae ABC104 described earlier from the UAE (GenBank Accession No. JX275775)6 and in E. cloacae SA4/2 from Saudi Arabia, the qacED1-sul1-orf5 structure was present downstream of the ISPa21. In the two Omani isolates, the integron lacked the dfrA1 and DaadA1 cassettes, and the 3′ CS was present downstream of ISPa21 (GenBank Accession No. MF178139; Table 1 and Figure 2C).
Discussion
VIM-producing Enterobacteriaceae have only been sporadically encountered in countries of the Arabian Peninsula.4,6,19,20 Between April 2009 and February 2011, a higher prevalence of mostly unrelated VIM-4 producer Enterobacteriaceae strains was observed in Kuwait.8 As shown by our current data, this increased rate of blaVIM-carrying strains was mostly due to local horizontal gene transmission, leading to the uniform presence of the same blaVIM-containing In416 integron in all Kuwaiti isolates, irrespective of the species and plasmids carried, and to the wide distribution of a 162 kb IncA/C2-type plasmid. It is noteworthy that this type of integron and plasmid was characteristic to the Kuwaiti isolates, whereas in strains from other countries of the region, there was much heterogeneity of episomes and integrons (Table 1 and Figure 2C).
It is of interest that a plasmid (pKP-Gr642) very similar to the one spreading blaVIM-4 in Kuwait was found in Greece. It bore a single amino acid variant of this enzyme, i.e., blaVIM-19.18 The slight differences between this latter plasmid and pKKp4-VIM are highlighted in Figure 2. The RI-1 of pKP-Gr642 contains an additional ISCR2-driven floR gene (Figure 2B), and its RI-3 lacks the In-t4-like integron containing aadB, cmlA7, qacED1 and sul1 genes (Figure 2D). Furthermore, pKP-Gr642 carries two insertion elements (ISEc23 and ISVpa4) in the plasmid backbone.
Moreover, features of pCC416, the conjugative IncA/C plasmid shown to transfer blaVIM-4 between E. cloacae and K. pneumoniae clinical isolates of a patient in Italy,14,21 were closely similar to the endemic pKKp4-VIM of Kuwait. Both RI-2 and RI-3 of pKKp4-VIM were 99% identical to the two fragments sequenced of pCC416 (GenBank Accession Nos. AJ875405 and AJ704863). Furthermore, pCC416 was also reported to carry a sul2 gene, which is located on RI-1 of pKKp4-VIM. After detecting its in vivo transfer in a patient, it was speculated that this particular IncA/C plasmid could play a role in the spread of carbapenem resistance.14,21 Our study confirmed this hypothesis by showing that such plasmids are indeed able to spread, over a year-long period, between strains carried by different patients, not even respecting species’ barriers.
A limitation of our study is the lack of epidemiological data linking the cases to each other. No details were available to us regarding possible routes of transmission. Furthermore, only eight strains (seven of the originally described outbreak set of 11 plus one further isolate) were available for the investigation. However, even the data on this set of strains showed the heterogeneity of strains being in sharp contrast with the near uniformity of plasmids and the complete identity of blaVIM-4 containing integrons in all Kuwaiti isolates. Importantly, this increased rate of VIM-positive isolates seems to be a temporary event, as strains collected subsequently from Kuwait and even from the same hospital expressed mostly NDM- and OXA-type enzymes, otherwise characteristic of the region.4,22
Our results also highlight the importance of the detailed molecular typing of CPE to obtain a realistic picture of the complexity of the spread of carbapenem resistance. The spread of plasmids and integrons represents a considerable challenge to infection control. Horizontal gene transfer is difficult to prevent by routine infection control measures, and only limiting the selective antibiotic pressure in the human body and in the environment may possibly mitigate its efficacy.
Supplementary materials
PFGE comparison of VIM-producing Enterobacteriaceae.
Abbreviation: PFGE, pulsed field gel electrophoresis.
rep-PCR comparison of Kuwaiti Klebsiella pneumoniae strains.
Abbreviation: rep-PCR, repetitive element sequence-based polymerase chain reaction.
Plasmid profiles of VIM-producing Enterobacteriaceae.
Notes: (A) Plasmid gel. (B) Membrane hybridized with blaVIM-4 probe. (C) Membrane hybridized with blaCMY-4 probe. (D) Membrane hybridized with IncA/C probe. Lane 1, Escherichia coli 39R861; Lane 2, E. coli J53RAZ; Lane 3, Klebsiella pneumoniae KKp1; Lane 4, K. pneumoniae KKp2; Lane 5, K. pneumoniae KKp4; Lane 6, K. pneumoniae KKp6; Lane 7, K. pneumoniae KKp8; Lane 8, K. pneumoniae KW11; Lane 9, E. coli KEc7; Lane 10, Enterobacter cloacae KEcl3; Lane 11, E. cloacae OM63; Lane 12, E. cloacae OM69; Lane 13, E. cloacae SA4/2; Lane 14, E. cloacae ABC104; Lane 15, E. coli 39R861.
Fusion of IncA/C-VIM and IncN plasmids.
Notes: (A) Plasmid gel. (B) Membrane hybridized with VIM probe. (C) Membrane hybridized with Inc A/C probe. (D) Membrane hybridized with Inc N probe. Lane 1, Klebsiella pneumoniae KKp1; Lane 2, K. pneumoniae KKp2; Lane 3, Escherichia coli J53RAZ(pKKp1-VIM); Lane 4, E. coli J53RAZ(pKKp2-VIM).
Table S1.
Primers used in sequencing the molecular structures carrying the blaVIM gene
| Primer name | 5′-3′ sequence | Annealing to AJ704863 | Size of products (bp) | Comment |
|---|---|---|---|---|
| AS_ClassIint_L | TGT CGT TTT CAG AAG ACG GCT GC | 5250 | 434 | For amplification and sequencing the 5′ end of class I integron |
| AS_ClassIint_R | CAA ACG TGC CGT AGA ACA AG | 5683C | ||
| AS_intI1_L | GGG AGG ACT TTC CGC AAC CG | 5363 | 1084 | For amplification and sequencing the blaVIM upstream region |
| AS_VIM_R | CGT TAC CAC CGC TGC GTT CG | 6446C | ||
| AS_VIM_GS_S1 | GCC TTG ATG TTA CCC GAG AG | 5937C | NA | |
| AS-VIM4GS-f | GAT GCG TGG AGA CCG AAA CC | 6228 | 1295 | For amplification and sequencing the blaVIM and its immediate surroundings |
| AS-VIM4GS-r | TGC CTA ACG CCT GAG TTG AG | 7522C | ||
| AS_VIM_L | AAT CGC TCA GTC GCC GAG TA | 7412 | 3750 | For amplification and sequencing the blaVIM downstream region |
| AS_ISPa21_R | CTA TAA GAC ACG AGG TGT CTG | 11161C | ||
| AS_ISPa21_L | CAC CAC AAC CGC AAG AAA TA | 10034 | NA | |
| AS_ISPa21_seq | CGC GCA TCG ATT GTT CGT AG | 10549 | NA | |
| AS_smr_f | GCT GGA CTC TTT GAG ATT GG | 9507 | NA | |
| AS_dhfrI_R | ACC CTT TTG CCA GAT TTG GT | 8597C | NA | |
| AS_aacA7_R | GAG CAA CCT CCG TGA ATC CA | 7955C | NA | |
| AS_VIMdn_LS1 | TTC GTT CAA GCC GAA CTT GC | 8010 | NA | |
| AS_VIMdn_LS2 | AAT AGA CAT CGA GCC GGA AG | 8477 | NA | |
| AS_VIMdn_LS3 | ACA TAG CGT TGC CTT GGT AG | 9030 | NA | |
| AS_orf5_R | TTA GAT TTC GAG TTC TAG GCG TTC TG | NA | 3647 bp (if class I integron classical 3′ end is present) | Primer AS_orf5_R anneals to the 3′ end of the class I integron; the two primers amplify the 3′ region of class I integron if present |
| AS_smr_f | GCT GGA CTC TTT GAG ATT GG | 9507 | ||
| AS_ISPa21_R | CTA TAA GAC ACG AGG TGT CTG | 11161C | NA | Sequencing the amplicon produced by the PCR above |
| AS_ISPa21_L | CAC CAC AAC CGC AAG AAA TA | 10034 | NA | |
| AS_ISPa21_seq | CGC GCA TCG ATT GTT CGT AG | 10549 | NA | |
| AS_sul1_R1 | TTG CCG ATC GCG TGA AGT TC | 13000C | NA | Sequencing the amplicon produced by the PCR using primer AS_orf5_R and AS_smr_f |
| AS_sul1_R2 | CACAACCTGGTCGATATCAC | 13311C | NA | |
| AS_orf5_L | ATGGACAGCGAGGAGC | 13249 | NA | |
| AS_qacED1_L | GCG AAG TAA TCG CAA CAT CC | 11978 | NA | |
| AS_VIMdn_LS4 | GAT CAG ATG CAC CGT GTT TC | 12501 | NA |
Abbreviations: NA, not applicable; PCR, polymerase chain reaction.
Table S2.
MIC values of antibiotics against VIM-producing strains and their derivatives
| Strain | Type | Ceftazidime | Cefotaxime | Azt | Ertapenem | Imipenem | Meropenem | Ciprofloxacin | Gn | Ak | Co-trimoxazole | Tet | Chl | Col | Tig | Fos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KKp1 | W | >128 | >128 | >128 | >64 | 128 | 128 | 2 | 256 | 32 | >256/4864 | >256 | >256 | <0.5 | 2 | 64 |
| J53RAZ(pKKp1-VIM) | TC | >128 | 128 | 128 | 16 | 8 | 2 | <0.125 | 32 | 8 | 128/2432 | 32 | 256 | <0.5 | 0.25 | 0.5 |
| KKp2 | W | >128 | >128 | >128 | >64 | 128 | 128 | 2 | 256 | 32 | >256/4864 | >256 | >256 | <0.5 | 2 | 64 |
| J53RAZ(pKKp2-VIM) | TC | 128 | 128 | 128 | 4 | 2 | 2 | <0.125 | 32 | 8 | 128/2432 | 32 | 256 | <0.5 | 0.25 | 0.5 |
| KEcl3 | W | >128 | >128 | >128 | >64 | 64 | 32 | 2 | >256 | 16 | >256/4864 | 4 | 16 | 4 | 0.5 | 32 |
| KKp4 | W | 128 | 128 | 32 | 64 | 16 | 16 | 0.5 | 64 | 16 | >256/4864 | >256 | 256 | <0.5 | 8 | 16 |
| J53RAZ(pKKp4-VIM) | TC | 64 | 32 | 32 | 4 | 1 | <0.25 | <0.125 | 16 | 4 | 128/2432 | 32 | 16 | <0.5 | 0.25 | 2 |
| KKp6 | W | 128 | >128 | >128 | 64 | 16 | 8 | 16 | 128 | 8 | >256/4864 | >256 | >256 | <0.5 | 8 | 4 |
| J53RAZ(pKKp6-VIM) | TC | 128 | 64 | 64 | 4 | 2 | 1 | <0.125 | 16 | 8 | 128/2432 | 64 | 128 | <0.5 | 0.25 | 2 |
| KEc7 | W | >128 | >128 | 64 | 64 | 8 | 8 | >64 | 64 | 16 | >256/4864 | 128 | 256 | <0.5 | 0.5 | 0.5 |
| J53RAZ(pKEc7-VIM) | TC | 64 | 64 | 64 | 16 | 2 | 0.5 | <0.125 | 16 | 8 | 256/4864 | 128 | 256 | <0.5 | 0.25 | 1 |
| KKp8 | W | >128 | >128 | >128 | 64 | 16 | 8 | >64 | 128 | 16 | >256/4864 | >256 | >256 | <0.5 | 0.5 | 16 |
| J53RAZ(pKKp8-VIM) | TC | 64 | 64 | 32 | 4 | 2 | <0.25 | <0.125 | 16 | 8 | 128/2432 | 64 | 128 | <0.5 | 0.25 | 2 |
| KW11 | W | >128 | >128 | >128 | >64 | >128 | 128 | >64 | 256 | 32 | >256/4864 | >256 | 16 | 64 | 2 | 128 |
| SA4/2 | W | 64 | >128 | 1 | 64 | 64 | 16 | 1 | 2 | 32 | >256/4864 | 4 | 8 | <0.5 | 1 | 4 |
| J53RAZ(pSA4/2-VIM) | TC | 32 | 64 | 0.5 | 32 | 4 | 2 | 0.25 | 1 | 8 | <0.5/9.5 | 2 | 8 | <0.5 | <0.125 | 1 |
| OM63 | W | 128 | >128 | >128 | 32 | 4 | 4 | 64 | 2 | 16 | >256/4864 | >256 | 8 | <0.5 | 0.5 | 32 |
| DH5α(pOM63-VIM) | TF | 16 | 32 | <0.25 | 2 | 0.5 | <0.25 | <0.125 | 1 | 8 | <0.5/9.5 | <0.5 | 1 | <0.5 | <0.125 | <0.25 |
| OM69 | W | 128 | >128 | 128 | 64 | 64 | 4 | 32 | 2 | 16 | >256/4864 | 256 | 8 | <0.5 | 0.5 | 32 |
| DH5α(pOM69-VIM) | TF | 16 | 32 | <0.25 | 2 | 0.5 | <0.25 | <0.125 | 1 | 8 | <0.5/9.5 | <0.5 | 1 | <0.5 | <0.125 | <0.25 |
| ABC104 | W | >128 | >128 | >128 | >64 | 32 | 4 | 32 | 64 | 8 | >256/4864 | >256 | 128 | <0.5 | 8 | 64 |
| J53RAZ | R | <0.25 | <0.25 | <0.25 | <0.125 | <0.25 | <0.25 | <0.125 | 1 | <0.5 | <0.5/9.5 | <0.5 | 8 | <0.5 | <0.125 | 0.5 |
| DH5α | R | <0.25 | <0.25 | <0.25 | <0.125 | <0.25 | <0.25 | <0.125 | 1 | 1 | <0.5/9.5 | <0.5 | 1 | <0.5 | <0.125 | <0.25 |
Abbreviations: MIC, minimal inhibitory concentration; Azt, aztreonam; Gn, gentamicin; Ak, amikacin; Tet, tetracycline; Chl, chloramphenicol; Col, colistin; Tig, tigecycline; Fos, fosfomycin; W, wild; TC, transconjugant; TF, transformant; R, recipient.
Acknowledgments
The kind help of Ms Geraldine Kershaw (CMHS, UAE University) in language editing the manuscript is highly appreciated. This work was supported by grants UPAR-25143 (31R061) and UAEU FMHS grants NP/12/13 and NP-10-11/1019 awarded to TP and UAEU UPAR-31M235 grant awarded to AS.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Diene SM, Rolain JM. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2014;20(9):831–838. doi: 10.1111/1469-0691.12655. [DOI] [PubMed] [Google Scholar]
- 2.Falcone M, Paterson D. Spotlight on ceftazidime/avibactam: a new option for MDR Gram-negative infections. J Antimicrob Chemother. 2016;71(10):2713–2722. doi: 10.1093/jac/dkw239. [DOI] [PubMed] [Google Scholar]
- 3.Zowawi HM, Sartor AL, Balkhy HH, et al. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother. 2014;58(6):3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonnevend A, Ghazawi AA, Hashmey R, et al. Characterization of carbapenem-resistant Enterobacteriaceae with high rate of autochthonous transmission in the Arabian Peninsula. PLoS One. 2015;10(6):e0131372. doi: 10.1371/journal.pone.0131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnevend A, Ghazawi A, Darwish D, AlDeesi Z, Kadhum AF, Pal T. Characterization of KPC-type carbapenemase-producing Klebsiella pneumoniae strains isolated in the Arabian Peninsula. J Antimicrob Chemother. 2015;70(5):1592–1593. doi: 10.1093/jac/dku576. [DOI] [PubMed] [Google Scholar]
- 6.Sonnevend A, Ghazawi A, Yahfoufi N, et al. VIM-4 carbapenemase-producing Enterobacter cloacae in the United Arab Emirates. Clin Microbiol Infect. 2012;18(12):E494–E496. doi: 10.1111/1469-0691.12051. [DOI] [PubMed] [Google Scholar]
- 7.Sonnevend A, Al Baloushi A, Ghazawi A, et al. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J Med Microbiol. 2013;62(pt 7):1044–1050. doi: 10.1099/jmm.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 8.Jamal W, Rotimi VO, Albert MJ, Khodakhast F, Nordmann P, Poirel L. High prevalence of VIM-4 and NDM-1 metallo-beta-lactamase among carbapenem-resistant Enterobacteriaceae. J Med Microbiol. 2013;62(pt 8):1239–1244. doi: 10.1099/jmm.0.059915-0. [DOI] [PubMed] [Google Scholar]
- 9.CLSI . Performance Standard for Antimicrobial Susceptibility Testing; M100–S24. Wayne, PA: Clinical and Laboratory Standard Institute; 2014. [Google Scholar]
- 10.EUCAST. European Committee on Antimicrobial Susceptibility Testing. [homepage on the Internet] [Accessed November 10, 2017]. http://www.eucast.org/
- 11.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 12.Xavier BB, Lammens C, Ruhal R, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27) doi: 10.2807/1560-7917.ES.2016.21.27.30280. [DOI] [PubMed] [Google Scholar]
- 13.Miriagou V, Tzouvelekis LS, Villa L, et al. CMY-13, a novel inducible cephalosporinase encoded by an Escherichia coli plasmid. Antimicrob Agents Chemother. 2004;48(8):3172–3174. doi: 10.1128/AAC.48.8.3172-3174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colinon C, Miriagou V, Carattoli A, Luzzaro F, Rossolini GM. Characterization of the IncA/C plasmid pCC416 encoding VIM-4 and CMY-4 beta-lactamases. J Antimicrob Chemother. 2007;60(2):258–262. doi: 10.1093/jac/dkm171. [DOI] [PubMed] [Google Scholar]
- 15.Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- 16.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Johnson TJ, Bielak EM, Fortini D, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68(1):43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Papagiannitsis CC, Dolejska M, Izdebski R, et al. Characterisation of IncA/C2 plasmids carrying an In416-like integron with the blaVIM-19 gene from Klebsiella pneumoniae ST383 of Greek origin. Int J Antimicrob Agents. 2016;47(2):158–162. doi: 10.1016/j.ijantimicag.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Yezli S, Shibl AM, Memish ZA. The molecular basis of beta-lactamase production in Gram-negative bacteria from Saudi Arabia. J Med Microbiol. 2015;64(pt 2):127–136. doi: 10.1099/jmm.0.077834-0. [DOI] [PubMed] [Google Scholar]
- 20.Memish ZA, Assiri A, Almasri M, et al. Molecular characterization of carbapenemase production among Gram-negative bacteria in Saudi Arabia. Microb Drug Resist. 2015;21(3):307–314. doi: 10.1089/mdr.2014.0121. [DOI] [PubMed] [Google Scholar]
- 21.Luzzaro F, Docquier JD, Colinon C, et al. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-beta-lactamase encoded by a conjugative plasmid. Antimicrob Agents Chemother. 2004;48(2):648–650. doi: 10.1128/AAC.48.2.648-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamal WY, Albert MJ, Rotimi VO. High prevalence of New Delhi metallo-beta-lactamase-1 (NDM-1) producers among carbapenem-resistant Enterobacteriaceae in Kuwait. PLoS One. 2016;11(3):e0152638. doi: 10.1371/journal.pone.0152638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PFGE comparison of VIM-producing Enterobacteriaceae.
Abbreviation: PFGE, pulsed field gel electrophoresis.
rep-PCR comparison of Kuwaiti Klebsiella pneumoniae strains.
Abbreviation: rep-PCR, repetitive element sequence-based polymerase chain reaction.
Plasmid profiles of VIM-producing Enterobacteriaceae.
Notes: (A) Plasmid gel. (B) Membrane hybridized with blaVIM-4 probe. (C) Membrane hybridized with blaCMY-4 probe. (D) Membrane hybridized with IncA/C probe. Lane 1, Escherichia coli 39R861; Lane 2, E. coli J53RAZ; Lane 3, Klebsiella pneumoniae KKp1; Lane 4, K. pneumoniae KKp2; Lane 5, K. pneumoniae KKp4; Lane 6, K. pneumoniae KKp6; Lane 7, K. pneumoniae KKp8; Lane 8, K. pneumoniae KW11; Lane 9, E. coli KEc7; Lane 10, Enterobacter cloacae KEcl3; Lane 11, E. cloacae OM63; Lane 12, E. cloacae OM69; Lane 13, E. cloacae SA4/2; Lane 14, E. cloacae ABC104; Lane 15, E. coli 39R861.
Fusion of IncA/C-VIM and IncN plasmids.
Notes: (A) Plasmid gel. (B) Membrane hybridized with VIM probe. (C) Membrane hybridized with Inc A/C probe. (D) Membrane hybridized with Inc N probe. Lane 1, Klebsiella pneumoniae KKp1; Lane 2, K. pneumoniae KKp2; Lane 3, Escherichia coli J53RAZ(pKKp1-VIM); Lane 4, E. coli J53RAZ(pKKp2-VIM).
Table S1.
Primers used in sequencing the molecular structures carrying the blaVIM gene
| Primer name | 5′-3′ sequence | Annealing to AJ704863 | Size of products (bp) | Comment |
|---|---|---|---|---|
| AS_ClassIint_L | TGT CGT TTT CAG AAG ACG GCT GC | 5250 | 434 | For amplification and sequencing the 5′ end of class I integron |
| AS_ClassIint_R | CAA ACG TGC CGT AGA ACA AG | 5683C | ||
| AS_intI1_L | GGG AGG ACT TTC CGC AAC CG | 5363 | 1084 | For amplification and sequencing the blaVIM upstream region |
| AS_VIM_R | CGT TAC CAC CGC TGC GTT CG | 6446C | ||
| AS_VIM_GS_S1 | GCC TTG ATG TTA CCC GAG AG | 5937C | NA | |
| AS-VIM4GS-f | GAT GCG TGG AGA CCG AAA CC | 6228 | 1295 | For amplification and sequencing the blaVIM and its immediate surroundings |
| AS-VIM4GS-r | TGC CTA ACG CCT GAG TTG AG | 7522C | ||
| AS_VIM_L | AAT CGC TCA GTC GCC GAG TA | 7412 | 3750 | For amplification and sequencing the blaVIM downstream region |
| AS_ISPa21_R | CTA TAA GAC ACG AGG TGT CTG | 11161C | ||
| AS_ISPa21_L | CAC CAC AAC CGC AAG AAA TA | 10034 | NA | |
| AS_ISPa21_seq | CGC GCA TCG ATT GTT CGT AG | 10549 | NA | |
| AS_smr_f | GCT GGA CTC TTT GAG ATT GG | 9507 | NA | |
| AS_dhfrI_R | ACC CTT TTG CCA GAT TTG GT | 8597C | NA | |
| AS_aacA7_R | GAG CAA CCT CCG TGA ATC CA | 7955C | NA | |
| AS_VIMdn_LS1 | TTC GTT CAA GCC GAA CTT GC | 8010 | NA | |
| AS_VIMdn_LS2 | AAT AGA CAT CGA GCC GGA AG | 8477 | NA | |
| AS_VIMdn_LS3 | ACA TAG CGT TGC CTT GGT AG | 9030 | NA | |
| AS_orf5_R | TTA GAT TTC GAG TTC TAG GCG TTC TG | NA | 3647 bp (if class I integron classical 3′ end is present) | Primer AS_orf5_R anneals to the 3′ end of the class I integron; the two primers amplify the 3′ region of class I integron if present |
| AS_smr_f | GCT GGA CTC TTT GAG ATT GG | 9507 | ||
| AS_ISPa21_R | CTA TAA GAC ACG AGG TGT CTG | 11161C | NA | Sequencing the amplicon produced by the PCR above |
| AS_ISPa21_L | CAC CAC AAC CGC AAG AAA TA | 10034 | NA | |
| AS_ISPa21_seq | CGC GCA TCG ATT GTT CGT AG | 10549 | NA | |
| AS_sul1_R1 | TTG CCG ATC GCG TGA AGT TC | 13000C | NA | Sequencing the amplicon produced by the PCR using primer AS_orf5_R and AS_smr_f |
| AS_sul1_R2 | CACAACCTGGTCGATATCAC | 13311C | NA | |
| AS_orf5_L | ATGGACAGCGAGGAGC | 13249 | NA | |
| AS_qacED1_L | GCG AAG TAA TCG CAA CAT CC | 11978 | NA | |
| AS_VIMdn_LS4 | GAT CAG ATG CAC CGT GTT TC | 12501 | NA |
Abbreviations: NA, not applicable; PCR, polymerase chain reaction.
Table S2.
MIC values of antibiotics against VIM-producing strains and their derivatives
| Strain | Type | Ceftazidime | Cefotaxime | Azt | Ertapenem | Imipenem | Meropenem | Ciprofloxacin | Gn | Ak | Co-trimoxazole | Tet | Chl | Col | Tig | Fos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KKp1 | W | >128 | >128 | >128 | >64 | 128 | 128 | 2 | 256 | 32 | >256/4864 | >256 | >256 | <0.5 | 2 | 64 |
| J53RAZ(pKKp1-VIM) | TC | >128 | 128 | 128 | 16 | 8 | 2 | <0.125 | 32 | 8 | 128/2432 | 32 | 256 | <0.5 | 0.25 | 0.5 |
| KKp2 | W | >128 | >128 | >128 | >64 | 128 | 128 | 2 | 256 | 32 | >256/4864 | >256 | >256 | <0.5 | 2 | 64 |
| J53RAZ(pKKp2-VIM) | TC | 128 | 128 | 128 | 4 | 2 | 2 | <0.125 | 32 | 8 | 128/2432 | 32 | 256 | <0.5 | 0.25 | 0.5 |
| KEcl3 | W | >128 | >128 | >128 | >64 | 64 | 32 | 2 | >256 | 16 | >256/4864 | 4 | 16 | 4 | 0.5 | 32 |
| KKp4 | W | 128 | 128 | 32 | 64 | 16 | 16 | 0.5 | 64 | 16 | >256/4864 | >256 | 256 | <0.5 | 8 | 16 |
| J53RAZ(pKKp4-VIM) | TC | 64 | 32 | 32 | 4 | 1 | <0.25 | <0.125 | 16 | 4 | 128/2432 | 32 | 16 | <0.5 | 0.25 | 2 |
| KKp6 | W | 128 | >128 | >128 | 64 | 16 | 8 | 16 | 128 | 8 | >256/4864 | >256 | >256 | <0.5 | 8 | 4 |
| J53RAZ(pKKp6-VIM) | TC | 128 | 64 | 64 | 4 | 2 | 1 | <0.125 | 16 | 8 | 128/2432 | 64 | 128 | <0.5 | 0.25 | 2 |
| KEc7 | W | >128 | >128 | 64 | 64 | 8 | 8 | >64 | 64 | 16 | >256/4864 | 128 | 256 | <0.5 | 0.5 | 0.5 |
| J53RAZ(pKEc7-VIM) | TC | 64 | 64 | 64 | 16 | 2 | 0.5 | <0.125 | 16 | 8 | 256/4864 | 128 | 256 | <0.5 | 0.25 | 1 |
| KKp8 | W | >128 | >128 | >128 | 64 | 16 | 8 | >64 | 128 | 16 | >256/4864 | >256 | >256 | <0.5 | 0.5 | 16 |
| J53RAZ(pKKp8-VIM) | TC | 64 | 64 | 32 | 4 | 2 | <0.25 | <0.125 | 16 | 8 | 128/2432 | 64 | 128 | <0.5 | 0.25 | 2 |
| KW11 | W | >128 | >128 | >128 | >64 | >128 | 128 | >64 | 256 | 32 | >256/4864 | >256 | 16 | 64 | 2 | 128 |
| SA4/2 | W | 64 | >128 | 1 | 64 | 64 | 16 | 1 | 2 | 32 | >256/4864 | 4 | 8 | <0.5 | 1 | 4 |
| J53RAZ(pSA4/2-VIM) | TC | 32 | 64 | 0.5 | 32 | 4 | 2 | 0.25 | 1 | 8 | <0.5/9.5 | 2 | 8 | <0.5 | <0.125 | 1 |
| OM63 | W | 128 | >128 | >128 | 32 | 4 | 4 | 64 | 2 | 16 | >256/4864 | >256 | 8 | <0.5 | 0.5 | 32 |
| DH5α(pOM63-VIM) | TF | 16 | 32 | <0.25 | 2 | 0.5 | <0.25 | <0.125 | 1 | 8 | <0.5/9.5 | <0.5 | 1 | <0.5 | <0.125 | <0.25 |
| OM69 | W | 128 | >128 | 128 | 64 | 64 | 4 | 32 | 2 | 16 | >256/4864 | 256 | 8 | <0.5 | 0.5 | 32 |
| DH5α(pOM69-VIM) | TF | 16 | 32 | <0.25 | 2 | 0.5 | <0.25 | <0.125 | 1 | 8 | <0.5/9.5 | <0.5 | 1 | <0.5 | <0.125 | <0.25 |
| ABC104 | W | >128 | >128 | >128 | >64 | 32 | 4 | 32 | 64 | 8 | >256/4864 | >256 | 128 | <0.5 | 8 | 64 |
| J53RAZ | R | <0.25 | <0.25 | <0.25 | <0.125 | <0.25 | <0.25 | <0.125 | 1 | <0.5 | <0.5/9.5 | <0.5 | 8 | <0.5 | <0.125 | 0.5 |
| DH5α | R | <0.25 | <0.25 | <0.25 | <0.125 | <0.25 | <0.25 | <0.125 | 1 | 1 | <0.5/9.5 | <0.5 | 1 | <0.5 | <0.125 | <0.25 |
Abbreviations: MIC, minimal inhibitory concentration; Azt, aztreonam; Gn, gentamicin; Ak, amikacin; Tet, tetracycline; Chl, chloramphenicol; Col, colistin; Tig, tigecycline; Fos, fosfomycin; W, wild; TC, transconjugant; TF, transformant; R, recipient.