Abstract
Justin et al. suggested that in the crystal structure of a polycomb repressive complex 2 from Chaetomium thermophilum (ctPRC2) a flexible linker region but not the H3K27M cancer mutant peptide better fits the electron density. Based on our new data we agree with this alternative interpretation and provide the crystal structure of ctPRC2 bound to a bona fide H3K27M sequence.
Although displaying obvious sequence diversity, ctPRC2 is functionally and structurally similar to human PRC2 in both basal and stimulated states (1–3). In our initial crystallization efforts, the H3K27M cancer mutant peptide, which also inhibited the enzymatic activity of ctPRC2 in solution, was strictly required to generate ctPRC2 crystals of sufficient quality for structural determination (1). In addition, electron density that clearly corresponded to a short peptide was observed within the substrate-binding groove of the Ezh2 active site. We assigned the H3K27M peptide to the electron density primarily based on the arginine residue unambiguously found to occupy the lysine access channel, although we also noted that the mutated methionine residue was not fully resolved. However, according to Justin et al. a linker region located at the N-terminus of Suz12(VFES) from a neighboring asymmetric unit in the crystal lattice better fits the electron density. Our own re-examination of the electron density map prompted by Justin et al. also indicated this possibility.
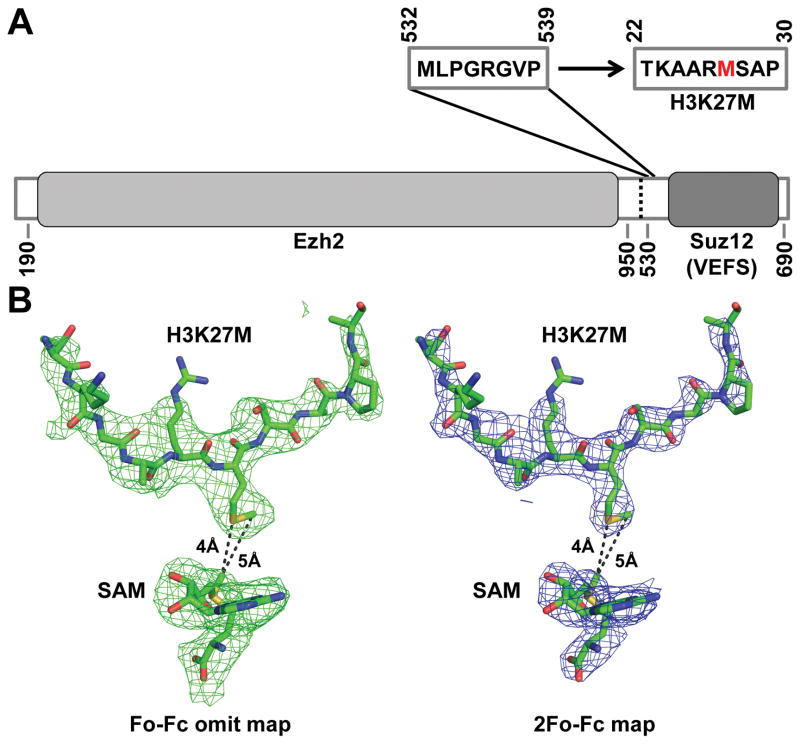
To clarify this situation and, more importantly, to reveal the mechanism of H3K27M recognition by ctPRC2, we replaced the corresponding linker region with an H3K27M peptide sequence in the context of the same Ezh2-Suz12(VEFS) fusion protein that was used for crystallization previously (Fig. 1). We obtained crystals of the sequence-modified ctPRC2 in complex with SAM and the H3K27me3 stimulating peptide in a similar crystal lattice under the same crystallization condition. We were not able to generate crystals of sufficient quality when SAM was replaced with SAH. We determined the new crystal structure to 2.95Å resolution, which showed that a methionine but not an arginine residue in the replaced H3K27M sequence from a neighboring asymmetric unit was inserted into the active site, contacting SAM directly (Fig. 1). Only the aliphatic portion of residue R26 side chain is ordered under the current condition. The remainder of the structure remained essentially identical to the published ctPRC2 structure in the stimulated state. H3K27M thus appears to use the same structural mechanism to inhibit ctPRC2 catalysis as previously suggested for human PRC2 (2, 4).
Figure 1. Crystal structure of ctPRC2 bound to H3K27M.
(A) Residues 532–539 of Suz12(VEFS) are replaced by residues 22–30 of histone H3K27M. Ezh2 and Suz12(VEFS) are indicated by grey boxes.
(B) The Fo−Fc omit (left) and 2Fo−Fc (right) electron density maps contoured at 2.0σ and 1.0σ respectively that correspond to H3K27M and SAM are indicated by the green and blue meshes. Structural models are shown in sticks. The side chain of the mutated methionine residue in H3K27M is clearly defined to occupy the lysine access channel and contact SAM.
The new crystal structure of SAM and H3K27M-bound ctPRC2 in the stimulated state was deposited in the Protein Data Bank under the PDB accession code 5KKL. We also updated the previously deposited PDB entries 5CH1 and 5CH2 to 5KJH and 5KJI, respectively. All other results and analyses of our original publication remain unaffected. We apologize for any confusion that this misinterpretation may have caused.
Acknowledgments
Preparation of this response was supported by Welch Foundation research grant I-1790, CPRIT research grant R1119, Rita Allen Foundation research grant, University of Texas Southwestern Medical Center Endowed Scholar fund, and NIH grant GM114576 to X.L. L.J. was supported by American Heart Association postdoctoral fellowship 16POST30700004. X.L. is a W. W. Caruth, Jr. Scholar in Biomedical Research. This research also received support from the Cecil H. and Ida Green Center Training Program in Reproductive Biology Sciences Research. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. DOE under contract no. DE-AC02-05CH11231. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
References
- 1.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. published online EpubOct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Justin N, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nature communications. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooun A, et al. Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance. Nature communications. 2016;7:11384. doi: 10.1038/ncomms11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. published online EpubMay 17. [DOI] [PMC free article] [PubMed] [Google Scholar]