Abstract
Myostatin (MSTN) is a powerful negative regulator of skeletal muscle mass in mammalian species that is primarily expressed in skeletal muscles, and mutations of its encoding gene can result in the double-muscling trait. In this study, the CRISPR/Cas9 technique was used to edit MSTN in Shaanbei Cashmere goats and generate knockout animals. RNA sequencing was used to determine and compare the transcriptome profiles of the muscles from three wild-type (WT) goats, three fibroblast growth factor 5 (FGF5) knockout goats (FGF5+/- group) and three goats with disrupted expression of both the FGF5 and MSTN genes (FM+/- group). The sequence reads were obtained using the Illumina HiSeq 2000 system and mapped to the Capra hircus reference genome using TopHat (v2.0.9). In total, 68.93, 62.04 and 66.26 million clean sequencing reads were obtained from the WT, FM+/- and FGF5+/- groups, respectively. There were 201 differentially expressed genes (DEGs) between the WT and FGF5+/- groups, with 86 down- and 115 up-regulated genes in the FGF5+/- group. Between the WT and FM+/- groups, 121 DEGs were identified, including 81 down- and 40 up-regulated genes in the FM+/- group. A total of 198 DEGs were detected between the FGF5+/- group and FM+/- group, with 128 down- and 70 up-regulated genes in the FM+/- group. At the transcriptome level, we found substantial changes in genes involved in fatty acid metabolism and the biosynthesis of unsaturated fatty acids, such as stearoyl-CoA dehydrogenase, 3-hydroxyacyl-CoA dehydratase 2, ELOVL fatty acid elongase 6 and fatty acid synthase, suggesting that the expression levels of these genes may be directly regulated by MSTN and that these genes are likely downstream targets of MSTN with potential roles in lipid metabolism in goats. Moreover, five randomly selected DEGs were further validated with qRT-PCR, and the results were consistent with the transcriptome analysis. The present study provides insight into the unique transcriptome profile of the MSTN knockout goat, which is a valuable resource for studying goat genomics.
Introduction
Myostatin (MSTN) is a secreted growth factor and a member of the TGF-β superfamily that functions as a critical autocrine/paracrine inhibitor and negatively regulates skeletal muscle growth and development through the regulation of anabolic and catabolic pathways in skeletal muscles [1]. Myostatin is expressed almost exclusively in skeletal muscle [2]. Mutations in the coding region of MSTN result in a double-muscling phenotype in many species, including cattle [3, 4], mice [5–7] and humans [8]. Myostatin both regulates lean tissue mass and affects body fat content in mice [9]. In MSTN knockout mice (MSTN-/-), the absence of MSTN results in increased skeletal muscle mass, reduced fat tissue, increased insulin sensitivity, enhanced fatty acid oxidation, and an elevated resistance to obesity [6, 10], whereas overexpression of myostatin induces muscle atrophy [11]. Consequently, a number of strategies to block the effects of myostatin have been developed and tested in various models of neuromuscular disorders, muscle-wasting conditions or metabolic disturbances [12–14]. Mice with Lewis Lung carcinoma treated with ActRIIB-Fc, a soluble myostatin receptor that binds myostatin, showed increased body weight and muscle weight as well as significantly increased grip strength [15]. In mdx mice, a model for Duchenne muscular dystrophy, an antibody-mediated myostatin blockade was found to ameliorate the pathophysiology and muscle weakness associated with this condition [16]. However, these strategies have less efficiency in blocking the effects of myostatin.
Natural myostatin gene mutations occur in cattle breeds such as the Belgian Blue, which exhibits obviously increased muscle mass [17]. Muscular hypertrophy, or the double-muscle phenotype, is a heritable condition in cattle [3]. The Shaanbei Cashmere goat is a local breed in China that produces both fiber and meat products. The meat is renowned for its lower fat content compared to that of beef and lamb [18], and thus, it is recommended as a healthy food for consumers. To increase meat production in goats, efforts aimed at the development of strategies to significantly increase muscle growth by manipulating MSTN gene expression have been intensively undertaken [19].
High-throughput mRNA sequencing (RNA-seq) offers the ability to discover new genes and transcripts and to measure both transcript abundance and expression in a single experiment [20, 21]. Based on RNA-seq technology, we measured the integrated global gene expression and signaling pathway activities in goats after MSTN gene knockout. Microarray technology enables a broad overview of the impact of treatments on the expression of all known genes, and this technology has been already been used to examine the effects of constitutive MSTN deficiency in mice and cattle [22–25]. However, high-throughput mRNA sequencing (RNA-Seq) offers the ability to discover new genes and transcripts and to measure transcript expression levels in a single assay [26–28]. Many studies have shown that RNA-Seq is more accurate over a greater dynamic range of gene expression than expression microarrays [29, 30]. Recently, genome-wide gene expression profiling has gained ground due to the development and application of large-scale sequencing techniques to obtain gene sequences and develop molecular markers, especially in less researched species [31–34].
Wang et al. recently reported the successful application of the CRISPR/Cas9 system to manipulate the goat genome [35]. They also demonstrated the utility of this approach by disrupting MSTN, which resulted in enhanced body weight and larger muscle fibers in goats with Cas9-mediated genetic modifications. They further characterized the effects of these genome modifications by hematoxylin and eosin (H&E) staining, quantitative PCR, Western blotting, and immunofluorescence staining (Animal Genetics, under review). However, a transcriptomic analysis of the changes in either gene expression or lipid metabolism in MSTN knockout goats has not been reported.
MSTN was initially characterized as a potent inhibitor of skeletal muscle growth and development [36]. However, its role in the regulation of glucose and lipid metabolism has gained significant attention [9, 37–40]. The goals of this study were to identify important candidate genes related to muscle, glucose and lipid metabolism and to further determine the transcriptomic changes using RNA-Seq. These results will provide valuable information about key genes in this species and improve our understanding of the molecular mechanisms regulating muscle, glucose and lipid metabolism in goats.
Materials and methods
Ethics statement
All animal experiments and procedures were carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals by the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of Northwest A&F University (Approval ID: 2014ZX08008-002).
Animal samples
The animals used in the present study were previously generated by Wang et al. [35]. The genotypes of three MSTN- and FGF5-disrupted goats and three FGF5-disrupted goats are summarized in S1 Table. All animals were raised in the same way under natural light with free access to water and food at the Shaanbei Cashmere Goat Farm at Yulin University. Longissimus dorsi muscles were obtained from nine one-year-old Shaanbei Cashmere goats: three wild-type (WT) goats (each was determined to possess an unedited genome), three goats with only the fibroblast growth factor 5 (FGF5) gene knocked out (FGF5+/- group) and three goats with disruptions in both the FGF5 and MSTN genes (FM+/- group). The transgenic goats were generated using the CRISPR/Cas9 system. Fresh longissimus dorsi muscle samples were harvested immediately from goats after surgery under local anesthesia using 3 mL of procaine (1%) injected into the skin/muscle tissues. All efforts were made to minimize animal suffering and to reduce the number of animals used. Obtained fresh longissimus dorsi muscle samples were washed with sterile normal saline three times, frozen in RNAlater (Takara, Dalian, China) using liquid nitrogen, and sent to Novogene Bioinformatics Technology Co. Ltd. (Beijing, China).
RNA extraction and preparation
Genome-wide transcriptome libraries were constructed from the three sets of muscle samples from the WT, FGF5+/- and FM+/- goats. Longissimus dorsi muscle tissue blocks containing approximately 80–100 mg of tissue were dissolved in TRIzol reagent (Invitrogen, USA) for total RNA extraction. RNA degradation and contamination were monitored by electrophoresis in 1% agarose gels. The quality and quantity of RNA were checked using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and a Qubit® RNA Assay Kit in a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using an RNA Nano 6000 Assay Kit and a Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
mRNA-Seq library preparation and sequencing
Illumina® (NEB, USA) mRNA-Seq libraries were prepared with 3 μg of total RNA using the NEBNext® Ultra™ RNA Library Prep Kit according to the manufacturer’s instructions, and index codes were added to attribute the sequences to each sample. Briefly, RNA was purified from total RNA using Poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature conditions in NEBNext First Strand Synthesis Reaction Buffer (5x). First-strand cDNA was synthesized using random hexamer primers and M-MLV Reverse Transcriptase (RNase H minus). Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of the 3’ ends of cDNA fragments, a NEBNext Adaptor with a hairpin loop structure was ligated to prepare the cDNA for hybridization. To preferentially select cDNA fragments of 150~200 bp in length, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA). Then, 3 μL of USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. Finally, the PCR products were purified (AMPure XP system), and the library quality was assessed using the Agilent Bioanalyzer 2100 system.
Clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina HiSeq 2000 platform, and 100-bp paired-end reads were generated.
Quality control and read mapping to the reference genome
Raw data (raw reads) in the FASTQ format were first processed using in-house Perl scripts and the Q20, Q30 and GC contents of the clean data were calculated.
For mapping, an index of the reference genome was built using Bowtie v2.0.6, and paired-end clean reads were aligned to the reference genome using TopHat v2.0.9.
Gene expression
The counts of the read numbers mapped to each gene were processed by HTSeq v0.6.1, and the FPKM (expected number of fragments per kilobase sequence per million base pairs sequenced) of each gene was calculated based on the length of the gene and the reads count mapped to that gene [41]. The FPKM considers both the effect of sequencing depth and gene length for the read count and is currently the most commonly used method for estimating gene expression levels [42].
DEGs were identified using the R package “DESeq” (1.10.1) with raw gene counts as the input, and quantile normalization was applied for variable library sizes. DESeq provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate [43]. Differentially expressed genes (DEGs) were determined by DEGseq with a cutoff threshold of P-value < 0.05 [44].
Differential gene functional annotation
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes databases (KEGG, http://www.genome.jp/kegg) enrichment was analyzed using differential genes as the foreground and all genes as the background for the identified differential transcripts. GO enrichment analysis of the DEGs was performed using the GOseq R package [45], in which gene length bias was corrected. GO terms with corrected P-values < 0.05 were considered significantly enriched by DEGs.
The KEGG is a database resource for understanding high-level functions and utilities of the biological system [46]. KEGG Orthology Based Annotation System (KOBAS, 2.0) software was used to test the statistical enrichment of differentially expression genes in the KEGG pathways.
qRT-PCR analysis for the validation of RNA-Seq data
Real-time quantitative reverse transcription PCR (qRT-PCR) was used to validate the RNA-Seq data. The total RNA isolated for RNA sequencing was used to perform qRT-PCR. Reverse transcription was performed with 1 μg of total RNA using the PrimeScript RT Reagent Kit (Invitrogen, USA). Real-time quantitative PCR analyses were then performed with a Bio-Rad IQ5 Optical System; individual reactions were prepared with 5 ng of cDNA and SYBR Green PCR master mix (Takara, Dalian, China) in a final volume of 20 μL. All reactions were performed in triplicate. Cycle threshold (Ct) values were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and comparative quantification of mRNA was performed using the 2-ΔΔCt method. The primer sequences used in the qPCR assay are provided in S2 Table [47–49].
Statistical analysis
All data are presented as the means ± standard deviation (SD), and comparisons were performed by analysis of variance (ANOVA) (SAS). A probability of less than 0.05 was considered to be statistically significant.
Results
Summary of the raw sequence reads and DEGs
To understand the gene expression profiles and differences between the three samples at transcript resolution, we performed RNA-Seq on three cDNA sequencing libraries constructed using muscle tissues from WT, FGF5+/- and FM+/- goats. We then mapped the filtered clean reads to the reference goat genome CHIR-1.0 (Table 1). In total, we obtained 68.93, 62.04 and 66.26 million reads for the sequencing libraries constructed from the WT, FGF5+/- and FM+/- goat muscle tissues, respectively. We found that in each sample, ~73% of reads could be mapped to the reference genome, which is comparable to the reports for two non-model organisms, swine [50] and bovine [51], in which 61.4–75.0% of reads are mapped to the reference genome. Thus, our mapping percentage suggests good sequence quality.
Table 1. Statistics of the Illumina RNA-Seq reads in the normal and transgenic goat muscle libraries that were mapped to the goat reference genome CHIR-1.0.
| Summary statistics | Number | Percentage | Number | Percentage | Number | Percentage |
|---|---|---|---|---|---|---|
| WT | FM+/- | FGF5+/- | ||||
| Total raw reads | 70553433 | 63677463 | 67796573 | |||
| Total mapped | 51626789 | 74.99% | 45847205 | 73.90% | 48958298 | 73.90% |
| Total clean reads | 68933544 | 62036347 | 66257588 | |||
| Total clean base pairs (Gb) | 10.34 Gb | 9.30 Gb | 9.94 Gb | |||
| Read map to '+' | 25346942 | 36.81% | 22469249 | 36.22% | 24050354 | 36.30% |
| Read map to '-' | 25053573 | 36.39% | 22207247 | 35.80% | 23732089 | 35.83% |
| Multiply mapped | 1226274 | 1.79% | 1170709 | 1.89% | 1175855 | 1.77% |
| Uniquely mapped | 50400515 | 73.00% | 44676496 | 72.01% | 47782443 | 72.13% |
| Non-splice reads | 25925041 | 37.59% | 22021886 | 35.52% | 24005700 | 36.24% |
| Splice reads | 24475474 | 35.61% | 22654610 | 36.49% | 23776743 | 35.88% |
Identification of DEGs
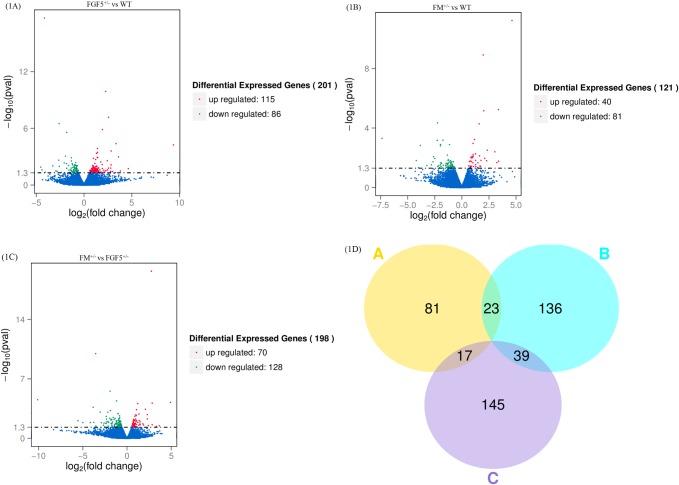
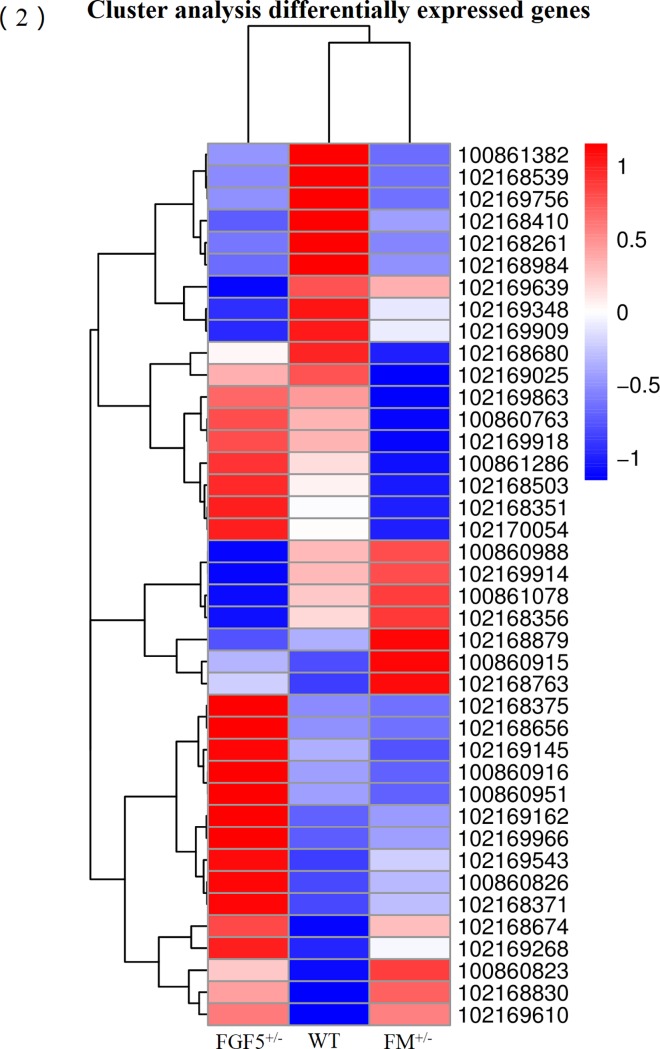
The read count data obtained from the transcriptome were used to analyze differences in gene expression. We obtained 201 DEGs, including 115 up-regulated genes and 86 down-regulated genes, between the FGF5+/- group and the WT group. Moreover, 121 DEGs, including 40 up-regulated genes and 81 down-regulated genes, were identified between the FM+/- group and the WT group. A total of 198 DEGs, including 70 up-regulated genes and 128 down-regulated genes, were identified between the FM+/- group and the FGF5+/- group. We used a false discovery rate (FDR) ≤ 0.001 and an absolute value of the log2 ratio ≥ 1 as the threshold to judge the significance of differences in gene expression. Library size-normalized counts for the three samples were generated, and volcano plots for the DEGs are shown in Fig 1A, 1B and 1C. A Venn diagram showing the number of DEGs among the three pairwise comparisons is shown in Fig 1D. Hierarchical clustering showed the expression profiles of the top 40 DEGs (Fig 2). The full names of the top 20 DEGs in the WT, FM+/- and FGF5+/- goats and their functional characteristics are shown in Table 2 [52–70].
Fig 1. Differentially expressed genes in the RNA-Seq data.
Volcano plot of statistically significant differentially expressed genes at P ≤ 0.05 identified from the RNA-Seq libraries of normal and transgenic goat muscle (1A, 1B, 1C). A Venn diagram showing the DEGs identified from comparisons of FGF5+/- vs WT goats, FM+/- vs WT goats and FM+/- vs FGF5+/- goats (1D).
Fig 2. A hierarchically clustered heatmap showing the expression patterns of the 40 most DEGs.
The red blocks represent the overexpressed genes, and the blue blocks represent genes with the lowest expression levels. Colored bars indicate the expression levels.
Table 2. Biological functions of the 20 most differentially expressed genes in the longissimus dorsi muscle identified by pairwise comparisons of FGF5+/- vs WT, FM+/- vs WT and FM+/- vs FGF5+/-.
| Gene ID | Symbol | Summary |
|---|---|---|
| 100861382 | KRT27 | This gene encodes a member of the type I (acidic) keratin family, which belongs to the superfamily of intermediate filament (IF) proteins that is essential for the proper assembly of type I and type II keratin protein complexes and the formation of keratin intermediate filaments in the inner root sheath (IRS) [52]. |
| 102168539 | ROR2 | The protein encoded by this gene is a receptor protein tyrosine kinase and type I transmembrane protein that belongs to the ROR subfamily of cell surface receptors. The protein may be involved in the early formation of the chondrocytes and may be required for cartilage and growth plate development [53]. |
| 102169758 | NUDT2 | This gene encodes a member of the MutT family of nucleotide pyrophosphatases, a subset of the larger NUDIX hydrolase family. The gene may be a candidate tumor suppressor gene [54]. |
| 102168410 | GJC3 | This gene encodes a gap junction protein. The encoded protein is a connexin that plays a role in the formation of gap junctions, which provide direct connections between adjacent cells [55]. |
| 102168261 | EPHA7 | Activation of the protein encoded by this gene results in activating phosphorylation of components of the ERK signaling pathway, including MAP2K1, MAP2K2, MAPK1 and MAPK3 [56]. |
| 102168984 | TNMD | The gene tenomodulin (TNMD) is a tendon-specific marker known to be important for tendon maturation, with key implications for the residing tendon stem/progenitor cells and for the regulation of endothelial cell migration in chordae tendineae cordis in the heart and in experimental tumor models [57]. Myostatin has a potential role in the induction of tenogenic differentiation of C2C12 cells [58]. |
| 102169639 | MIB1 | This gene encodes a protein that contains multiple ankyrin repeats and RING finger domains and functions as an E3 ubiquitin ligase. The encoded protein positively regulates Notch signaling by ubiquitinating Notch receptors, thereby facilitating Notch receptor endocytosis. This protein may also promote the ubiquitination and degradation of death-associated protein kinase 1 (DAPK1) [59]. |
| 102169348 | CDH19 | This gene is one of three related type II cadherin genes situated in a cluster on chromosome 18. The encoded protein is a calcium-dependent cell-cell adhesion glycoprotein containing five extracellular cadherin repeats. CDH19 plays important roles in cell adhesion and in the formation of adherens junctions, which bind cells together within tissues [60]. |
| 102169909 | ARL15 | The function of this gene has yet to be established. |
| 102168680 | LOC102168680 | The function of this gene has yet to be established. |
| 102169025 | SLC24A3 | Plasma membrane sodium/calcium exchangers play important roles in intracellular calcium homeostasis and electrical conduction. Potassium-dependent sodium/calcium exchangers such as SLC24A3 are believed to exchange 1 intracellular calcium ion and 1 potassium ion for 4 extracellular sodium ions [61]. |
| 102169863 | EGFL6 | This gene encodes a member of the epidermal growth factor (EGF) repeat superfamily. Members of this superfamily are characterized by the presence of EGF-like repeats and are often involved in the regulation of the cell cycle, proliferation, and developmental processes [62]. |
| 100860763 | SCD | Stearoyl-CoA desaturase (Δ-9-desaturase) is an endoplasmic reticulum enzyme that catalyzes the rate-limiting step in the formation of monounsaturated fatty acids (MUFAs), specifically oleate and palmitoleate, from stearoyl-CoA and palmitoyl-CoA [63]. |
| 102169918 | HECW2 | This gene encodes a member of a family of E3 ubiquitin ligases that plays an important role in the proliferation, migration and differentiation of neural crest cells via the regulation of glial cell line-derived neurotrophic factor (GDNF)/Ret signaling. This gene also plays an important role in angiogenesis by stabilizing endothelial cell-to-cell junctions via the regulation of angiomotin-like 1 stability [64]. |
| 100861286 | FASN | Fatty acid synthase is a multi-enzyme protein that catalyzes fatty acid synthesis. Its main function is to catalyze the synthesis of palmitate (C16:0), a long-chain saturated fatty acid, from acetyl-CoA and malonyl-CoA in the presence of NADPH [65]. |
| 102168503 | DOK6 | DOK6 is a member of the DOK (see DOK1; MIM 602919) family of intracellular adaptors that plays a role in the RET (MIM 164761) signaling cascade [66]. |
| 102168351 | TIAM2 | This gene encodes a guanine nucleotide exchange factor. A highly similar mouse protein specifically activates ras-related C3 botulinum substrate 1, converting this Rho-like guanosine triphosphatase (GTPase) from an inactive, guanosine diphosphate-bound state to an active, guanosine triphosphate-bound state. The encoded protein may also play a role in neural cell development [67]. |
| 102170054 | MERTK | This gene encodes a member of the MER/AXL/TYRO3 receptor kinase family. The encoded transmembrane protein contains two fibronectin type-III domains, two Ig-like C2-type (immunoglobulin-like) domains, and one tyrosine kinase domain. Mutations in this gene have been associated with disruption of the retinal pigment epithelium (RPE) phagocytosis pathway and the onset of autosomal recessive retinitis pigmentosa [68]. |
| 100860988 | TPMT | Thiopurine methyltransferase methylates thiopurine compounds. The methyl donor is S-adenosyl-L-methionine, which is converted to S-adenosyl-L-homocysteine. This enzyme uses S-adenosyl-L-methionine as an S-methyl donor to metabolize thiopurine drugs, producing S-adenosyl-L-homocysteine as a byproduct [69]. |
| 102169914 | KLHL23 | The protein encoded by this gene is a member of the kelch family of proteins, which is characterized by a 44–56 amino acid repeat motif. The kelch motif appears in many different polypeptide contexts and contains multiple potential protein-protein contact sites. Members of this family have diverse activities and are present both throughout the cell and extracellularly [70]. |
Functional enrichment of DEGs
The identified DEGs were further analyzed using GO and KEGG enrichment to determine their potential functions and metabolic pathways.
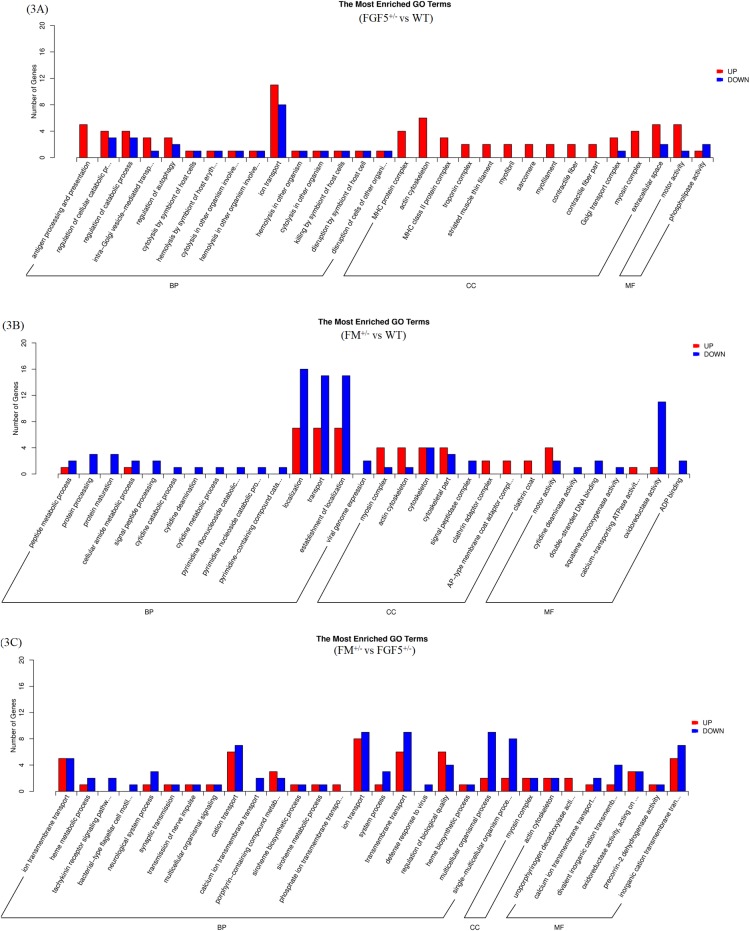
GO analysis based on biological process (BP) enrichment was performed for sets of DEGs with significant cluster profiles. Only significant GO categories with P-values < 0.05 were chosen for analysis. The results of the GO enrichment analysis of DEGs were classified into three categories: BP, cellular component (CC) and molecular function (MF). Fig 3 shows the GO classifications of the DEGs from the comparisons between FGF5+/- and WT goats, FM+/- and WT goats, and FM+/- and FGF5+/- goats.
Fig 3. Gene ontology analysis summary.
Classification of the annotated amino-acid sequences. Amino-acid sequences were grouped into different functional subcategories: cellular component (CC), molecular function (MF) and biological process (BF). 3A: FGF5+/- vs WT; 3B: FM+/- vs WT; and 3C: FM+/- vs FGF5+/-.
The top 20 up- and down-regulated gene-enriched GO terms are shown in Tables 3 and 4. By comparing the gene list of FGF5+/- goats with that of WT goats, two major GO terms relevant to muscle contraction [(actin cytoskeleton and myosin heavy chain (MHC) protein complex)] were identified (Fig 3A). The machinery that powers cell migration is built from the actin cytoskeleton, which is larger than any organelle, and the assembly or disassembly of the actin cytoskeleton can easily change cell morphology [71]. The distribution of enriched GO terms between gene lists of FM+/- and WT goats is shown in Fig 3B. Moreover, the myosin complex and actin cytoskeleton were significantly enriched in the CC. Myosin comprises a superfamily of ATP-dependent motor proteins and is best known for its roles in muscle contraction and its involvement in a wide range of other motility processes in eukaryotes [72]. In terms of BPs, several processes in the fatty acid metabolic system, including peptide metabolism, protein processing and cellular amide metabolism, were found to be enriched in the genes down-regulated in FM+/- goats compared with their expression in WT goats. There were several major enriched GO terms in the comparison of gene lists from FM+/- and FGF5+/- goats (Fig 3C).
Table 3. The top 20 GO terms of significantly up-regulated genes.
| Treatments | GO ID | Category | GO description | P-Value |
|---|---|---|---|---|
| FGF5+/- vs WT | GO:0015629 | cellular component | actin cytoskeleton | 4.29E-06 |
| GO:0042611 | cellular component | MHC protein complex | 4.45E-05 | |
| GO:0019882 | biological process | antigen processing and presentation | 7.64E-05 | |
| GO:0016459 | cellular component | myosin complex | 1.45x10-4 | |
| GO:0042613 | cellular component | MHC class II protein complex | 2.97 x10-4 | |
| GO:0003774 | molecular function | motor activity | 3.38 x10-4 | |
| GO:0005861 | cellular component | troponin complex | 6.65 x10-4 | |
| GO:0005865 | cellular component | striated muscle thin filament | 6.65 x10-4 | |
| GO:0030016 | cellular component | myofibril | 6.65 x10-4 | |
| GO:0030017 | cellular component | sarcomere | 6.65 x10-4 | |
| GO:0036379 GO:0043292 GO:0044449 |
cellular component cellular component cellular component |
myofilament contractile fiber contractile fiber part |
6.65 x10-4 6.65 x10-4 6.65 x10-4 |
|
| GO:0005886 | cellular component | plasma membrane | 1.31x10-3 | |
| GO:0044281 | biological process | small molecule metabolic process | 1.44x10-3 | |
| GO:0032182 | molecular function | mall conjugating protein bindings | 1.72 x10-3 | |
| GO:0043130 | molecular function | ubiquitin binding | 1.72 x10-3 | |
| GO:0017119 | cellular component | Golgi transport complex | 1.96 x10-3 | |
| GO:0006206 | biological process | pyrimidine nucleobase metabolic process | 2.78 x10-3 | |
| GO:0019856 | biological process | pyrimidine nucleobase biosynthetic process | 2.78 x10-3 | |
| FM+/- vs WT | GO:0016459 | cellular component | myosin complex | 2.68E-06 |
| GO:0015629 | cellular component | actin cytoskeleton | 1.46E-05 | |
| GO:0003774 | molecular function | motor activity | 6.82E-05 | |
| GO:0030131 | cellular component | clathrin adaptor complex | 1.14 x10-3 | |
| GO:0030119 | cellular component | AP-type membrane coat adaptor complex | 1.39 x10-3 | |
| GO:0030118 | cellular component | clathrin coat | 1.85 x10-3 | |
| GO:0016192 | biological process | vesicle-mediated transport | 3.12 x10-3 | |
| GO:0044430 | cellular component | cytoskeletal part | 3.62 x10-3 | |
| GO:0009755 | biological process | hormone-mediated signaling pathway | 4.8 x10-3 | |
| GO:0032870 | biological process | cellular response to hormone stimulus | 4.8 x10-3 | |
| GO:0071495 | biological process | cellular response to endogenous stimulus | 4.97 x10-3 | |
| GO:0005575 | cellular component | cellular component | 5.13 x10-3 | |
| GO:0005388 | molecular function | calcium-transporting ATPase activity | 5.18 x10-3 | |
| GO:0071310 | biological process | cellular response to organic substance | 5.29 x10-3 | |
| GO:0009725 | biological process | response to hormone stimulus | 5.29 x10-3 | |
| GO:0009719 | biological process | response to endogenous stimulus | 5.45 x10-3 | |
| GO:0005856 | cellular component | cytoskeleton | 5.95 x10-3 | |
| GO:0070887 | biological process | cellular response to chemical stimulus | 6.49 x10-3 | |
| GO:0010033 | biological process | response to organic substance | 6.67 x10-3 | |
| GO:0009399 | biological process | nitrogen fixation | 6.89 x10-3 | |
| FM+/- vs FGF5+/- | GO:0004853 | molecular function | uroporphyrinogen decarboxylase activity | 4.70E-05 |
| GO:0034220 | biological process | ion transmembrane transport | 1.72 x10-3 | |
| GO:0031090 | cellular component | organelle membrane | 2.2 x10-3 | |
| GO:0065008 | biological process | regulation of biological quality | 2.72 x10-3 | |
| GO:0035435 | biological process | phosphate ion transmembrane transport | 3.82 x10-3 | |
| GO:0019725 | biological process | cellular homeostasis | 4.22 x10-3 | |
| GO:0015672 | biological process | monovalent inorganic cation transport | 5.14 x10-3 | |
| GO:0006779 | biological process | porphyrin-containing compound biosynthetic process | 5.56 x10-3 | |
| GO:0042592 | biological process | homeostatic process | 7.41 x10-3 | |
| GO:0006778 | biological process | porphyrin-containing compound metabolic process | 7.66 x10-3 | |
| GO:0006811 | biological process | ion transport | 8.43 x10-3 | |
| GO:0051701 | biological process | interaction with host | 9.33 x10-3 | |
| GO:0004427 | molecular function | inorganic diphosphatase activity | 9.39 x10-3 | |
| GO:0033014 | biological process | tetrapyrrole biosynthetic process | 0.01 | |
| GO:0005388 | molecular function | calcium-transporting ATPase activity | 0.01 | |
| GO:0006812 | biological process | cation transport | 0.01 | |
| GO:0015114 | molecular function | phosphate ion transmembrane transporter activity | 0.01 | |
| GO:0033013 | biological process | tetrapyrrole metabolic process | 0.01 | |
| GO:0045454 | biological process | cell redox homeostasis | 0.01 | |
| GO:0019048 | biological process | modulation by virus of host morphology or physiology | 0.01 |
Table 4. The top 20 GO terms of significantly down-regulated genes.
| Treatments | GO ID | Category | GO description | P-Value |
|---|---|---|---|---|
| FGF5+/- vs WT | GO:0009055 | molecular function | electron carrier activity | 1.95 x10-3 |
| GO:0019911 | molecular function | structural constituent of myelin sheath | 3.11 x10-3 | |
| GO:0015109 | molecular function | chromate transmembrane transporter activity | 3.89 x10-3 | |
| GO:0015703 | biological process | chromate transport | 3.89 x10-3 | |
| GO:0005198 | molecular function | structural molecule activity | 7.78 x10-3 | |
| GO:0045502 | molecular function | dynein binding | 9.59 x10-3 | |
| GO:0008813 | molecular function | chorismate lyase activity | 0.01 | |
| GO:0005515 | molecular function | protein binding | 0.01 | |
| GO:0004620 | molecular function | phospholipase activity | 0.01 | |
| GO:0042803 | molecular function | protein homodimerization activity | 0.01 | |
| GO:0019015 | cellular component | viral genome | 0.01 | |
| GO:0007156 | biological process | homophilic cell adhesion | 0.01 | |
| GO:0016833 | molecular function | oxo-acid-lyase activity | 0.02 | |
| GO:0016337 | biological process | cell-cell adhesion | 0.02 | |
| GO:0003913 | molecular function | DNA photolyase activity | 0.02 | |
| GO:0005521 | molecular function | lamin binding | 0.02 | |
| GO:0046983 | molecular function | protein dimerization activity | 0.02 | |
| GO:0015099 | molecular function | nickel cation transmembrane transporter activity | 0.03 | |
| GO:0015675 | biological process | nickel cation transport | 0.03 | |
| GO:0044699 | biological process | single-organism process | 0.03 | |
| FM+/- vs WT | GO:0016485 | biological process | protein processing | 7.3 x10-4 |
| GO:0051604 | biological process | protein maturation | 7.3 x10-4 | |
| GO:0016491 | molecular function | oxidoreductase activity | 2.62 x10-3 | |
| GO:0005787 | cellular component | signal peptidase complex | 2.94 x10-3 | |
| GO:0006465 | biological process | signal peptide processing | 2.94 x10-3 | |
| GO:0055114 | biological process | oxidation-reduction process | 4.44 x10-3 | |
| GO:0004126 | molecular function | cytidine deaminase activity | 7.06 x10-3 | |
| GO:0006216 | biological process | cytidine catabolic process | 7.06 x10-3 | |
| GO:0009972 | biological process | cytidine deamination | 7.06 x10-3 | |
| GO:0046087 | biological process | cytidine metabolic process | 7.06 x10-3 | |
| GO:0046133 | biological process | pyrimidine ribonucleoside catabolic process | 7.06 x10-3 | |
| GO:0046135 | biological process | pyrimidine nucleoside catabolic process | 7.06 x10-3 | |
| GO:0072529 | biological process | pyrimidine-containing compound catabolic process | 7.06 x10-3 | |
| GO:0003690 | molecular function | double-stranded DNA binding | 7.11 x10-3 | |
| GO:0019080 | biological process | viral genome expression | 7.86 x10-3 | |
| GO:0006518 | biological process | peptide metabolic process | 9.37 x10-3 | |
| GO:004353 | molecular function | ADP binding | 0.01 | |
| GO:0004506 | molecular function | squalene monooxygenase activity | 0.01 | |
| GO:0005135 | molecular function | interleukin-3 receptor binding | 0.02 | |
| GO:0043566 | molecular function | structure-specific DNA binding | 0.02 | |
| FM+/- vs FGF5+/- | GO:0007217 | biological process | tachykinin receptor signaling pathway | 1.27 x10-3 |
| GO:0071973 | biological process | bacterial-type flagellar cell motility | 4.24 x10-3 | |
| GO:0070838 | biological process | divalent metal ion transport | 4.79 x10-3 | |
| GO:0070588 | biological process | calcium ion transmembrane transport | 5.67 x10-3 | |
| GO:0072511 | biological process | divalent inorganic cation transport | 8.87 x10-3 | |
| GO:0051607 | biological process | defense response to virus | 9.86 x10-3 | |
| GO:0032501 | biological process | multicellular organismal process | 0.01 | |
| GO:0042168 | biological process | heme metabolic process | 0.01 | |
| GO:0072509 | molecular function | divalent inorganic cation transmembrane transporter activity | 0.01 | |
| GO:0007601 | biological process | visual perception | 0.01 | |
| GO:0030553 | molecular function | cGMP binding | 0.01 | |
| GO:0050953 | biological process | sensory perception of light stimulus | 0.01 | |
| GO:0003856 | molecular function | 3-dehydroquinate synthase activity | 0.01 | |
| GO:0044707 | biological process | single-multicellular organism process | 0.01 | |
| GO:0030416 | biological process | methylamine metabolic process | 0.01 | |
| GO:1901160 | biological process | primary amino compound metabolic process | 0.01 | |
| GO:0006816 | biological process | calcium ion transport | 0.01 | |
| GO:0050877 | biological process | neurological system process | 0.01 | |
| GO:0008113 | molecular function | peptide-methionine (S)-S-oxide reductase activity | 0.01 | |
| GO:0030551 | molecular function | cyclic nucleotide binding | 0.01 |
Pathway analysis
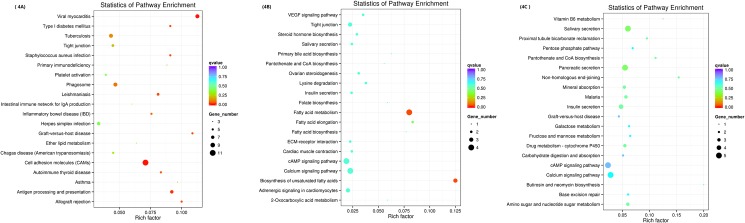
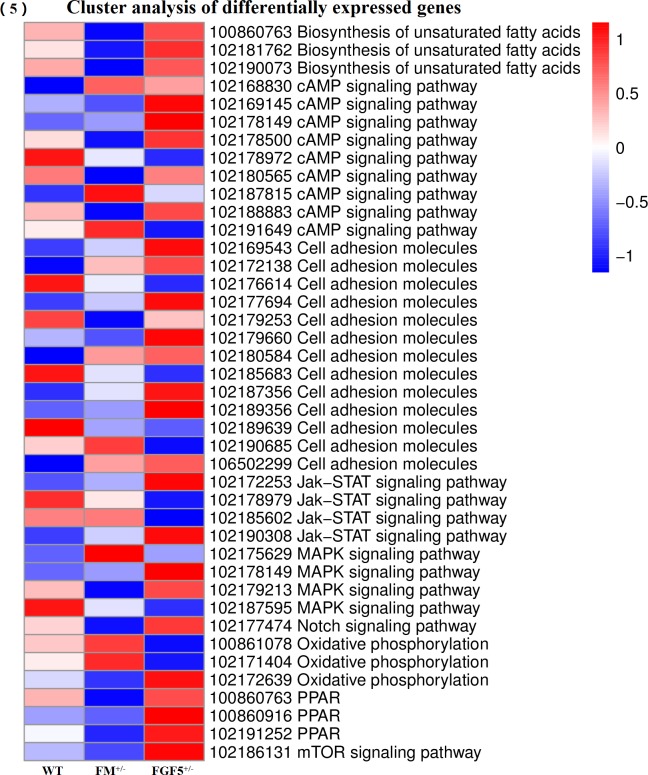
As shown in Fig 4, the top 20 pathways were identified by pairwise comparisons of complete DEGs lists from every group. The results from the KEGG analysis revealed many genes related to fatty acid metabolism, the peroxisome proliferator-activated receptor (PPAR) signaling pathway, the mitogen-activated protein kinase (MAPK) signaling pathway and cell adhesion molecules. Additionally, the KEGG analysis revealed several genes involved in Notch signaling, AMP-activated protein kinase (AMPK) signaling, mTOR-signaling, oxidative phosphorylation, and Jak-STAT signaling. Many genes significantly down-regulated in the FM+/- goats compared to their expression in WT and FGF5+/- goats were associated with fatty acid biosynthesis (stearoyl-CoA desaturase, fatty acid synthase, ELOVL fatty acid elongase 6 and 3-hydroxyacyl-CoA dehydratase 2) and the biosynthesis of unsaturated fatty acids (stearoyl-CoA desaturase, fatty acid synthase and 3-hydroxyacyl-CoA dehydratase 2). Genes involved in the PPAR signaling pathway (stearoyl-CoA desaturase), MAPK signaling pathway [(mitogen-activated protein kinase 3, MAPK3 or ERK1), (ras-related C3 botulism toxin substrate 2, RAC2), cell division cycle 25B (CDC25B), phospholipase A2 group IVE (PLA2G4E)], biosynthesis of unsaturated fatty acids, Notch signaling (hes family bHLH transcription factor 1 and HES 1), AMPK signaling (calcium binding protein 39-like and fructose-bisphosphatase 1) and mTOR-signaling (calcium binding protein 39-like) were also down-regulated. On the other hand, along with an increased percentage of differentially expressed gene sets between FGF5+/- goats and WT goats, the DEGs clustered in the pathways of viral myocarditis, cell adhesion molecules (CAMs), antigen processing and presentation, graft-versus-host disease, leishmaniosis, allograft rejection, staphylococcus aureus infection and type I diabetes mellitus were up-regulated. Our KEGG pathway enrichment analysis of the DEGs showed that genes involved in oxidative phosphorylation (pyro phosphatase and ATPase H+ transporting V0 subunit a4) and Jak-STAT signaling (colony stimulating factor 3) were up-regulated after knockout of the MSTN gene (Fig 5).
Fig 4. KEGG pathway enrichment analysis.
4A: FM+/- vs WT; 4B: FGF5+/- vs WT. 4C: FM+/- vs FGF5+/-.
Fig 5. Cluster analysis of DEGs.
Heat map of the expression levels of DEGs related to lipid, glucose, protein and oxidative phosphorylation metabolism pathways in the muscles of the goats.
Verification of DEGs by qRT-PCR
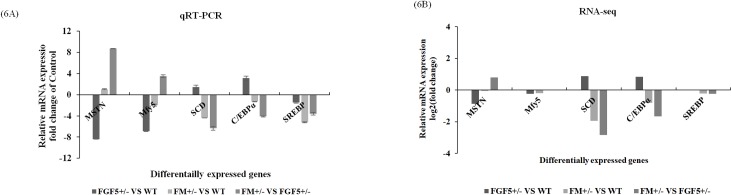
To further confirm and validate the transcriptome analysis results, the RNA samples isolated for RNA-Seq were used in the qRT-PCR analysis. The DEGs were selected based on the expression profiles and the following criteria: DEGs with a fold-change (log2) ≥ 1 or fold-change (log2) ≤ -1 and a P-value < 0.05. A subset of 5 genes selected from the list of DEGs was selected for qRT-PCR analysis. These genes were associated with muscle structure [myogenic actor 5 (Mfy5)], muscle fat content [stearoyl coenzyme A dehydrogenase (SCD), CCAAT-enhancer-binding protein α (C/EBPα) and sterol-regulatory element binding proteins (SREBP)] and muscle growth/differentiation (MSTN). Overall, the qPCR results showed good correspondence with the transcriptome analysis results, indicating that the RNA-Seq data were reliable and accurate (Fig 6). The discrepancies with respect to the ratio should be attributed to the different algorithms and sensitivity of the two techniques [73, 74].
Fig 6. Relative expression levels of MSTN, SCD, C/EBPα, SREBP and Mfy5 determined by qRT-PCR and RNA-Seq.
A: qRT-PCR; and B: RNA-Seq.
Discussion
Myostatin is a powerful negative regulator of skeletal muscle growth and development. Although myostatin inhibition in skeletal muscles is valuable for agricultural applications, the molecules downstream of myostatin in skeletal muscle have not been fully identified [75]. Transcriptome analysis is an important method of exploring functional genes and is both the foundation and starting point for studying gene functions and structures [76]. Previous studies have not explored the transcriptome profile of the MSTN knockout goat. To our knowledge, this is the first study to investigate the effects of MSTN knockout on muscle hypertrophy and functionality as well as whole-body lipid metabolism in a goat model.
FGF5 resides on chromosome 4q21.21, directly within the region of homozygosis [77]. FGF5, which is a member of the FGF family and has 23 related genes, regulates hair length in humans and a variety of other animals. The FGF5 family is involved in the control of numerous physiological processes, including embryonic development, neuronal survival, wound repair and angiogenesis, but is also involved in a number of pathological responses [78–80]. In postnatal muscles, satellite cells are the myogenic precursors and are situated beneath the myofiber basement membrane. FGF family proteins can maintain the proliferative state of satellite cells in rat myofiber cultures; specifically, FGF1, FGF4, and FGF6 enhance satellite cell proliferation, whereas FGF5 and FGF7 are ineffective [81]. Mice that are homozygous null for FGF5 display no obvious defects in limb or axial muscle development but are characterized by substantial hair overgrowth. This finding, which is an important demonstration that FGF5 is not required for normal limb development [82], provides evidence that FGF5 is not involved in muscle development. Recent studies have suggested that the FGF5 gene is associated with hair length and in controls the cessation of the anagen stage. FGF5 signaling pathway is binding of canonical FGFs to FGFR with heparin sulfate as a cofactor induces the formation of ternary FGF—FGFR-HS complex, which activates the FGFR intracellular tyrosine kinase domain by phosphorylation of specific tyrosine residues. The activated receptor is coupled to intracellular RAS-MAPK signaling pathway [83, 84]. However, MSTN is a negative regulator of skeletal muscle development and growth, and it is expressed mainly in muscle. The myostatin-Smad pathway alters the activity of protein kinase AKT, thereby inhibiting mTOR pathway and protein synthesis [85]. Therefore, hitherto, no works have yet addressed the possible interaction between MSTN and FGF5. Taken together, our results support that FGF5 has not affect the function of MSTN. We assessed the function and molecular mechanism of introducing myostatin dysfunction in FGF5 knockout goats in order to compare the differences between FGF5/MSTN dual knockout and FGF5 single knockout goats.
Myostatin dysfunction results in a dramatic increase of animal muscle mass due to increases in both the numbers and cross-sectional areas of myofibrils [7, 86]. In the present study, we were the first to use transcriptome analysis to identify genes expression changes caused by the knockout of MSTN in goats. In total, 68.93 Gb of clean data were obtained. Several DEGs involved in muscle development, fatty acid metabolism, glucose metabolism and oxidative phosphorylation were identified following MSTN and FGF5 gene knockout; these results may benefit studies of the function and molecular mechanism of myostatin in goats. Changes in the downstream molecules of MSTN, including the increased expression levels of MYH15 (myosin heavy chain 15), MAPKAPK3 (mitogen-activated protein kinase-active protein kinase) and MYOZ3 (myozenin 3) [87–89], indicated that functional MSTN activity or expression was enhanced in FM +/- goats compared to that in the FGF5 +/- and WT goats. Myostatin expression is restricted to developing skeletal muscles, but myostatin protein is still expressed and secreted by skeletal muscles in adulthood [90, 91]. Though the effects of MSTN knockout varies by age, no differences were found in body size and weight at birth among homozygous KO lambs and their WT counterparts. However, KO lambs were 20–30% heavier than WT lambs 60 days later despite having a similar body size [92, 93]. The causes of these results are not entirely understood, but it is clear that these results are dependent on the genetic background.
This study showed that genes related to the immune system were up-regulated in the FGF5+/- group compared with the WT and FM+/- groups. Overexpression of KRT27 has been reported in the inner root sheath [52], and KRT27 has been shown to regulate innate immune functions [94]. Furthermore, cell adhesion pathway enrichment analysis of the DEGs revealed changes in the intercellular adhesion molecule 3 (ICAM-3), human leukocyte antigen (HLA) class II histocompatibility antigen, BOLA class I histocompatibility antigen and HLA class II histocompatibility antigen genes. ICAM-3, a member of the ICAM immunoglobulin family of adhesion molecules, binds to leukocyte function antigen and mediates the initial localization of neutrophils to sites of tissue injury and inflammation in autoimmune diseases [95]. CAMs of the immunoglobulin superfamily nucleate and maintain groups of cells at key sites during early development and in the adult stage. In addition to their adhesive properties, the binding of CAMs can affect intracellular signaling and developmental events, including cell migration, proliferation, and differentiation [96].
Our KEGG pathway enrichment analysis of the DEGs showed that the skeletal muscle growth, fatty acid metabolism, glucose metabolism and oxidative phosphorylation pathways were enriched in DEGs after MSTN knockout. The DEG HES1, which is involved in Notch signaling, may act as a regulator of myogenesis by inhibiting the function of MyoD1 and ASH1. It is well known that AMPK acts as a key energy sensor that balances anabolism and catabolism by monitoring either cellular ATP levels or glycogen content.
The MAPK signaling pathway was significantly enriched in DEGs (MAPKAPK3, RAC2, CDC25B and PLA2G4E) after knockout of the MSTN and FGF5 genes. Myostatin activates Erk1/2 MAPK in both proliferating and differentiating C2C12 cells [97]; thus, MSTN may affect muscle structures in goats. The MAPK/ERK pathway is reported to be associated with cell proliferation, differentiation, migration, senescence and apoptosis [98, 99]. A wealth of data has revealed a cross-talk between myostatin and the intracellular AKT/mTOR signaling pathway, strongly supporting the notion that myostatin affects the muscle protein balance by regulating protein synthesis and degradation [100]. Adult muscle growth is primarily due to the increase of protein content through activation of the AKT/mTOR pathway, which regulates protein synthesis [101]. Inhibition of myostatin, a negative growth modulator in muscles, functionally enhances muscle mass and improves glucose and fat metabolism in myostatin propeptide transgenic mice [102]. Published studies have clearly shown that adipogenesis is decreased in MSTN KO mice. Moreover, the absence of MSTN results in enhanced peripheral tissue fatty acid oxidation and increased thermogenesis, culminating in increased fat utilization and reduced adipose tissue mass [103]. Consistent with previous studies, our study showed that fatty acid synthesis is decreased in heterozygous MSTN goats, and our KEGG pathway enrichment analysis of the DEGs showed that the fatty acid metabolism pathway was significantly enriched in DEGs after MSTN knockout in goats.
Both adipocytes and myocytes are derived from the same mesodermal precursor cells during development. C3H10T(1/2) cells, a mesenchymal fibroblast-like cell line of embryonic origin, have the capacity to undergo differentiation into multiple cell lineages, such as myoblasts, chondrocytes, and adipocytes, after incubation in different media in vitro [104–106]. Later, differentiation controlled by different transcriptional pathways yields individual tissues, suggesting that the myostatin gene is involved in regulating both adiposity and muscularity [92]. In a previous study, we used CRISPR/Cas9 technology and single cell embryo injection and showed that these two methods differ in terms of gene editing efficiency. Specifically, single cell embryo injection could result in abnormal fetus development and high fetal mortality, whereas CRISPR/Cas9 technology did not affect the health of the animals [107]. More importantly, the present study identified the response of some important genes, including TNMD, SCD and FANS, to MSTN disruption. These results will be beneficial for breeding purposes.
Conclusions
In this study, we assembled, characterized, and evaluated the muscle tissue transcriptome and quantified the gene expression levels in MSTN and FGF5 gene-modified goats. Through the development of a rigorous multistep bioinformatics approach, the sequencing reads obtained from MSTN knockout goats were successfully cleaned of host sequences and were found to contain high numbers of important genes. The results presented here and the associated assembly will help improve our understanding of the MSTN gene expression profile and molecular mechanisms. These data are valuable resources for future studies of goat genomics and will be beneficial for breeding applications that target multiple genes with strong effects on economically important traits in animals. Finally, the results of this study suggest that myostatin disruption might be a promising strategy for the treatment of type 2 diabetes and related metabolic diseases.
Supporting information
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This study was carried out using the animals from the Shaanbei Cashmere Goat Farm at Yulin University. The authors would like to thank all participants who were involved in our study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is supported by National Natural Science Foundation of China (31372279, 31171377, 31572369) to YC, and China Agriculture Research System (CARS-39-12) to YC, as well as the Major Projects for New Varieties of Genetically Modified Organisms of China (2014ZX08008-002) to YC, National Natural Science Foundation of China (31402038) and the key Research Program of Shaanxi Province (2017NY-072) to XW, and the Special Fund for Agro-scientific Research in the Public Interest (201303059) to YC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lee S. Regulation of muscle mass by myostatin. Annual Review of Cell and Developmental Biology. 2004;20(1):61–86. [DOI] [PubMed] [Google Scholar]
- 2.Mcpherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387(6628):83 doi: 10.1038/387083a0 [DOI] [PubMed] [Google Scholar]
- 3.Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Research. 1997;7(9):910 [DOI] [PubMed] [Google Scholar]
- 4.Mcpherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proceedings of the National Academy of Sciences. 1997;94(23):12457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakatani M, Takehara Y, Sugino H, Matsumoto M, Hashimoto O, Hasegawa Y, et al. Transgenic expression of a myostatin inhibitor derived from follistatin increases skeletal muscle mass and ameliorates dystrophic pathology in mdx mice. Faseb Journal Official Publication of the Federation of American Societies for Experimental Biology. 2008;22(2):477–87. doi: 10.1096/fj.07-8673com [DOI] [PubMed] [Google Scholar]
- 6.Mcpherron AC, Lee SJ. McPherron A.C. & Lee S.J. Suppression of body fat accumulation in myostatin-deficient mice. J. Clin. Invest. 109, 595–601. Journal of Clinical Investigation. 2002;109(5):595–601. doi: 10.1172/JCI13562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SJ. Quadrupling Muscle Mass in Mice by Targeting TGF-ß Signaling Pathways. Plos One. 2007;2(8):: e789 doi: 10.1371/journal.pone.0000789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuelke M, Wagner KR, Stolz LE, Hübner C, Riebel T, Kömen W, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. New England Journal of Medicine. 2004;350(26):2682–8. doi: 10.1056/NEJMoa040933 [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Mcfarlane C, Lokireddy S, Masuda S, Ge X, Gluckman PD, et al. Inhibition of myostatin protects against diet-induced obesity by enhancing fatty acid oxidation and promoting a brown adipose phenotype in mice. Diabetologia. 2012;55(1):183–93. doi: 10.1007/s00125-011-2304-4 [DOI] [PubMed] [Google Scholar]
- 10.Guo T, Jou W, Chanturiya T, Portas J, Gavrilova O, Mcpherron AC. Myostatin inhibition in muscle, but not adipose tissue, decreases fat mass and improves insulin sensitivity. Plos One. 2009;4(3):e4937 doi: 10.1371/journal.pone.0004937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, et al. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296(5572):1486–8. doi: 10.1126/science.1069525 [DOI] [PubMed] [Google Scholar]
- 12.Breitbart A, Auger-Messier M, Molkentin JD, Heineke J. Myostatin from the heart: local and systemic actions in cardiac failure and muscle wasting. American Journal of Physiology Heart & Circulatory Physiology. 2011;300(6):1973–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kainulainen H, Papaioannou K, Silvennoinen M, Autio R, Saarela J, Oliveira BM, et al. Myostatin/activin blocking combined with exercise reconditions skeletal muscle expression profile of mdx mice. Molecular and Cellular Endocrinology. 2015;399:131–42. doi: 10.1016/j.mce.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 14.Heineke J, Augermessier M, Xu J, Sargent MA, York AJ, Welle S, et al. Genetic Deletion of Myostatin From the Heart Prevents Skeletal Muscle Atrophy in Heart Failure. Circulation. 2010;121(3):419–25. doi: 10.1161/CIRCULATIONAHA.109.882068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sílvia B, Míriam T, Marcel O, David M, Maria P, Eva C, et al. Myostatin blockage using actRIIB antagonism in mice bearing the Lewis lung carcinoma results in the improvement of muscle wasting and physical performance. Journal of Cachexia Sarcopenia & Muscle. 2012;3(1):37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, et al. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420(6914):418–21. doi: 10.1038/nature01154 [DOI] [PubMed] [Google Scholar]
- 17.Dominique JE, Gérard C. Myostatin regulation of muscle development: Molecular basis, natural mutations, physiopathological aspects. Experimental Cell Research. 2006;312(13):2401–14. doi: 10.1016/j.yexcr.2006.04.012 [DOI] [PubMed] [Google Scholar]
- 18.James NA, Berry BW. Use of chevon in the development of low-fat meat products. Journal of Animal Science. 1997;75(2):571–7. [DOI] [PubMed] [Google Scholar]
- 19.Patel UA, Patel AK, Joshi CG. Stable suppression of myostatin gene expression in goat fetal fibroblast cells by lentiviral vector-mediated RNAi. Biotechnology Progress. 2015;31:452–9. doi: 10.1002/btpr.2022 [DOI] [PubMed] [Google Scholar]
- 20.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold BJ. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 21.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The Transcriptional Landscape of the Yeast Genome Defined by RNA Sequencing. Science. 2008;320(5881):1344–9. doi: 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cassarmalek I, Passelaigue F, Bernard C, Léger J, Hocquette JF. Target genes of myostatin loss-of-function in muscles of late bovine fetuses. BMC Genomics. 2007;8(1):423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sadkowski T, Jank M, Zwierzchowski L, Siadkowska E, Oprządek J, Motyl T. Gene expression profiling in skeletal muscle of Holstein-Friesian bulls with single-nucleotide polymorphism in the myostatin gene 5’-flanking region. Journal of Applied Genetics. 2008;49(3):237–50. doi: 10.1007/BF03195620 [DOI] [PubMed] [Google Scholar]
- 24.Steelman CA, Recknor JC, Nettleton D, Reecy JM. Transcriptional profiling of myostatin-knockout mice implicates Wnt signaling in postnatal skeletal muscle growth and hypertrophy. Faseb Journal Official Publication of the Federation of American Societies for Experimental Biology. 2006;20(3):580–2. doi: 10.1096/fj.05-5125fje [DOI] [PubMed] [Google Scholar]
- 25.Welle S, Cardillo A, Zanche M, Tawil R. Skeletal muscle gene expression after myostatin knockout in mature mice. Physiological Genomics. 2009;38(3):342–50. doi: 10.1152/physiolgenomics.00054.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 27.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nature Methods. 2008;5(7):613–9. doi: 10.1038/nmeth.1223 [DOI] [PubMed] [Google Scholar]
- 28.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881): 1344–9. doi: 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Research. 2008;18(9):1509–17. doi: 10.1101/gr.079558.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10(1):161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strickler SR, Bombarely A, Mueller LA. Designing a transcriptome next-generation sequencing project for a nonmodel plant species. American Journal of Botany. 2012;99(2):257–66. doi: 10.3732/ajb.1100292 [DOI] [PubMed] [Google Scholar]
- 32.Calduchginer JA, Bermejonogales A, Beneditopalos L, Estensoro I, Ballesterlozano G, Sitjàbobadilla A, et al. Deep sequencing for de novo construction of a marine fish (Sparus aurata) transcriptome database with a large coverage of protein-coding transcripts. BMC Genomics. 2013;14(1):178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panserat S. Differential gene expression after total replacement of dietary fish meal and fish oil by plant products in rainbow trout (Oncorhynchus mykiss) liver. Aquaculture. 2009;294(1–2):123–31. [Google Scholar]
- 34.Chen QL, Luo Z, Huang C, Pan YX, Wu K. De novo characterization of the liver transcriptome of javelin goby Synechogobius hasta and analysis of its transcriptomic profile following waterborne copper exposure. Fish Physiology and Biochemistry. 2016;42(3):979–94. doi: 10.1007/s10695-015-0190-2 [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Yu H, Lei A, Zhou J, Zeng W, Zhu H, et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Scientific Reports. 2015;5:13878 doi: 10.1038/srep13878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirk SP, Oldham JM, Kambadur R, Sharma M, Dobbie PM, Bass JJ. Myostatin regulation during skeletal muscle regeneration. Journal of Cellular Physiology. 2000;184(3):356–63. doi: 10.1002/1097-4652(200009)184:3<356::AID-JCP10>3.0.CO;2-R [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Mcfarlane C, Lokireddy S, Bonala S, Ge X, Masuda S, et al. Myostatin-deficient mice exhibit reduced insulin resistance through activating the AMP-activated protein kinase signalling pathway. Diabetologia. 2011;54(6):1491–501. doi: 10.1007/s00125-011-2079-7 [DOI] [PubMed] [Google Scholar]
- 38.Mcpherron AC, Lee SJ. Suppression of body fat accumulation in myostatin-deficient mice. Journal of Clinical Investigation. 2002;109(5):595–601. doi: 10.1172/JCI13562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo T, William J, Tatyana C, Jennifer P, Oksana G, Mcpherron AC. Myostatin Inhibition in Muscle, but Not Adipose Tissue, Decreases Fat Mass and Improves Insulin Sensitivity. Plos One. 2009;4(3):e4937 doi: 10.1371/journal.pone.0004937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao B, Wall RJ, Yang J. Transgenic expression of myostatin propeptide prevents diet-induced obesity and insulin resistance. Biochemical & Biophysical Research Communications. 2005;337(1):248–55. [DOI] [PubMed] [Google Scholar]
- 41.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28(5):511–5. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035 doi: 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. Controlling The False Discovery Rate—A Practical And Powerful Approach To Multiple Testing. Journal of the Royal Statistical Society. 1995;57(57):289–300. [Google Scholar]
- 44.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology. 2010;11(2):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Research. 2008;36(Database issue):D480–D4. doi: 10.1093/nar/gkm882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripathi AK, Ramani UV, Patel AK, Rank DN, Joshi CG. Short Hairpin RNA-Induced Myostatin Gene Silencing in Caprine Myoblast Cells In Vitro. Applied Biochemistry & Biotechnology. 2013;169(2):688–94. [DOI] [PubMed] [Google Scholar]
- 48.Patel UA, Patel AK, Joshi CG. Stable suppression of myostatin gene expression in goat fetal fibroblast cells by lentiviral vector-mediated RNAi. Biotechnology Progress. 2015;31(2):452 doi: 10.1002/btpr.2022 [DOI] [PubMed] [Google Scholar]
- 49.Ohsaki H, Sawa T, Sasazaki S, Kano K, Taniguchi M, Mukai F, et al. Stearoyl-CoA desaturase mRNA expression during bovine adipocyte differentiation in primary culture derived from Japanese Black and Holstein cattle. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2007;148(3):629–34. [DOI] [PubMed] [Google Scholar]
- 50.Chen C, Ai H, Ren J, Li W, Li P, Qiao R, et al. A global view of porcine transcriptome in three tissues from a full-sib pair with extreme phenotypes in growth and fat deposition by paired-end RNA sequencing. Bmc Genomics. 2011;12(1):448–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wickramasinghe S, Rincon G, Islas-Trejo A, Medrano JF. Transcriptional profiling of bovine milk using RNA sequencing. Bmc Genomics. 2012;13(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Langbein L, Rogers MA, Praetzelwunder S, Helmke B, Schirmacher P, Schweizer J. K25 (K25irs1), K26 (K25irs2), K27 (K25irs3), and K28 (K25irs4) represent the type I inner root sheath keratins of the human hair follicle. Journal of Investigative Dermatology. 2006;126(11):2377–86. doi: 10.1038/sj.jid.5700494 [DOI] [PubMed] [Google Scholar]
- 53.Ma SSQ, Henry C, Llamosas E, Higgins R, Daniels B, Hesson LB, et al. Validation of specificity of antibodies for immunohistochemistry: the case of ROR2. Virchows Archiv. 2016:1–10. [DOI] [PubMed] [Google Scholar]
- 54.Ogawa T, Ueda Y, Yoshimura K, Shigeoka S. Comprehensive Analysis of Cytosolic Nudix Hydrolases in Arabidopsis thaliana. Journal of Biological Chemistry. 2005;280(26):25277–83. doi: 10.1074/jbc.M503536200 [DOI] [PubMed] [Google Scholar]
- 55.Marsh A, Caseygreen K, Probert F, Withall DM, Mitchell DA, Dilly SJ, et al. Simvastatin Sodium Salt and Fluvastatin Interact with Human Gap Junction Gamma-3 Protein. PLOS ONE. 2016;11(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, et al. The Eph-Receptor A7 Is a Soluble Tumor Suppressor for Follicular Lymphoma. Cell. 2011;147(3):554–64. doi: 10.1016/j.cell.2011.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shukunami C, Takimoto A, Oro M, Hiraki Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Developmental Biology. 2006;298(1):234–47. doi: 10.1016/j.ydbio.2006.06.036 [DOI] [PubMed] [Google Scholar]
- 58.Uemura K, Hayashi M, Itsubo T, Oishi A, Iwakawa H, Komatsu M, et al. Myostatin promotes tenogenic differentiation of C2C12 myoblast cells through Smad3. Febs Open Bio. 2017;7(4):522–32. doi: 10.1002/2211-5463.12200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon DY, Dimitriadi M, Terzic B, Cable C, Hart AC, Chitnis AB, et al. The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Molecular Biology of the Cell. 2013;24(12):1863–71. doi: 10.1091/mbc.E13-01-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin J, Luo J, Redies C. Cadherin-19 expression is restricted to myelin-forming cells in the chicken embryo. Neuroscience. 2010;165(1):168–78. doi: 10.1016/j.neuroscience.2009.10.032 [DOI] [PubMed] [Google Scholar]
- 61.Kraev A, Quednau BD, Leach SA, Li X, Dong H, Winkfein R, et al. Molecular Cloning of a Third Member of the Potassium-dependent Sodium-Calcium Exchanger Gene Family,NCKX3. Journal of Biological Chemistry. 2001;276(25):23161–72. doi: 10.1074/jbc.M102314200 [DOI] [PubMed] [Google Scholar]
- 62.Wouters MA, Rigoutsos I, Chu CK, Feng LL, Sparrow DB, Dunwoodie SL. Evolution of distinct EGF domains with specific functions. Protein Science. 2005;14(4):1091–103. doi: 10.1110/ps.041207005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinemann FS, Ozols J. Stearoyl-CoA desaturase, a short-lived protein of endoplasmic reticulum with multiple control mechanisms. Prostaglandins Leukotrienes and Essential Fatty Acids. 2003;68(2):123–33. [DOI] [PubMed] [Google Scholar]
- 64.Choi K, Choi H, Lee J, Im S, Zhang H, Jeong Y, et al. The endothelial E3 ligase HECW2 promotes endothelial cell junctions by increasing AMOTL1 protein stability via K63-linked ubiquitination. Cellular Signalling. 2016;28(11):1642–51. doi: 10.1016/j.cellsig.2016.07.015 [DOI] [PubMed] [Google Scholar]
- 65.Loftus TM, Jaworsky DE, Frehywot GL, Townsend CA, Ronnett GV, Lane MD, et al. Reduced Food Intake and Body Weight in Mice Treated with Fatty Acid Synthase Inhibitors. Science. 2000;288(5475):2379–81. [DOI] [PubMed] [Google Scholar]
- 66.Crowder RJ, Enomoto H, Yang M, Johnson EM, Milbrandt J. Dok-6, a novel p62 Dok family member, promotes Ret-mediated neurite outgrowth. Journal of Biological Chemistry. 2004;279(40):42072–81. doi: 10.1074/jbc.M403726200 [DOI] [PubMed] [Google Scholar]
- 67.Chiu C, Leng S, Martin KA, Kim EHJ, Gorman SE, Duhl D. Cloning and Characterization of T-Cell Lymphoma Invasion and Metastasis 2 (TIAM2), a Novel Guanine Nucleotide Exchange Factor Related to TIAM1 ☆. Genomics. 1999;61(1):66–73. doi: 10.1006/geno.1999.5936 [DOI] [PubMed] [Google Scholar]
- 68.Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, et al. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nature Genetics. 2000;26(3):270–1. doi: 10.1038/81555 [DOI] [PubMed] [Google Scholar]
- 69.Wang L, Pelleymounter LL, Weinshilboum RM, Johnson JA, Hebert JM, Altman RB, et al. Very important pharmacogene summary: thiopurine S-methyltransferase. Pharmacogenetics and Genomics. 2010;20(6):401–5. doi: 10.1097/FPC.0b013e3283352860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dhanoa BS, Cogliati T, Satish AG, Bruford EA, Friedman JS. Update on the Kelch-like (KLHL) gene family. Human Genomics. 2013;7(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uzman A. Molecular Cell Biology (4th edition). Biochemistry & Molecular Biology Education. 2001;29(3):126–8. [Google Scholar]
- 72.Pollard TD, Korn ED. Acanthamoeba myosin. I. Isolation from Acanthamoeba castellanii of an enzyme similar to muscle myosin. Journal of Biological Chemistry. 1973;248(13):4682–90. [PubMed] [Google Scholar]
- 73.Li PH, Ponnala L, Gandotra N, Wang L, Si YQ, Tausta SL, et al. The developmental dynamics of the maize leaf transcriptome. Nature Genetics. 2010;42(12):1060–7. doi: 10.1038/ng.703 [DOI] [PubMed] [Google Scholar]
- 74.Shi T, Gao Z, Wang L, Zhang Z, Zhuang W, Sun H, et al. Identification of differentially-expressed genes associated with pistil abortion in Japanese apricot by genome-wide transcriptional analysis. Plos One. 2012;7(10):17302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Egerman MA, Cadena SM, Gilbert JA, Meyer A, Nelson HN, Swalley SE, et al. GDF11 Increases with Age and Inhibits Skeletal Muscle Regeneration. Cell Metabolism. 2015;22(1):164–74. doi: 10.1016/j.cmet.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glenn TC. Field guide to nextextA, Meyer A, Nelson HN, Swalley SE, et al. GDF11 Increases with Age and I
- 77.Higgins CA, Petukhova L, Harel S, Ho YY, Drill E, Shapiro L, et al. FGF5 is a crucial regulator of hair length in humans. Proceedings of the National Academy of Sciences. 2014;111(29):10648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerwins P, Sköldenberg E, Claesson-Welsh L. Function of fibroblast growth factors and vascular endothelial growth factors and their receptors in angiogenesis. Critical Reviews in Oncology/hematology. 2000;34(3):185–94. [DOI] [PubMed] [Google Scholar]
- 79.Braun S, Auf dKU, Beer HD, Krampert M, Müller M, S, et al. Meeting report: growth factors in development, repair and disease. European Journal of Cell Biology. 2002;81(7):375–82. [DOI] [PubMed] [Google Scholar]
- 80.Giavazzi R, Sennino B, Coltrini D, Garofalo A, Dossi R, Ronca R, et al. Distinct role of fibroblast growth factor-2 and vascular endothelial growth factor on tumor growth and angiogenesis. American Journal of Pathology. 2003;162(6):1913 doi: 10.1016/S0002-9440(10)64325-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kästner S, Elias MC, Rivera AJ, Yablonkareuveni Z. Gene expression patterns of the fibroblast growth factors and their receptors during myogenesis of rat satellite cells. Journal of Histochemistry & Cytochemistry Official Journal of the Histochemistry Society. 2000;126(8):1079. [DOI] [PubMed] [Google Scholar]
- 82.Hébert JM, Rosenquist T, Götz J, Martin GR. FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell. 1994;78(6):1017 [DOI] [PubMed] [Google Scholar]
- 83.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdisciplinary Reviews-Developmental Biology. 2015;4(3):215–66. doi: 10.1002/wdev.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belov AA, Mohammadi M. Molecular Mechanisms of Fibroblast Growth Factor Signaling in Physiology and Pathology. Cold Spring Harbor Perspectives in Biology. 2013;5(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodriguez J, Vernus B, Chelh I, Cassarmalek I, Gabillard J, Sassi AH, et al. Myostatin and the skeletal muscle atrophy and hypertrophy signaling pathways. Cellular and Molecular Life Sciences. 2014;71(22):4361–71. doi: 10.1007/s00018-014-1689-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SJ, Mcpherron AC. Regulation of myostatin activity and muscle growth. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(16):9306–11. doi: 10.1073/pnas.151270098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, et al. Mitogen-Activated Protein Kinase Kinase 3 Regulates Seed Dormancy in Barley. Current Biology. 2016;26(6):775–81. doi: 10.1016/j.cub.2016.01.024 [DOI] [PubMed] [Google Scholar]
- 88.Rossi AC, Mammucari C, Argentini C, Reggiani C, Schiaffino S. Two novel/ancient myosins in mammalian skeletal muscles: MYH14/7b and MYH15 are expressed in extraocular muscles and muscle spindles. The Journal of Physiology. 2010;588(2):353–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takada F, Woude DLV, Tong H, Thompson TG, Watkins SC, Kunkel LM, et al. Myozenin: An α-actinin- and γ-filamin-binding protein of skeletal muscle Z lines. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(4):1595–600. doi: 10.1073/pnas.041609698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mcpherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387(6628):83–90. doi: 10.1038/387083a0 [DOI] [PubMed] [Google Scholar]
- 91.Elkasrawy MN, Hamrick MW. Myostatin (GDF-8) as a key factor linking muscle mass and bone structure. Journal of Musculoskeletal & Neuronal Interactions. 2010;10(1):56–63. [PMC free article] [PubMed] [Google Scholar]
- 92.Lin J, Arnold HB, Della-Fera MA, Azain MJ, Hartzell DL, Baile CA. Myostatin knockout in mice increases myogenesis and decreases adipogenesis. Biochemical & Biophysical Research Communications. 2002;291(3):701–6. [DOI] [PubMed] [Google Scholar]
- 93.Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, Santosneto PC, et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. Plos One. 2014;10(8):e0136690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lessard JC, Pinapaz S, Rotty JD, Hickerson RP, Kaspar RL, Balmain A, et al. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(48):19537–42. doi: 10.1073/pnas.1309576110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang XL, YN, Zhou JK, Zhu HJ, Ma BH, Chen YL, et al. CRISPR/Cas9-mediated MSTN disruption and heritable mutagenesis in goats causes increased body mass 2017;under review. [DOI] [PubMed] [Google Scholar]
- 96.Crossin KL, Krushel LA. Cellular signaling by neural cell adhesion molecules of the immunoglobulin superfamily. Developmental Dynamics. 2000;218(2):260–79. doi: 10.1002/(SICI)1097-0177(200006)218:2<260::AID-DVDY3>3.0.CO;2-9 [DOI] [PubMed] [Google Scholar]
- 97.Rodriguez JB, Vernus B, Chelh I, Cassarmalek I, Gabillard JC, Sassi AH, et al. Myostatin and the skeletal muscle atrophy and hypertrophy signaling pathways. Cellular and Molecular Life Sciences. 2014;71(22):4361–71. doi: 10.1007/s00018-014-1689-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu Q, Hofmann PA. Protein phosphatase 2A-mediated cross-talk between p38 MAPK and ERK in apoptosis of cardiac myocytes. American Journal of Physiology-Heart and Circulatory Physiology. 2004;286(6):H2204–H12. doi: 10.1152/ajpheart.01050.2003 [DOI] [PubMed] [Google Scholar]
- 99.Sun Y, Liu W-Z, Liu T, Feng X, Yang N, Zhou H-F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. Journal of Receptors and Signal Transduction. 2015;35(6):600–4. doi: 10.3109/10799893.2015.1030412 [DOI] [PubMed] [Google Scholar]
- 100.Elliott B, Renshaw D, Getting SJ, Mackenzie R. The central role of myostatin in skeletal muscle and whole body homeostasis. Acta Physiologica. 2012;205(3):324–40. doi: 10.1111/j.1748-1716.2012.02423.x [DOI] [PubMed] [Google Scholar]
- 101.Rodriguez J, Vernus B, Chelh I, Cassarmalek I, Gabillard JC, Hadj SA, et al. Myostatin and the skeletal muscle atrophy and hypertrophy signaling pathways. Cellular & Molecular Life Sciences Cmls. 2014;71(22):4361–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qiao C, Li J, Zhu X, Wang B, Li J, Xiao X. Myostatin propeptide gene delivery by adeno-associated virus serotype 8 vectors enhances muscle growth and ameliorates dystrophic phenotypes in mdx mice. Human Gene Therapy. 2008;19(3):241–54. doi: 10.1089/hum.2007.159 [DOI] [PubMed] [Google Scholar]
- 103.Mcpherron AC, Lee S. Suppression of body fat accumulation in myostatin-deficient mice. Journal of Clinical Investigation. 2002;109(5):595–601. doi: 10.1172/JCI13562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feldman BJ, Streeper RS, Farese RV, Yamamoto KR. Myostatin modulates adipogenesis to generate adipocytes with favorable metabolic effects. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(42):15675–80. doi: 10.1073/pnas.0607501103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Artaza JN, Bhasin S, Magee TR, Reiszporszasz S, Shen R, Groome NP, et al. Myostatin Inhibits Myogenesis and Promotes Adipogenesis in C3H 10T(1/2) Mesenchymal Multipotent Cells. Endocrinology. 2011;146(8):3547–57. [DOI] [PubMed] [Google Scholar]
- 106.Muruganandan S, Roman A, Sinal CJ. Adipocyte differentiation of bone marrow-derived mesenchymal stem cells: Cross talk with the osteoblastogenic program. Cellular and Molecular Life Sciences. 2009;66(2):236–53. doi: 10.1007/s00018-008-8429-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nature Biotechnology. 2013;31(8):681 doi: 10.1038/nbt.2661 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.